Abstract
Autosomal recessive cerebellar ataxia (ARCA) comprises a large and heterogeneous group of neurodegenerative disorders. For many affected patients, the genetic cause remains undetermined. Through whole-exome sequencing, we identified compound heterozygous mutations in ubiquitin-like modifier activating enzyme 5 gene (UBA5) in two Chinese siblings presenting with ARCA. Moreover, copy number variations in UBA5 or ubiquitin-fold modifier 1 gene (UFM1) were documented with the phenotypes of global developmental delays and gait disturbances in the ClinVar database. UBA5 encodes UBA5, the ubiquitin-activating enzyme of UFM1. However, a crucial role for UBA5 in human neurological disease remains to be reported. Our molecular study of UBA5-R246X revealed a dramatically decreased half-life and loss of UFM1 activation due to the absence of the catalytic cysteine Cys250. UBA5-K310E maintained its interaction with UFM1, although with less stability, which may affect the ability of this UBA5 mutant to activate UFM1. Drosophila modeling revealed that UBA5 knockdown induced locomotive defects and a shortened lifespan accompanied by aberrant neuromuscular junctions (NMJs). Strikingly, we found that UFM1 and E2 cofactor knockdown induced markedly similar phenotypes. Wild-type UBA5, but not mutant UBA5, significantly restored neural lesions caused by the absence of UBA5. The finding of a UBA5 mutation in cerebellar ataxia suggests that impairment of the UFM1 pathway may contribute to the neurological phenotypes of ARCA.
Introduction
Autosomal recessive cerebellar ataxias (ARCAs) are a large group of neurodegenerative disorders that manifest mainly in children and young adults. Most ARCAs are heterogeneous with respect to the age at onset, severity of the disease progression, and frequency of extracerebellar and systemic signs. [1,2] Five main pathogenic mechanisms are distinguishable: defective DNA repair, abnormal protein folding and degradation, channelopathies, and mitochondrial and metabolic defects. [3] Although a growing list of rare molecular defects associated with ARCAs have been identified, as yet many affected families and individuals have an unknown etiology. [4,5,6]
We applied whole-exome sequencing to DNA from two siblings with progressive cerebellar ataxia who had non-consanguineous parents. Consequently, compound heterozygous variants were found in UBA5. UBA5, also known as ubiquitin-like modifier activating enzyme 5, is an ubiquitin-activating enzyme (E1) of the ubiquitin-fold modifier 1 (UFM1) pathway. UFM1 is an ubiquitin-like protein (UBL) that, upon activation by a dedicated E1 (UBA5), forms a thioester bond with an E2 cofactor (UFC1), resulting in the tagging of reactive ubiquityl units to substrates by an E3 ligase (UFL1).[7–11] In ischemic myocardial cells and pancreatic islet beta cells, endoplasmic reticulum (ER) stress can specifically induce the expression of UFM1 cascade members, thus suggesting the protection of the UFM1 cascade during cellular homeostasis.[12,13] In UBA5, the catalytic cysteine (Cys250) is part of the adenylation domain within the helical motif through which the ubiquityl-enzyme thioester is formed.[14] Our cellular studies revealed that both of the two mutants (p.R246X and p.K310E) became less stable which caused reduced enzymatic activity of UBA5. A role for UBA5 in human neurological disease has yet to be identified. Here we studied a cohort of patients with ARCA and identified UBA5 mutations in these patients.
Drosophila is a powerful model organism with which to study neural development and neuronal maintenance. A Drosophila model containing a loss-of-function of the UBA5 homologue confirmed the role of this in disease pathogenesis. In particular, we found that UBA5 knockdown resulted in remarkably reduced locomotor activity, a shortened lifespan, and neuromuscular junction (NMJ) defects. Furthermore, similar phenotypes were observed with UFM1 and UFC1 knockdown, although UFM1 knockdown resulted in a more severe phenotype. Drosophila NMJs provide a simpler and genetically more amenable system with which to explore the molecular mechanisms underlying synapse development and maturation.[15] Both Drosophila and human wild type UBA5, but not UBA5 mutations, significantly rescue the neuromuscular junction (NMJ) defects. Our data therefore establish a novel ARCA syndrome by a mutation in UBA5 and thus shed light on the biological function of the corresponding protein.
Materials and Methods
Ethic statement
This study protocol was approved by the Ethic Committee of the Xiangya Hospital of Central South University in China (equivalent to an Institutional Review Board). The individual in this manuscript have given written informed consent.
Patients
Clinical data and blood samples were obtained from two Chinese siblings who presented with progressive ataxia during childhood and had non-consanguineous parents. Both patients underwent a standardized neurologic examination conducted by two neurological specialists. We used DNA sequencing and capillary electrophoresis to exclude mutations and repeat expansions in known ataxia genes. In addition, 500 unaffected, healthy Chinese individuals were analyzed as controls. The relevant ethical authorities approved this study, and written informed consent was obtained from all subjects.
