Abstract
Accumulating evidence has recently supported the association of bacterial infection with the growth and development of cancers, particularly in organs that are constantly exposed to bacteria such as the lungs, colon, cervical cancer etc. Our in silico study on the proteome of Chlamydia pneumoniae suggests an unprecedented idea of the etiology of lung cancer and have revealed that the infection of C. pneumoniae is associated with lung cancer development and growth. It is reasonable to assume that C. pneumoniae transports its proteins within host-intracellular organelles during infection, where they may work with host-cell proteome. The current study was performed for the prediction of nuclear targeting protein of C. pneumoniae in the host cell using bioinformatics predictors including ExPASy pI/Mw tool, nuclear localization signal (NLS) mapper, balanced sub cellular localization predictor (BaCeILo), and Hum-mPLoc 2.0. We predicted 47/1112 nuclear-targeting proteins of C. pneumoniae connected with several possible alterations in host replication and transcription during intracellular infection. These nuclear-targeting proteins may direct to competitive interactions of host and C. pneumoniae proteins with the availability of same substrate and may be involved as etiological agents in the growth and development of lung cancer. These novel findings are expected to access in better understanding of lung cancer etiology and identifying molecular targets for therapy.
Introduction
Lungs cancer is the most common cause of death worldwide [1]. In 2013, according to CA cancer reports, lung cancer is second most prevalent cancer in US as there were an estimated 159,480 (118,080 males and 110,110 females) deaths from lung cancer and 228,190 (male 87,260 and 72,220) new cases of lung cancer reported [2]. The process of carcinogenesis of lung cancer is still not completely understood. Beside smoking, there are other potential genetic and environmental factors such as exposure to asbestos and radon definite metals, coal smoke, various hormones, and air pollution as well as genetic incompatibility and chronic infections of bacteria and parasites have been connected to lung carcinogenesis including C. Pneumoniae, [3–6]. The equivocal association of infectious agents in the etiology of cancer has focused the interest of scientists in recent year.
The role of C. pneumoniae as an infectious carcinogen in lung cancer has been studied since more than 10 years ago [7, 8]. Epidemiological associations indicated that C. pneumoniae is potentially associated with the growth and development of lungs cells carcinoma [9]. Various studies were then performed to analyze the possible connection between C. pneumoniae infection and risk of lung cancer, but the results have not been consistent [10]. It is proposed that the C. pneumoniae acts as a cofactor with other causes for the progression and development of lungs caner [7, 8, 11, 12]. It has been observed that the titers of C. pneumoniae antibody elevated in lung cancer patients. Among witch, patients with high titers of anti-C. pneumoniae IgA antibody have ten-time risk of adenocarcinomas and small cell carcinomas of the lung [9]. This possibility is enhanced specifically in male smoker patients with chronic infection of C. pneumoniae [11]. One more finding have demonstrated an important connection between elevated Chlamydia Hsp-60 seropositivity and the chance of lung cancer, which may suggest the etiological role of C. pneumoniae in the growth and development of lung cancer [3].
C. pneumoniae is a common intracellular respiratory pathogen which requires regulating the host cell for their survival and growth. The following mechanisms have been suggested to elucidate how chronic infection of C. pneumoniae could enhance the possibility of lung cancer. One potential mechanism is mediated through the generation of reactive oxygen species during inflammation, which may contribute to DNA damage [7]. Moreover, inflammation results in cell injury and subsequent repair that may enhance the rate of cell division. The multiplication of cells will increase the risk of a mutation through a fixed rate of DNA damage, which may lead to cancer [13]. Collecting evidence proposes that immunological events contribute in part in the carcinogenic action of C. pneumoniae. Earlier in vitro studies have in fact shown that TNF-α, IL-1β, IL8, and superoxide oxygen radicals released by alveolar macrophages from healthy persons play a crucial role in lung tissue and DNA damage [14]. C. pneumoniae is also effective inducer of TNF-α, IL-1β, and IL-6 in host monocytic cells that may potentially contribute in carcinogenesis. In this paper, we are trying to predict the nuclear targeted proteins of C. pneumoniae due to their potential role in the host cells regulation and involvement in the progression and development of lung cancer.
Results
Selection of protein database
We selected TW-183 strain of C. pneumoniae as it contains highest number (1112) of protein in complete proteome G/11222. However, the rest of the four strains include AR39, LPCoLN, CWL029 and J138 have 1109, 1105, 1052, and 1069 proteins, respectively [15–18].
Prediction of nuclear localization signal
The cNLS mapper predicted the location of protein in cytoplasm, both nucleus and cytoplasm, partially in nucleus, and localized in the nucleus with cut of value 1–2, 3–5, 7–8, and 8–10, respectively. The results were illustrated in Supplementary data (S1 Table).
