Abstract
Olfactory receptors (ORs) are chemosensors that are responsible for one’s sense of smell. In addition to this specialized role in the nose, recent evidence suggests that ORs are also found in a variety of additional tissues including the kidney. As this list of renal ORs continues to expand, it is becoming clear that these ORs play important roles in renal and whole-body physiology, including a novel role in blood pressure regulation. In this review, we highlight important considerations that are crucial when studying ORs, and present the current literature on renal ORs and their emerging relevance in maintaining renal function.
Keywords: olfactory receptors, blood pressure, kidney, adenylate cyclase 3, odorant receptor, ligand
Introduction
Olfactory receptors (ORs) are 7 transmembrane domain G protein-coupled chemosensors that mediate the sense of smell in the nose. Localized to the cilia on the tips of olfactory sensory neurons (OSNs), they exhibit exquisite sensitivity to a diverse range of volatile odorants [1]. First identified in 1991 by Linda Buck and Richard Axel, it is now known that there are more than 1000 OR genes in mice and roughly 350 in humans making them the largest gene family in the genome [2–4]. Despite the diversity of molecules that activate these receptors, they all signal through a conserved downstream signaling pathway whereby stimulation by a volatile odorant leads to increased intracellular cAMP which depolarizes the OSN [5–7].
In the seminal paper that first described ORs, an assumption was made that OR expression would be restricted to OSNs given their sensory function [1]. While it is true that the expression level of ORs is highest within the olfactory epithelium (OE), it is now clear that at least some ORs are expressed in a variety of additional tissues. With new techniques such as whole transcriptome sequencing, the list of ‘ectopic ORs’ has greatly expanded in the last few years, and these sites now include the GI tract, muscle, heart, pancreas, liver, lung and the skin [8–19]. While the physiological functions that these ORs play in non-sensory tissues is an ongoing area of research, it is evident that ORs are serving important roles in physiology. Notably, murine and human ORs have been found in sperm and play a key role in sperm chemotaxis [10;16]. In addition, MOR23 is found in muscle myocytes where it regulates migration and adhesion and contributes to muscle regeneration [11]. The functional importance of these ORs clearly indicates that these receptors are likely playing novel roles in a variety of different tissues.
Given its sensory functions, the kidney appears to be an ideal organ to take advantage of OR signaling. The epithelial lining of the kidney comes in contact with a diverse range of chemicals, and it must keep careful track of the composition of the tubular fluid as it moves through the different segments of the nephron. The kidney adjusts its filtration, reabsorption and secretion rates in response to changes in electrolytes and metabolites; thus, it is well-suited to take advantage of specialized chemosensors such as ORs, in order to maintain homeostasis. In this review, we will begin by outlining challenges specific to OR biology which shape these investigations, and subsequently will focus on the identification of individual renal ORs and their roles in renal physiology.
Special considerations and challenges of studying ORs
The ‘ectopic’ expression of sensory receptors such as ORs is an exciting and emerging field. However, there are a number of experimental challenges that are specific to the study of ORs, and it is imperative that one understands these challenges in order to properly evaluate and understand data regarding OR expression in the kidney and elsewhere. Therefore, we will begin by briefly reviewing OR-specific ‘best practices’. First, very few antibodies for ORs are available. While G protein-coupled receptors in general tend not to be particularly antigenic, ORs are persistently difficult to generate antibodies against (despite enormous efforts in this area). Out of the 1000 murine ORs and 350 human ORs, there are only a few antibodies which appear reliable: MOR28 [20–22], M50 [23], M71 [23], MOR256-17 [24], MOR11-4 [22], mOR37A-E [24], I7 [21], mOR-EG [21], MOR10 [25], MOR18-2 [26], MOR256-50 [27] and MOR42-3 [27]. Of these, the only OR expressed in the kidney is MOR18-2, although this antibody strongly cross-reacts with a mitochondrial protein when used outside the OE [26]. It is important to note that although numerous additional OR antibodies are readily available for purchase via multiple vendors and companies, to our knowledge only two commercially available OR antibodies have been successfully validated (MOR256-50 and MOR42-3 [27]) – note that an antibody for MOR127-1 could not be validated in this same reference). We and others have attempted to validate a great number of additional commercial antibodies, to no avail. Therefore, if one wishes to employ an OR antibody which has not been previously published, proper validation is absolutely critical – typically, at a minimum, a demonstration that at least one (preferably both) of the following are true: (a) the antibody gives the expected staining pattern in the OE (if the expression pattern for that particular OR is not known, in situ hybridization coupled with immunohistochemistry would be the proper demonstration of specificity), and (b) the antibody recognizes cells transfected with the target protein (as in [26;27]). However, even with this validation, it is worth bearing in mind that a positive result would provide key evidence that the antibody recognizes its intended target, but would not mean that the antibody is specific for the intended target - as opposed to the other ~1000 ORs.
In the absence of antibodies, one often turns to other methods, including transgenic mice (i.e., with GFP or LacZ reporters) and RNA expression. ORs have an ‘ancient’ genomic structure – all mammalian ORs have single-exon coding sequences (although introns can be present in OR genes upstream of the coding exon [28; 29]). Therefore, when using RT-PCR it is important to include mock controls for ORs (where no reverse transcriptase enzyme is added to the RT reaction) as genomic DNA and RNA will yield the same size bands for primers directed against the coding sequence. Likewise, simply because the OR gene family is so large, it is important to sequence RT-PCR products to assure identity – with 1000 murine ORs which all have significant sequence similarity, even the most carefully designed primers may be able to cross-react with a ‘sibling’ of the intended target. Similarly, in situ hybridization is often employed for OR localization, and once again one must take care when designing probes that they are specific for the intended OR. When available, knockout mice can be utilized to confirm specificity of in situ or antibody signal. When knockout mice are not available, the number of glomeruli in the olfactory bulb which express the OR is a decent surrogate to gauge specificity of an in situ probe or antibody (if the probe is specific for a single OR, one would expect to see signal in 2–4 glomeruli [30]; it is imperative that an in situ looking at OR expression in glomeruli be isotopic due to the low level of OR mRNA expression in axons).
The high sequence similarity within this large gene family presents an additional challenge – identifying orthologs across species. When comparing rat and murine ORs, for example, both gene families have ~1000 members, and all of them have similarity to one another. Identifying an ortholog based on sequence similarity alone becomes quite difficult. Once a putative ortholog is identified based on sequence, to prove functional relevance one ought to demonstrate that the ORs are functional orthologs (i.e., respond to the same ligand).
