Abstract
Atomically detailed computer simulations of complex molecular events attracted the imagination of many researchers in the field as providing comprehensive information on chemical, biological, and physical processes. However, one of the greatest limitations of these simulations is of time scales. The physical time scales accessible to straightforward simulations are too short to address many interesting and important molecular events. In the last decade significant advances were made in different directions (theory, software, and hardware) that significantly expand the capabilities and accuracies of these techniques. This perspective describes and critically examines some of these advances.
INTRODUCTION
Atomically detailed computer simulations of molecular processes have proven useful in many areas of chemical physics, material science, and molecular biophysics (for books see Ref. 1). Of interest to the present article is the technique of classical Molecular Dynamics (MD) which is loosely defined as the numerical solution of the classical equations of motion for molecular systems. We denote by Γ ∈ R6L the phase space vector of a system of L particles and by the sequence of phase points as a function of time t (the MD trajectory). Trajectories produced by the MD method are used to investigate molecular kinetics and thermodynamics.
As the title suggests, we are primarily interested in the study of kinetics, which is the time evolution of an ensemble of trajectories between predetermined states. This is to be contrasted with the sampling of an ensemble of configurations for equilibrium averages and for calculations of thermodynamic observables. An ensemble representing equilibrium can be computed with or without a time series. For kinetics, at least one time series is necessary to determine correlation functions and transition rates between states.
The most widely used numerical realizations of MD trajectories are solutions of initial value problems. Indeed, initial value integration algorithms, such as Verlet,2 dominate the field and form the core of a number of molecular modeling software packages. It is amusing to note that later in his life Verlet became interested in the history of science and discovered that “his” algorithm was published in Newton’s Principia in 1687 to prove Kepler’s second law.3 The wide use and successes of the initial value formulation underline the significant drawback of MD, namely, the limitation of time scales. To appreciate the source of this limitation and its importance consider an explicit implementation of the Verlet algorithm in which the coordinates and velocities are solved in small time steps. We call this and related algorithms Initial Value Solvers (IVSs)
| (1) |
To ensure numerical accuracy and stability of the time series, the time step, Δt, must be small relative to the time scale of the fastest coordinate changes in the system. Bond vibrations are typical examples for fast motions that require small time steps, indeed, periods of vibrations of bonds with hydrogen atoms are of a few tens of femtoseconds which makes it necessary to set Δt ∼ 1 fs to follow these vibrations accurately. The overall time scale studied is the sum of many steps computed numerically. Without special hardware4 and/or massively parallel computers, it is difficult to find simulations of condensed phase systems that exceed 109 steps or a few microseconds.
However, even this time scale is far too short to investigate many interesting problems in molecular biophysics. Rapid events of protein folding take place over tens of microseconds to milliseconds,5,6 and so do many conformational transitions, changing the structure of the protein from inactive to active forms.7 Simulations of microsecond length are a significant feat on readily accessible computers. Many enzymatic reactions that couple chemical reactions with major conformational transitions occur on the millisecond to second time scale, or longer, making it necessary to further extend the time scales of simulation to new uncharted territories. Furthermore, observing and sampling a single event are not sufficient to study kinetics and sampling multiple transitions is required. Statistically meaningful observation requires the measurement of the change of population and not of a single trajectory. A single event, even if measured experimentally with the tools of “single-molecule” spectroscopy,8 is hard to assess quantitatively.
Given that MD is a general method appropriate for a wide range of tasks with a minimal set of approximations (the most obvious ones are the use of classical mechanics, and an empirical force field) it is desirable to extend its applicability to kinetics beyond the microsecond time scale. The present Perspective discusses different approaches to reach long time scales in molecular dynamics simulations. In brief, the advances can be divided into three categories: numerical and physics-based analysis, special purpose hardware, and physical modeling. From a methodological viewpoint, we consider calculations of individual trajectories, sampling of trajectories, and the computation and use of trajectory fragments.
NUMERICAL AND PHYSICS-BASED ANALYSIS
In one direction of research, techniques from numerical analysis and applied mathematics are used to seek an effective increase in the time step and hence significant speed up in MD simulations. One of the earliest approaches of this category is SHAKE9 in which constraining bond lengths, dij, in molecules to their ideal values, dij,0 ( bonded pairs) removes fast motions and enables the increase of efficiency of the calculations by about a factor of two. Since bond lengths vary a little in normal circumstances, the physics of the system is only slightly altered, allowing for sound physical representation of atomically detailed models at reduced costs and larger time steps.
A factor of two is significant and essentially all major molecular dynamics software packages include the SHAKE option (or variants) in their arsenal. However, it is not the factor of thousands and millions that we mentioned in the Introduction. Can we increase the time step further given that the fastest motions are removed?
Simulations in condensed phases have another type of fast processes that is difficult to eliminate during simulations. These extra fast processes make it challenging to expand the scope of MD and limit the benefits of SHAKE. In condensed phases, we find hard-core collisions of particles that are frequently modeled by the stiff parts of the Lennard Jones terms (A/r12) where r is the distance between the two particles and A an empirical constant. Pairs of atoms that enter the domain in which this interaction is significant accelerate until they leave the collision zone. The velocity changes in the collision zone can be particularly dramatic and require accurate, and small-step integration. In contrast to gas phase and typical of solvated phases of biological molecules, collisions are frequent and sequential configurations in numerical integration of midsize and large systems are rarely collision-free. How to model collision events with SHAKE like algorithms is, however, not known. Collisions are transient events, a pair of atoms interacts only a fraction of time of the total trajectory, and the identity of colliding pairs is changing rapidly. In current applications of SHAKE, we have a list of fixed fast degrees of freedom (covalently bonded atoms). No such list is available for condensed phase collisions.
A conceptually similar approach to SHAKE (in the sense that fast motions are treated separately in IVS) is proposed by multi-stepping algorithms in which rapidly changing degrees of freedom are integrated with small time steps while coordinates or forces that are slow to vary are integrated with larger time steps. A leading algorithm of this type is RESPA (REference System Propagation Algorithm)10 based on a Trotter expansion of the time propagator, ensuring time reversibility. The forces are split into a sum of long range and short-range interactions. Short range and repulsive Lennard Jones forces are integrated with smaller time steps. The long-range interactions that are more numerous are integrated less frequently. The splitting leads to significant computational gains and an increase in the size of the time step used for the slower degrees of freedom. Unfortunately, the hierarchical integration adds significant complexity to the calculations and reduces the benefits. The increase in the time step is also moderate (a factor of several). It is bounded not only by fast vibrations but also by rapid collision events. The overall efficiency for deterministic dynamics in complex systems rarely exceeds a factor of 2. This is of course useful but not a solution for the extensive time scale gap mentioned in the Introduction.
The above formulations are rigorous and no compromise on accuracy and details of the modeling is made. Is it possible to obtain useful, but approximate solutions at lower cost? Unfortunately, the popular Verlet or similar algorithms are not a convenient starting point for approximations. If the time step describing fast motions in atomically detailed simulations increases significantly from 1 fs to say ∼10 fs the solution develops instabilities and “blows up.” Different formulations are required in order to obtain sensible approximate solutions. An intriguing numerical approach to generate approximate trajectories with a large step is the backward Euler algorithm,11 in which an exceptionally large time step can be taken (tens of femtoseconds) and a correction step (minimization) follows. The algorithm was shown to “leak” energy from high frequency modes. A fix was proposed in conjunction with a Langevin formulation using random noise to supply back lost energy.12
Stochastic dynamics (like the Langevin “fix” mentioned in the above paragraph) can use time steps larger than deterministic algorithms. If the noise and friction are gentle (e.g., they are added only to a few external variables such as thermostat degrees of freedom) then correlation functions were shown to be reproduced accurately.13,14 The efficiency in this case is found to be less than a factor of 6.15,16 Further research on gentle stochastic algorithms is desired. Moreover, in the context of algorithms with multiple time steps and Nose-Hoover thermostats,17 it is possible to sample from the correct ensembles efficiently with a very large time step. While the dynamic content of these trajectories is not clear, they may provide initial conditions for the large number of short trajectories that are used to enhance sampling of kinetics (see section titled Physical Modeling).
Filtering of high frequency modes and focusing on slow motions with the largest amplitudes at thermal conditions was the focus of another research direction.18 Rather than solving an initial value problem like in Eq. (1) an action is defined and boundary value formulation is used. Of interest was the Onsager-Machlup action18 at the zero friction limit, , which is discretized and minimized to obtain an approximate classical trajectory (depending on the size of the time step Δt)
| (2) |
where the are constraints on overall translations and rotations of the images of the system xi.19 The coordinates of the edges of the path x0 and xN are fixed. After the discretization, the action is a function of the complete trajectories and a solution can be found by setting the gradient of the discrete action to zero . An algorithm to optimize the action using the functional derivative (or the derivatives of the discrete action) is the following.