Prediction of subcellular localization in eukaryotic cell organelles
The results of BaCeILo assessment of total protein of C. pneumoniae summarized in Table 1. The result showed that BaCeILo predicted total cytoplasmic (515), mitochondrial (183), nuclear (98) and Secretory (318) proteins.
Table 1. Possible sub-cellular localization of C. pneumoniae proteins in host cell as per BaCeILo and Hum-mPLoc 2.0.
| Type of Protein | Number of Proteins predicted by BaCeILo | Number of proteins by predicted Hum-mPLoc 2.0 |
|---|---|---|
| Cytoplasm | 513 | 250 |
| Mitochondria | 183 | 196 |
| Nucleus | 98 | 292 |
| Secretory | 318 | - |
| Plasma Membrane | - | 94 |
| Endoplasmic reticulum | - | 63 |
| Golgi apparatus | - | 05 |
| Peroxisome | - | 11 |
| Microsome | - | 01 |
| Lysosome | - | 11 |
| Extracell | - | 179 |
| Centrosome | - | 05 |
| Cytoskeleton | - | 01 |
| Unknown | 04 |
Prediction of subcellular localization in human cell organelles
The results of Hum-mPLoc 2.0 shows in Table 1 and compared with the result of BaCeILo. The result showed that Hum-mPLoc 2.0 predicted total cytoplasmic (250), mitochondrial (196), nuclear (292) plasma membrane 94, endoplasmic reticulum 63, Golgi apparatus 05, peroxisome 11, microsome 01, lysosome 11, extracell 179, centrosome 05, cytoskeleton 01 and unknown (04) proteins.
Synchronization of BaCeILo predicted proteins with Hum-mPLoc 2.0 predictor
The synchronization results indicated that among the BaCeILo predicted total cytoplasmic (515), mitochondrial (183), nuclear (98) and secretory (318) proteins, not all follow the same prediction results by Hum-mPLoc 2.0 (Table 2). When these BaCeILo proteins further compared with Hum-mPLoc 2.0 results, only 47 proteins were found consistent with BaCeILo results and showed nuclear localization through in silico prediction by both computational tools.
Table 2. Possible sub-cellular localization prediction of C. pneumoniae proteins with BaCeILo and their synchronization with Hum-mPLoc 2.0 predictions.
| S. No | Intracellular distribution of BaCeILo predicted proteins as per Hum-mPLoc 2.0 | |||
|---|---|---|---|---|
| Cytoplasmic (513) | Nuclear (98) | Mitochondrial (183) | Secretory (318) | |
| 1. | Cytoplasm: 167 | Cytoplasm: 13 | Cytoplasm: 32 | Cytoplasm: 38 |
| 2. | Mitochondrion: 111 | Mitochondrion: 18 | Mitochondrion: 47 | Mitochondrion: 19 |
| 3. | Nucleus: 131 | Nucleus: 47 | Nucleus: 42 | Nucleus: 73 |
| 4. | Centrosome: 03 | Centrosome: 0 | Centrosome: 02 | Centrosome |
| 5. | Cytoskeleton: 01 | Cytoskeleton: 0 | Cytoskeleton | Cytoskeleton |
| 6. | Endoplasmic reticulum: 15 | Endoplasmic reticulum: 03 | Endoplasmic reticulum: 12 | Endoplasmic reticulum: 33 |
| 7. | Extracell: 46 | Extracell: 12 | Extracell: 19 | Extracell: 102 |
| 8. | Golgi apparatus: 1 | Golgi apparatus: 0 | Golgi apparatus: 02 | Golgi apparatus: 02 |
| 9. | Lysosome: 2 | Lysosome: 0 | Lysosome: 02 | Lysosome: 07 |
| 10. | Microsome: 1 | Microsome: 0 | Microsome | Microsome |
| 11. | Plasma Membrane: 25 | Plasma Membrane: 05 | Plasma Membrane: 23 | Plasma Membrane: 41 |
