Abstract
The purpose of the dbNSFP is to provide a one-stop resource for functional predictions and annotations for human non-synonymous single-nucleotide variants (nsSNVs) and splice site variants (ssSNVs), and to facilitate the steps of filtering and prioritizing SNVs from a large list of SNVs discovered in an exome-sequencing study. A list of all potential nsSNVs and ssSNVs based on the human reference sequence were created, functional predictions and annotations were curated and compiled for each SNV. Here we report a recent major update of the database to version 3.0. The SNV list has been rebuilt based on GENCODE 22 and currently the database includes 82,832,027 nsSNVs and ssSNVs. An attached database dbscSNV, which compiled all potential human SNVs within splicing consensus regions and their deleteriousness predictions, add another 15,030,459 potentially functional SNVs. Eleven prediction scores (MetaSVM, MetaLR, CADD, VEST3, PROVEAN, 4× fitCons, fathmm-MKL and DANN) and allele frequencies from the UK10K cohorts and the Exome Aggregation Consortium (ExAC), among others, have been added. The original seven prediction scores in v2.0 (SIFT, 2× Polyphen2, LRT, MutationTaster, MutationAssessor and FATHMM) as well as many SNV and gene functional annotations have been updated. dbNSFP v3.0 is freely available at http://sites.google.com/site/jpopgen/dbNSFP.
Keywords: dbNSFP, dbscSNV, non-synonymous mutation, splice site mutation, functional prediction, database
INTRODUCTION
With the advancement of technologies and the drop of the associated expenses, DNA sequencing is increasingly used as a research as well as diagnostic tool for human diseases. Among all the sequencing strategies, whole exome sequencing (WES) is probably the most popular for identifying novel genes and mutations causing genetic diseases. Currently, the cost of WES is roughly on par with targeted sequencing of a few genes while delivering the genotypes of the whole exome. Compared to whole genome sequencing with the same depth, with only a fraction of the cost WES is able to discover some of the most important candidates for disease causing mutations, including presumably functional single-nucleotide variants (SNVs): stop-gain, stop-loss, missense, splice site, and those within splicing consensus regions (−3 to +8 at the 5’ splice site and −12 to +2 at the 3’ splice site).
The major aim of dbNSFP is to facilitate the process of filtering and prioritizing the above mentioned presumably functional SNVs from a long list of SNVs identified in a typical WES study. To make it truly scalable to large WES studies and avoid security concerns, dbNSFP was designed to work as a local and self-sustaining database without need for internet connection. This database compiled all potential non-synonymous SNVs (nsSNVs, including stop-gain, stop-loss and missense), splice site SNVs (ssSNVs) and SNVs in splicing consensus regions (scSNVs, via attached database dbscSNV; see below) based on a human reference sequence. Functional predictions and annotations for each SNV from many methods and resources were exhaustively curated. Searching the database using the companion Java program can be accomplished by a single command line call, therefore it is easy to operate for researchers with minimum bioinformatics training.
dbNSFP has expanded since its first release in 2011. dbNSFP v1.0 (Liu et al. 2011) was based on the human reference sequence version hg18 and the gene model of Consensus Coding Sequence (CCDS) version 20090327 (Pruitt et al. 2009). It included 75,931,005 nsSNVs and four functional prediction scores: SIFT (Ng and Henikoff 2001), Polyphen2 (Adzhubei et al. 2010), LRT (Chun and Fay 2009) and MutationTaster (Schwarz et al. 2010), and one conservation score: phyloP (Siepel et al. 2006) for each nsSNV. dbNSFP v2.0 (Liu et al. 2013) was rebuilt based on the human reference sequence version hg19 and the gene model of GENCODE 9 (Harrow et al. 2012). It compiled 87,347,043 nsSNVs and 2,270,742 ssSNVs. It added two functional prediction scores, MutationAssessor (Reva et al. 2011) and FATHMM (Shihab et al. 2013), two conservation scores, GERP++ (Davydov et al. 2010) and SiPhy (Garber et al. 2009; Lindblad-Toh et al. 2011), and allele frequencies from the 1000 Genomes Project phase 1 data (The 1000 Genomes Project Consortium 2012) and the NHLBI Exome Sequencing Project data (Fu et al. 2013). Rich functional annotations for human genes were also added to dbNSFP v2.0. dbNSFP has gained popularity among human geneticists and has been adopted by mainstream annotation tools/resources, including the UCSC Genome Browser’s Variant Annotation Integrator (http://genome.ucsc.edu/cgi-bin/hgVai), Ensembl’s Variant Effect Predictor (McLaren et al. 2010), ANNOVAR (Wang et al. 2010), SnpEff/SnpSift (Cingolani et al. 2012) and HGMD (Stenson et al. 2014), among others.
Here we report a recent major update of dbNSFP to v3.0. The core SNVs have been rebuilt based on the human reference sequence version hg38. It now includes 82,832,027 nsSNVs and ssSNVs. An attached database called dbscSNV (Jian et al. 2014) which compiled all potential human scSNVs (15,030,459 in total) is distributed along with dbNSFP, and can be searched using the same companion search program of dbNSFP. Compared to v2.0, the new version added eleven new prediction scores: MetaSVM and MetaLR (Dong et al. 2015), CADD (Kircher et al. 2014), VEST3 (Carter et al. 2013), PROVEAN (Choi et al. 2012), 4× fitCons scores (Gulko et al. 2015), fathmm-MKL (Shihab et al. 2015) and DANN (Quang et al. 2015), two conservation scores: 2× phastCons (Siepel et al. 2005), and allele frequencies from the UK10K cohorts (The UK10K Consortium 2015) and the Exome Aggregation Consortium (ExAC, http://exac.broadinstitute.org/), among others. Many prediction scores and resources have been updated. Details of the updates and preliminary analyses of the functional prediction scores and conservation scores are reported in the following sections.