Indeed, when studying any G protein-coupled receptor, one typically would consider potential roles for the ligand – however, most ORs are ‘orphan’ receptors, with no known ligand. This is certainly a challenge for the field. There are several well-developed assays that can be used to screen ORs for potential ligands – for example, calcium signaling in HEK cells [31], cAMP-mediated CFTR activation in oocytes [32], or cAMP-mediated luciferase activation in HEK cells [33]. However, all of these assays absolutely require, as a prerequisite, that the OR be expressed on the plasma membrane. For reasons which remain somewhat unclear, although ORs appear to efficiently traffic to the cell surface in vivo, they typically remain trapped in the endoplasmic reticulum when expressed exogenously in cell lines [34–36]. This frustrating fact means that screening an OR for potential ligands in a high throughput in vitro fashion is not as straightforward as one would hope. Given this, there are several approaches that one could take: first of all, the clearest solution is to screen ORs which Nature has expressed for us – that is, using native OE to test for potential ligands. Typically, potential ligands are tested while calcium responses are monitored, and if an OSN exhibits a calcium response to a particular ligand, then single-cell RT-PCR will be used to identify the receptor expressed in that OSN [37] (dogma holds that each OSN expresses one, and only one, receptor, in a monoallelic fashion [38]). Although in many ways this approach is the most straightforward, it is technically demanding, is not high-throughput, and without the use of GFP or another marker it is difficult to focus research on a particular OR. A second possible approach to screening ORs is to use a variety of aids to help the OR achieve surface expression when exogenously expressed in vitro. A number of studies have identified either chaperones or N-terminal tags which aid in OR surface expression [39–53], and when successful this allows for a high-throughput approach focused on a specific OR.
Finally, careful and elegant work has found that the expression of ORs exhibits strong developmental regulation in the OE – that is, different ORs may be strategically expressed at different developmental time points [54]. Furthermore, even in an OSN, there is a 4-day lag in the onset of OR expression [55]. Therefore, although it is unknown whether temporal regulation of expression takes place in tissues beyond the OE, it is worth noting that the ‘absence’ of OR expression does not necessarily mean that the OR is never expressed in that tissue - but rather that it is not expressed at that time point.
Characterizing OR signaling in the kidney
Despite the challenges of studying ORs, significant progress has been made in the characterization of renal ORs. In this section of the review, we will begin by discussing the identification of renal ORs and components of their downstream signaling, and then explore their relevance to both renal and whole body physiology.
AC3 and Golf
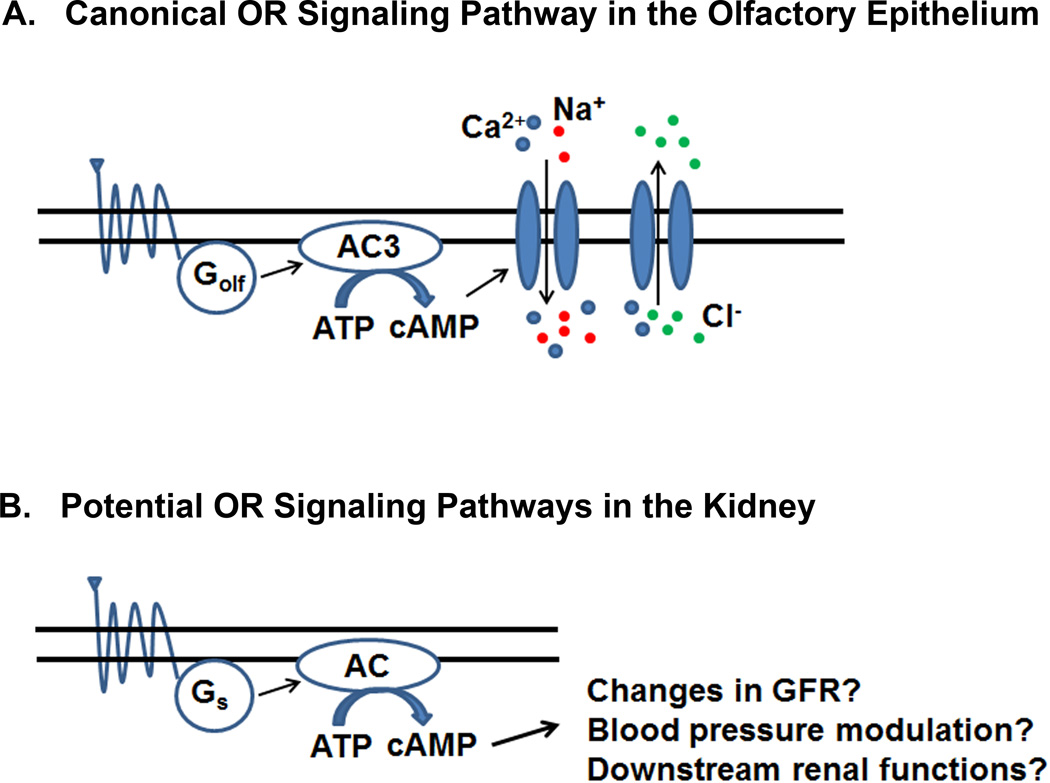
Despite the fact that there are more than 1000 ORs (mice), all mammalian ORs - at least in the nose - signal through a conserved downstream signaling pathway. Upon OR-odorant binding, the trimeric olfactory G protein (Golfactory or Golf) is activated, triggering the stimulation of membrane-bound adenylate cyclase 3 (AC3) (Fig. 1A) [5–7]. While this signaling pathway is obligatory for olfaction (mice null for AC3 cannot smell), the components of this pathway are also expressed outside of the nose [13; 56–58], and one of these sites is the kidney. AC3 and Golf are both expressed in the murine kidney at the transcript and protein level [59], and antibodies have localized them to the distal convoluted tubule and to an extremely specialized structure known as the macula densa (MD). The MD is a group of tightly packed sensory cells that are localized at the junction between the thick ascending limb and the distal convoluted tubule, where the nephron loops back to the afferent arteriole feeding its own parent glomerulus. MD cells take advantage of this unique placement to monitor the composition of the tubular fluid in the distal nephron, and then communicate this information back to the afferent arteriole feeding the parent glomerulus. For example, if the MD cells detect a decrease in the NaCl concentration in the filtrate, they will ‘tell’ the afferent arteriole/glomerulus to increase both glomerular filtration rate (GFR) and renin secretion. Ultimately, both of these actions serve to increase NaCl concentration in the filtrate back toward normal. This fine-tuning of GFR regulation allows for constant and precise regulation at the single nephron level, a process known as tubuloglomerular feedback (TGF).
Figure 1.
Olfactory receptor (OR) signaling pathways. A) The canonical OR signaling pathway in the olfactory epithelium. OR-ligand binding activates Golfactory (Golf) leading to increased cAMP via Adenylate Cyclase 3 (AC3) and the subsequent depolarization of olfactory sensory neurons. B) The potential OR signaling pathways in the kidney. As Golf and AC3 are not ubiquitous in the nephron, it is likely that an activated renal OR binds a stimulatory G protein (Gs) leading to cAMP production by different adenylate cyclases and changes in renal function.