| 12. | Peroxisome: 7 | Peroxisome: 0 | Peroxisome: 02 | Peroxisome: 02 |
| 14. | Unknown: 3 | Unknown: 0 | Unknown | Unknown: 01 |
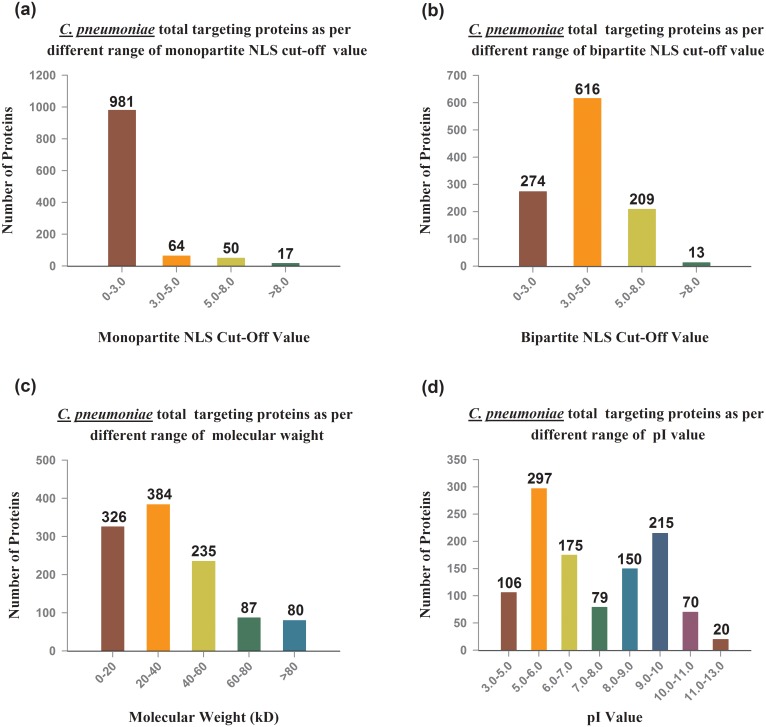
These 47 host nuclear-targeted proteins illustrated in Table 3 and arranged as per their NLS cutoff value. Increasing the cutoff value of monopartite NLS are linked with decreased nuclear targeting, whereas the reverse pattern is observed with bipartite NLS values, where the high cutoff value enhances the percentage of protein targeting to nucleus except cutoff value >8 (Table 3). The S1 Table provides details about predicted proteins target to nucleus during in silico analysis.
Table 3. Computationally prediction of C. pneumoniae proteins targeting to nucleus of host cell and their relation with all proteins with similar NLS.
| NLS | NLS cutoff | Number of proteins targeting to nucleus | Total number of proteins in this range | Percentage |
|---|---|---|---|---|
| Monopartite | 0–3.0 | 36 | 981 | 3.66 |
| 3.0–5.0 | 5 | 64 | 7.81 | |
| 5.0–8.0 | 5 | 50 | 10 | |
| >8.0 | 1 | 17 | 5.88 | |
| Bipartite | 0–3.0 | 8 | 274 | 2.91 |
| 3.0–5.0 | 17 | 616 | 2.75 | |
| 5.0–8.0 | 21 | 209 | 10.04 | |
| >8.0 | 1 | 13 | 7.69 |
Nevertheless, not any accurate relation was found between nuclear targeting protein and molecular weight, but the increased molecular weight consistently increased nuclear targeting except the one range of molecular weight 60–80 kDa, the highest molecular weight proteins (>80 kDa) were observed most targeted proteins to nucleus of host cell (Table 4).
Table 4. Computationally prediction of C. pneumoniae proteins targeting to nucleus of host cell and their relation with proteins with similar molecular weight.
| Molecular weight | Number of proteins Targeting to Nucleus | Total number of proteins | Percentage |
|---|---|---|---|
| 0–20 kD | 9 | 326 | 2.76 |
| 20–40 kD | 16 | 384 | 4.16 |
| 40–60 kD | 13 | 235 | 5.53 |
| 60–80 kD | 1 | 87 | 1.14 |
| >80 kD | 8 | 80 | 10 |
Furthermore, the value of isoelectric point (pI) did not show any constant pattern for mitochondrial targeting (Table 5).
Table 5. Computationally prediction of C. pneumoniae proteins targeting to nucleus of host cell, and their relation with all proteins with similar pI value.
| Range of pI value | Number of proteins targeting to Nucleus | Total number of proteins | Percentage |
|---|---|---|---|
| 3.0–5.0 | 3 | 106 | 2.83 |
| 5.0–6.0 | 11 | 297 | 3.70 |
| 6.0–7.0 | 10 | 175 | 5.71 |
| 7.0–8.0 | 4 | 79 | 5.06 |
| 8.0–9.0 | 8 | 150 | 5.33 |
| 9.0–10 | 10 | 215 | 4.65 |
| 10.0–11.0 | 1 | 70 | 1.42 |
| 11.0–13.0 | 0 | 20 | 0.0 |
The patterns of C. pneumoniae protein targeting in host cell nucleus with different parameter shows in Fig 1, whereas the all proteins targeting of C. pneumoniae in host cells components with different parameters illustrates in Fig 2. The supplementary data provides details regarding proteins predicted to target nucleus of host cell during our analysis (S1 Table).
Fig 1. Computationally prediction of C. pneumoniae proteins targeting to nucleus of host cells and their relation with various parameters.