NEW AND UPDATED FUNCTIONAL ANNOTATIONS
To keep up with the updates of new gene models, we have rebuilt our backbone nsSNVs and ssSNVs using the GENCODE 22, which is based on human reference sequence version hg38. As described previously (Liu et al. 2013), we artificially “mutated” each non-N reference allele to the three alternative alleles. Then we checked the “mutations” against the gene models and collected all those nsSNVs or ssSNVs (on the first two and last two nucleotide sites of an intron) into our database. To balance false positives and false negatives of the gene models, we included putative genes but excluded genes with incomplete 5′ ends. Genes on the mitochondrial genome has been included for the first time. This resulted in 80,622,428 nsSNVs and 2,209,599 ssSNVs in the database. Genome positions were converted to corresponding coordinates in hg19 (no missing) and then in hg18 (0.09% missing) using the liftOver tool of the UCSC Genome Browser (Rosenbloom et al. 2015). Please note that there are a few SNVs whose coordinates in hg38 and hg19 (hg18) have inconsistent chromosome numbers.
Two new nsSNV-focused prediction scores, PROVEAN and VEST 3.0 have been added, which were kindly provided by Drs. Yongwook Choi and Rachel Karchin, respectively. PROVEAN scores range from −14 to 14 in dbNSFP, with a lower score indicating a higher likelihood to be deleterious. PROVEAN also provides binary predictions (Neutral versus Damaging) with a score cut-off of −2.5. Multiple scores and predictions corresponding to multiple transcripts of the same gene are separated by “;” and the transcript IDs are presented in the Ensembl_proteinid column. VEST 3.0 scores range from 0 to 1 with a higher score indicating a higher likelihood to be deleterious. VEST does not provide binary predictions. Multiple scores are separated by “;” and the corresponding transcript IDs are presented in the Transcript_id_VEST3 column.
Recently, several “general” prediction scores have been proposed, which incorporated DNA/protein sequence features as well as epigenomic signals and provide deleteriousness predictions for any SNV in the human genome, coding or non-coding. Examples of such scores include CADD, fitCons, fathmm-MKL and DANN. Among them CADD, fathmm-MKL and DANN provide predictions for a SNV while fitCons is more coarse-grained and has predictions at the genome position level as a conservation score. We included the above mentioned four “general” prediction scores in dbNSFP v3.0 to provide more choices for our users. fathmm-MKL separated their scores for coding and non-coding SNVs and we included those designed for coding SNVs.
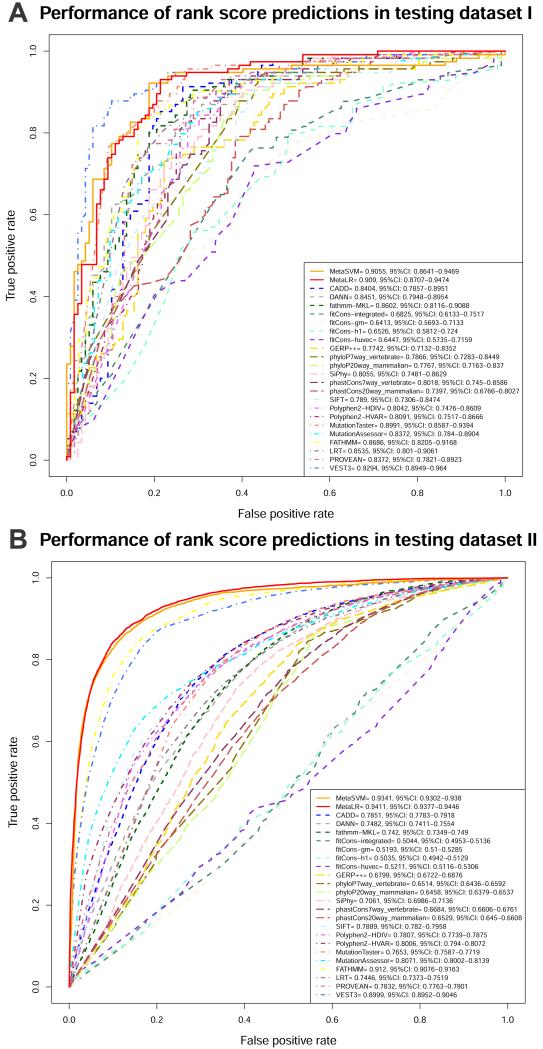
Although having more prediction scores for an nsSNV has an advantage of providing additional perspectives, sometime a consensus prediction is also useful in practice. We recently developed two ensemble scores, MetaLR and MetaSVM, based on 10 component scores (SIFT, PolyPhen-2 HDIV, PolyPhen-2 HVAR, GERP++, MutationTaster, Mutation Assessor, FATHMM, LRT, SiPhy, PhyloP) and the maximum frequency observed in the 1000 genomes populations (Dong et al. 2015). Based on our comparison, the two ensemble scores outperform all their component scores. MetaLR achieved the highest separation power (AUC = 0.92 and 0.94 for testing dataset I and II, respectively) followed by MetaSVM (AUC = 0.91 and 0.93 for testing dataset I and II, respectively).