Consistent with localization to the MD, AC3 knockout mice were found to present with both decreased GFR and decreased plasma renin. Therefore, as AC3 and Golf both localize to the MD and AC3 knockout mice exhibit an altered GFR, it was hypothesized that that the decreased GFR is due to an altered TGF response. However, when TGF was measured in these mice using stop-flow pressure with artificial tubular fluid, the response was normal. It should be noted that, to date, the individual ORs that are upstream of Golf and AC3 in the MD have not been identified. As ORs often respond to individual metabolites (see below), it is likely that the absence of an altered TGF response in the AC3 KO mice may due to the fact that the artificial perfusate was lacking key OR ligands/metabolites. Nonetheless, it is clear that AC3 and Golf do play important functional roles within the renal nephron.
Olfactory receptors
Given the expression of both AC3 and Golf in the murine kidney, it is reasonable to assume that individual ORs are also found in the kidney. Indeed, using a degenerate primer screening approach that takes advantage of the incredible sequence similarity amongst ORs, 6 ORs were initially found to be expressed in whole murine kidney [59]. Further analysis in kidneys from multiple independent mice determined that 4 of these original 6 ORs are consistently expressed in every sample examined. As such, this initial screen revealed 4 novel renal ORs (Olfr78, Olfr90, Olfr1392 and Olfr1393). As degenerate primer screens are likely to pick out only the most highly expressed ORs, more recent targeted RT-PCR screens have also been performed to identify additional renal ORs [60]. Using this strategy, an additional 6 ORs were identified in whole kidney (Olfr99, Olfr545, Olfr691, Olfr693, Olfr31 and Olfr1426). All 10 of these murine renal ORs have been cloned from the kidney from start to stop confirming full-length expression of the coding mRNA (Table 1). In addition to targeted screens, renal ORs have also been identified in mice by microarray studies. Notably, several ORs were found to be expressed by microarray analysis in isolated Juxtaglomerular cells or in renin-containing cells, and their expression may be developmentally regulated (Table 1) [61]. In the future, it will be important to confirm these microarray data by sequencing and to examine their potential developmental regulation in more detail. There are also a number of intriguing RNA sequencing screens that have found renal OR expression in different segments of the nephron. While this data is currently unpublished, the raw datasets are readily available in the GenitoUrinary Development Molecular Anatomy Project (GUDMAP.org).
Table I.
Identified Renal Olfactory Receptors (ORs)
| OR | Species | Method of Detection |
Localization | Ligands | References |
|---|---|---|---|---|---|
| Olfr78 | Mouse | RT-PCR Microarray |
Afferent arteriole Isolated JG Cells |
Short chain fatty acids | [59; 64] [61] |
| (Ortholog of Olr59 and OR51E2) | |||||
| Olr59 | Rat | RNA-seq | Glomeruli | - | [63] |
| (Ortholog of Olfr78 and OR51E2) | |||||
| OR51E2 | Human | RNA-seq | - | Propionate | [62; 86] |
| (Ortholog of Olfr78 and Olr59) | Acetate β-ionone Steroid hormones |
||||
| Olfr90 | Mouse | RT-PCR | Macula Densa | - | [59] |
| Olfr1392 | Mouse | RT-PCR | - | - | [59] |
| Olfr1393 | Mouse | RT-PCR | - | - | [59] |
| Olfr99 | Mouse | RT-PCR | - | - | [59] |
| Olfr545 | Mouse | RT-PCR | - | Sebacic acid* | [60; 87] |
| Olfr691 | Mouse | RT-PCR | - | Short and medium chain fatty acids |
[60; 68] |
| Olfr693 | Mouse | RT-PCR | - | - | [60] |
| Olfr31 | Mouse | RT-PCR | - | - | [60] |
| Olfr1426 | Mouse | RT-PCR | - | - | [60] |
| Olfr95 | Mouse | Microarray | Isolated JG Cells | - | [61] |
| Olfr796 | Mouse | Microarray | Isolated JG Cells | - | [61] |
| Olfr1303 | Mouse | Microarray | Isolated JG Cells | - | [61] |
| Olfr239 | Mouse | Microarray | Isolated JG Cells | - | [61] |
| Olfr1200 | Mouse | Microarray | Isolated JG Cells | - | [61] |
| Olfr373 | Mouse | Microarray | Isolated JG Cells | - | [61] |
| Olfr558 | Mouse | Microarray | Isolated JG Cells | - | [61] |
| Olr1104 | Rat | RNA-seq | DCT | - | [63] |
| Olr1694 | Rat | RNA-seq | S3, LDLIM, tAL, IMCD, Glomeruli |
- | [63] |
| Olr560 | Rat | RNA-seq | Entire nephron, Glomeruli |
- | [63] |
| Olr63 | Rat | RNA-seq | Glomeruli | - | [63] |
| OR10A2 | Human | RNA-seq | - | - | [62] |
| OR1L8 | Human | RNA-seq | - | - | [62] |
| OR2A1 | Human | RNA-seq | - | - | [62] |
| OR2A42 | Human | RNA-seq | - | - | [62] |
| OR2A7 | Human | RNA-seq | - | - | [62] |
| OR2AG1 | Human | RNA-seq | - | Amylbutyrate | [62; 88] |
| OR2T10 | Human | RNA-seq | - | Maltyl isobutyrate, cinnamaldehyde, vanillin, terpinyl acetate and α- damascone |
[62; 89] |
| OR51E1 | Human | RNA-seq | - | Methyl valeric acid, valeric acid and isovaleric acid |
[62; 90] |
| OR51I1 | Human | RNA-seq | - | - | [62] |
| OR51I2 | Human | RNA-seq | - | - | [62] |
| OR56B1 | Human | RNA-seq | - | - | [62] |
| OR9A4 | Human | RNA-seq | - | - | [62] |
JG Cells, Juxtaglomerular cells; DCT, distal convoluted tubule; S3, third segment of proximal tubule; LDLIM, long descending limb of the loop of Henle in the inner medulla; tAL, thin ascending limb of the loop of Henle; IMCD, inner medullary collecting duct
Renal OR expression is not unique to the mouse – indeed, ORs are also known to be expressed in the rat and human (Table 1). A recent RNA sequencing study on human tissues found that ORs are broadly expressed in different tissues and at least 13 different ORs are expressed in human kidney (excluding pseudogenes) [62]. Similar findings have emerged in the rat. Using hand-dissected nephron segments and isolated glomeruli, the Knepper lab has localized 5 different ORs within the rat nephron [63]. It is important to keep in mind that identification of functional orthologs based on sequence identity alone is usually not possible. Thus, it will be crucial to determine the localization and ligand profiles of this expanding list of renal ORs in order to determine their common functions. To date, a handful of ORs have been characterized and we will explore what is known about these ORs in the next few sections.