Fig 2. Computationally prediction of whole proteome of C. pneumoniae proteins (UniProt data base) and their relation with various parameters.
Discussion
Epidemiological reports have founded several well-defined potential factors for the growth and development of cancer such as heredity, age, use of tobacco, diet, inflammation, and chronic infections with pathogens. Infection is the cause for approximately 16% of all malignancies worldwide [19]. A number of bacteria have shown the capacity to alter many pathways and molecules of host cells for their intracellular survival in the host. The thought that an infection of bacteria could promote to carcinogenesis disregards initially. However, landmark studies in the early 1990s established C. pneumoniae as a causative agent of different lung cancers, resulting in a new orientation of the scientific focus toward patterns of bacterial association with cancers (5, 6). C. pneumoniae bacterial derived effector molecules can change the internal environment of host cell through the production of chronic inflammation, inhibition of tumor suppressor mechanisms, induction of immunosuppression, and transformation of cells by transfer of oncogenes [7, 13, 14, 20–22]. The proteins of C. pneumoniae will be existed in the host cell during chronic infection and some proteins may migrate to the many organelles of the host cell such as nucleus, endoplasmic reticulum, Golgi apparatus, mitochondria etc. Proteins enter into host nuclei have many adverse effects that may inhibit or promote certain important biological activities leading to the development of cancer.
Subcellular protein targeting may be predicted by various tools, that works on different principles and different parameters. These parameters included composite motifs through the artificial neural feed-forward network, different binding grooves of importin α, simple Hidden Markov model, identity/alignment search, linear motifs and their role in cell signaling and regulation, support vector machine (SVM), and functional domain information along with sequential evolution information [23–28]. We predicted sub cellular proteins targeting using cNLS mapper, BaCeILo and Hum-mPLoc 2.0 predictors in order to achieve more accurate and consistent results.
Nuclear localization signals (NLSs) are very critical to ensure the selective transport of proteins into the nucleus [29]. The cNLS Mapper tool correctly locates the nuclear localization signals (NLSs) particularly to the importin α/β pathway by predicting NLS scores. The calculated NLSs are divided into two classes, monopartite (1 basic cluster) and bipartite (2 basic cluster) NLSs, according to the existence of cluster of rich basic amino acid residues. Furthermore, the scores of NLS are evaluated with four classes of profiles with specified cut off values. Higher values of NLS score show more NLS activities. Proteins with cut off value 8–10 were predicted as localized to the nucleus, 8–10 as partially in the nucleus, 3–5 as both in nucleus and cytoplasm, and 1–2 as particularly in cytoplasm. The proteins of C. pneumoniae indicating intermediary cut off value was included in a specific range of cut off value in protein list such as a cut off value 7.5 or above was rounded up to 8 while 7.4 or below was rounded down to 7. cNLS mapper was used to compute NLS activity instead of NLS sequence because NLS sequences are not stringent enough [26]. However, it must be remembered that the NLS profiles of cNLS mapper were produced by nuclear import assays using the data of yeast. Therefore, the prediction of NLS for other species may not be as accurate as in yeast although the importin α/β pathway of eukaryotes is highly conserved. Recent study identified a new bacterial protein SINC that targets the nuclear envelope in the infected and non-infected neighboring host cell with the potential of modifying nuclear envelope functions. These capabilities of C. psittaci bacteria may promote the process of destructive pathogenesis [30].
BaCelLo is a computational predictor used in our study for the subcellular localization of proteins in eukaryotes. Animals, plants, and fungi predictors were implemented in BaCelLo; hence, we used animal specific predictor. It is based on diverse SVMs for the prediction of nuclear, cytoplasmic, mitochondrial, secretory, and chloroplast targeting proteins [24]. BaCelLo predicts subcellular targeting based on residue sequence information and evolutionary information included in alignment profiles within the N and C termini and entire protein sequence.
Prediction of subcellular localization of proteins in human is a more challenging task. Another subcellular localization prediction tool Hum-mPLoc 2.0 was used to deal with the nuclear targeting of protein in human system. The Hum-mPLoc 2.0 tool predicts the proteins targeting on the basis of domain information and the sequential evolution information. The predictor computes 14 subcellular locations including nucleus, cytoplasm, mitochondrion, plasma membrane, endoplasmic reticulum, extracell, Golgi apparatus, cytoskeleton, endosome, lysosome, peroxisome, microsome, synapse, and centriole. Although the comparative results of BaCeILo and Hum-mPLoc 2.0 were demonstrated slight difference in subcellular localization of C. pneumoniae proteins in host organelles, the slight difference in results of BaCeILo and Hum-mPLoc 2.0 may be due to the existence of different data in their respective datasets used during prediction. Therefore, the little variations in outcomes obtained from different tools can be accordingly justified.