Whole-exome sequencing
Genomic DNA was extracted from the peripheral blood of the two affected individuals (II:2 and II:3) using standard methods (QIAGEN, Valencia, CA). Whole-exome sequencing was performed using a Genome Analyser II platform. Sequencing data were aligned to the human genome reference (UCSC hg 18 version). The variants were confirmed using Sanger sequencing.
Plasmid construction, cell culture, and transfection
Human wild-type UBA5 cDNA was PCR amplified from a human cDNA library and inserted in-frame into p3xFlag-CMV-24 (Sigma, St. Louis, MO, USA) at the EcoRI/SalI sites. Mutants were generated via QuikChange site-directed mutagenesis according to the manufacturer’s protocol (Stratagene, La Jolla, CA, USA). All constructs were confirmed by sequencing. Human embryonic kidney 293A cells (HEK293A, Invitrogen, R705-07) and human cervical carcinoma HeLa cells (Chinese Academy of Sciences, TCHu187) were cultured in Dulbecco’s Modified Eagle Medium (Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS) at 37°C and 5% CO2. Plasmids were transfected into cells using Lipofectamine 2000 (Invitrogen).
Degradation assay, immunoprecipitation, and westernblotting
After transfection, HEK293A cells expressing the indicated plasmids were treated with 100 μg/ml cycloheximide (CHX; Sigma). The cells were harvested after 0, 12, 24, and 36 h of CHX treatment. For co-immunoprecipitation, the following antibodies were used: monoclonal anti-Flag (Sigma); monoclonal anti-UFM1 (Epitomics/Abcam, USA); monoclonal anti-GAPDH (Sigma); polyclonal anti-UBA5 (Epitomics/Abcam), and sheep anti-rabbit antibody (Amersham Pharmacia Biotech).
Immunocytochemical analyses
Cells transfected with the indicated plasmids were grown on cover slides and fixed with 4% paraformaldehyde (PFA) for 5 min at room temperature; the cells were subsequently incubated with 0.25% Triton X-100 for 5 min and blocked with 0.1% FBS in phosphate-buffered saline (PBS). DAPI (Sigma) was used for nuclear staining. Anti-UBA5 (Epitomics/Abcam) and anti-GM130 antibodies (Sigma) were also used for staining. All cells were imaged using a fluorescence microscope equipped with a cooled charge-coupled device camera (CTR MIC; Leica, Wetzlar, Germany).
Third-instar larval muscles were dissected and stained using a modified protocol described by Dr. Wei Xie.[16] Larval muscles were fixed in 4% PFA and incubated with anti-DLG antibody (DSHB) at a dilution of 1:50. The NMJs from muscle 4 of abdominal segments 2, 3, and 4 were imaged using a Leica TCS SP5 confocal station and ImageJ software (National Institutes of Health, Bethesda, MD, USA) to quantify the bouton numbers and sizes.
Drosophila genetics and fly stocks
Fly cultures and crosses were performed according to standard procedures. The DA-GAL4, UBA5 RNAi, UFM1 RNAi, w1118 and attP2 fly lines were obtained from the Bloomington Drosophila Stock Center at Indiana University (Bloomington, IN, USA). UFC1 RNAi and UFL1 RNAi fly lines were obtained from the Vienna Drosophila RNAi Center (Vienna, Austria). The OK6-GAL4 fly line was a gift from Dr. Wei Xie at Southeast University. Wild-type UBA5 cDNA was amplified from Drosophila cDNA. Human wild-type and mutant UBA5 cDNA sequences were HA-tagged and subcloned into the pUAST vector at the EcoRI/XhoI restriction sites. P-element–mediated germ line transformations were performed via microinjection into w1118 background flies using P-element vectors (pUAST-HA-dUBA5-WT, pUAST-HA-hUBA5-WT, pUAST-HA-hUBA5-R246X, and pUAST-HA-hUBA5-K310E). F1 transformants were selected on the basis of white eye-color rescue.
Quantification of wing phenotypes and light microscopy
The percentage of 3-day-old male flies that exhibited unfolded vertical-turned wings was measured (n ≥ 300). Whole flies were analyzed using an OLYMPUS DP72 microscope (Olympus Corporation, Tokyo, Japan) to obtain light microscopy (LM) images.
Behavioral and longevity assays
For climbing assays, groups of ten 3-day-old male flies were transferred into 1.25-cm-diameter and 28-cm-height plastic tubes for a 30-min incubation at room temperature. The time at which the fifth fly arrived at the 15-cm finish line was recorded and analyzed. For flight assays, a vial of twenty 3-day-old male flies was guided into a 500-ml measuring cylinder. In this test, normal flies run into the inner wall of the measuring cylinder and are glued to the top, whereas disabled flies fall straight to the bottom. For longevity assays, 20 male flies were placed in a vial maintained at 25°C and supplied with fresh food every 3 days. Five vials were recorded per genotype. The above assays were repeated at least 3 times per genotype.