OR localization
The renal nephron is comprised of a number of specialized cell types, all with important and unique functions. Therefore, it is important to know where these renal ORs are expressed in order to understand potential physiological functions. As noted above, antibodies for these receptors are consistently unreliable and very few exist for renal ORs. Therefore, alternative methods are often used to determine OR localization. Using hand-dissected nephron segments and isolated glomeruli, all 5 rat identified renal ORs were localized within the kidney [63]. One of these, OR1104, is highly expressed in a single nephron segment – the distal convoluted tubule – while others are found more widely distributed along the length of the nephron (Table 1). Two ORs are exclusively expressed in the rat glomeruli, although the individual cell types are not yet known. In addition, mouse renal Olfr90 is expressed in an MD cell line [59]. While not yet confirmed in vivo, this data suggests that Olfr90 may be one of the ORs that signals via AC3 and Golf and could be participating in the AC3-mediated renin release/GFR response. Finally, using a lacZ reporter from a knockout mouse, Olfr78 localizes to the renal afferent arteriole, as well as large renal vessels (i.e. renal artery), and the physiological function of this OR is expanded on in detail below.
While the majority of renal ORs have not yet been localized, it is likely (based on both published and unpublished data) that they are widely distributed throughout the nephron. However, AC3 and Golf display a much more restricted localization pattern as they are both found primarily in the distal tubule and MD [59]. While AC3 and Golf are thought to be the obligate downstream signaling molecules in OSNs, it does not appear to be the case in the kidney. Since ORs have been found in cell types that lack AC3 and Golf (i.e. Olfr78 in the afferent arteriole [64]), these receptors clearly must be signaling through a different G protein. Consistent with this, ORs in vitro can also couple to Gs (or, under specific conditions, Gq) [31; 32; 39; 60; 64; 65]. In addition, a handful of ORs have been found to be expressed in the vomeronasal organ, the site of pheromone sensing in the mouse, where they are also thought to couple to other G proteins [66; 67]. Additionally, it is also possible that the renal ORs would signal through AC3 but not Golf, or vice versa. This is supported by the fact that AC3 can be stimulated by other Gs proteins [6; 58]. Clearly additional studies are required to determine the complete expression pattern of ORs within in the kidney and to delineate their downstream signaling (Fig. 1B).
OR ligands
Most ORs are orphan receptors with no known ligands, and the subset of renal ORs are no exception (Table 1). Deorphanizing these receptors is a critical step in determining the function of these renal ORs, and to date, a few of these renal receptors now have known ligands. As mentioned above, functional heterologous expression must be achieved before ligand screening can occur in vitro. To date, with the help of various tags and chaperone proteins, 9 different renal murine ORs have been found to traffic to the cell surface in HEK293T cells [39; 60]. Ligand screens have been performed on these ORs, leading to the identification of several different OR-ligand pairs. Olfr78 and its human ortholog hOR51E2 both respond to short chain fatty acids (SCFAs) and the functional relevance of this is discussed in detail below. Olfr691 responds to a wide range of short and medium chain fatty acids [60; 68] (Table 1). Notably, one of these ligands, valproate, has been used as an antiepileptic drug and patients have been shown to develop Fanconi syndrome [69; 70]. This link could potentially reveal a novel mechanism by which proximal tubule transport is regulated and highlights the clear importance of understanding the ligands and functional roles for ORs in the kidney.
Olfactory Receptor 78: A novel mechanism of blood pressure regulation
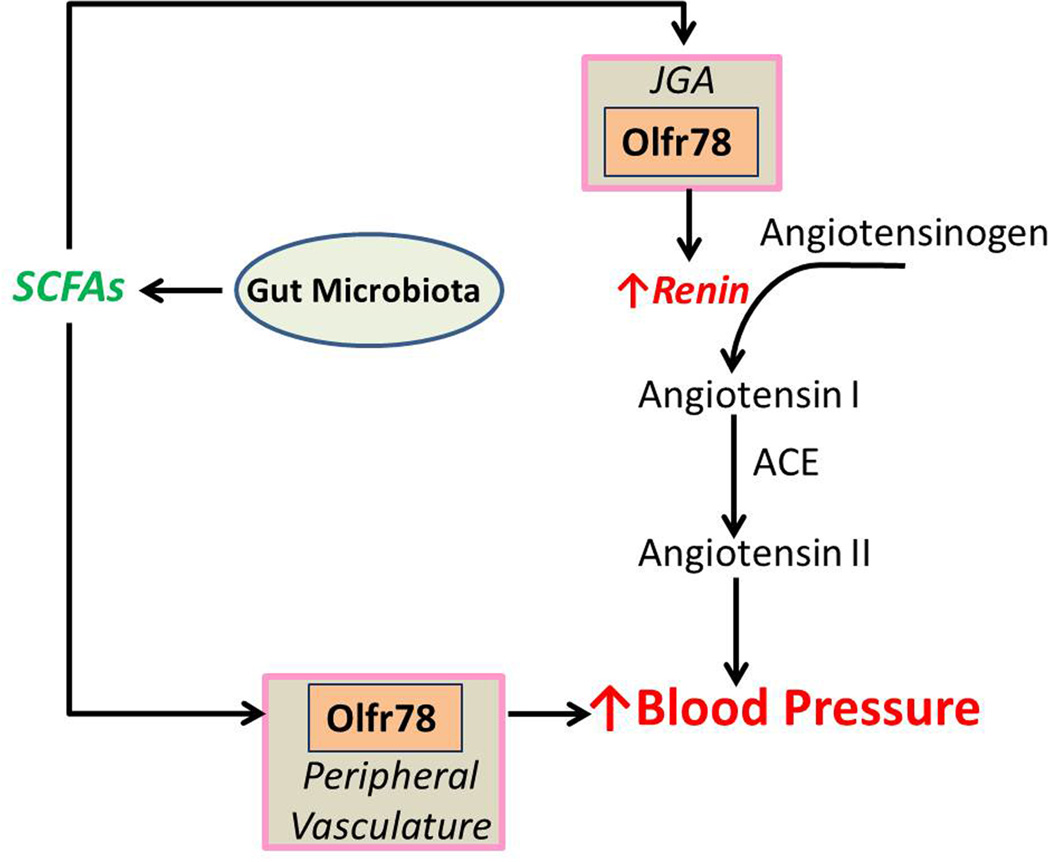
Olfr78 (MOR18-2) was initially identified as being expressed in whole kidney in 2009 [59]. Although an antibody against Olfr78 was successfully generated [26], it strongly cross-reacts with a mitochondrial protein in all other tissues tested – including the kidney. Therefore, a transgenic mouse model in which LacZ expression is driven by the Olfr78 promoter was used to identify cell types which normally express Olfr78. Using this tool, it was found that Olfr78 is found in the renal afferent arteriole [71] in the renal juxtaglomerular apparatus (JGA). Interestingly, the rat ortholog for Olfr78 (Olr59) was also found in whole glomeruli (Table 1) [63], suggesting that this localization pattern may be conserved across species. In addition, LacZ staining of non-renal tissues revealed that Olfr78 also localizes to small resistance vessels in a variety of vascular beds in the peripheral vasculature (Fig. 2). It is intriguing to note that both the JGA and the peripheral vasculature are classical sites known to play key roles in blood pressure regulation, and that the human ortholog of Olfr78 (hOR51E2) is also expressed at the RNA level in tissues consistent with these findings [62] (kidney, skeletal muscle, heart, etc.).