In this study, we present a systematic computational prediction of C. pneumoniae proteins using differently functioning predictors: NLS mapper, BaCeILo, and Hum-mPLoc 2.0 that works on different datasets. The prediction of BaCeILo based on animal dataset, whereas the Hum-mPLoc 2.0 worked on human specific datasets which includes 3,681 human proteins classified into 14 different human sub cellular locations. Therefore, the results were further narrow down and scrutinized after using human dataset specific predictor. It has been reported that many proteins can localize in the nucleus in the absence of NLS [24, 31]. Moreover, proteins less than 40kD can freely diffuse to nucleus [32]. According to our predictions, little variation in the results expected due to the use of different tools. Therefore, these results of in silico prediction require further experimental verification prior to any final conclusion. Furthermore, we focused on the potential effects of these nuclear targeting proteins in tumorigenesis and development of cancer.
DNA replication and DNA binding proteins
The genomic instability is a crucial factor in cancer. Nonetheless, the mechanisms of its growth and development remain not fully understood. A frequently stated assumption is that anomalies in translesion DNA synthesis or error-prone phenotypes in DNA replication participate in genomic DNA instability and are prominent cause of the development of cancer. Such, erroneous DNA replication mechanisms have been implicated as an etiological factor in many cancers [33–35].
For instance, DNA polymerase beta protein is involved in approximate 30% all human tumors reported to date due to mutations [33, 34]. The bacterial DNA polymerase III subunit beta has a homolog of eukaryotic proliferating cell nuclear antigen (PCNA). PCNA is identified as a molecular marker for cell proliferation during replication [36, 37]. In our study, we found nuclear localization of DNA polymerase III subunit beta protein. PCNA was characterized as a potential antigen that is expressed during the phase of DNA synthesis in cell cycle and involved in carcinogenesis [38]. Therefore, during infection, the possible existence of two homologs proteins in same cell with unique enzymatic action alters the relative activity of host protein. As DNA polymerase III subunit beta is a DNA-replication connected proteins, the anomaly in DNA replication can also act as a factor for the growth and development of cancer.
Another chaperone protein DnaJ a homolog of HSP40 is also predicted as nuclear target that may change the activity of HSP40 and involved in carcinogenesis [39, 40]. In addition, certain DNA replication and binding proteins are also predicted as nuclear targeting proteins such as DNA gyrase subunit A, single-stranded DNA-binding protein (SSB), and primosomal protein, which may also be involved in the development of cancer.
Gene expression associated proteins
Translational regulation is a crucial process in the progression and development of cancer. It manages both the overall expression of protein synthesis and the specific translation of selective mRNAs that may support various oncogenic properties including cell transformation, tumor cell survival, invasion, metastasis, and angiogenesis. The nuclear targeting of these gene expression proteins implies their potential roles in the growth and development of lung cancer. Consequently, alteration in the gene expression is connected with the growth and development of cancer through the dysregulation of many critical genes. Dysregulation may direct the activation of proto-oncogenes and suppression of anti-oncogenes [41].
The results of our study show that DNA-directed RNA polymerase β and β’ subunits of C. pneumoniae are targeted to host cell nucleus. This is consistent with other reports that demonstrated an alteration in the levels of gene expression in many hosts, including human and other eukaryotes, as an action of bacterial RNA polymerase. For instance, various human genes may be transcribed through the involvement bacterial transcription regulators using E. coli DNA-directed RNA polymerase II [42, 43]. These predicted transcription-associated proteins may efficiently bind to host DNA and consequently hinder the binding affinity of the host transcription regulators and ultimately deregulate gene expression [42]. Although it has been confirmed that C. pneumoniae is associated with alteration of host gene expression [21], its involvement in the progression and development of lung cancer in human requires further experimental assessment. Our result encompasses important findings that can contribute to this emerging field.
DNA damage and repair proteins
Previous study showed that C. pneumoniae has the ability to induce DNA damage through the induction of reactive oxygen species (ROS) [13]. Our findings have demonstrated nuclear targeting of DNA-damaging proteins including exonuclease V subunit RecB, ribonuclease R, exodeoxyribonuclease VII small subunit, and exodeoxyribonuclease VII large subunit. Moreover, it is found that DNA mismatch repair is essential for enhancing the fidelity of replication in most of the organisms including bacteria, yeast, and humans etc. MutS has been identified as a protein of the ABC ATPase superfamily, which is involved in unpaired and mispaired bases in double stranded DNA that initiates mismatch repair. Mutation in MutS may be a possible cause of the growth and development of cancer [44, 45]. We predicted nuclear localization of DNA mismatch repair proteins MutS and MutL during the analysis. Alteration in mismatch repair proteins is potentially associated with various types of human cancers including lung.