-
1.
Provide a guess for a trajectory of N steps: .
-
2.
Compute action of the current trajectory (iteration n) and its derivatives: and .
-
3.
Test if where ε is a small positive number used as convergence criterion. If the test is true the process is terminated. If not continue. Note that the check is for S being stationary, not necessarily a minimum.
-
4.
Generate a step Δ in discrete trajectory space to find a new trajectory that reduces the value of the target function Θ. If the derivatives are known, use derivative information to “quench” the trajectory following the direction opposite to the gradient: where a is a scalar chosen to minimize Θ. Alternatively, an optimization step can be chosen stochastically. A random displacement Δ is accepted or rejected according to Metropolis criterion: , where is a fictitious Boltzmann factor that increases during the optimization. Optimal action and trajectories are obtained when .
-
5.
Return to step 2 to evaluate the current trajectory.
It was shown that a global minimum of Eq. (2) filters out motions with frequencies ω ≥ π/Δt, so one effect is the removal of high frequency motions. Another effect is the indirect impact of the removal of these motions on the slower degrees of freedom, which is harder to assess. The creation of a numerically stable (but approximate) trajectory uses a similar motivation as the backward Euler integration algorithm. The numerical stability of a trajectory as a function of the time step generated with action optimization is going far beyond the stability of initial value solvers. This observation makes it possible to produce approximate trajectories at highly extended time scales. We call this class of numerical approaches Boundary Value Solvers (BVSs).
While initial value solvers of the equations of motion are restricted in practice to time as a variable, more diversity is found in action optimization. Besides solution as a function of time one also finds practical solutions in the arc length that gained in popularity.20–25 These trajectory calculations were conducted for alternative types of dynamics, from Hamiltonian dynamics19,20,25,26 to overdamped Langevin.21–24,27 Optimization of actions and determination of trajectories as a function of the arc-length were used to find order of events in protein folding20,25,28 and to suggest reaction coordinates for more elaborate rate calculations.29 This type of study has recently gained considerable momentum following rigorous mathematical analysis of path integral formulation of stochastic trajectories, in both time19,26,30,31 and arc-length.21–24,27 It should be noted that the conversion between time and arc-length is far from trivial. The trajectories computed with large steps (either in time or in the arc-length) are based on filtering high frequency motions that add up noisy and diffusive contributions to the trajectories, retaining the more deterministic and aimed displacements. The part that is left out can be long in time. The filtering was discussed extensively in the past.18 As such, the time estimated directly from filtered trajectories is bound to be too short, depending on the extent of the filtering. Nevertheless, the set of structures generated by these calculations can be used as a starting point for more elaborate calculations of the kinetics, such as Milestoning. The structures provide seeds for coarse and importance sampling of the relevant reaction space. For example, in the method of Milestoning, the seeds can be made centers of Voronoi cells32,33 that are used in the calculations of short trajectories between the cells and eventually in the studies of kinetics and thermodynamics (for more information see section titled Enhancing Sampling of Reactive Trajectories by “Celling”).
In general, IVS approaches to molecular dynamics are much more popular than BVS. Nevertheless, the use of BVS is still significant. A comparison between the two approaches is therefore in place. When we compare the costs of calculations of a single trajectory of the same accuracy computed with initial value formulation or with an optimization of an action the initial value formulation is a clear winner. It is faster and more accurate. In initial value formulation, we generate one phase space point at a time, and after it is computed (e.g., using the Verlet algorithm) the phase space point remains fixed and is no longer considered computationally. In the action optimization, we adjust each of the structures along the trajectory many times until the convergence criteria are satisfied (e.g., the functional derivative of the action or the derivatives of the discretized action are sufficiently close to zero). Hence, the optimization of the action for the same accuracy and length will be slower by a factor of the number of optimization cycles.
In practice the optimization of the action, starting from an initial guess for a trajectory is considerably more difficult than producing an IVS solution. The action minimization is a global optimization problem for a sequence of structures along a discrete approximation of a curve. Hence, the dimensionality of the optimization problem is extremely high. If protein folding is a hard problem to solve, the problem of finding a pathway for folding, which is much larger, is obviously harder.
Most frequently only local optimizations of the action were computed, refining initial guesses. For example, the focus was on estimates of trajectories in the neighborhood of minimum energy pathways.20,25 The trajectories so obtained allow the incorporation of thermal (kinetic) energy into the ensemble of pathways. These calculations improve the value of the action and the pathway. However, the path remains correlated with the initial guess. It is therefore desired to develop more efficient global optimization schemes for pathways, to make the BVS approach more attractive. Bai and Elber26 examined a global optimization relaxation algorithm to determine trajectories from the classical action. They illustrate their algorithm for an isomerization problem in a Lennard-Jones cluster of 38 atoms. However, more could be done to address the global optimization problem of pathways and we hope to see more future research in this direction.
Another non-trivial complication for BVS use is the treatment of explicit solvent. Solvent molecules are mobile and permutable. It is therefore not advisable to label them explicitly and to optimize action using the Cartesian coordinate of each water molecules. Averages over solvent coordinate to produce densities are likely to be effective (though expensive). Averages of this type were employed already in the study of hydrophobic collapse by Miller and Vanden-Eijnden34 using the string method, and by Cardenas and Elber in the investigation of membrane fluctuations35 using Milestoning.
A hybrid approach36 combines the generation of IVS trajectories with a bias towards a desired product state with the re-weighting of the trajectories using an action. The advantage is the efficiency and simplicity of the protocol. The bias, however, may tilt the ensemble towards restricted sampling missing mechanisms that are inconsistent with the functional choice of the bias.
The use of much larger time or length steps in these protocols is a necessity to make the efficiency of these computational techniques competitive with the initial value formulation. However, they clearly cannot compete in accuracy. Despite the lower efficiency, action-based trajectory calculations are attractive for a number of reasons. First, they are typically computed between known end points. If the task at hand is the elucidation of reaction mechanisms between two known molecular structures or states, the action optimization is using all the information at hand. A solution of an initial value problem uses only local information (typically one of the end states) and cannot guarantee that the trajectory will end at the desired point for reasonable values of trajectory lengths. Many trials may be necessary adding significantly to the complexity of the calculation. If the transition is particularly rare (i.e., the number of trajectory trials before transition is detected is large) then the efficiency of calculations can shift from IVS to BVS. Typical biological examples that can benefit from the boundary value formulation include conformational transitions in proteins24 and transport through membranes.37
Simulation of protein folding is not a natural candidate for BVS since the unfolded state is not a single or even the neighborhood of a single structure. A plausible workaround is to sample unfolded configurations and conduct action optimization from the sampled set to folded conformations.20,25 In this case, we sample widely different starting configurations while converging to a set of similar structures. A large step (in the arc-length) can still be used.
The action formulation is interesting for stochastic dynamics because it assigns variable weights to different trajectories or pathways. The usual IVS formulation of Equation (1) assigns the same weight for all trajectories with the same energy. The BVS formulation makes it possible to determine most probable pathways,18,21,23,24,38 or pathways of maximum weight. Such pathways are attractive for qualitative analysis. Individual BVS trajectories are therefore easier to interpret, assign weight, and understand than a single constant-energy IVS path.
Correct weights of pathways in BVS formulation make it also possible to use standard statistical sampling of Markov chains to generate an ensemble of trajectories with the BVS formulation. Sampling of pathways using BVS has the additional advantage that new trajectories can be obtained from perturbations to existing pathways simplifying the refinement of an initial guess to the trajectory. This argument must be done with extra care. For molecular trajectories with hundreds (or more) of particles that interact via a non-linear potential, we expect chaotic trajectories. The deviation between IVS trajectories that start from (almost) the same initial condition can rapidly develop to large values. In BVS, we expect the trajectories to be more stable to perturbation. The optimization is global for a whole trajectory and some (noisy) high frequency motions are removed. Hence in contrast to IVS, we ask if we can find a nearby whole trajectory once the original trajectory is perturbed. The answer to this question, using local minimization on the rough energy landscape of pathways, is typically yes.
Nevertheless optimizing whole trajectories, especially if they are quite long, is an expensive task that requires special hardware like massively parallel computing systems. The use of large time steps makes the trajectories approximate and the optimization is costly. It is therefore no surprise that IVS are considerably more popular. This brings us to the next set of approaches for computing long time dynamics: Exploitation of modern, and task specific hardware.