Results
Clinical description of the UBA5 family
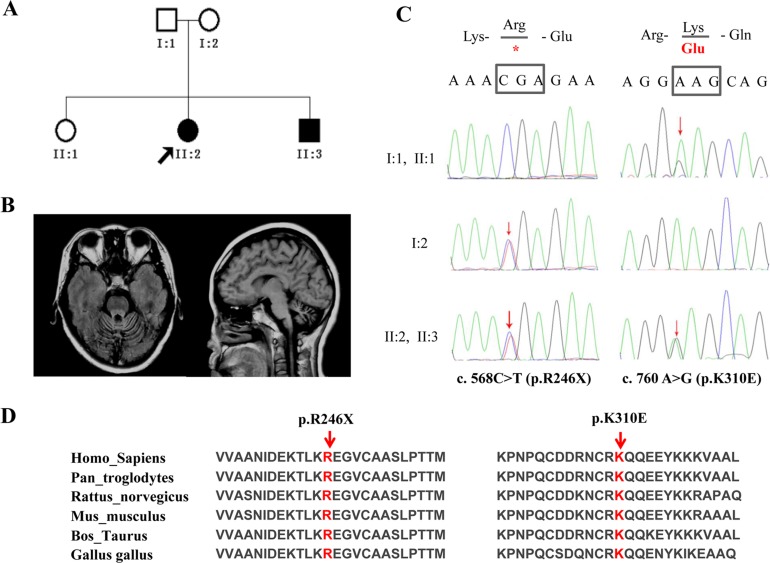
The affected family originated from the Shanxi province in China (Fig 1A). The parents (I:1 and I:2) were non-consanguineous. The father of the affected siblings (II:2 and II:3) reported no gait difficulties at the age of 80, and the mother presented with an essential tremor of the head but no gait difficulties. Neurologic examinations of the parents (I:1 and I:2) yielded normal findings. The two affected siblings, one female and one male, reported gait instability since childhood. Both siblings exhibited markedly delayed growth during childhood but had achieved a normal body size in adulthood. Both developed cataracts while young and had undergone cataract-removal surgeries in 2010. Neither sibling exhibited signs of cognitive involvement. The disease progressed insidiously in the proband (II:2), who presented with marked cerebellar atrophy as determined by brain magnetic resonance imaging (MRI; Fig 1B), leading to a loss of the ability to walk and caregiver dependency at 39 years of age. The disease course was apparently stable in the younger brother (II:3), who was 36 years old, worked full-time, and displayed a mildly spastic gait that did not interfere with his daily activities, and also exhibited mild cerebellar atrophy on MRI. Neurophysiologic studies indicated no other central or peripheral system impairments in the proband. The brother (II:3) exhibited partial peripheral nerve impairment with a demyelinating sensory-motor peripheral neuropathy. Phenotypic details are given in Table 1.
Fig 1. Genetic findings in a family with UBA5 mutations.
(A) The pedigree of family 1 with autosomal recessive spinocerebellar ataxia. (B) Brain magnetic resonance imaging of patient II:2. Panel (left): axial T1-weighted image showing atrophy of the cerebellar vermis. Panel (right): midline sagittal T1-weighted image showing cerebellar atrophy, particularly in the superior vermis, with enlargement of the fourth ventricle. (C) The UBA5 variants [c. 568C > T; c. 760A > G], [p. Arg246X (R246X); p. Lys310Glu (K310E)] segregated in this family. Red arrows indicate the mutation sites. (D) Two mutations (red) affected amino acids that are highly conserved across species.
Table 1. The phenotypic characteristics of patients with intermediate.
| Patient II:2 | Patient II:3 | |
|---|---|---|
| Age (y) | 40 | 36 |
| Sex | Female | Male |
| Age at onset (y) | 5 | 8 |
| Symptoms | Gait instability, speech difficulties | Gait instability, speech difficulties |
| Clinical signs | Gait and limb ataxia, dysarthria, horizontal nystagmus | Gait and limb ataxia, dysarthria, horizontal nystagmus |
| Other symptoms | Cataract | Cataract |
| EMG and NCV | Normal | EMG: Normal; NCV: partial abnormality |
| Evoked potentials | VEP: normal; AEP: normal;SEP: increased latency in RLL | VEP: normal; AEP: increased latency; SEP: normal |
| ICARS | 46 | 29 |
| SARA | 17 | 11 |
| MMSE | 27 | 30 |
| Brain MRI | Cerebellar atrophy | Cerebellar atrophy |
Whole-exome sequencing identified UBA5 mutations in siblings with ARCA
The non-consanguineous pedigree of the index family includes two siblings (patient II:2 and II:3) with ataxia. DNA from both affected individuals was subjected to whole-exome sequencing. Variants were prioritized according to the presence of compound heterozygosity and homozygosity, based on recessive inheritance. Only two variants in the UBA5 (NM_198329.2) cosegregated completely in the conserved domain of the UBA5 protein: c.568C > T; p. R246X and c.760A > G; p. K310E (Fig 1C and 1D). These variants were not identified in any of the 500 unaffected controls.