Figure 2.
Short chain fatty acids (SCFAs) produced by the gut microbiota activate Olfr78 in the both the renal juxtaglomerular apparatus (JGA) and peripheral vasculature. Activation of Olfr78 in the JGA leads to increased renin release. In the peripheral vasculature, activation of Olfr78 modulates a hypotensive response mediated by other SCFA receptors. In both cases, activation of Olfr78 leads to an increased blood pressure response.
Although Olfr78 was initially an orphan receptor, an in vitro ligand screen revealed that it is a receptor for SCFAs – specifically, acetate and propionate [71] – and that the human ortholog of this receptor (hOR51E2) responds to the same two compounds. Although identifying OR orthologs across species is typically problematic, this OR appears to be uniquely well-conserved amongst mammalian species (with a clear ortholog in mice, rats, rabbits, elephants, horses and 5 species of primates [72]). As Golf is not expressed in the afferent arteriole [59], Olfr78 in the afferent arteriole likely couples to Gs.
Intriguingly, SCFAs are present in the bloodstream primarily due to production by the gut microbiota [73]; bacteria in the gut produce SCFAs in large quantities (such that the concentration in the colonic lumen is ~100mM [73]), and then they are absorbed into the bloodstream in the low mM range (0.1–10mM [74–77]). The half maximal effective concentration (EC50) of Olfr78 for acetate (2.35mM) and propionate (0.92mM) falls within this plasma range. SCFAs produced by the gut microbiota have been shown to play important roles in influencing several aspects of host physiology, including inflammatory responses and metabolism [75; 78].
As mentioned, Olfr78 localizes to two important sites of blood pressure control. One of these sites is the afferent arteriole. Here, renin - the rate-limiting step in the renin-angiotensin-aldosterone pathway - is stored and secreted into the bloodstream. Renin is responsible for the hydrolysis of angiotensinogen to angiotensin I, which is further cleaved to angiotensin II, a potent vasoconstrictor (Fig. 2). Because Olfr78 localizes to the renal afferent arteriole, it was investigated whether Olfr78 might mediate an increase in renin secretion in response to SCFAs. Indeed, in an ex vivo preparation it was demonstrated that SCFAs induced renin release, and that this effect was dependent on Olfr78 [71]. Furthermore, in support of this, it was found that Olfr78 null animals had lowered plasma renin, and lowered baseline blood pressure [71] (Fig. 2). Thus, these findings present a novel pathway whereby activation of a sensory receptor, Olfr78, mediates changes in plasma renin and in blood pressure. It should be noted that some ORs have been shown to be mechanosensitive [79; 80]; however, it is unknown whether Olfr78 may be responding to mechanical (changes in blood flow?) as well as chemical stimuli.
Because Olfr78 also localizes to the peripheral vasculature, it was also determined whether SCFAs might also have an acute effect on vascular resistance, another important aspect of blood pressure control. Previous studies had reported that SCFAs can indeed induce vasodilation in isolated vessels [81–83]; to determine if there was an in vivo consequence of this vasodilation, SCFAs were delivered exogenously (intravenously) and blood pressure was measured. It was found that SCFA delivery caused a rapid, reproducible and dose-dependent hypotensive response. However, subsequent studies showed that this response is primarily due to activation of another SCFA receptor (Gpr41), whereas Olfr78 acts as a ‘brake’ on this response. Therefore, Olfr78 activation by SCFAs serves to increase blood pressure through two independent methods: increased renin release from the JGA and changes in in peripheral vascular resistance (Fig. 2). (The opposing actions of Olfr78 and Gpr41 on blood pressure regulation have been reviewed in detail: [84; 85]).
Conclusion
In conclusion, recent studies have highlighted novel and unexpected roles that sensory receptors, including ORs, play in a variety of tissues. As a key regulator of homeostasis, the kidney is a particularly intriguing location to employ such receptors, and recent studies have shown that ORs, taste receptors and other sensory GPRs are found in the kidney. Although there are a number of challenges to studying to ORs, it will be important for future studies to carefully explore and elucidate the novel roles that these receptors play in renal function.
Acknowledgements
The authors are grateful to Dr. Diego Rodriguez-Gil (Yale Univ.) for a critical reading of portions of this manuscript and for valued insights and discussions, and to members of the Pluznick Lab for helpful discussions.
Funding Jennifer L. Pluznick: Carl W. Gottschalk Research Scholar Grant from the American Society of Nephrology; Blythe D. Shepard: National Institutes of Health (F32-DK096780).