DESIGNING AND EXPLOITING SPECIAL HARDWARE
The idea of designing special purpose hardware to conduct long time molecular dynamics simulations has been explored for a long time.4,39–41 One focus was on special computer boards for rapid calculation of non-bonded interactions. The boards or the machine exploit the observation that the functional form of empirical force fields in molecular dynamic simulations has remained essentially the same for a long time. It is a sum of covalent interactions and non-covalent terms
| (3) |
where the b, θ, ϕ, Φ are the bond lengths, bond angles, torsions, and improper torsions, respectively. Subscript “0” means an ideal equilibrium value of that variable. The periodicity of a torsion n is denoted by ln. The coefficients (ki, ai, δi, κi, Ai, and Bi) are constants of the force fields, and qi are the atomic charges.
The computational cost of the forces is dominated by the last two summations of the non-bonded interactions. The cost of electrostatic and van der Waals interactions scales in the worst-case scenario as L2 where L is the number of atoms. Usually the long range interactions are either truncated or summed up in Fourier space (particle meshed Ewald42) which improves the scaling with the number of particles to L or L log(L). Even with the improved scaling the non-bonded interaction term is the most expensive component of the whole calculation. Special computer hardware was designed39,40 to reduce the time of computing the non-bonded interactions and hence speeding up the integration in Equation (1). The basic concept is straightforward. Given the current coordinate set xi, the summations in Equation (3) can be divided between different cores or threads speeding up the computations of the forces, ideally by the number of threads. The list of the covalent interactions (bonds, angles, torsions, and improper torsions) is fixed during the simulations and is precisely divided between the processors at the beginning of the run. The parallelization of the non-bonded interactions is more challenging since the list of interactions is dynamic. Interacting pairs may be added or subtracted as the particles are getting closer or farther away. Updates of the list of non-bonded interactions are computed periodically and stored in arrays that can be accessed rapidly.43
Until the last decade special purpose hardware did not make significant impact on the field. There are multiple reasons for the limited influence. First, progress of widely available commercial Central Processing Units (CPUs) was exceptionally fast. In the past, the time from the design of the special hardware to production was too long to compete effectively with general-purpose machines. Special hardware appeared late in the market and as a result was not sufficiently attractive to potential buyers. Companies that produced this hardware did not show profit in the limited time allowed by investors and the companies failed. If the performance difference between a general-purpose machine and specialized hardware is not very large then a general purpose machine is the natural choice to buy and use.
A second reason that reduced the impact of early special hardware is the focus on the non-bonded interactions. Non-bonded interactions typically require 90% of the total computation time. If the remaining ten percent are left “as is” it is impossible to speed up the original code by more than a factor of 10. The cloak time (time that passes in the real world between initiation to termination of the program) can be expressed as t = a/p + b also called Amdahl’s law.44 The parameter a is the time component that is amenable for parallelization and b is the time required for running the part of the code that is executed serially. It is necessary to create a balanced optimization scheme of all the components of the MD algorithm (i.e., make b as close as possible to zero). Small b makes it possible obtain speedup factors that are linear in the number of processors and are in the thousands in many molecular biophysics applications on readily accessible parallel computers. The significant change in the impact of special purpose machines occurs when Anton was introduced.4 With significant investment and comprehensive approach for parallelization, the Anton project provided a special purpose machine that is about 1000 times faster than alternatives. This intriguing development opened the way for a direct study of new molecular processes and at present is leading the field in computational speed of individual trajectories. However, given the exceptionally long time scales of many molecular processes in biophysics, the necessity of averaging over initial conditions and the cost of a single machine Anton is not a complete answer for many intriguing questions in molecular biophysics.
An intermediate approach runs molecular dynamics applications directly on general purpose parallel computers and includes programs like NAMD,45 GROMACS,46 and Desmond47 that scales well with the number of processors (roughly linear for typical systems) and allow for significant speed up. Several tens or hundreds of cores can be used effectively in supercomputer centers. Parallelization at national centers makes it possible to conduct research by a broad community of investigators (and on a diverse set of problems). Parallelization also makes it possible to study exceptionally large systems by dividing the system between nodes.
Another important avenue of supercomputing is growing; the use of Graphic Processing Units (GPUs)48–52 in molecular dynamics simulations. The GPUs enable significant speed up of simulations using a similar concept to the original special purpose boards (massive parallelism that primarily reduces the calculation time of the non-bonded interactions). Considerable re-thinking of the original code is required, since the GPU bottle-neck tends to be memory-access and not numerical operations. In contrast to special purpose boards, the design of the GPU for the game industry made the cost to the end customer significantly cheaper. It is possible to build up today the equivalent of tens of parallel cores using GPUs on a desktop machine at modest cost. Though the investment in software writing and optimization for the GPU is significant the payoff is considerable as well and a number of MD software packages take advantage of the GPU.48,49,51–53 A concern is of stability of computing models and retention of software. No one would like to write a significant code that has to be re-written in another year. The GPU is still an evolving architecture. Nevertheless, stable programming models for GPU architecture are emerging which encourages investments in more general development of portable software.54 Given the price performance ratio and the wide market of GPU processors (beyond the use in MD simulation) it is likely that the GPU architecture is here to stay. It is also likely to be merged with other architectures to provide more versatile systems.
Another concern is the accuracy and consistency of the numerical integration across different computing platforms. While the standard are either single or double precision arithmetic, a single precision accuracy (32 bits) is usually insufficient to ensure energy conservation of mechanical systems in extended simulations. Earlier GPU models support efficient calculations of 32 bits arithmetic. This restriction was removed, however, in more recent products. Care must be used to ensure correct mechanical behavior of trajectories produced if single precision mode is exploited (on CPU or GPU). Frequently, compromises are sought between numerical efficiency and accuracy considered sufficient for the task at hand. For sampling configurations from the appropriate ensemble (e.g., the canonical ensemble) it is necessary to know the distribution from which the structures were sampled. Without energy conservation and/or accurate integration of the equations of motion it is not known what is this sampled distribution. Of particular concern are simulations of long time dynamics in which “energy drift” significantly affect the system motions. Strategies were developed to ensure accurate integration at the microsecond time scale which is probably the upper bound of straightforward MD simulations that are run routinely on widely accessible computing platforms. Integration using fixed point arithmetic is used to ensure exact reproducibility of the simulations.4
PHYSICAL MODELING
Processes with a clear transition domain
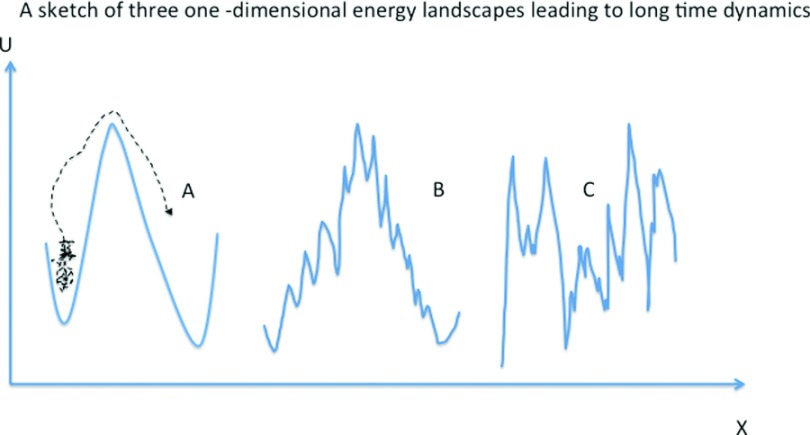
Here, we consider theoretical approaches to investigate and predict long time scale processes and kinetic. The first question is what is the physical mechanism that makes molecular processes so slow compared to the fastest motion of molecules (femtoseconds). The common answer to that question is activation. Many molecular processes require passage over a large (free) energy barrier for completion. This transition is rare and the system spends a long time in a metastable state before climbing up the barrier and rapidly transitioning to another metastable configuration (Fig. 1). Hence, while the “waiting” time at the metastable states is long the actual transition time is very short.
FIG. 1.