Conclusion
We proposed a new and integrative in silico approach for identifying the suspicious role of C. pneumoniae proteins in the growth and development of lung cancer in human. The results of in silico prediction revealed 47 candidates proteins. Out of which, various proteins may have the potential to trigger cancer growth through the alteration in replication, transcription, and DNA damage repair mechanism. It is confirmed that various proteins of C. pneumoniae can target to different organelles including nucleus and other parts of host cells, which may be an etiological cause of lung cancer. Our prediction data demonstrated more accuracy of computational prediction due to the use of different prediction tool based on different datasets, which may suggest that nuclear targeting proteins of C. pneumoniae can be potential targets for lung cancer management. Therefore, the outcome of this in silico study can open the new avenue for lung cancer research. Although the oncogenic potential and significant contribution of this nuclear-targeted protein of C. pneumoniae in growth and development of cancer was suggested by our knowledge and computational analysis, the confirmatory roles and specificity of these predicted proteins in carcinogenesis process require further experimental validation.
Materials and Methods
Selection of protein database
C. pneumoniae is an obligate intracellular gram negative pathogen, infects humans and suspiciously involved as an etiological agent of lung cancer [5, 46]. The proteome of C. pneumoniae TW-183 were downloaded from Uniprot database. Five proteomes of different strains of C. pneumoniae were available [15–18]. The proteome of TW-183 strain of C. pneumoniae was analyzed for the prediction of the nuclear localization signal and human cell subcellular localization using different computational tools.
Prediction of nuclear localization signal
cNLS mapper tool for eukaryotic cells was used for the prediction of nuclear localization signal in TW-183 protein of C. pneumoniae [23]. The complete sequence of each C. pneumoniae protein was used for the prediction of monopartite and bipartite NLS sequence.
Prediction of subcellular localization in eukaryotic cell organelles
The balanced subcellular localization predictor (BaCeILo) was used to predict the subcellular localization of TW-183 protein of C. pneumoniae in eukaryotic cell compartments. BaCelLo has based on three specific predictors for eukaryotic kingdoms including animals, plants, and fungi [24]. BaCeILo predict five classes of sub cellular localization including nuclear, mitochondrial, cytoplasmic, secretory, and chloroplast. We were done the prediction with animal specific predictor using proteins of TW-183 strain of C. pneumoniae.
Prediction of subcellular localization in human cell organelles
Furthermore, human protein subcellular localization Hum-mPLoc 2.0 (Hum-mPLoc 2.0) predictor was used to predict the subcellular localization of TW-183 of C. pneumoniae proteins in nucleus and other cell organelles in human [25]. Hum-mPLoc 2.0 predict the fourteen classes of subcellular localization that includes nucleus, cytoplasm, mitochondrion, endoplasmic reticulum, centriole, cytoskeleton, endosome, extracell, Golgi apparatus, lysosome, microsome, peroxisome, plasma membrane, and synapse.
Synchronization of BaCeILo predicted proteins with Hum-mPLoc 2.0 predictor
Moreover synchronization was preformed for the predication of nuclear targeting protein in human using Hum-mPLoc 2.0. The results of BaCelLo was used to narrow down the sub cellular localization of C. pneumoniae proteins.
Supporting Information
(DOC)
Acknowledgments
This project was funded by the Research Groups Program (Research Group number RG-1436-027), Deanship of Scientific Research, King Saud University, Riyadh, Saudi Arabia.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This project was funded by the Research Groups Program (Research Group number RG-1436-027), Deanship of Scientific Research, King Saud University, Riyadh, Saudi Arabia.