Characterization of UBA5 and its mutant protein
Immunofluorescence was used to visualize exogenously expressed WT and mutant UBA5 in HEK293A cells. As shown in Fig 2A, wild-type and K310E UBA5 were localized in the cytoplasm, whereas the truncated R246X protein was mainly localized in the nucleus. We also found that endogenous UBA5 co-localized with GM130 (Golgi marker) (Fig 2B). We next aimed to investigate the degradation of UBA5. After overexpressing UBA5 in HEK293A cells, MG132 or bafilomycin A1 (BafAl) was added to the cells, followed by a 12-h incubation. The results demonstrated a marked increase in UBA5 protein levels after the addition of BafA1 (autophagy inhibitor), but not MG132 (proteasome inhibitor), indicating that UBA5 was degraded through autophagy. K310E protein was degraded in the same manner as wild-type UBA5 (Fig 2C). However, levels of the truncated R246X protein increased significantly in the presence of MG132 but not BafA1, suggesting that the R246X protein was degraded through the ubiquitin proteasome pathway (Fig 2C). To examine whether UBA5 mutations impaired the stability of the protein product, a CHX-chase analysis was performed. The results indicated that wild-type UBA5 protein was quite stable with a half-life of more than 36 h, whereas K310E was less stable and R246X protein had a half-life of less than 30 min (Fig 2D).
Fig 2. The subcellular localization, manner of degradation, and stability of mutant UBA5 and its interaction with UFM1.
(A) Immunostaining of overexpressed UBA5 and its mutants, as well as endogenous UBA5. (B) Immunostaining of endogenous UBA5 in HEK293A and HeLa cells. (C) Flag-UBA5 or related mutants were overexpressed via plasmid transfection in HEK293A cells. After 24 h, the cells were treated with MG132 (100μg/ml) or Bafilomycin A1 (BafAl, 100μg/ml). (D) UBA5 stability analysis. (E) Interactions of UBA5 mutants with Ufm1.
Activity of UBA5 mutants with regard to UFM1
UBA5 is among the least characterized of all human E1 enzymes. In a previous report, UBA5 was shown to activate UFM1 and SUMO2. [8] However, we and others demonstrated a lack of interaction between UBA5 and SUMO2 (data not shown). Next, we focused on UFM1. A co-immunoprecipitation study demonstrated that R246X protein failed to interact with endogenous UFM1, indicating that this mutation impaired the ability of UBA5 to activate UFM1 (Fig 2E). K310E remained able to interact with UFM1, similar to wild-type UBA5; thus, the pathogenesis associated with this mutation will require further investigation (Fig 2E).
Impairment of the UFM1 pathway causes an abnormal Drosophila phenotype
Drosophila UBA5 (dUBA5, CG1749) is highly homologous to human UBA5 and shares 67% identity with human UBA5 at the amino acid level. Similarly, the respective UFM1 and E2 homologues, ubiquitin-like protein and UFC1, share 88% and 79% identity, respectively. Therefore, we used a fly model to determine whether the identified mutations in our patients would impair the function of the UFM1 pathway in the nervous system. As a loss-of-function of UBA5 is associated with ARCA, FRT UBA5, and hs-flp were crossed to generate UBA5 null mutant fly lines. Unfortunately, UBA5 null mutant flies of both sexes died during embryonic development. UBA5 RNAi flies were found to exhibit an 80% reduction in dUBA5 mRNA expression. Here, dUBA5 knockdown was applied to model UBA5-associated ARCA and explore the in vivo functions of the UFM1 system. The resulting flies were analyzed at 3 days of age.