Sketches of three energy landscapes (U) along a coordinate (X). The dynamics on these three potentials is of long time scales which are difficult to access with straightforward Molecular Dynamics simulations. A—Potential energy with large and smooth energy barrier separating two metastable states, which is significantly larger than the thermal energy. Crossing this barrier is a rare event. However, Transition State Theory (TST)55,56 and the calculations of transmission coefficient57 are likely to provide adequate estimate of the rate. B—A rough energy landscape with a dominant barrier separating metastable states. Transitions between the metastable states are rare. However transient trajectories are expected to be short. The precise interface of the transition state may be hard to identify, and the transmission coefficient is slow to converge due to the local roughness and small barrier distribution. Methods like transition path sampling (TPS)58 that probe short transient trajectories between reactants and products are likely to provide adequate estimate of the rate. C—A rough energy landscape without a single dominant barrier and/or multiple metastable states. The motion is a mixed of activated processes and diffusion. Methods insensitive to the shape of the potential energy are appropriate for these types of systems and include weighted ensemble (WE),59 partial path transition interface sampling (PPTIS),60 non-equilibrium umbrella sampling (NEUS),61 and Milestoning.62
For activated processes with a single dominating barrier (which is significantly larger than the typical thermal energy kT) the TST55,56 is appropriate. TST is not trajectory based and therefore does not provide a dynamical picture. Supplementing TST with dynamics aspects of the process can be done with the transmission coefficient.57 Trajectories are sampled and run starting from the neighborhood of the barrier. Some of these trajectories will return to the reactants and some to the products. The transmission coefficient is the fraction of trajectories that were initiated at the transition state and continue to the product state before returning to the reactant. The basic form of the TST assumes that all the trajectories that made it to the dividing surface (Transition State) are reactive. Both the transmission coefficient and the more recent approach of the TPS58 build on the idea that the transitional trajectories are short and accessible to straightforward molecular dynamics simulations.
Sampling short (but rare) transitional trajectories
Individual short trajectories that are passing over barriers can be computed explicitly with straightforward molecular dynamics. However, to study long time kinetics, there remains the following two issues that were addressed by TPS:58,63 (i) how to find them (these trajectories are rare and are difficult to sample directly) and (ii) how to analyze the ensemble of trajectories to obtain experimentally measured quantities such as rate constants. The generation of TPS trajectories is done by a Monte Carlo procedure in trajectory space. A single reactive trajectory is provided and new reactive trajectories are generated from it. For example, micro-canonical sampling selects an intermediate point along the trajectory and samples a displacement to the momentum direction, keeping the kinetics energy (and of course the total energy) fixed. Once a new momentum is assigned a trajectory is computed backward and forward to termination in the metastable states of the “reactants” and “products.”
Some of the trajectories will end up at the same basin they started from and are non-reactive. Losing some of the sampled points is the price we need to pay for using the more convenient IVS approach. If these trajectories were computed in BVS formulation then the acceptance probability of “trial” trajectories would be one. However, keep in mind that the calculation of each of the BVS trajectories is significantly more expensive than IVS solutions if the same accuracy is desired in both formulations.
Activated processes on corrugated energy landscapes
In many molecular processes, the experimental kinetic is described by a single exponential as a function of time. In this case, it is tempting to interpret an ensemble of reactive trajectories using a simple two state kinetics. For example, if is the reactant population that survived after time t then exponential kinetics implies that where N0 is the population at time zero and k is the rate coefficient which is time independent. Alternatively we can focus only on reactive trajectories. Let the number of product molecules be B, the phenomenological rate equation for the appearance of the product (starting from zero product concentration) takes the form . For sufficiently small rate constant, k (or equivalently short times) the fraction of trajectories that makes it to state B at time t is
| (4) |
We can determine the rate constant from a slope of the above ratio as a function of time even if the trajectory duration is significantly shorter than .
The above procedure, which requires only the sampling of reactive transitional trajectories, avoids the use of a transition state or a reaction coordinate and is still able to determine a rate constant. This is an important advance that enables the study of kinetics of complex molecular systems by simulations. It is important to remember, however, the basic requirements. The calculation is conducted assuming that only two states are under consideration. The existence of metastable states can make the analysis considerably more complicated. It is also assumed that the states are well defined and separated by a significant (free) energy barrier. The assumption of large barrier implies that the transitional trajectories are short, which is exploited by the TPS algorithm. What can we do if the separation into distinct metastable states is not obvious? For example, if the rugged energy landscape includes a number of metastable states with comparable barriers separating them (Fig. 1). Note that the energy landscape can be rugged and still have only two metastable states if separation of scales of barrier heights is present. However, the transitional trajectories will no longer be short.
Enhancing sampling of reactive trajectories by “celling”
In typical energy landscapes of condensed phase systems, even with a dominant barrier, the remaining barriers can be relatively high (Fig. 1, B) and transitions inaccessible on the usual MD time scale. The remaining local corrugation can significantly slow down the dynamics to the extent that direct trajectories passing over the barrier are hard to sample. Broad distributions of barrier heights were illustrated for realistic systems in the past.64–66 A recent numerical study of a two-dimensional random energy model illustrates some of the issues mentioned above.67
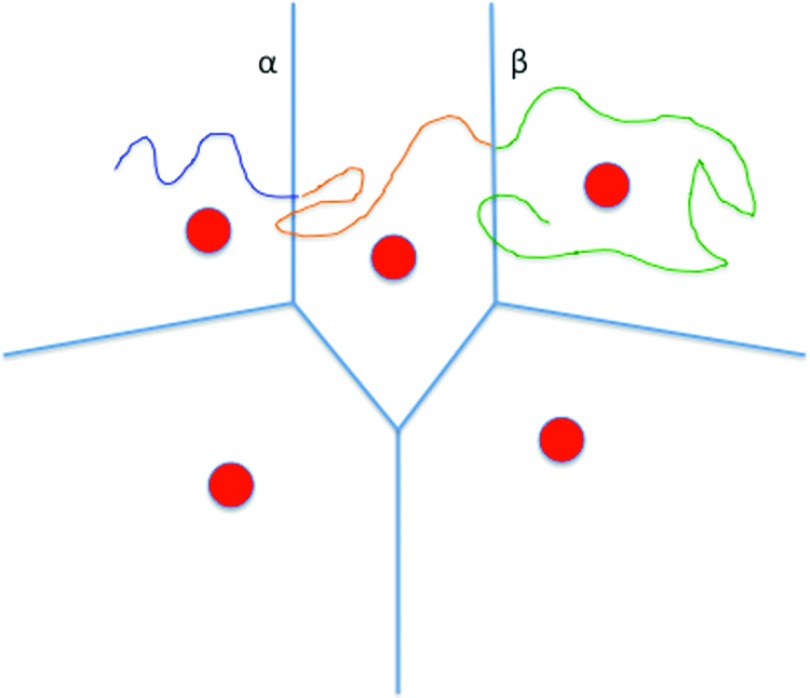
An alternative technology is required to sample reactive trajectories under these conditions. An early idea that is used extensively in recent methods splits phase space into “cells,” and estimates local kinetics between cells. In Fig. 2, we illustrate the concepts of cells for the method of Milestoning.
FIG. 2.
A schematic drawing of Milestoning cells in two dimensions. The milestones are the blue lines between the anchors (red circles). A trajectory is a curve that alternates colors when the last milestone crossed is modified. In the illustration, it crossed milestone α and then β.
The parameters of the local kinetics (e.g., rate coefficients, the probability of transition between cells, the distribution of transition times between cells) are collected to obtain global properties of the system. We call this concept in the present manuscript “celling.” How to choose the cells in an effective and efficient way is an important open question that has attracted considerable research. One possibility is to create cells along one dimensional reaction coordinate. Choices of reaction coordinates in complex molecular system vary from minimum energy paths,68 maximum flux pathways,69 most probable or dominant pathways,24 minimum free energy paths,70 and iso-committors.71–74 In the context of the present discussion (identifying optimal cells), iso-committors are special, since they make a straightforward connection to kinetics.74,75 The choice of the order parameter (as long it is sensible and follows a few guidelines listed below for different methods) is not expected to affect the accuracy of the calculations, but it will certainly affect the efficiency.
The first implementation of the celling idea was in the WE approach for stochastic dynamics along a one-dimensional reaction coordinate.76 This method was revisited and enhanced significantly more recently.59 In WE, particles in cells along the reaction coordinate, executing stochastic dynamics, are created and destroyed as they transition between cells, and their statistical weights are modified. The process is repeated until stationary distributions in all the cells are obtained. The final distributions of particles and their weights determine the free energy and the flux. The fluxes between the cells determine the rate.
The same concept of “celling” along one dimensional reaction coordinates was also used in the approaches of Transition Interface Sampling (TIS),77 PPTIS,60 Milestoning,62,78 Markovian Milestoning,32 Non-Equilibrium Umbrella Sampling,79 Forward Flux Sampling (FFS),80 and others.81,82 The sheer number of these approaches underlines the importance of the topic for current computational sciences but can also be confusing for the user that wishes to pick up the most appropriate technology for his/her applications.