To make the functional prediction scores and conservation scores in the dbNSFP more comparable to each other, we created a rank score for each of them. First, we converted scores if necessary to make them monotonic in the same direction (a higher score indicating more likely to be damaging, see Suppporting Information for details). Then for each type of score (such as a converted SIFT score) we ranked all the (converted SIFT) scores in the dbNSFP and the rank score is the ratio of the rank (or tied rank) of the (converted SIFT) score over the total number of (converted SIFT) scores in the dbNSFP. In the case when an nsSNV has multiple scores due to multiple transcripts, only the most deleterious one was used in ranking. Therefore, a rank score is always between 0 and 1 and a score of 0.9 means it is more likely to be damaging than 90% of all potential nsSNVs predicted by that method.
Many prediction scores and conservation scores have been updated from the dbNSFP v2.0 to v3.0: SIFT to the version based on Ensembl 66; MutationTaster to MutationTaster2 (Schwarz et al. 2014); FATHMM to v2.3; phyloP to phyloP7way_vertebrate and phyloP20way_mammalian (both based on hg38); phastCons to phastCons7way_vertebrate and phastCons20way_mammalian (both based on hg38). As many prediction scores provide multiple (often different) scores or predictions for the same nsSNV due to multiple transcripts of the same gene, we included those transcript specific predictions in this new version.
Besides prediction scores and conservation scores, many annotation resources have been added or updated. Noticeably, allele frequencies from the UK10K cohorts and the Exome Aggregation Consortium (ExAC) have been added; those of human populations in the 1000 Genomes Project have been updated to the phase 3 data set. Clinvar (Landrum et al. 2014), dbSNP (Sherry et al. 2001) 142 and phenotypes of mouse and zebra fish homologs have been added. More details on the resources and their version in dbNSFP can be found in the Supporting Information and the readme file distributed with the database file.
The dbNSFP v3.0 is provided in two branches: v3.0a and v3.0c. The former includes all the prediction scores and annotation resources while the latter excludes prediction scores that require licenses for commercial usages, such as VEST, CADD and DANN. The whole database is in plain text format. No database management system is needed. A Java companion search program along with the database files are freely available at https://sites.google.com/site/jpopgen/dbNSFP. Alternatively, dbNSFP can be queried via MyVariant.info web service (http://myvariant.info/), either calling its API directly or using its Python client (Mark 2015a) and R client from Bioconductor (Mark 2015b).
A COMPARISON OF FUNCTIONAL PREDICTION SCORES AND CONSERVATION SCORES
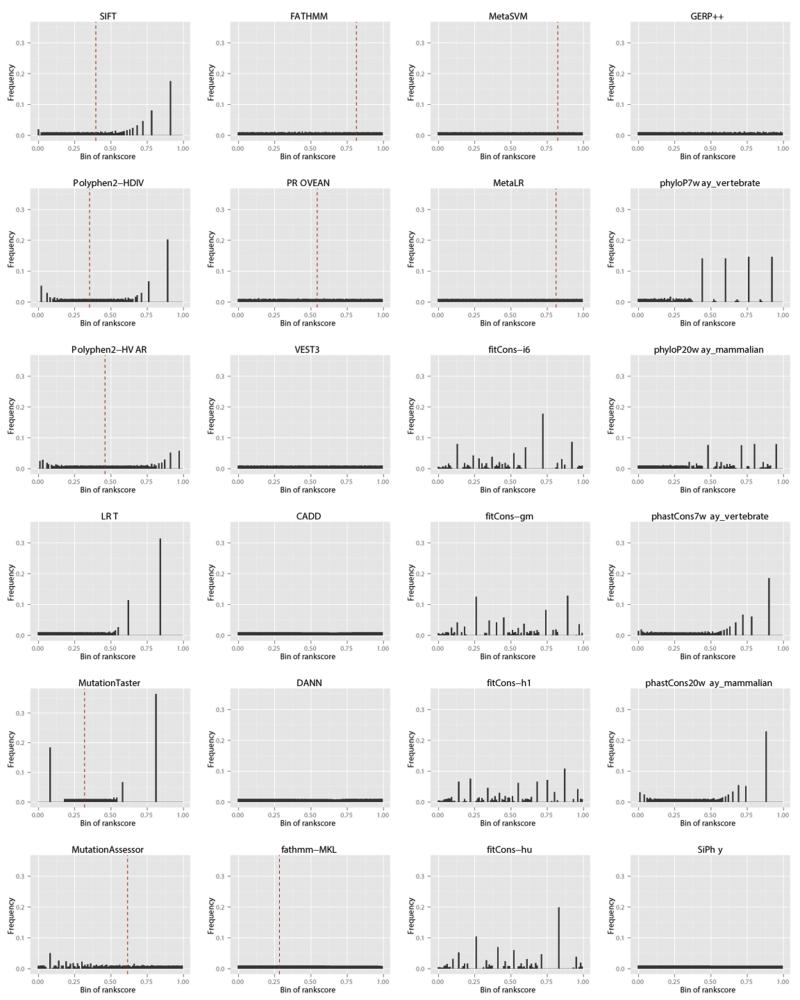
We conducted some preliminary analyses comparing the 24 functional prediction scores and conservation scores based on the 80,622,428 nsSNVs in dbNSFP v3.0. A summary of the 24 scores is presented in Table 1. nsSNV-focused scores typically have a higher missingness percentage in the dbNSFP (a minimum of 2.15% for MutationTaster to a maximum of 16.68% for LRT) compared to “general” prediction scores or conservation scores (a minimum of <0.01% for CADD to a maximum of 3.97% for fitCons), largely due to gene model inconsistency (Table 2). As to the distributions of the rank scores (Figure 1), while some rank scores are more or less evenly distributed, such as MutationAssessor, FATHMM, PROVEAN, VEST3, CADD, DANN, fathmm-MKL, MetaSVM, MetaLR, GERP++ and SiPhy, others are more sparse and have high spikes, suggesting a large amount of raw scores having tied ranks in the database.