References
- 1.(WHO) WHO. GLOBOCAN 2012: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012. International Agency for Research on Cancer; 2014. [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013; 63:11–30. 10.3322/caac.21166 [DOI] [PubMed] [Google Scholar]
- 3.Chaturvedi AK, Gaydos CA, Agreda P, Holden JP, Chatterjee N, Goedert JJ, et al. Chlamydia pneumoniae infection and risk for lung cancer. Cancer Epidemiol Biomarkers Prev 2010; 19:1498–505. 10.1158/1055-9965.EPI-09-1261 [DOI] [PubMed] [Google Scholar]
- 4.Lin TY, Huang WY, Lin JC, Lin CL, Sung FC, Kao CH, et al. Increased lung cancer risk among patients with pneumococcal pneumonia: a nationwide population-based cohort study. Lung 2014; 192:159–65. 10.1007/s00408-013-9523-z [DOI] [PubMed] [Google Scholar]
- 5.Zhan P, Suo LJ, Qian Q, Shen XK, Qiu LX, Yu LK, et al. Chlamydia pneumoniae infection and lung cancer risk: a meta-analysis. Eur J Cancer 2011; 47:742–7. 10.1016/j.ejca.2010.11.003 [DOI] [PubMed] [Google Scholar]
- 6.Chu DJ, Guo SG, Pan CF, Wang J, Du Y, Lu XF, et al. An experimental model for induction of lung cancer in rats by Chlamydia pneumoniae. Asian Pac J Cancer Prev 2012; 13:2819–22. [DOI] [PubMed] [Google Scholar]
- 7.Laurila AL, Anttila T, Laara E, Bloigu A, Virtamo J, Albanes D, et al. Serological evidence of an association between Chlamydia pneumoniae infection and lung cancer. Int J Cancer 1997; 74:31–4. [DOI] [PubMed] [Google Scholar]
- 8.Littman AJ, White E, Jackson LA, Thornquist MD, Gaydos CA, Goodman GE, et al. Chlamydia pneumoniae infection and risk of lung cancer. Cancer Epidemiol Biomarkers Prev 2004; 13:1624–30. [PubMed] [Google Scholar]
- 9.Littman AJ, Jackson LA, Vaughan TL. Chlamydia pneumoniae and lung cancer: epidemiologic evidence. Cancer Epidemiol Biomarkers Prev 2005; 14:773–8. [DOI] [PubMed] [Google Scholar]
- 10.Smith JS, Kumlin U, Nyberg F, Fortes C, Zaridze D, Ahrens W, et al. Lack of association between serum antibodies of Chlamydia pneumoniae infection and the risk of lung cancer. Int J Cancer 2008; 123:2469–71. 10.1002/ijc.23814 [DOI] [PubMed] [Google Scholar]
- 11.Kocazeybek B. Chronic Chlamydophila pneumoniae infection in lung cancer, a risk factor: a case-control study. J Med Microbiol 2003; 52:721–6. [DOI] [PubMed] [Google Scholar]
- 12.Koyi H, Branden E, Gnarpe J, Gnarpe H, Steen B. An association between chronic infection with Chlamydia pneumoniae and lung cancer. A prospective 2-year study. Apmis 2001; 109:572–80. [DOI] [PubMed] [Google Scholar]
- 13.Jackson LA, Wang SP, Nazar-Stewart V, Grayston JT, Vaughan TL. Association of Chlamydia pneumoniae immunoglobulin A seropositivity and risk of lung cancer. Cancer Epidemiol Biomarkers Prev 2000; 9:1263–6. [PubMed] [Google Scholar]
- 14.Redecke V, Dalhoff K, Bohnet S, Braun J, Maass M. Interaction of Chlamydia pneumoniae and human alveolar macrophages: infection and inflammatory response. Am J Respir Cell Mol Biol 1998; 19:721–7. [DOI] [PubMed] [Google Scholar]
- 15.Shirai M, Hirakawa H, Kimoto M, Tabuchi M, Kishi F, Ouchi K, et al. Comparison of whole genome sequences of Chlamydia pneumoniae J138 from Japan and CWL029 from USA. Nucleic Acids Res 2000; 28:2311–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalman S, Mitchell W, Marathe R, Lammel C, Fan J, Hyman RW, et al. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat Genet 1999; 21:385–9. [DOI] [PubMed] [Google Scholar]
- 17.Myers GS, Mathews SA, Eppinger M, Mitchell C, O'Brien KK, White OR, et al. Evidence that human Chlamydia pneumoniae was zoonotically acquired. J Bacteriol 2009; 191:7225–33. 10.1128/JB.00746-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, et al. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res 2000; 28:1397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gisserot O, Romeo E, Boudin L, Tsitsi Nding Tsogou P, Abed S, Blade JS, et al. [Can we prevent or cure infection-related cancers?]. Rev Med Interne 2014; 35:259–63. [DOI] [PubMed] [Google Scholar]
- 20.Eickhoff M, Thalmann J, Hess S, Martin M, Laue T, Kruppa J, et al. Host cell responses to Chlamydia pneumoniae in gamma interferon-induced persistence overlap those of productive infection and are linked to genes involved in apoptosis, cell cycle, and metabolism. Infect Immun 2007; 75:2853–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hess S, Peters J, Bartling G, Rheinheimer C, Hegde P, Magid-Slav M, et al. More than just innate immunity: comparative analysis of Chlamydophila pneumoniae and Chlamydia trachomatis effects on host-cell gene regulation. Cell Microbiol 2003; 5:785–95. [DOI] [PubMed] [Google Scholar]