“Celling” for diffusive and activated processes
Below an attempt is made to differentiate and classify the different technologies that use the concept of “celling” along a one-dimensional reaction coordinate. Extensions to more than a single reaction coordinate are possible and are discussed later. The first classification is to methods that are aimed at activated processes (called class A) and to methods with a focus on diffusive dynamics, called class B. In class A, the reactive trajectories tend to be short but they are rare. In class B, the trajectories are not necessarily rare but can be long individually. Class A methods include TIS,77 FFS,80 and hyperdynamics.83 Class B includes PPTIS,60 Milestoning,84 Markovian Milestoning,32 Non-Equilibrium Umbrella Sampling,79 trajectory tilting,81 and exact Milestoning.62 All the class A techniques compute complete trajectories from reactants to products. A Monte Carlo algorithm in trajectory space, enhancing and building on TPS, enables the calculations of the weights of trajectories and makes it possible to compute kinetic and equilibrium.85 The use of complete trajectories is in principle exact but it can also be a weakness, since individual trajectories cannot be longer than the time range accessible to ordinary MD (microseconds at most, considering the need to produce an ensemble of trajectories). Intermediate metastable states that trap the system for a substantial length of time may prevent it from reaching the end product at the MD time scale and/or complicate the analysis based on the argument in Eq. (4).
If class A technologies are used in type B systems two things may happen: (i) The trajectories will not reach the product and/or the reactant, since they are designed to be short. As such they will fail to connect the two metastable states and no rate could be estimated. The partially good news is that the failure will be easily detected by the user. (ii) Some short time trajectories connecting the reactant and product will be found and sampled. Since typical trajectories in these systems are longer than those sampled, the statistics will be biased towards short, and atypical trajectories, potentially leading to incorrect results. That is, even though class A approach is in principle exact, it is hard to detect and avoid the second problem mentioned above.
In PPTIS and Milestoning (class B), the emphasis is on diffusive behavior. Instead of computing complete trajectories, only trajectory fragments between interfaces (milestones) are computed (Fig. 2). These trajectory fragments are used to calculate transition probabilities and typical times of transitions between the cells or cell interfaces which we call milestones.78 The overall kinetics and thermodynamics are computed with these intermediate entities.60,86
The use of trajectory fragments in calculations of kinetics is more complex to manage than the use of complete trajectories from reactants to products. It is necessary to design a statistical model that will make it possible to “patch” the information extracted from the fragments to obtain a complete transition. These statistical models are at the core of the methods of WE,59,76 Milestoning,84 PPTIS,60 non-equilibrium umbrella sampling,79 and trajectory tilting.81 They can be approximate or exact. The approximate version is highly efficient and the exact formulation is significantly more expensive but still profoundly cheaper than straightforward MD. Below we describe in details the framework of trajectory fragments in the language of Milestoning. Related concepts are used in other approaches.
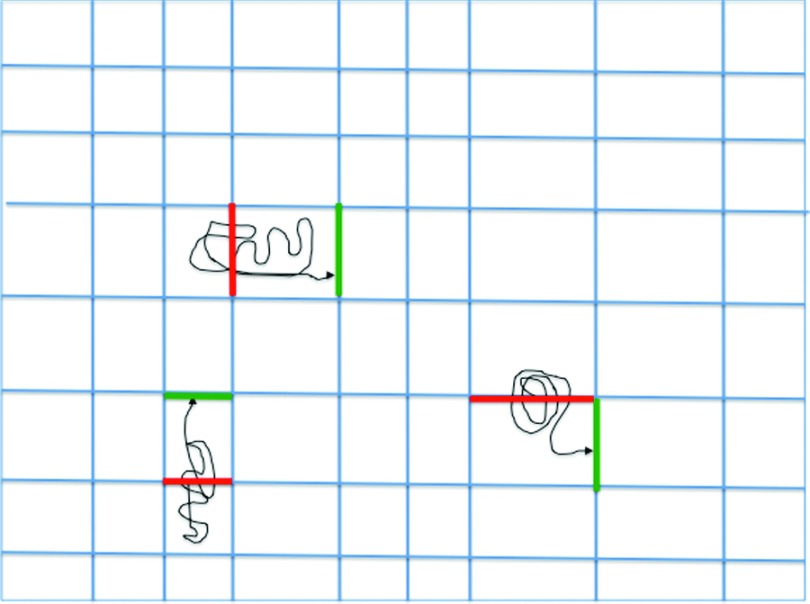
The trajectory fragments in Milestoning are conducted between milestones that are boundaries separating phase space cells (Fig. 3). In one realization we consider stationary processes close to equilibrium. To retain a stationary state we have a source that generates new trajectories (reactant state) and a sink that absorbs them (product state). For every trajectory absorbed by the sink, we have a new trajectory initiated at the source keeping the total number of trajectories fixed. The system is stationary since at the long time limit the number of trajectories per unit time that crosses any milestone in the system is fixed.
FIG. 3.
A schematic drawing describing the use of trajectory fragments (thin black lines) in Milestoning. Trajectories are initiated at a milestone (a boundary of a cell, red thick lines) and are integrated until they hit for the first time another milestone (green think line). From the trajectories the probability for a direct transition between two milestones (red and green) is estimated and so is the average time of the transition.
Consider the transition probability density or the kernel. Given the end points, Xα and Xβ, it is the probability density of a trajectory fragment that connects the two. In other words, given that a trajectory fragment last crosses milestone α at a phase space point Xα, the kernel is the probability that the trajectory fragment will cross next, before any other milestone, milestone β at phase space point Xβ. The end points are set closed in space, or at least close from kinetic perspective. This means that the kernel, or elements of the kernel, can be sampled and estimated directly by short molecular dynamic trajectories between the milestones. To obtain adequate statistics a large number of short trajectories are needed. Because the trajectories are typically of picosecond lengths, they can be computed using massive parallelism. Furthermore, other considerations87 further illustrate that this approach is profoundly more efficient than straightforward MD.
A probability density of interest is the number of trajectories that crosses milestone α at phase space point Xα in unit time. This function is the absolute stationary flux . This probability density can be normalized since the total number of trajectories in the system in unit time remains fixed under stationary conditions. In principle, running long time trajectories and enumerating the instances in which they cross milestones can estimate this function. This procedure, however, is expensive and is equivalent to an exact calculation of the dynamics. This is where the short trajectory information is very useful. The stationary flux is the solution of the vector-matrix equation where the integration over Mα is over an interface (milestone) between two cells. This equation is written in a more compact form as qt = qtK and can be solved exactly by iterations: qt(n+1) = qt(n)K where n is the iteration number62 or as an eigenvalue problem where q is the eigenvector of K with eigenvalue of one. The same procedure of iteratively sample short trajectories between cells is used in non-equilibrium umbrella sampling79 and in trajectory tilting.81 In the first version of Milestoning, we are using only one iteration to solve for the flux vector.84 We estimate the zero order iteration of the flux from the Boltzmann distribution assuming that the system is at equilibrium.86
The flux vector contains information needed to compute the thermodynamics and kinetics of the system. From the short trajectories, we extract another vector t. An element of t say tα is the average time that it takes a trajectory after crossing milestone α to cross any other milestone. We also call it the lifetime of a milestone and it can be estimated from the same short trajectories that sample elements of the kernel. With t and q, we can compute the probability that the last milestone that was crossed is . The free energy associated with milestone α is . Finally, the Mean First Passage Time, MFPT, to an absorbing milestone f in stationary conditions is
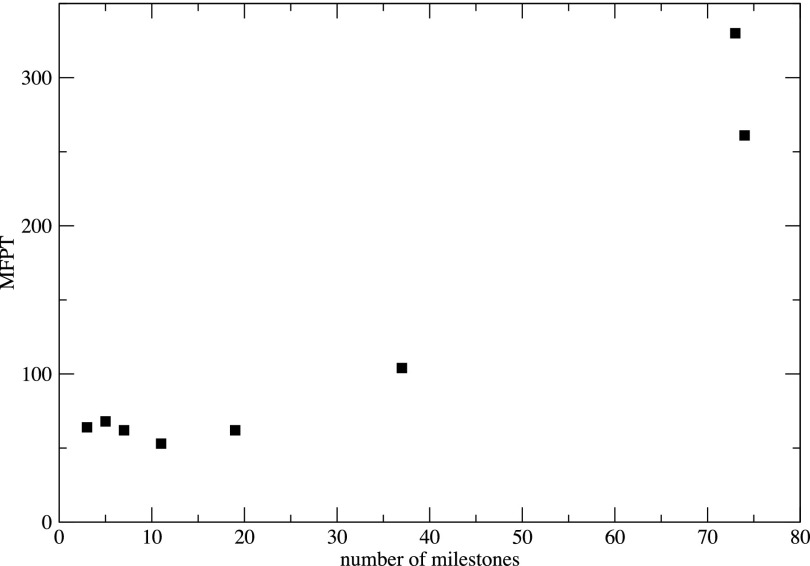
The approximation of estimating the flux from an equilibrium distribution at the milestone and using a single iteration to estimate the weight of the milestone62 is cheaper than the exact calculation by a factor of about one hundred. It is therefore worth exploring and was examined a number of times in the past, conceptually and empirically. We expect the approximate calculation to be accurate if the system has enough time to relax to a local equilibrium while it transitions from one milestone to the next. In that case the sampling from the canonical distribution for initial conditions at the milestone will be appropriate. A way of making the local transition time longer is by increasing the spatial separation between the milestones. In Fig. 4 (a reproduction from Ref. 86), the MFPT as a function of the number of milestones is plotted.