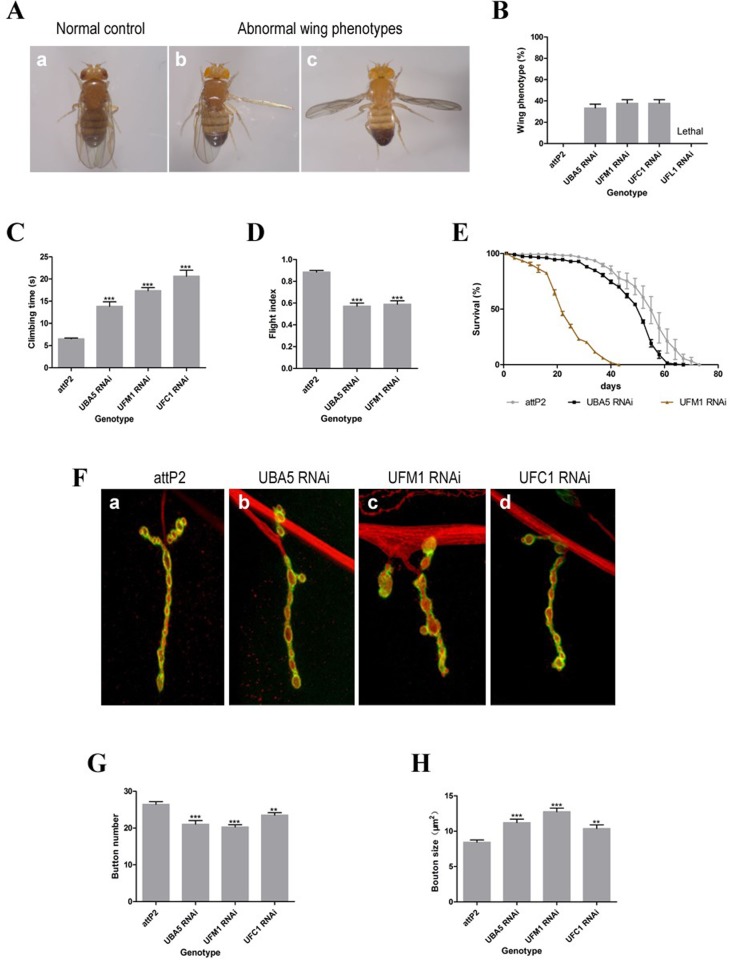
UBA5 knockdown flies (both male and female) exhibited abnormal vertical-turned wings that failed to fold under resting conditions at 25°C. The wing postures indicated unilateral or bilateral wing abnormalities, and the percentages of abnormal wing phenotypes are shown in Fig 3A and 3B. Similar phenotypes were observed in dUFM1 RNAi and dUFC1 RNAi lines, whereas UFL1 knockdown was found to be lethal. In contrast, the control flies always held their wings parallel to the body axis. The penetrance of this phenotype increased when flies were raised at 29°C; specifically, 56.9% of the dUBA5 RNAi lines were found to exhibit wing defects. The climbing and flight abilities and lifespans of dUFM1 RNAi, dUBA5 RNAi, and dUFC1 RNAi lines were greatly reduced, compared with control flies (Fig 3C and 3D). In an evaluation of the climbing abilities of these flies after removing the wings to exclude wing involvement in climbing movements, a similar tendency was observed (data not shown).
Fig 3. RNAi-mediated knockdown of UBA5 and other molecules of UFM1 pathway induce neurodegeneration.
(A) Knockdown of UBA5 and other crucial molecules of UFM1 pathway, UFM1 and UFC1, in Drosophila exhibit abnormal vertical-turned wings that fail to fold, whereas knockdown of UFL1 is fatal. The wing phenotypes are: normal (a), unilateral wing abnormality (b), bilateral wing abnormality (c). (B) Percentage of abnormal wing phenotypes. (C) Comparison of climbing abilities shows climbing disabilities of the RNAi flies. (D) Comparison of flight abilities presents flight declines of the RNAi flies. (E) The life spans of the RNAi flies are significantly shortened. (F) The confocal images of larval muscle exhibit reduced type Ib bouton number and increased bouton size of the RNAi flies. (G) The statistical graph of bouton numbers. (H) The statistical graph of bouton sizes. All the indicated genotypes in panel B-E are: DA-GAL4>attP2, DA-GAL4>UBA5 RNAi, DA-GAL4>UFM1 RNAi, DA-GAL4>UFC1 RNAi, DA-GAL4>UFL1 RNAi. All the indicated genotypes in panel F-H are: OK6-GAL4>attP2, OK6-GAL4>UBA5 RNAi, OK6-GAL4>UFM1 RNAi, OK6-GAL4>UFC1 RNAi. *** = p < 0.0001 and ** = p < 0.01, Student’s t-test and Mann–Whitney test.
A histological analysis revealed no disruption in muscle morphology in the indirect flight muscles of Da-Gal4 > dUFM1 RNAi and Da-Gal4 > dUBA5 RNAi flies. Defective NMJs remained another anatomical explanation for the observed abnormal wing posture. Confocal images of larval muscles revealed a reduced number of type Ib boutons and increased bouton size in UFM1 RNAi, UBA5 RNAi and UFC1 RNAi flies relative to WT controls; this effect was particularly pronounced in the motor neurons controlled by an OK6-Gal4 driver (Fig 3E–3H). We expressed dUFM1 RNAi and dUBA5 RNAi specifically in the nervous system using the Elav-Gal4 driver and in muscle cells using the MHC-Gal4 driver. The above-described bouton changes were also observed with neuron-specific Elav-Gal4, but not MHC-Gal4, indicating that the UFM1 pathway is active in neurons. We noticed that all UFM1 knockdown phenotypes were highly reminiscent of those associated with UBA5, albeit stronger.