- 22.Kuper H, Adami HO, Trichopoulos D. Infections as a major preventable cause of human cancer. J Intern Med 2000; 248:171–83. [DOI] [PubMed] [Google Scholar]
- 23.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci U S A 2009; 106:10171–6. 10.1073/pnas.0900604106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pierleoni A, Martelli PL, Fariselli P, Casadio R. BaCelLo: a balanced subcellular localization predictor. Bioinformatics 2006; 22:e408–16. [DOI] [PubMed] [Google Scholar]
- 25.Shen HB, Chou KC. A top-down approach to enhance the power of predicting human protein subcellular localization: Hum-mPLoc 2.0. Anal Biochem 2009; 394:269–74. 10.1016/j.ab.2009.07.046 [DOI] [PubMed] [Google Scholar]
- 26.Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, Tomita M, et al. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J Biol Chem 2009; 284:478–85. 10.1074/jbc.M807017200 [DOI] [PubMed] [Google Scholar]
- 27.Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics 2009; 10:202 10.1186/1471-2105-10-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diella F, Haslam N, Chica C, Budd A, Michael S, Brown NP, et al. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front Biosci 2008; 13:6580–603. [DOI] [PubMed] [Google Scholar]
- 29.Su EC, Chang JM, Cheng CW, Sung TY, Hsu WL. Prediction of nuclear proteins using nuclear translocation signals proposed by probabilistic latent semantic indexing. BMC Bioinformatics 2012; 13 Suppl 17:S13 10.1186/1471-2105-13-S17-S13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mojica SA, Hovis KM, Frieman MB, Tran B, Hsia RC, Ravel J, et al. SINC, a type III secreted protein of Chlamydia psittaci, targets the inner nuclear membrane of infected cells and uninfected neighbors. Mol Biol Cell 2015; 26:1918–34. 10.1091/mbc.E14-11-1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.la Cour T, Gupta R, Rapacki K, Skriver K, Poulsen FM, Brunak S. NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res 2003; 31:393–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorlich D, Mattaj IW. Nucleocytoplasmic transport. Science 1996; 271:1513–8. [DOI] [PubMed] [Google Scholar]
- 33.Tan X, Wang H, Luo G, Ren S, Li W, Cui J, et al. Clinical significance of a point mutation in DNA polymerase beta (POLB) gene in gastric cancer. Int J Biol Sci 2015; 11:144–55. 10.7150/ijbs.10692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dalal S, Chikova A, Jaeger J, Sweasy JB. The Leu22Pro tumor-associated variant of DNA polymerase beta is dRP lyase deficient. Nucleic Acids Res 2008; 36:411–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki M, Takahashi T. Aberrant DNA replication in cancer. Mutat Res 2013; 743–744:111–7. 10.1016/j.mrfmmm.2012.07.003 [DOI] [PubMed] [Google Scholar]
- 36.Jeruzalmi D, Yurieva O, Zhao Y, Young M, Stewart J, Hingorani M, et al. Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell 2001; 106:417–28. [PubMed] [Google Scholar]
- 37.Wang SC. PCNA: a silent housekeeper or a potential therapeutic target? Trends Pharmacol Sci 2014; 35:178–86. 10.1016/j.tips.2014.02.004 [DOI] [PubMed] [Google Scholar]
- 38.Leonardi E, Girlando S, Serio G, Mauri FA, Perrone G, Scampini S, et al. PCNA and Ki67 expression in breast carcinoma: correlations with clinical and biological variables. J Clin Pathol 1992; 45:416–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hattori H, Liu YC, Tohnai I, Ueda M, Kaneda T, Kobayashi T, et al. Intracellular localization and partial amino acid sequence of a stress-inducible 40-kDa protein in HeLa cells. Cell Struct Funct 1992; 17:77–86. [DOI] [PubMed] [Google Scholar]
- 40.Sterrenberg JN, Blatch GL, Edkins AL. Human DNAJ in cancer and stem cells. Cancer Lett 2011; 312:129–42. 10.1016/j.canlet.2011.08.019 [DOI] [PubMed] [Google Scholar]
- 41.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med 2004; 10:789–99. [DOI] [PubMed] [Google Scholar]
- 42.Karin M. Too many transcription factors: positive and negative interactions. New Biol 1990; 2:126–31. [PubMed] [Google Scholar]
- 43.Mote J Jr., Reines D. Recognition of a human arrest site is conserved between RNA polymerase II and prokaryotic RNA polymerases. J Biol Chem 1998; 273:16843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature 2000; 407:711–7. [DOI] [PubMed] [Google Scholar]
- 45.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature 2000; 407:703–10. [DOI] [PubMed] [Google Scholar]
- 46.Yang J, Hooper WC, Phillips DJ, Tondella ML, Talkington DF. Induction of proinflammatory cytokines in human lung epithelial cells during Chlamydia pneumoniae infection. Infect Immun 2003; 71:614–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.