FIG. 4.
The Mean First Passage Time (MFPT) as a function of the number of milestones M. The calculations are for a conformational transition in solvated alanine dipeptide from an alpha helix to a beta sheet configuration; see Ref. 86 for more details.
Interestingly the use of three or nineteen milestones did not impact significantly the value of the MFPT. This observation suggests that the MFPT is insensitive to the distance between the milestones as long as the distance is large enough to allow for local equilibration during the time the system travels between the two milestones. This insensitivity to the number of milestones is a useful consistency check of the approximate Milestoning calculations. The more Milestoning we introduce, the smaller is the separation between the milestones and the calculation is more efficient. However, the assumption of local equilibration may not be satisfied if approximate Milestoning is used.
Other approaches to verify and enhance the memory loss of trajectories were proposed.87 One is the retention of partial history of previous interfaces or milestones that were crossed by the trajectory fragment.88 Without diminishing from the seriousness of the physical assumption of de-correlation, practice and adjustments mentioned above seem to keep the errors at bay. For practical purposes, the above computational methods can be made accurate by proper choice of simulation conditions (distances between Milestones).
The remarkable thing about class B methods is the huge computational gain that they provide for a broad range of processes. A simple illustration of required computational resources follows. The trajectory fragments are of a few to hundreds of picoseconds long. The total number of trajectory fragments rarely exceeds one hundred thousand. The accumulated computing time is therefore of order of less than 10 μs. The predicted and actual physical time scales in the system studied routinely exceed milliseconds89 and for membrane transport it was found to be hours.90 Hence the speed up compared to straightforward MD is at least thousands.
It is useful to discuss the differences between class A and B methods for diffusive and activated processes. We already mentioned that methods for activated processes do not handle efficiently diffusive dynamics since individual diffusive trajectories can be long. Care must also be exercised when using the methods designed for diffusive dynamics in the investigation of activated processes. The requirement of memory loss and local equilibration time poses a challenge for the short time trajectories typical of passages over large barriers. The problem is less significant for trajectory fragments that climb up the barrier, since the probability of going up is low and many (uncorrelated) climbing trials are expected. Memory loss and local equilibration is less likely for trajectories that are (rapidly) moving downhill from the top of the barrier. The solution in this case is to keep the milestones that point downhill as far as possible from each other. Since the trajectories are rapid, the calculation is not significantly more expensive and memory loss can be achieved with sufficient separation. This is an example for directional Milestoning, in which the separation between milestones depends on the direction in which they are crossed.33
It is also useful to discuss the technologies within a class, since some of the differences are significant and should impact selection. The prime attraction of the forward flux sampling (of the class A category) is that only forward trajectories are stored and considered. Trajectories are initiated at the reactants and are “pushed” towards the product along an appropriate reaction coordinate. This selection simplifies considerably the book keeping of trajectory events compared to approaches that count both forward and backward trajectories (TIS). It also makes it possible to investigate non-equilibrium dynamics,91 since the trajectories are initiated from a particular state at fixed time. This is to be contrasted with trajectory sampling in TPS and TIS calculations from a known statistical ensemble in which trajectories are launched anywhere “in between” and integrated forward and backward in time. The disadvantages of forward-only integration are somewhat less stable calculations due to “initiation” from the reactant position only. The FFS is probably restricted to one dimensional reaction coordinate since the concept of “forward” is harder to define in more than one-dimensional reaction space.
The methods of PPTIS and Milestoning focus on running short trajectories between the interfaces/milestones to estimate local transition times, rates, and transition probability. For example, in Milestoning, the transition probability between milestones α and β is estimated using trajectories as , where nα is the number of trajectories initiated at interface α and is the number of trajectories that were initiated at α and made to β exactly at time t. Both technologies were proposed for one-dimensional systems. Milestoning was extended to reaction space of several coarse variables,33 making it possible to address problems with multiple coupled and slow degrees of freedom. An alternative approach to deal with coarse space with more than one degree of freedom is the use of several pathways (a network) and stochastic sampling to switch between pathways. Extension to the TIS approach makes this type of calculation possible.92 Celling has made it possible to simulate in atomic details many complex and interesting systems. For example, conformational transitions in proteins,70,93 and folding.94,95
Finally, the approach of Markov State Model (MSM)96–98 made it possible to partition long time trajectories between cells and therefore to suggest another view of “celling” which can be analyzed to yield significantly long time scales. The MSM was introduced as a method to analyze long time trajectories and to rigorously build, based on this analysis, a coarse grained description of the system including a relatively small number of metastable states. Shalloway formulated an elegant way to dissect conformation space to several macrostates and estimate transition rates between them.99,100 Schuette and co-workers101 put the foundation of MSM and showed how a transition matrix can be constructed from detailed trajectories, using cells in the space of coarse variables. The time may be discrete or continuous and MSM is expected to accurately describe the long time behavior. In the simplest terms, the dynamics is being mapped to a master equation of transitions between cells where α and β denote cells that are assumed to be in local thermal equilibrium. is the probability of being at cell α at time t, and kαβ is a matrix of rate coefficients. While the above model is approximate for general dynamics (the generalized master equation is exact102), it is simple and attractive. Therefore, a great deal of work was published on the design of and estimates of the rate coefficients.96,97,101,103–106 The prime goal is to reproduce the long time behavior. Efficient and accurate estimates of the rate coefficients will continue to be a topic of intense debate.
It should be noted that WE, non-equilibrium umbrella sampling, and Milestoning are viable alternatives to the MSM model by providing kinetic description at broader scales and being exact for their corresponding dynamics. This is achieved at the additional cost of more involved and complex modeling. In Milestoning, for example, two approaches are possible: (i) exact calculations retaining the phase space points at the crossing milestones62 or (ii) an approximate calculation that is going beyond Markovian approximation in coarse space and model the dynamics following the generalized master equation.78,107
SUMMARY
Atomically detailed molecular simulations continue to attract the interest and imagination of many scientists. They offer a tool that explores molecular processes at great detail and makes it possible to analyze and focus at the smallest component. A drawback of simulations is their approximate nature. The force field is not exact, and the sampling for kinetics or thermodynamics is not comprehensive. Nevertheless, significant progress has been made to make these simulations quantitative, predictive, and subject to rigorous tests by a wide range of experiments. The present perspective focuses on extending the scope of simulations of kinetics.
Kinetics is a critical aspect of molecular processes that has so far received less attention compared to the calculations of thermodynamic equilibrium data. In biology, many processes are determined by kinetics and can be far from global equilibrium. Emphasizing only thermodynamics can therefore be misleading. Study of kinetics (as opposed to equilibrium) suggests new challenges. The calculation of a single reactive trajectory can be a demanding task if the process under consideration is slow. Obviously the calculation of an ensemble of such trajectories is even more challenging. In this perspective, we consider different approaches to address these problems. The most straightforward approaches focus on speeding up the computation of time steps either by special hardware4,51,52 or numerical analysis aiming to increase the fundamental time step13 and make the straightforward calculations of trajectories more rapid. Other approaches focus on the physical characteristics of the problem, exploiting the short length in time of transitional and rare trajectories.58 Other physical characteristics that are exploited in long time algorithms are memory loss in which de-correlation simplifies the mechanical equations of motions and makes it possible to use short trajectories and local kinetic operators77,78,108 to extrapolate to much longer times.
In the future, we expect more interactions between the three directions. For example, the physically motivated approaches are likely to be subject to more rigorous analysis and mathematical verifications of the type that led to methods like SHAKE. Special-purpose machines may be built to run ensemble of trajectories that are computed and analyzed on the fly to decide where and when to launch new trajectory fragments. Trajectory fragment approaches are particularly suitable for massively parallel computing systems. Hence approaches for quantification of kinetic calculations are likely to be automated, not only at the level of software (as is partially done with geometric clustering in Markov State Models109) and more extensively in the Copernicus package110 but also at the level of hardware.
Finally, we comment on a new direction that is not fully explored yet with enhanced sampling techniques. It is the time dependent approach of a system to a stationary non-equilibrium state. Coupling of time dependent relaxation process to stationary phenomena is relevant to many cellular processes. An approach to the problem has been formulated by Ciccotti,111 and applications using enhanced sampling techniques for kinetics that were discussed in this perspective are anticipated.
Acknowledgments
The writing of this perspective was supported by NIH Grant No. GM 059796. The author is grateful to two anonymous reviewers and Juan M. Bello-Rivas for their comments that improved the original version significantly.