Table 1. A summary of functional prediction scores and conservation scores.
| Score | Training data | Information used | Prediction model |
|---|---|---|---|
| PolyPhen2- HDIV |
5564 Mendelian disease mutations and 7539 divergence SNVs from close mammalian homolog proteins |
eight sequence-based and three structure-based predictive features |
naive Bayes classifier |
| PolyPhen2- HVAR |
22196 disease associated SNVs and 21119 common SNVs |
same as above | same as above |
| SIFT | 1750 deleterious and 2254 tolerant nsSNVs of E. coli LacI gene |
sequence homology based on PSI-BLAST |
position specific scoring matrix |
| Mutation Taster |
SNVs from 1000 G (1000 Genomes Project), HGMD |
conservation, splice site, mRNA features, protein features; regulatory features |
naive Bayes classifier |
| LRT | coding sequences of 32 vertebrate species |
sequence homology | likelihood ratio test of codon neutrality |
| Mutation Assessor |
SNVs from COSMIC database | sequence homology of protein families and sub- families within and between species |
combinatorial entropy formalism |
| FATHMM | SNVs from HGMD and UniProt | sequence homology | hidden Markov models |
| PROVEAN | SNVs from UniProt/HUMSAVAR | sequence homology | Delta alignment score |
| VEST3 | SNVs from HGMD and the Exome Sequencing Project |
86 sequence features | Random Forest |
| fathmm-MKL coding |
SNVs from HGMD and 1000G | conservation, epigenomic signals |
multiple kernel learning |
| MetaSVM | 36,192 SNVs from UnPprot | 9 prediction scores and allele frequencies in 1000G |
radial kernel support vector machine |
| MetaLR | same as above | same as above | logistic regression |
| CADD | 16,627,775 “observed” variants and 49,407,057 “simulated” variants |
63 annotations (949 features) | linear kernel support vector machine |
| DANN | same as above | same as above | deep neural network |
| fitCons-i6 | genomes of 54 unrelated human individuals |
epigenomic signals of GM12878, H1-hESC and HUVEC |
INSIGHT (Inference of Natural Selection from Interspersed Genomically coHerent elemenTs) |
| fitCons-gm | same as above | epigenomic signals of GM12878 |
same as above |
| fitCons-h1 | same as above | epigenomic signals of H1-hESC |
same as above |
| fitCons-hu | same as above | epigenomic signals of HUVEC |
same as above |
| SiPhy | genomes of 29 mammals | multiple alignments | inferring nucleotide substitution pattern per site |
| GERP++ | genomes of 34 mammals | multiple alignments and phylogenetic tree |
maximum likelihood evolutionary rate estimation |
| phyloP7way _vertebrate |
genomes of 7 vertebrates | same as above | distributions of the number of substitutions based on a phylogenetic hidden Markov model |
| phyloP20way _mammalian |
genomes of 20 mammals | same as above | same as above |
| phastCons7way _vertebrate |
genomes of 7 vertebrates | same as above | two-state phylogenetic hidden Markov model |
| phastCons20way _mammalian |
genomes of 20 mammals | same as above | same as above |
Table 2. Number of nsSNVs in each chromosome and the percentages of missingness of functional prediction scores and conservation scores.
| Chr | nsSNV | SIFT | Poly phen2 |
LRT | Mutation Taster |
Mutation Assessor |
FATHMM | PROVEAN | VEST3 | CADD | DANN | fathmm -MKL |
MetaSVM MetaLR |
fitCons | GERP++ | phyloP 7way |
phyloP 20way |
Phast Cons 7way |
Phast Cons 20way |
SiPhy |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | 23145 | 64.21 | 100.00 | 100.00 | 6.35 | 100.00 | 13.07 | 13.04 | 100.00 | 1.06 | 100.00 | 100.00 | 100.00 | 100.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 100.00 |
| 1 | 8085329 | 10.97 | 10.22 | 15.11 | 1.53 | 12.53 | 15.40 | 10.51 | 7.21 | 0.00 | 0.00 | 0.00 | 7.56 | 0.00 | 0.60 | 0.53 | 0.24 | 0.53 | 0.24 | 1.42 |