Overexpression of wild-type UBA5 significantly restored neural lesions in UBA5 RNAi flies, whereas mutant UBA5 could not confer effective restoration
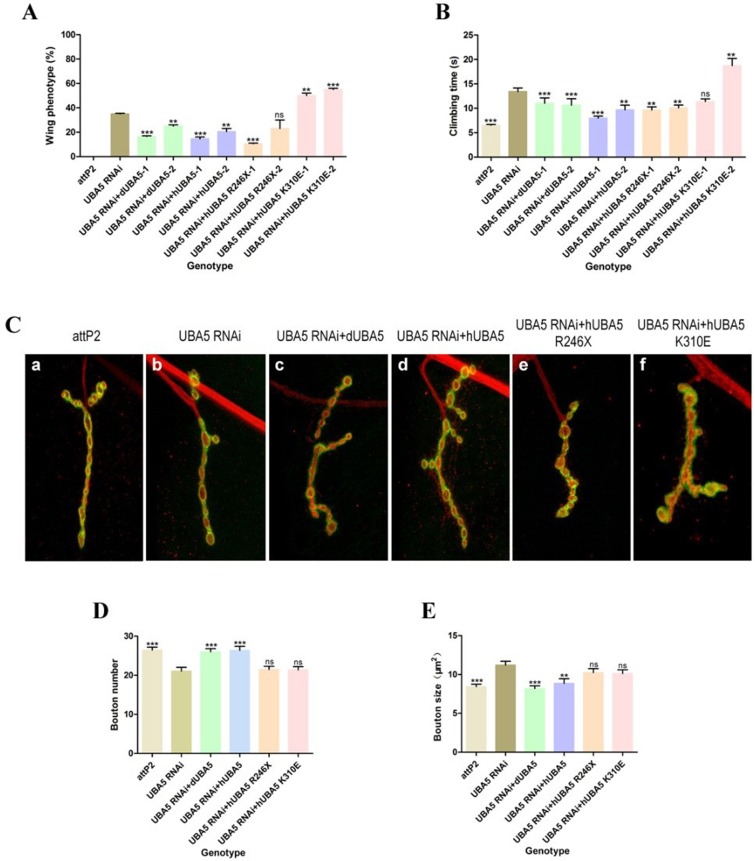
We generated transgenic fly lines that would overexpress Drosophila UBA5, wild-type human UBA5, and mutated or deleted human UBA5 to investigate whether the identified mutations would impair the function of UBA5. Quantitative real-time PCR was performed to confirm similar expression levels. Rescue experiments were performed to determine whether mutated UBA5 could rescue the neurodegenerative defect caused by the loss of the Drosophila ortholog UBA5. The expression of wild-type hUBA5 and dUBA5 in the UBA5 knockdown fly significantly reduced the observed neurodegeneration but was unable to completely convert the phenotype to wild-type (Fig 4). The rescued flies exhibited an increased climbing speed. Consistent with these findings, significant NMJ restoration was observed with UBA5 expression, and the size within these regions was also markedly restored (Fig 4C and 4E). As shown in Fig 4A and 4B, the expression of mutant UBA5 could not rescue the wing phenotype and climbing defect.
Fig 4. Overexpression of wild-type UBA5 significantly restores neural lesions of UBA5 RNAi flies while mutant UBA5 could not confer effective rescue.
(A) Percentage of abnormal wing phenotypes. (B) Comparison of climbing abilities. (C) The confocal images of larval muscle. (D) The statistical graph of bouton numbers. (E) The statistical graph of bouton sizes. All the indicated genotypes in panel A-B are: DA-GAL4>attP2, DA-GAL4>UBA5 RNAi, DA-GAL4>UBA5 RNAi+dUBA5-1, DA-GAL4>UBA5 RNAi+dUBA5-2, DA-GAL4>UBA5 RNAi+hUBA5-1, DA-GAL4>UBA5 RNAi+hUBA5-2, DA-GAL4>UBA5 RNAi+hUBA5 R246X-1, DA-GAL4>UBA5 RNAi+hUBA5 R246X-2, DA-GAL4>UBA5 RNAi+hUBA5 K310E-1, DA-GAL4>UBA5 RNAi+hUBA5 K310E-2. All the indicated genotypes in panel C-E are: OK6-GAL4>attP2, OK6-GAL4>UBA5 RNAi, OK6-GAL4>UBA5 RNAi+dUBA5-1, OK6-GAL4>UBA5 RNAi+hUBA5-1, OK6-GAL4>UBA5 RNAi+hUBA5 R246X-1, OK6-GAL4>UBA5 RNAi+hUBA5 K310E-1. *** = p < 0.0001, ** = p < 0.01 and ns = no significance, Student’s t-test and Mann–Whitney test.
Discussion
ARCAs are genetically heterogeneous. At present, a large proportion of ARCA cases remain unexplained by mutations in known genes. Here we uncovered the genetic basis for a new subtype of ARCA. This report demonstrates that ataxia can be caused by mutations in UBA5 (NM_198329.2). Both affected individuals carried a mixture of two mutant alleles (c. 568 C > T; p. R246X and c. 760A > G; p. K310E), whereas their parents were heterozygous mutation carriers [the mother (I:2) carried c. 568 C > T (p. R246X) and the father (I:1) carried c.760A > G (p. K310E)]. Both variants were located in the conserved region and cosegregated completely (Fig 1D). Cases with copy number variations in the UBA5 and UFM1 region have been documented in the ClinVar database. A 0.4-Mb heterozygous loss in 3q22.1, which includes UBA5 and NPHP3, was previously detected in a patient with global developmental delays, impaired hearing, muscular hyptonia, and seizures. Because NPHP3 functions in the renal tubular development process, the loss of UBA5 may have resulted in these phenotypes. A 1-Mb duplication in 13q13.3, including UFM1, was also identified in a case with global developmental delays, gait disturbance, and other abnormalities. Our molecular characterization of UBA5-R246X revealed a loss in UFM1 activation in the absence of the catalytic cysteine Cys250 (Fig 2). The activity of UBA5-K310E was not clear; the interaction between this mutant protein and UFM1 was maintained but less stable, which may affect its ability to activate UFM1 (Fig 2).