REFERENCES
- 1.Frenkel D. and Berend S., Understanding Molecular Simulation: From Algorithms to Applications (Academic Press, San Diego, 1996). [Google Scholar]
- 2.Verlet L., Phys. Rev. 159(1), 98–103 (1967). 10.1103/PhysRev.159.98 [DOI] [Google Scholar]
- 3.Hairer E., Lubich C., and Wanner G., Acta Numer. 12, 399–450 (2003). 10.1017/S0962492902000144 [DOI] [Google Scholar]
- 4.Shaw D. E., Deneroff M. M., Dror R. O., Kuskin J. S., Larson R. H., Salmon J. K., Young C., Batson B., Bowers K. J., Chao J. C., Eastwood M. P., Gagliardo J., Grossman J. P., Ho C. R., Ierardi D. J., Kolossvary I., Klepeis J. L., Layman T., Mcleavey C., Moraes M. A., Mueller R., Priest E. C., Shan Y. B., Spengler J., Theobald M., Towles B., and Wang S. C., Commun. ACM 51(7), 91–97 (2008). 10.1145/1364782.1364802 [DOI] [Google Scholar]
- 5.Eaton W. A., Munoz V., Hagen S. J., Jas G. S., Lapidus L. J., Henry E. R., and Hofrichter J., Annu. Rev. Biophys. Biomol. Struct. 29, 327–359 (2000). 10.1146/annurev.biophys.29.1.327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prigozhin M. B. and Gruebele M., Phys. Chem. Chem. Phys. 15(10), 3372–3388 (2013). 10.1039/c3cp43992e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elber R. and Kirmizialtin S., Curr. Opin. Struct. Biol. 23(2), 206–211 (2013). 10.1016/j.sbi.2012.12.002 [DOI] [PubMed] [Google Scholar]
- 8.Makarov D. E., Single Molecule Science: Physical Principles and Models (CRC Press, Boca Raton, FL, 2015). [Google Scholar]
- 9.Ryckaert J. P., Ciccotti G., and Berendsen H. J. C., J. Comput. Phys. 23(3), 327–341 (1977). 10.1016/0021-9991(77)90098-5 [DOI] [Google Scholar]
- 10.Tuckerman M., Berne B. J., and Martyna G. J., J. Chem. Phys. 97(3), 1990–2001 (1992). 10.1063/1.463137 [DOI] [Google Scholar]
- 11.Peskin C. S. and Schlick T., Commun. Pure Appl. Math. 42, 1001–1031 (1989). 10.1002/cpa.3160420706 [DOI] [Google Scholar]
- 12.Zhang G. H. and Schlick T., J. Chem. Phys. 101(6), 4995–5012 (1994). 10.1063/1.467422 [DOI] [Google Scholar]
- 13.Leimkuhler B. and Matthews C., Molecular Dynamics with Deterministic and Stochastic Numerical Methods (Springer, Switzerland, 2015). [Google Scholar]
- 14.Leimkuhler B., Noorizadeh E., and Theil F., J. Stat. Phys. 135(2), 261–277 (2009). 10.1007/s10955-009-9734-0 [DOI] [Google Scholar]
- 15.Morrone J. A., Markland T. E., Ceriotti M., and Berne B. J., J. Chem. Phys. 134(1), 014103 (2011). 10.1063/1.3518369 [DOI] [PubMed] [Google Scholar]
- 16.Ma Q. and Izaguirre J. A., Multiscale Model. Simul. 2(1), 1–21 (2003). 10.1137/S1540345903423567 [DOI] [Google Scholar]
- 17.Leimkuhler B., Margul D. T., and Tuckerman M. E., Mol. Phys. 111(22-23), 3579–3594 (2013). 10.1080/00268976.2013.844369 [DOI] [Google Scholar]
- 18.Olender R. and Elber R., J. Chem. Phys. 105(20), 9299–9315 (1996). 10.1063/1.472727 [DOI] [Google Scholar]
- 19.Elber R., Meller J., and Olender R., J. Phys. Chem. B 103(6), 899–911 (1999). 10.1021/jp983774z [DOI] [Google Scholar]
- 20.Ghosh A., Elber R., and Scheraga H. A., Proc. Natl. Acad. Sci. U. S. A. 99(16), 10394–10398 (2002). 10.1073/pnas.142288099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elber R. and Shalloway D., J. Chem. Phys. 112(13), 5539–5545 (2000). 10.1063/1.481131 [DOI] [Google Scholar]
- 22.Faccioli P., Sega M., Pederiva F., and Orland H., Phys. Rev. Lett. 97(10), 108101 (2006). 10.1103/PhysRevLett.97.108101 [DOI] [PubMed] [Google Scholar]
- 23.Beccara S. A., Faccioli P., Sega M., Pederiva F., Garberoglio G., and Orland H., J. Chem. Phys. 134(2), 024501 (2011). 10.1063/1.3514149 [DOI] [PubMed] [Google Scholar]
- 24.Faccioli P., Lonardi A., and Orland H., J. Chem. Phys. 133(4), 045104 (2010). 10.1063/1.3459097 [DOI] [PubMed] [Google Scholar]
- 25.Cardenas A. E. and Elber R., Proteins: Struct., Funct., Genet. 51(2), 245–257 (2003). 10.1002/prot.10349 [DOI] [PubMed] [Google Scholar]
- 26.Bai D. and Elber R., J. Chem. Theory Comput. 2(3), 484–494 (2006). 10.1021/ct060028m [DOI] [PubMed] [Google Scholar]
- 27.Olender R. and Elber R., J. Mol. Struct.: THEOCHEM 398, 63–71 (1997). 10.1016/S0166-1280(97)00038-9 [DOI] [Google Scholar]
- 28.Cardenas A. E. and Elber R., Biophys. J. 85(5), 2919–2939 (2003). 10.1016/s0006-3495(03)74713-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elber R., Biophys. J. 92(9), L85–L87 (2007). 10.1529/biophysj.106.101899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller T. F. and Predescu C., J. Chem. Phys. 126(14), 144102 (2007). 10.1063/1.2712444 [DOI] [PubMed] [Google Scholar]
- 31.Eastman P., Gronbech-Jensen N., and Doniach S., J. Chem. Phys. 114(8), 3823–3841 (2001). 10.1063/1.1342162 [DOI] [Google Scholar]
- 32.Vanden-Eijnden E. and Venturoli M., J. Chem. Phys. 130(19), 194101 (2009). 10.1063/1.3129843 [DOI] [PubMed] [Google Scholar]
- 33.Majek P. and Elber R., J. Chem. Theory Comput. 6(6), 1805–1817 (2010). 10.1021/ct100114j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller T. F., Chandler D., and Vanden-Eijnden E., “Solvent coarse-graining and the string method applied to the hydrophobic collapse of a hydrated chain,” Proc. Natl. Acad. Sci. U. S. A. 104, 14559 (2007). 10.1073/pnas.0705830104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cardenas A. E. and Elber R., J. Chem. Phys. 141(5), 054101 (2014). 10.1063/1.4891305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beccara S., Tatjana S., Covino R., and Faccioli P., Proc. Natl. Acad. Sci. U. S. A. 109(7), 2330–2335 (2012). 10.1073/pnas.1111796109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siva K. and Elber R., Proteins: Struct., Funct., Bioinf. 50(1), 63–80 (2003). 10.1002/prot.10256 [DOI] [PubMed] [Google Scholar]
- 38.Passerone D. and Parrinello M., Phys. Rev. Lett. 87(10), 108302 (2001). 10.1103/PhysRevLett.87.108302 [DOI] [PubMed] [Google Scholar]
- 39.Fine R., Dimmler G., and Levinthal C., Proteins: Struct., Funct., Genet. 11(4), 242–253 (1991). 10.1002/prot.340110403 [DOI] [PubMed] [Google Scholar]
- 40.Toyoda S., Miyagawa H., Kitamura K., Amisaki T., Hashimoto E., Ikeda H., Kusumi A., and Miyakawa N., J. Comput. Chem. 20(2), 185–199 (1999). [DOI] [Google Scholar]
- 41.Weinbach Y. and Elber R., J. Comput. Phys. 209(1), 193–206 (2005). 10.1016/j.jcp.2005.03.015 [DOI] [Google Scholar]
- 42.Darden T., York D., and Pedersen L., J. Chem. Phys. 98(12), 10089–10092 (1993). 10.1063/1.464397 [DOI] [Google Scholar]
- 43.Allen M. P. and Tildesley D. J., Computer Simulation of Liquids (Oxford University Press, New York, 1987). [Google Scholar]
- 44.Amdahl G. M., presented at the AFIPS Conference Proceeding,1967.