| 2 | 5960951 | 9.20 | 10.87 | 19.41 | 1.84 | 11.82 | 14.01 | 8.76 | 7.11 | 0.00 | 0.00 | 0.00 | 7.04 | 0.00 | 0.38 | 0.04 | 0.05 | 0.04 | 0.05 | 0.77 |
| 3 | 4647575 | 8.30 | 8.47 | 12.87 | 1.34 | 11.14 | 13.09 | 8.01 | 6.74 | 0.00 | 0.00 | 0.00 | 6.67 | 0.00 | 0.35 | 0.06 | 0.04 | 0.06 | 0.04 | 0.65 |
| 4 | 3238883 | 8.96 | 11.67 | 12.24 | 2.04 | 11.54 | 13.60 | 8.52 | 7.14 | 0.00 | 0.00 | 0.00 | 7.23 | 0.00 | 0.39 | 0.20 | 0.03 | 0.20 | 0.03 | 2.19 |
| 5 | 3718178 | 8.67 | 8.76 | 16.17 | 0.74 | 10.92 | 12.36 | 8.17 | 6.95 | 0.00 | 0.00 | 0.00 | 7.67 | 0.00 | 0.12 | 0.09 | 0.07 | 0.09 | 0.07 | 0.54 |
| 6 | 4123833 | 9.43 | 9.77 | 12.46 | 2.92 | 12.06 | 12.89 | 8.58 | 6.70 | 0.00 | 0.00 | 0.00 | 7.75 | 0.02 | 0.16 | 0.01 | 0.13 | 0.01 | 0.13 | 0.70 |
| 7 | 3797070 | 12.30 | 11.09 | 19.95 | 4.36 | 14.63 | 17.17 | 11.64 | 8.05 | 0.00 | 0.00 | 0.00 | 9.35 | 0.00 | 1.02 | 0.24 | 0.06 | 0.24 | 0.06 | 2.34 |
| 8 | 2706162 | 11.11 | 10.03 | 13.94 | 3.13 | 13.95 | 17.21 | 10.98 | 7.09 | 0.00 | 0.00 | 0.00 | 8.23 | 0.00 | 0.50 | 0.15 | 0.08 | 0.15 | 0.08 | 1.05 |
| 9 | 3168302 | 9.69 | 9.38 | 13.34 | 1.87 | 10.95 | 14.77 | 9.24 | 7.31 | 0.00 | 0.00 | 0.00 | 7.08 | 0.00 | 0.14 | 0.02 | 0.00 | 0.02 | 0.00 | 0.48 |
| 10 | 3114019 | 9.98 | 9.88 | 12.36 | 2.01 | 12.67 | 14.86 | 9.68 | 7.51 | 0.00 | 0.00 | 0.00 | 8.01 | 0.00 | 0.54 | 0.04 | 0.00 | 0.04 | 0.00 | 1.05 |
| 11 | 4735763 | 9.68 | 9.83 | 16.46 | 2.62 | 12.41 | 14.25 | 9.10 | 7.49 | 0.00 | 0.00 | 0.00 | 7.90 | 0.00 | 0.08 | 0.00 | 0.00 | 0.00 | 0.00 | 0.59 |
| 12 | 4223205 | 8.71 | 9.62 | 12.89 | 0.57 | 12.23 | 13.74 | 8.05 | 6.41 | 0.00 | 0.00 | 0.06 | 6.36 | 0.00 | 0.06 | 0.03 | 0.03 | 0.03 | 0.03 | 0.59 |
| 13 | 1470936 | 12.14 | 11.00 | 10.19 | 2.87 | 12.86 | 16.69 | 11.83 | 7.01 | 0.00 | 0.00 | 0.00 | 8.91 | 0.00 | 0.12 | 0.01 | 0.00 | 0.01 | 0.00 | 0.91 |
| 14 | 2546323 | 8.74 | 10.61 | 13.94 | 1.51 | 12.55 | 14.04 | 8.01 | 7.12 | 0.00 | 0.00 | 0.00 | 7.68 | 0.00 | 0.23 | 0.16 | 0.11 | 0.16 | 0.11 | 0.59 |
| 15 | 2790630 | 9.17 | 11.82 | 15.92 | 2.22 | 13.26 | 13.30 | 8.63 | 7.05 | 0.00 | 0.00 | 0.00 | 9.99 | 0.00 | 1.28 | 0.11 | 0.07 | 0.11 | 0.07 | 2.00 |
| 16 | 3434017 | 11.11 | 11.91 | 17.86 | 2.81 | 15.81 | 15.34 | 9.85 | 7.37 | 0.00 | 0.00 | 0.00 | 10.00 | 0.00 | 2.24 | 1.44 | 0.37 | 1.44 | 0.37 | 3.36 |
| 17 | 4608227 | 17.72 | 10.19 | 16.23 | 1.32 | 12.57 | 17.14 | 17.43 | 7.28 | 0.00 | 0.00 | 0.00 | 8.85 | 0.00 | 0.50 | 0.08 | 0.03 | 0.08 | 0.03 | 1.17 |
| 18 | 1286209 | 12.95 | 11.07 | 17.43 | 0.59 | 11.82 | 12.44 | 12.37 | 8.02 | 0.00 | 0.00 | 0.00 | 8.76 | 0.00 | 0.08 | 0.06 | 0.02 | 0.06 | 0.02 | 1.01 |
| 19 | 5373215 | 16.72 | 11.65 | 35.61 | 2.56 | 14.12 | 14.97 | 16.36 | 9.63 | 0.00 | 0.00 | 0.00 | 10.25 | 0.00 | 0.22 | 0.04 | 0.02 | 0.04 | 0.02 | 1.73 |
| 20 | 1930545 | 8.16 | 9.28 | 10.60 | 0.98 | 11.73 | 13.47 | 7.77 | 7.61 | 0.00 | 0.00 | 0.00 | 7.08 | 0.00 | 0.19 | 0.04 | 0.04 | 0.04 | 0.04 | 0.55 |
| 21 | 790792 | 10.06 | 9.03 | 16.10 | 0.46 | 13.34 | 12.77 | 8.05 | 7.21 | 0.00 | 0.00 | 0.00 | 6.59 | 0.00 | 0.24 | 0.11 | 0.22 | 0.11 | 0.22 | 1.35 |
| 22 | 1668348 | 10.95 | 9.63 | 13.39 | 1.41 | 12.17 | 17.38 | 10.76 | 6.91 | 0.00 | 0.00 | 0.00 | 6.64 | 0.00 | 0.46 | 0.14 | 0.03 | 0.14 | 0.03 | 1.25 |
| X | 3010269 | 11.94 | 11.13 | 16.72 | 1.27 | 13.59 | 14.48 | 11.70 | 7.03 | 0.00 | 0.00 | 0.00 | 7.39 | 100.00 | 0.42 | 0.12 | 0.06 | 0.12 | 0.06 | 2.76 |
| Y | 170502 | 15.38 | 24.85 | 96.11 | 100.00 | 22.95 | 20.52 | 13.93 | 11.34 | 0.00 | 0.00 | 0.00 | 88.33 | 100.00 | 38.26 | 3.78 | 2.81 | 3.78 | 2.81 | 100.00 |
|
| ||||||||||||||||||||
| Total | 80622428 | 10.88 | 10.37 | 16.68 | 2.15 | 12.66 | 14.64 | 10.36 | 7.37 | 0.00 | 0.03 | 0.03 | 8.18 | 3.97 | 0.54 | 0.19 | 0.08 | 0.19 | 0.08 | 1.50 |
Figure 1. Distributions of the rank scores of the prediction and conservation scores based on 100 bins between 0 and 1.