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
References
- 1.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 2.Glusman G, Yanai I, Rubin I, Lancet D. The complete human olfactory subgenome. Genome Res. 2001;11:685–702. doi: 10.1101/gr.171001. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- 4.Zozulya S, Echeverri F, Nguyen T. The human olfactory receptor repertoire. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-6-research0018. RESEARCH0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belluscio L, Gold GH, Nemes A, Axel R. Mice deficient in G(olf) are anosmic. Neuron. 1998;20:69–81. doi: 10.1016/s0896-6273(00)80435-3. [DOI] [PubMed] [Google Scholar]
- 6.Wong ST, Trinh K, Hacker B, Chan GC, Lowe G, Gaggar A, Xia Z, Gold GH, Storm DR. Disruption of the type III adenylyl cyclase gene leads to peripheral and behavioral anosmia in transgenic mice. Neuron. 2000;27:487–497. doi: 10.1016/s0896-6273(00)00060-x. [DOI] [PubMed] [Google Scholar]
- 7.Lowe G, Nakamura T, Gold GH. Adenylate cyclase mediates olfactory transduction for a wide variety of odorants. Proc Natl Acad Sci U S A. 1989;86:5641–5645. doi: 10.1073/pnas.86.14.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Busse D, Kudella P, Gruning NM, Gisselmann G, Stander S, Luger T, Jacobsen F, Steinstrasser L, Paus R, Gkogkolou P, Bohm M, Hatt H, Benecke H. A Synthetic Sandalwood Odorant Induces Wound-Healing Processes in Human Keratinocytes via the Olfactory Receptor OR2AT4. J Invest Dermatol. 2014;134:2823–2832. doi: 10.1038/jid.2014.273. [DOI] [PubMed] [Google Scholar]
- 9.Feldmesser E, Olender T, Khen M, Yanai I, Ophir R, Lancet D. Widespread ectopic expression of olfactory receptor genes. BMC Genomics. 2006;7:121. doi: 10.1186/1471-2164-7-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukuda N, Yomogida K, Okabe M, Touhara K. Functional characterization of a mouse testicular olfactory receptor and its role in chemosensing and in regulation of sperm motility. J Cell Sci. 2004;117:5835–5845. doi: 10.1242/jcs.01507. [DOI] [PubMed] [Google Scholar]
- 11.Griffin CA, Kafadar KA, Pavlath GK. MOR23 promotes muscle regeneration and regulates cell adhesion and migration. Dev Cell. 2009;17:649–661. doi: 10.1016/j.devcel.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang N, Koo J. Olfactory receptors in non-chemosensory tissues. BMB Rep. 2012;45:612–622. doi: 10.5483/BMBRep.2012.45.11.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang N, Bahk YY, Lee N, Jae Y, Cho YH, Ku CR, Byun Y, Lee EJ, Kim MS, Koo J. Olfactory receptor Olfr544 responding to azelaic acid regulates glucagon secretion in alpha-cells of mouse pancreatic islets. Biochem Biophys Res Commun. 2015;460:616–621. doi: 10.1016/j.bbrc.2015.03.078. [DOI] [PubMed] [Google Scholar]
- 14.Kim SH, Yoon YC, Lee AS, Kang N, Koo J, Rhyu MR, Park JH. Expression of human olfactory receptor 10J5 in heart aorta, coronary artery, and endothelial cells and its functional role in angiogenesis. Biochem Biophys Res Commun. 2015;460:404–408. doi: 10.1016/j.bbrc.2015.03.046. [DOI] [PubMed] [Google Scholar]
- 15.Pluznick JL, Caplan MJ. Novel sensory signaling systems in the kidney. Curr Opin Nephrol Hypertens. 2012;21:404–409. doi: 10.1097/MNH.0b013e328354a6bd. [DOI] [PubMed] [Google Scholar]
- 16.Spehr M, Gisselmann G, Poplawski A, Riffell JA, Wetzel CH, Zimmer RK, Hatt H. Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science. 2003;299:2054–2058. doi: 10.1126/science.1080376. [DOI] [PubMed] [Google Scholar]
- 17.Wu C, Jia Y, Lee JH, Kim Y, Sekharan S, Batista VS, Lee SJ. Activation of OR1A1 suppresses PPAR-gamma expression by inducing HES-1 in cultured hepatocytes. Int J Biochem Cell Biol. 2015;64:75–80. doi: 10.1016/j.biocel.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Cui T, Tsolakis AV, Li SC, Cunningham JL, Lind T, Oberg K, Giandomenico V. Olfactory receptor 51E1 protein as a potential novel tissue biomarker for small intestine neuroendocrine carcinomas. Eur J Endocrinol. 2013;168:253–261. doi: 10.1530/EJE-12-0814. [DOI] [PubMed] [Google Scholar]
- 19.Ichimura A, Kadowaki T, Narukawa K, Togiya K, Hirasawa A, Tsujimoto G. In silico approach to identify the expression of the undiscovered molecules from microarray public database: identification of odorant receptors expressed in non-olfactory tissues. Naunyn Schmiedebergs Arch Pharmacol. 2008;377:159–165. doi: 10.1007/s00210-007-0255-6. [DOI] [PubMed] [Google Scholar]
- 20.Serizawa S, Miyamichi K, Takeuchi H, Yamagishi Y, Suzuki M, Sakano H. A neuronal identity code for the odorant receptor-specific and activity-dependent axon sorting. Cell. 2006;127:1057–1069. doi: 10.1016/j.cell.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 21.Kaneko-Goto T, Yoshihara S, Miyazaki H, Yoshihara Y. BIG-2 mediates olfactory axon convergence to target glomeruli. Neuron. 2008;57:834–846. doi: 10.1016/j.neuron.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 22.Barnea G, O’Donnell S, Mancia F, Sun X, Nemes A, Mendelsohn M, Axel R. Odorant receptors on axon termini in the brain. Science. 2004;304:1468. doi: 10.1126/science.1096146. [DOI] [PubMed] [Google Scholar]
- 23.Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 24.Strotmann J, Levai O, Fleischer J, Schwarzenbacher K, Breer H. Olfactory receptor proteins in axonal processes of chemosensory neurons. J Neurosci. 2004;24:7754–7761. doi: 10.1523/JNEUROSCI.2588-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeuchi H, Inokuchi K, Aoki M, Suto F, Tsuboi A, Matsuda I, Suzuki M, Aiba A, Serizawa S, Yoshihara Y, Fujisawa H, Sakano H. Sequential arrival and graded secretion of Sema3F by olfactory neuron axons specify map topography at the bulb. Cell. 2010;141:1056–1067. doi: 10.1016/j.cell.2010.04.041. [DOI] [PubMed] [Google Scholar]
- 26.Pluznick JL, Rodriguez-Gil DJ, Hull M, Mistry K, Gattone V, Johnson CA, Weatherbee S, Greer CA, Caplan MJ. Renal cystic disease proteins play critical roles in the organization of the olfactory epithelium. PLoS One. 2011;6:e19694. doi: 10.1371/journal.pone.0019694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang N, Kim H, Jae Y, Lee N, Ku CR, Margolis F, Lee EJ, Bahk YY, Kim MS, Koo J. Olfactory marker protein expression is an indicator of olfactory receptor-associated events in non-olfactory tissues. PLoS One. 2015;10:e0116097. doi: 10.1371/journal.pone.0116097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young JM, Shykind BM, Lane RP, Tonnes-Priddy L, Ross JA, Walker M, Williams EM, Trask BJ. Odorant receptor expressed sequence tags demonstrate olfactory expression of over 400 genes, extensive alternate splicing and unequal expression levels. Genome Biol. 2003;4:R71. doi: 10.1186/gb-2003-4-11-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sosinsky A, Glusman G, Lancet D. The genomic structure of human olfactory receptor genes. Genomics. 2000;70:49–61. doi: 10.1006/geno.2000.6363. [DOI] [PubMed] [Google Scholar]