UBA5 is a member of the ubiquitin-activating protein family and the only known E1 enzyme in the UFM1 cascade. In humans, UBA5 is transcribed as two distinct isoforms (1–404 and 57–404).The role of the additional N-terminal residues encoded by the longer splice transcript is not clear; however, these residues are not strongly conserved and are not required for ubiquitin-like modifier (UBL) activation. [8,14] UBA5 knockout mice were found to die in utero because of severe anemia associated with the defective differentiation of both megakaryocytes and erythrocytes. [17] However, there are no previous reports of a relationship between UBA5 and neurological disorders. The form of ARCA described herein is the new phenotype to be associated with impairment in the ubiquitin-like system. Emerging evidence of mutations in the ubiqutin system have been found in cerebellar ataxia patients, as was described in our report as well as others. [18,19]
UFM1 belongs to the UBL family, the members of which are present in nearly all eukaryotic organisms, with the exception of fungi. As described in detail previously, the UFM1 cascade, which includes the ligase UFL1, UFM1-specific protease UfSP2, and known target proteins (Ufbp1 and Cdk5rap3), has been implicated in ER functioning and cell cycle control. [20,21] A loss-of-function of the UFM1 cascade in mice leads to apoptosis in fetal liver cells and pancreatic beta cells. [13,17] In particular, the ligase UFL1 was shown to be involved in spinocerebellar ataxia type 1 (SCA1), a polyglutamine disease. Furthermore, UFL1 deficiency contributes to SCA1 pathology through a functional deficiency in Bergmann glia, which regulate cell proliferation through the regulation of CDK5RAP3. [22]
Drosophila was selected as a model with which to study the involvement of UBA5 in the disease phenotype. The Drosophila larval NMJ is a well-established synaptic model system that shares major features with central excitatory synapses in the mammalian brain and has been successfully used to investigate human neural disorders, as described in detail previously. [23] Our UBA5 knockdown Drosophila models exhibited abnormal wing phenotypes, a shortened life span and climbing disabilities. Neuron-specific UBA5 knockdown resulted in reduced NMJ growth and a consistent decrease in synaptic size, which presented as a reduced number of type Ib boutons and increased bouton size in the larval muscles, suggesting that the UFM1 cascade might play a role in the development of NMJs in Drosophila. Our data thus shed light on a role for the UFM1 cascade in neurological diseases.
In conclusion, we identified mutations in UBA5, an E1 enzyme, in patients with ARCA. A Drosophila model provided lines of evidence suggesting that a UFM1 pathway impairment might contribute to the neurological phenotypes of ARCA. Rescue experiments involving wild-type and mutated human UBA5 definitively demonstrated the deleterious nature of these mutations and shed light on the underlying molecular pathogenesis of this disease. An analysis of the UFM1 pathway should be performed in patients presenting with both sporadic and familial ARCA.
Acknowledgments
We are grateful to the family for their cooperation. We would like to thank Dr. Lei Xue at the Tongji University for valuable suggestions on Drosophila research. We also thank Dr. Wei Xie at the Southeast University for sharing fly line of OK6-GAL4.
Abbreviations
- ARCA
autosomal recessive cerebellar ataxia
- UBA5
ubiquitin-like modifier activating enzyme 5
- Ufm1
ubiquitin-like modifier
- UBLs
ubiquitin like molecules
- RT-PCR
Reverse transcription PCR
Data Availability
All relevant data are within the paper.
Funding Statement
The State Key Program of the National Natural Science Foundation of China (grant number: 81130021), National Natural Science Foundation of China (grant number: 81571253, 81172513, 81071028).