Dash lines indicate the cut-offs for binary predictions.
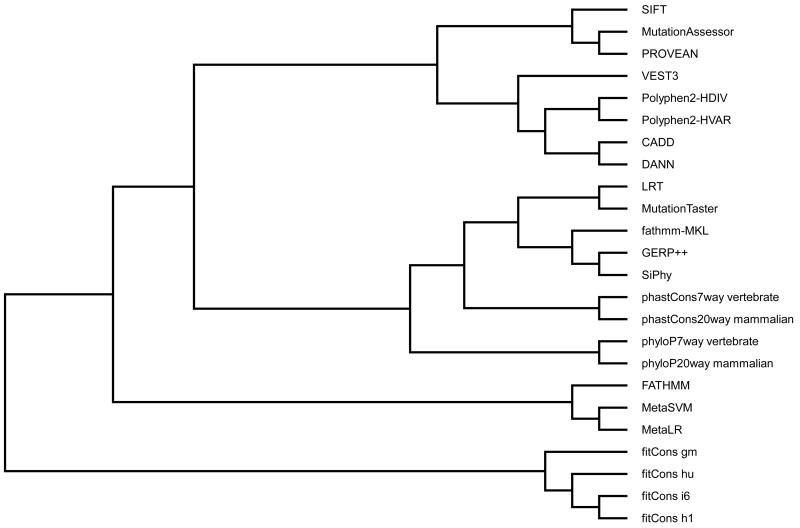
Knowing the correlation between scores helps researchers to weight the predictions from multiple methods. For each pair of scores, we calculated the Pearson’s correlation coefficient (r) between their rank scores as a measure of correlation (Table 3). Some of the highly correlated (r > 0.7) pairs either use the same training data or use the same method, such as Polyphen2-HVIR and Polyphen2-HVAR, MetaSVM and MetaLR, CADD and DANN, fitCons-i6 and fitCons-h1, phyloP7way_vertebrate and phyloP20way_mammalian, and phastCons7way_vertebrate and phastCons20way_mammalian. The others are less obvious, such as FATHMM and MetaLR (or MetaSVM), CADD and Polyphen2-HVAR (or Polyphen2-HDIV), CADD and VEST3, fathmm-MKL and GERP++, fathmm-MKL and SiPhy, and GERP++ and SiPhy. fitCons scores have low correlations (r < 0.3) with other scores. There is even a negative (though close to 0) correlation between fitCons-gm and FATHMM. To provide an entire perspective, we clustered the scores using UPGMA (Unweighted Pair Group Method with Arithmetic Mean) with 1-r as distance between scores (Figure 2). The scores fall in four clusters. The largest one includes LRT, MutationTaster, fathmm-MKL, GERP++, SiPhy, 2× phastCons scores and 2× phyloP scores. All conservation scores are in this cluster suggesting that the prediction scores in this cluster may put a heavy weight on conservation information. The second largest cluster includes SIFT, MutationAssessor, PROVEAN, VEST3, CADD, DANN and 2× Polyphen2 scores. The smallest cluster includes FATHMM, MetaSVM and MetaLR, which are highly correlated among themselves while having low to moderate correlation with other scores. Finally, the 4× fitCons scores form their own cluster and serve as an out group.
Table 3. Pearson’s correlation coefficients between rank scores (upper-triangle) and the ratio of binary predictions’ agreement between scores (lower-triangle).