- 30.Richard MB, Taylor SR, Greer CA. Age-induced disruption of selective olfactory bulb synaptic circuits. Proc Natl Acad Sci U S A. 2010;107:15613–15618. doi: 10.1073/pnas.1007931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kajiya K, Inaki K, Tanaka M, Haga T, Kataoka H, Touhara K. Molecular bases of odor discrimination: Reconstitution of olfactory receptors that recognize overlapping sets of odorants. J Neurosci. 2001;21:6018–6025. doi: 10.1523/JNEUROSCI.21-16-06018.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katada S, Nakagawa T, Kataoka H, Touhara K. Odorant response assays for a heterologously expressed olfactory receptor. Biochem Biophys Res Commun. 2003;305:964–969. doi: 10.1016/s0006-291x(03)00863-5. [DOI] [PubMed] [Google Scholar]
- 33.Zhuang H, Matsunami H. Evaluating cell-surface expression and measuring activation of mammalian odorant receptors in heterologous cells. Nat Protoc. 2008;3:1402–1413. doi: 10.1038/nprot.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu M, Echeverri F, Moyer BD. Endoplasmic reticulum retention, degradation, and aggregation of olfactory G-protein coupled receptors. Traffic. 2003;4:416–433. doi: 10.1034/j.1600-0854.2003.00097.x. [DOI] [PubMed] [Google Scholar]
- 35.McClintock TS, Sammeta N. Trafficking prerogatives of olfactory receptors. Neuroreport. 2003;14:1547–1552. doi: 10.1097/00001756-200308260-00001. [DOI] [PubMed] [Google Scholar]
- 36.Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci. 2004;5:263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- 37.Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 38.Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 39.Shepard BD, Natarajan N, Protzko RJ, Acres OW, Pluznick JL. A cleavable N-terminal signal peptide promotes widespread olfactory receptor surface expression in HEK293T cells. PLoS One. 2013;8:e68758. doi: 10.1371/journal.pone.0068758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaillard I, Rouquier S, Pin JP, Mollard P, Richard S, Barnabe C, Demaille J, Giorgi D. A single olfactory receptor specifically binds a set of odorant molecules. Eur J Neurosci. 2002;15:409–418. doi: 10.1046/j.0953-816x.2001.01871.x. [DOI] [PubMed] [Google Scholar]
- 41.Hague C, Uberti MA, Chen Z, Bush CF, Jones SV, Ressler KJ, Hall RA, Minneman KP. Olfactory receptor surface expression is driven by association with the beta2-adrenergic receptor. Proc Natl Acad Sci U S A. 2004;101:13672–13676. doi: 10.1073/pnas.0403854101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu M, Staszewski L, Echeverri F, Xu H, Moyer BD. Endoplasmic reticulum degradation impedes olfactory G-protein coupled receptor functional expression. BMC Cell Biol. 2004;5:34. doi: 10.1186/1471-2121-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall RA. Olfactory receptor interactions with other receptors. Ann N Y Acad Sci. 2009;1170:147–149. doi: 10.1111/j.1749-6632.2009.03879.x. [DOI] [PubMed] [Google Scholar]
- 44.Matsunami H, Mainland JD, Dey S. Trafficking of mammalian chemosensory receptors by receptor-transporting proteins. Ann N Y Acad Sci. 2009;1170:153–156. doi: 10.1111/j.1749-6632.2009.03888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belluscio L, Gold GH, Nemes A, Axel R. Mice deficient in G(olf) are anosmic. Neuron. 1998;20:69–81. doi: 10.1016/s0896-6273(00)80435-3. [DOI] [PubMed] [Google Scholar]
- 46.Kajiya K, Inaki K, Tanaka M, Haga T, Kataoka H, Touhara K. Molecular bases of odor discrimination: Reconstitution of olfactory receptors that recognize overlapping sets of odorants. J Neurosci. 2001;21:6018–6025. doi: 10.1523/JNEUROSCI.21-16-06018.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krautwurst D, Yau KW, Reed RR. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell. 1998;95:917–926. doi: 10.1016/s0092-8674(00)81716-x. [DOI] [PubMed] [Google Scholar]
- 48.Nara K, Saraiva LR, Ye X, Buck LB. A large-scale analysis of odor coding in the olfactory epithelium. J Neurosci. 2011;31:9179–9191. doi: 10.1523/JNEUROSCI.1282-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 50.Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. Odor coding by a Mammalian receptor repertoire. Sci Signal. 2009;2 doi: 10.1126/scisignal.2000016. ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Von Dannecker LE, Mercadante AF, Malnic B. Ric-8B promotes functional expression of odorant receptors. Proc Natl Acad Sci U S A. 2006;103:9310–9314. doi: 10.1073/pnas.0600697103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhuang H, Matsunami H. Synergism of accessory factors in functional expression of mammalian odorant receptors. J Biol Chem. 2007;282:15284–15293. doi: 10.1074/jbc.M700386200. [DOI] [PubMed] [Google Scholar]
- 53.Zhuang H, Matsunami H. Evaluating cell-surface expression and measuring activation of mammalian odorant receptors in heterologous cells. Nat Protoc. 2008;3:1402–1413. doi: 10.1038/nprot.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodriguez-Gil DJ, Treloar HB, Zhang X, Miller AM, Two A, Iwema C, Firestein SJ, Greer CA. Chromosomal location-dependent nonstochastic onset of odor receptor expression. J Neurosci. 2010;30:10067–10075. doi: 10.1523/JNEUROSCI.1776-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodriguez-Gil DJ, Bartel DL, Jaspers AW, Mobley AS, Imamura F, Greer CA. Odorant receptors regulate the final glomerular coalescence of olfactory sensory neuron axons. Proc Natl Acad Sci U S A. 2015;112:5821–5826. doi: 10.1073/pnas.1417955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Defer N, Marinx O, Poyard M, Lienard MO, Jegou B, Hanoune J. The olfactory adenylyl cyclase type 3 is expressed in male germ cells. FEBS Lett. 1998;424:216–220. doi: 10.1016/s0014-5793(98)00178-1. [DOI] [PubMed] [Google Scholar]
- 57.Ishikawa Y, Grant BS, Okumura S, Schwencke C, Yamamoto M. Immunodetection of adenylyl cyclase protein in tissues. Mol Cell Endocrinol. 2000;162:107–112. doi: 10.1016/s0303-7207(00)00210-0. [DOI] [PubMed] [Google Scholar]
- 58.Xia Z, Choi EJ, Wang F, Storm DR. The type III calcium/calmodulin-sensitive adenylyl cyclase is not specific to olfactory sensory neurons. Neurosci Lett. 1992;144:169–173. doi: 10.1016/0304-3940(92)90742-p. [DOI] [PubMed] [Google Scholar]