References
- 1.Anheim M, Tranchant C, Koenig M. The autosomal recessive cerebellar ataxias. N Engl J Med 2012; 366:636–646. 10.1056/NEJMra1006610 [DOI] [PubMed] [Google Scholar]
- 2.Vermeer S, van de Warrenburg BP, Willemsen MA, Cluitmans M, Scheffer H, Kremer BP, et al. Autosomal recessive cerebellar ataxias: the current state of affairs. J Med Genet 2011; 48:651–659. 10.1136/jmedgenet-2011-100210 [DOI] [PubMed] [Google Scholar]
- 3.De Michele G, Coppola G, Cocozza S, Filla A. A pathogenetic classification of hereditary ataxias: is the time ripe? J Neurol 2004; 251:913–922. [DOI] [PubMed] [Google Scholar]
- 4.Depondt C, Donatello S, Simonis N, Rai M, van Heurck R, Abramowicz M, et al. Autosomal recessive cerebellar ataxia of adult onset due to STUB1 Mutations. Neurology 2014; 82:1749–1750. 10.1212/WNL.0000000000000416 [DOI] [PubMed] [Google Scholar]
- 5.Sailer A, Houlden H. Recent advances in the genetics of cerebellar ataxias. Curr Neurol Neurosci Rep 2012; 12:227–236. 10.1007/s11910-012-0267-6 [DOI] [PubMed] [Google Scholar]
- 6.Hersheson J, Haworth A, Houlden H. The inherited ataxias: genetic heterogeneity, mutation databases, and future directions in research and clinical diagnostics. Hum Mutat 2012; 33:1324–1332. 10.1002/humu.22132 [DOI] [PubMed] [Google Scholar]
- 7.Komatsu M1, Chiba T, Tatsumi K, Iemura S, Tanida I, Okazaki N, et al. A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J 2004; 23:1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng M, Gu X, Zheng D, Yang Z, Li F, Zhao J, et al. UBE1DC1, an ubiquitin-activating enzyme, activates two different ubiquitin-like proteins. J Cell Biochem 2008; 104:2324–2334. 10.1002/jcb.21791 [DOI] [PubMed] [Google Scholar]
- 9.Tatsumi K, Sou YS, Tada N, Nakamura E, Iemura S, Natsume T, et al. A novel type of E3 ligase for the Ufm1 conjugation system. J Biol Chem 2010; 285:5417–5427. 10.1074/jbc.M109.036814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M, Zhu X, Zhang Y, Cai Y, Chen J, Sivaprakasam S, et al. RCAD/Ufl1, a Ufm1 E3 ligase, is essential for hematopoietic stem cell function and murine hematopoiesis. Cell Death Differ 201551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hertel P, Daniel J, Stegehake D, Vaupel H, Kailayangiri S, Gruel C, et al. The ubiquitin-fold modifier 1 (Ufm1) cascade of Caenorhabditis elegans. J Biol Chem 2013; 288:10661–10671. 10.1074/jbc.M113.458000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azfer A, Niu J, Rogers LM, Adamski FM, Kolattukudy PE. Activation of endoplasmic reticulum stress response during the development of ischemic heart disease. Am J Physiol Heart Circ Physiol 2006; 291:H1411–H1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lermaire K, Moura RF, Granvik M, Igoillo-Esteve M, Hohmeier HE, Hendrickx N, et al. Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic beta cells from ER stress-induced apoptosis. PLoS One 2011; 6:e18517 10.1371/journal.pone.0018517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bacik JP, Walker JR, Ali M, Schimmer AD, Dhe-Paganon S. Crystal structure of the human ubiquitin-activating enzyme 5 (UBA5) bound to ATP: mechanistic insights into a minimalistic E1 enzyme. J Biol Chem 2010; 285:20273–20280. 10.1074/jbc.M110.102921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins CA, DiAntonio A. Synaptic development: insights from Drosophila. Curr Opin Neurobiol 2007; 17:35–42. [DOI] [PubMed] [Google Scholar]
- 16.Xing G, Gan G, Chen D, Sun M, Yi J, Lv H, et al. Drosophila neuroligin3 regulates neuromuscular junction development and synaptic differentiation. J Biol Chem 2014; 289:31867–31877. 10.1074/jbc.M114.574897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tatsumi K, Yamamoto-Mukai H, Shimizu R, Waguri S, Sou YS, Sakamoto A, et al. The Ufm1-activating enzyme Uba5 is indispensable for erythroid differentiation in mice. Nat Commun 2011; 2:181 10.1038/ncomms1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi Y, Wang J, Li JD, Ren H, Guan W, He M, et al. Identification of CHIP as a novel causative gene for autosomal recessive cerebellar ataxia. PLoS One 2013; 8:e81884 10.1371/journal.pone.0081884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ronnebaum SM, Patterson C, Schisler JC . Emerging evidence of coding mutations in the ubiquitin–proteasome system associated with cerebellar ataxias. Human Genome Variation 2014; 14018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daniel J, Liebau E. The ufm1 cascade. Cells 2014; 3:627–638. 10.3390/cells3020627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ha BH, Jeon YJ, Shin SC, Tatsumi K, Komatsu M, Tanaka K, et al. Structure of ubiquitin-fold modifier 1-specific protease UfSP2. J Biol Chem 2011; 286:10248–10257 10.1074/jbc.M110.172171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shiwaku H, Yoshimura N, Tamura T, Sone M, Ogishima S, Watase K, et al. Suppression of the novel ER protein Maxer by mutant ataxin-1 in Bergman glia contributes to non-cell-autonomous toxicity. EMBO J 2010; 29:2446–24. 10.1038/emboj.2010.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willemsen MH, Nijhof B, Fenckova M, Nillesen WM, Bongers EM, Castells-Nobau A, et al. GATAD2B loss-of-function mutations cause a recognisable syndrome with intellectual disability and are associated with learning deficits and synaptic undergrowth in Drosophila. J Med Genet. 2013;50:507–14. 10.1136/jmedgenet-2012-101490 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.