| Score | SIFT | HDIV | HVAR | LRT | MT | MA | FAT | PROV | MKL | SVM | LR | VEST3 | CADD | DANN | i6 | gm | h1 | hu | GERP | phP7 | phP20 | phC7 | phC20 | SiPhy |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SIFT | - | 0.63 | 0.63 | 0.36 | 0.36 | 0.59 | 0.14 | 0.63 | 0.36 | 0.40 | 0.41 | 0.53 | 0.63 | 0.50 | 0.02 | 0.01 | 0.02 | 0.02 | 0.28 | 0.25 | 0.22 | 0.21 | 0.18 | 0.31 |
| HDIV | 0.75 | - | 0.97 | 0.50 | 0.48 | 0.62 | 0.14 | 0.64 | 0.48 | 0.46 | 0.50 | 0.66 | 0.72 | 0.62 | 0.07 | 0.05 | 0.07 | 0.06 | 0.42 | 0.31 | 0.28 | 0.31 | 0.28 | 0.46 |
| HVAR | 0.74 | 0.89 | - | 0.53 | 0.51 | 0.64 | 0.16 | 0.66 | 0.51 | 0.48 | 0.53 | 0.68 | 0.73 | 0.62 | 0.07 | 0.06 | 0.08 | 0.06 | 0.43 | 0.32 | 0.30 | 0.34 | 0.31 | 0.48 |
| LRT | 0.66 | 0.71 | 0.72 | - | 0.66 | 0.44 | 0.18 | 0.46 | 0.68 | 0.37 | 0.42 | 0.64 | 0.55 | 0.50 | 0.22 | 0.19 | 0.22 | 0.21 | 0.55 | 0.38 | 0.36 | 0.59 | 0.53 | 0.59 |
| MT | 0.66 | 0.72 | 0.70 | 0.80 | - | 0.43 | 0.22 | 0.45 | 0.70 | 0.38 | 0.45 | 0.65 | 0.66 | 0.54 | 0.24 | 0.18 | 0.23 | 0.21 | 0.59 | 0.41 | 0.41 | 0.58 | 0.55 | 0.62 |
| MA | 0.68 | 0.66 | 0.72 | 0.65 | 0.61 | - | 0.16 | 0.69 | 0.43 | 0.49 | 0.53 | 0.57 | 0.60 | 0.50 | 0.05 | 0.05 | 0.05 | 0.05 | 0.31 | 0.28 | 0.24 | 0.25 | 0.20 | 0.36 |
| FAT | 0.46 | 0.43 | 0.50 | 0.48 | 0.44 | 0.61 | - | 0.16 | 0.22 | 0.71 | 0.85 | 0.22 | 0.19 | 0.16 | 0.02 | −0.01 | 0.02 | 0.01 | 0.16 | 0.12 | 0.11 | 0.18 | 0.16 | 0.18 |
| PROV | 0.73 | 0.70 | 0.74 | 0.67 | 0.65 | 0.76 | 0.56 | - | 0.46 | 0.43 | 0.46 | 0.64 | 0.66 | 0.50 | 0.07 | 0.06 | 0.07 | 0.06 | 0.35 | 0.32 | 0.28 | 0.28 | 0.24 | 0.39 |
| MKL | 0.65 | 0.71 | 0.68 | 0.76 | 0.86 | 0.56 | 0.40 | 0.61 | - | 0.39 | 0.47 | 0.67 | 0.62 | 0.56 | 0.30 | 0.26 | 0.31 | 0.29 | 0.76 | 0.56 | 0.55 | 0.63 | 0.60 | 0.76 |
| SVM | 0.53 | 0.51 | 0.60 | 0.55 | 0.50 | 0.71 | 0.88 | 0.65 | 0.44 | - | 0.87 | 0.46 | 0.47 | 0.37 | 0.04 | 0.02 | 0.04 | 0.04 | 0.32 | 0.24 | 0.21 | 0.27 | 0.23 | 0.37 |
| LR | 0.52 | 0.50 | 0.58 | 0.53 | 0.48 | 0.69 | 0.90 | 0.63 | 0.44 | 0.96 | - | 0.51 | 0.52 | 0.44 | 0.08 | 0.04 | 0.08 | 0.07 | 0.39 | 0.29 | 0.27 | 0.34 | 0.30 | 0.43 |
| VEST3 | - | - | - | - | - | - | - | - | - | - | - | - | 0.73 | 0.57 | 0.18 | 0.14 | 0.18 | 0.16 | 0.60 | 0.45 | 0.44 | 0.51 | 0.47 | 0.61 |
| CADD | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.74 | 0.19 | 0.15 | 0.18 | 0.16 | 0.57 | 0.37 | 0.38 | 0.48 | 0.49 | 0.58 |
| DANN | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.19 | 0.15 | 0.18 | 0.16 | 0.52 | 0.34 | 0.36 | 0.45 | 0.46 | 0.53 |
| i6 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.67 | 0.74 | 0.68 | 0.23 | 0.15 | 0.17 | 0.26 | 0.27 | 0.22 |
| gm | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.57 | 0.60 | 0.20 | 0.13 | 0.14 | 0.21 | 0.22 | 0.18 |
| h1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.57 | 0.23 | 0.15 | 0.17 | 0.25 | 0.25 | 0.22 |
| hu | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.22 | 0.14 | 0.16 | 0.24 | 0.24 | 0.21 |
| GERP | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.61 | 0.64 | 0.57 | 0.55 | 0.80 |
| phP7 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.74 | 0.51 | 0.44 | 0.43 |
| phP20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.44 | 0.50 | 0.43 |
| phC7 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.82 | 0.54 |
| phC20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
HDIV: Polyphen2_HDIV; HVAR: Polyphen2_HVAR; MT: MutationTaster; MA: MutationAssessor; FAT: FATHMM; PROV: PROVEAN; MKL: fathmm-MKL; SVM: MetaSVM; LR: MetaLR; i6: fitCons-i6; gm: fitCons-gm; h1: fitCons-h1; hu: fitCons-hu; GERP: GERP++; phP7: phyloP7way_vertebrate; phP20: phyloP20way_mammalian; phC7: phastCons7way_vertebrate; phC20: phastCons20way_mammalian
Figure 2. UPGMA dendrogram of the prediction and conservation scores.
Among the 24 scores, eleven provide binary predictions (deleterious or tolerated) for nsSNVs. Comparison of their prediction agreement shows that majority of the pairs have low to moderate agreement rate (< 70%) (Table 3). The lowest agreement rate (40%) is between FATHMM and fathmm-MKL (coding score). The highest agreement is between MetaLR and MetaSVM (96%) followed by FATHMM and MetaLR (90%) and the two Polyphen2 scores (89%).