- 59.Pluznick JL, Zou DJ, Zhang X, Yan Q, Rodriguez-Gil DJ, Eisner C, Wells E, Greer CA, Wang T, Firestein S, Schnermann J, Caplan MJ. Functional expression of the olfactory signaling system in the kidney. Proc Natl Acad Sci U S A. 2009;106:2059–2064. doi: 10.1073/pnas.0812859106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rajkumar P, Aisenberg WH, Acres OW, Protzko RJ, Pluznick JL. Identification and Characterization of Novel Renal Sensory Receptors. PLoS One. 2014;9:e111053. doi: 10.1371/journal.pone.0111053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brunskill EW, Sequeira-Lopez ML, Pentz ES, Lin E, Yu J, Aronow BJ, Potter SS, Gomez RA. Genes that confer the identity of the renin cell. J Am Soc Nephrol. 2011;22:2213–2225. doi: 10.1681/ASN.2011040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Flegel C, Manteniotis S, Osthold S, Hatt H, Gisselmann G. Expression profile of ectopic olfactory receptors determined by deep sequencing. PLoS One. 2013;8:e55368. doi: 10.1371/journal.pone.0055368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee JW, Chou CL, Knepper MA. Deep sequencing in microdissected renal tubules identifies nephron segment-specific transcriptomes. J Am Soc Nephrol. 2015 doi: 10.1681/ASN.2014111067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J, Brunet I, Wan LX, Rey F, Wang T, Firestein SJ, Yanagisawa M, Gordon JI, Eichmann A, Peti-Peterdi J, Caplan MJ. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci U S A. 2013;110:4410–4415. doi: 10.1073/pnas.1215927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Offermanns S, Simon MI. G alpha 15 and G alpha 16 couple a wide variety of receptors to phospholipase C. J Biol Chem. 1995;270:15175–15180. doi: 10.1074/jbc.270.25.15175. [DOI] [PubMed] [Google Scholar]
- 66.Berghard A, Buck LB, Liman ER. Evidence for distinct signaling mechanisms in two mammalian olfactory sense organs. Proc Natl Acad Sci U S A. 1996;93:2365–2369. doi: 10.1073/pnas.93.6.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trinh K, Storm DR. Vomeronasal organ detects odorants in absence of signaling through main olfactory epithelium. Nat Neurosci. 2003;6:519–525. doi: 10.1038/nn1039. [DOI] [PubMed] [Google Scholar]
- 68.Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 69.Endo A, Fujita Y, Fuchigami T, Takahashi S, Mugishima H. Fanconi syndrome caused by valproic acid. Pediatr Neurol. 2010;42:287–290. doi: 10.1016/j.pediatrneurol.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 70.Knorr M, Schaper J, Harjes M, Mayatepek E, Rosenbaum T. Fanconi syndrome caused by antiepileptic therapy with valproic Acid. Epilepsia. 2004;45:868–871. doi: 10.1111/j.0013-9580.2004.05504.x. [DOI] [PubMed] [Google Scholar]
- 71.Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J, Brunet I, Wan LX, Rey F, Wang T, Firestein SJ, Yanagisawa M, Gordon JI, Eichmann A, Peti-Peterdi J, Caplan MJ. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci U S A. 2013;110:4410–4415. doi: 10.1073/pnas.1215927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Niimura Y, Matsui A, Touhara K. Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Res. 2014;24:1485–1496. doi: 10.1101/gr.169532.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bugaut M. Occurrence, absorption and metabolism of short chain fatty acids in the digestive tract of mammals. Comp Biochem Physiol B. 1987;86:439–472. doi: 10.1016/0305-0491(87)90433-0. [DOI] [PubMed] [Google Scholar]
- 74.Le PE, Loison C, Struyf S, Springael JY, Lannoy V, Decobecq ME, Brezillon S, Dupriez V, Vassart G, Van DJ, Parmentier M, Detheux M. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem. 2003;278:25481–25489. doi: 10.1074/jbc.M301403200. [DOI] [PubMed] [Google Scholar]
- 75.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, Xavier RJ, Teixeira MM, Mackay CR. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci U S A. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, Marsland BJ. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 78.Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, Hammer RE, Williams SC, Crowley J, Yanagisawa M, Gordon JI. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci U S A. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Connelly T, Yu Y, Grosmaitre X, Wang J, Santarelli LC, Savigner A, Qiao X, Wang Z, Storm DR, Ma M. G protein-coupled odorant receptors underlie mechanosensitivity in mammalian olfactory sensory neurons. Proc Natl Acad Sci U S A. 2015;112:590–595. doi: 10.1073/pnas.1418515112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grosmaitre X, Santarelli LC, Tan J, Luo M, Ma M. Dual functions of mammalian olfactory sensory neurons as odor detectors and mechanical sensors. Nat Neurosci. 2007;10:348–354. doi: 10.1038/nn1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nutting CW, Islam S, Daugirdas JT. Vasorelaxant effects of short chain fatty acid salts in rat caudal artery. Am J Physiol. 1991;261:H561–H567. doi: 10.1152/ajpheart.1991.261.2.H561. [DOI] [PubMed] [Google Scholar]
- 82.Mortensen FV, Nielsen H, Mulvany MJ, Hessov I. Short chain fatty acids dilate isolated human colonic resistance arteries. Gut. 1990;31:1391–1394. doi: 10.1136/gut.31.12.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nutting CW, Islam S, Ye MH, Batlle DC, Daugirdas JT. The vasorelaxant effects of acetate: role of adenosine, glycolysis, lyotropism, and pHi and Cai2+ Kidney Int. 1992;41:166–174. doi: 10.1038/ki.1992.23. [DOI] [PubMed] [Google Scholar]
- 84.Pluznick J. A novel SCFA receptor, the microbiota, and blood pressure regulation. Gut Microbes. 2013;5:202–207. doi: 10.4161/gmic.27492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pluznick JL. Renal and cardiovascular sensory receptors and blood pressure regulation. Am J Physiol Renal Physiol. 2013;305:F439–F444. doi: 10.1152/ajprenal.00252.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Neuhaus EM, Zhang W, Gelis L, Deng Y, Noldus J, Hatt H. Activation of an olfactory receptor inhibits proliferation of prostate cancer cells. J Biol Chem. 2009;284:16218–16225. doi: 10.1074/jbc.M109.012096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abaffy T, Matsunami H, Luetje CW. Functional analysis of a mammalian odorant receptor subfamily. J Neurochem. 2006;97:1506–1518. doi: 10.1111/j.1471-4159.2006.03859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Neuhaus EM, Mashukova A, Zhang W, Barbour J, Hatt H. A specific heat shock protein enhances the expression of mammalian olfactory receptor proteins. Chem Senses. 2006;31:445–452. doi: 10.1093/chemse/bjj049. [DOI] [PubMed] [Google Scholar]
- 89.Gonzalez-Kristeller DC, do Nascimento JB, Galante PA, Malnic B. Identification of agonists for a group of human odorant receptors. Front Pharmacol. 2015;6:35. doi: 10.3389/fphar.2015.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fujita Y, Takahashi T, Suzuki A, Kawashima K, Nara F, Koishi R. Deorphanization of Dresden G protein-coupled receptor for an odorant receptor. J Recept Signal Transduct Res. 2007;27:323–334. doi: 10.1080/10799890701534180. [DOI] [PubMed] [Google Scholar]