Finally, we compared the performance of the nsSNV prediction scores, “general” prediction scores and conservation scores in dbNSFP v3.0 using their rank scores. We re-used the testing dataset I and testing dataset II from Dong et al. (2015) after removing nsSNVs that causing different amino acid changes in different transcripts according to GENCODE 22, which resulting in 115 true positives and 117 true negatives in testing data set I (Supp. Table S1) and 5,979 true positives and 13,025 true negatives in testing data set II (Supp. Table S2), respectively. The performance of the scores was measured using receiver operating characteristic (ROC) curve and area under the curve (AUC) (Figure 3). We found that, the two ensemble rank scores, MetaSVM and MetaLR, achieved excellent prediction accuracy (AUC > 0.9) in both testing datasets. Two other scores that reached excellent prediction accuracy in either testing dataset include VEST3 (AUC=0.9294 in testing data set I) and FATHMM (AUC=0.912 in testing data set II). The results also showed that those recently proposed “general” scores did not stand out as to nsSNV deleteriousness prediction, although some of those, such as CADD, DANN and fathmm-MKL, showed comparable performance as popular nsSNV prediction scores Polyphen2 and SIFT.
Figure 3. ROC curves for the functional prediction scores and conservation scores in dbNSFP v3.0 with testing data set I (A) and II (B).
ATTACHED DATABASE
Recently we developed a method for predicting the splice-altering effect of a scSNV (a SNV located within splicing consensus regions) (Jian et al. 2014). The resulting two ensemble prediction scores (ada_score and rf_score) and predictions were pre-computed for all potential scSNVs in the human genome based on RefSeq release 62 and Ensembl release 73. Those scores along with related annotations were compiled into a plain text database called dbscSNV and serves as an attached database for the dbNSFP. It is freely available for download at https://sites.google.com/site/jpopgen/dbNSFP. The companion Java search program distributed with dbNSFP v3.0 supports searching dbscSNV and SPIDEX (Xiong et al. 2015), another prediction tool for splice-altering SNVs, along with dbNSFP using the “-s” option.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Yongwook Choi, Rachel Karchin and Dominik Seelow for kindly providing the PROVEAN/SIFT, VEST3 and MutationTaster2 scores, respectively. We thank Dr. CS (Jonathan) Liu for providing hosting space. We thank Jason J. Corneveaux, Chris Gillies, Mihail Halachev, Jocob Hsu, James Ireland, Xueqiu Jian, Seung-Tae Lee, Alexander Li, John McGuigan, Zena Ng, Adam Novak, Kirill Prusov, and Lishuang Shen, for reporting bugs and providing suggestions. We thank Sara Barton for copy editing this manuscript. This project was supported by the US National Institutes of Health (5RC2HL102419 and U54HG003273).
Footnotes
The authors declare that they have no conflict of interests.
REFERENCES
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter H, Douville C, Stenson PD, Cooper DN, Karchin R. Identifying Mendelian disease genes with the variant effect scoring tool. BMC Genomics. 2013;14(Suppl 3):S3. doi: 10.1186/1471-2164-14-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PloS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a High Fraction of the Human Genome to be under Selective Constraint Using GERP++ PLoS Comput Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Wei P, Jian X, Gibbs R, Boerwinkle E, Wang K, Liu X. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, O’Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, Gabriel S, Rieder MJ, Altshuler D, Shendure J, Nickerson DA, Bamshad MJ, et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M, Guttman M, Clamp M, Zody MC, Friedman N, Xie X. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics. 2009;25:i54–i62. doi: 10.1093/bioinformatics/btp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulko B, Hubisz MJ, Gronau I, Siepel A. A method for calculating probabilities of fitness consequences for point mutations across the human genome. Nat. Genet. 2015;47:276–283. doi: 10.1038/ng.3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian X, Boerwinkle E, Liu X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014;42:13534–13544. doi: 10.1093/nar/gku1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, Ward LD, Lowe CB, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jian X, Boerwinkle E. dbNSFP: A lightweight database of human nonsynonymous SNPs and their functional predictions. Hum. Mutat. 2011;32:894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum. Mutat. 2013;34:E2393–2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark A. MyVariant.py: MyVariant.info Python client. 2015a https://pypi.python.org/pypi/myvariant.
- Mark A. MyVariant.R: MyVariant.info R client. 2015b http://bioconductor.org/packages/myvariant/
- McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, Hart E, Suner M-M, et al. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quang D, Chen Y, Xie X. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinforma. Oxf. Engl. 2015;31:761–763. doi: 10.1093/bioinformatics/btu703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, Harte RA, Heitner S, et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015;43:D670–681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GLA, Edwards KJ, Day INM, Gaunt TR. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihab HA, Rogers MF, Gough J, Mort M, Cooper DN, Day INM, Gaunt TR, Campbell C. An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics. 2015;31:1536–1543. doi: 10.1093/bioinformatics/btv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Pollard KS, Haussler D. RECOMB 2006. LNCS (LNBI) Vol. 3909. Springer; Heidelberg: 2006. New methods for detecting lineage-specific selection; pp. 190–205. [Google Scholar]
- Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UK10K Consortium. The UK10K project identifies rare variants in health and disease. Nature advance online publication. 2015 Sep 14; doi: 10.1038/nature14962. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong HY, Alipanahi B, Lee LJ, Bretschneider H, Merico D, Yuen RKC, Hua Y, Gueroussov S, Najafabadi HS, Hughes TR, Morris Q, Barash Y, et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347:1254806. doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.