- 45.Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kale L., and Schulten K., J. Comput. Chem. 26(16), 1781–1802 (2005). 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hess B., Kutzner C., van der Spoel D., and Lindahl E., J. Chem. Theory Comput. 4(3), 435–447 (2008). 10.1021/ct700301q [DOI] [PubMed] [Google Scholar]
- 47.Bowers K. J., Chow E., Xu H., Dror R. O., Eastwood M. P., Gregersen B. A., Kelpeis J. L., Kolossvary I., Moraes M. A., D. S. F., Salmon J. K., Shan Y., and Shaw D. E., presented at the Scientific Computing, Tampa, Florida,2006.
- 48.Anderson J. A., Lorenz C. D., and Travesset A., J. Comput. Phys. 227(10), 5342–5359 (2008). 10.1016/j.jcp.2008.01.047 [DOI] [Google Scholar]
- 49.Harvey M. J., Giupponi G., and De Fabritiis G., J. Chem. Theory Comput. 5(6), 1632–1639 (2009). 10.1021/ct9000685 [DOI] [PubMed] [Google Scholar]
- 50.Ruymgaart A. P., Cardenas A. E., and Elber R., J. Chem. Theory Comput. 7(10), 3072–3082 (2011). 10.1021/ct200360f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stone J. E., Phillips J. C., Freddolino P. L., Hardy D. J., Trabuco L. G., and Schulten K., J. Comput. Chem. 28(16), 2618–2640 (2007). 10.1002/jcc.20829 [DOI] [PubMed] [Google Scholar]
- 52.van Meel J. A., Arnold A., Frenkel D., Zwart S. F. P., and Belleman R. G., Mol. Simul. 34(3), 259–266 (2008). 10.1080/08927020701744295 [DOI] [Google Scholar]
- 53.Ruymgaart A. P. and Elber R., J. Chem. Theory Comput. 8(11), 4624–4636 (2012). 10.1021/ct300324k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jaaskelainen P. O., Lama C. S. de La, Huerta P., and Takala J. H., presented at the 2010 International Conference on Embedded Computer Systems (SAMOS) (IEEE),2010.
- 55.Vanden-Eijnden E. and Tal F. A., J. Chem. Phys. 123(18), 184103 (2005). 10.1063/1.2102898 [DOI] [PubMed] [Google Scholar]
- 56.Truhlar D. G., Garrett B. C., and Klippenstein S. J., J. Phys. Chem. 100(31), 12771–12800 (1996). 10.1021/jp953748q [DOI] [Google Scholar]
- 57.Chandler D., J. Chem. Phys. 68(6), 2959–2970 (1978). 10.1063/1.436049 [DOI] [Google Scholar]
- 58.Bolhuis P. G., Chandler D., Dellago C., and Geissler P. L., Annu. Rev. Phys. Chem. 53, 291–318 (2002). 10.1146/annurev.physchem.53.082301.113146 [DOI] [PubMed] [Google Scholar]
- 59.Zhang B. W., Jasnow D., and Zuckerman D. M., J. Chem. Phys. 132(5), 054107 (2010). 10.1063/1.3306345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moroni D., Bolhuis P. G., and van Erp T. S., J. Chem. Phys. 120(9), 4055–4065 (2004). 10.1063/1.1644537 [DOI] [PubMed] [Google Scholar]
- 61.Dickson A., Warmflash A., and Dinner A. R., J. Chem. Phys. 130(7), 074104 (2009). 10.1063/1.3070677 [DOI] [PubMed] [Google Scholar]
- 62.Bello-Rivas J. M. and Elber R., J. Chem. Phys. 142(9), 094102 (2015). 10.1063/1.4913399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dellago C., Bolhuis P. G., and Geissler P. L., Advances in Chemical Physics (John Wiley & Sons, Inc., New York, 2002), Vol. 123, pp. 1–78. [Google Scholar]
- 64.Austin R. H., Beeson K. W., Eisenstein L., Frauenfelder H., and Gunsalus I. C., Biochemistry 14(24), 5355–5373 (1975). 10.1021/bi00695a021 [DOI] [PubMed] [Google Scholar]
- 65.Czerminski R. and Elber R., J. Chem. Phys. 92(9), 5580–5601 (1990). 10.1063/1.458491 [DOI] [Google Scholar]
- 66.Wales D. J., Energy Landscapes: Applications to Clusters, Biomolecules and Glasses (Cambridge University Press, Cambridge, 2003). [Google Scholar]
- 67.Bello-Rivas J. M. and Elber R., “Simulations of thermodynamics and kinetics on rough energy landscapes with milestoning,” J. Comput. Chem. (published online). 10.1002/jcc.24039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ulitsky A. and Elber R., J. Chem. Phys. 92(2), 1510–1511 (1990). 10.1063/1.458112 [DOI] [Google Scholar]
- 69.Berkowitz M., Morgan J. D., McCammon J. A., and Northrup S. H., J. Chem. Phys. 79(11), 5563–5565 (1983). 10.1063/1.445675 [DOI] [Google Scholar]
- 70.Ovchinnikov V., Karplus M., and Vanden-Eijnden E., J. Chem. Phys. 134(8), 085103 (2011). 10.1063/1.3544209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maragliano L., Fischer A., Vanden-Eijnden E., and Ciccotti G., J. Chem. Phys. 125(2), 024106 (2006). 10.1063/1.2212942 [DOI] [PubMed] [Google Scholar]
- 72.Peters B., J. Chem. Phys. 125(24), 241101 (2006). 10.1063/1.2409924 [DOI] [PubMed] [Google Scholar]
- 73.Ma A. and Dinner A. R., J. Phys. Chem. B 109(14), 6769–6779 (2005). 10.1021/jp045546c [DOI] [PubMed] [Google Scholar]
- 74.E W. and Vanden-Eijnden E., Annu. Rev. Phys. Chem. 61, 391–420 (2010). 10.1146/annurev.physchem.040808.090412 [DOI] [PubMed] [Google Scholar]
- 75.Vanden Eijnden E., Venturoli M., Ciccotti G., and Elber R., J. Chem. Phys. 129(17), 174102 (2008). 10.1063/1.2996509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huber G. A. and Kim S., Biophys. J. 70(1), 97–110 (1996). 10.1016/S0006-3495(96)79552-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Erp T. S., Moroni D., and Bolhuis P. G., J. Chem. Phys. 118(17), 7762–7774 (2003). 10.1063/1.1562614 [DOI] [Google Scholar]
- 78.Faradjian A. K. and Elber R., J. Chem. Phys. 120(23), 10880–10889 (2004). 10.1063/1.1738640 [DOI] [PubMed] [Google Scholar]
- 79.Warmflash A., Bhimalapuram P., and Dinner A. R., J. Chem. Phys. 127(15), 154112 (2007). 10.1063/1.2784118 [DOI] [PubMed] [Google Scholar]
- 80.Allen R. J., Frenkel D., and ten wolde P. R., J. Chem. Phys. 124(19), 194111 (2006). 10.1063/1.2198827 [DOI] [PubMed] [Google Scholar]
- 81.Vanden-Eijnden E. and Venturoli M., J. Chem. Phys. 131(4), 044120 (2009). 10.1063/1.3180821 [DOI] [PubMed] [Google Scholar]
- 82.Glowacki D. R., Paci E., and Shalashilin D. V., J. Chem. Theory Comput. 7(5), 1244–1252 (2011). 10.1021/ct200011e [DOI] [PubMed] [Google Scholar]
- 83.Voter A. F., Phys. Rev. Lett. 78(20), 3908–3911 (1997). 10.1103/PhysRevLett.78.3908 [DOI] [Google Scholar]
- 84.Kirmizialtin S. and Elber R., J. Phys. Chem. A 115(23), 6137–6148 (2011). 10.1021/jp111093c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moroni D., van Erp T. S., and Bolhuis P. G., Phys. Rev. E 71(5), 056709 (2005). 10.1103/physreve.71.056709 [DOI] [PubMed] [Google Scholar]
- 86.West A. M. A., Elber R., and Shalloway D., J. Chem. Phys. 126(14), 145104 (2007). 10.1063/1.2716389 [DOI] [PubMed] [Google Scholar]
- 87.Hawk A. T., “Milestoning with coarse memory,” J. Chem. Phys. 138(15), 154105 (2013). 10.1063/1.4795838 [DOI] [PubMed] [Google Scholar]
- 88.Hawk A. T. and Makarov D. E., J. Chem. Phys. 135(22), 224109 (2011). 10.1063/1.3666840 [DOI] [PubMed] [Google Scholar]
- 89.Elber R. and West A., Proc. Natl. Acad. Sci. U. S. A. 107, 5001–5005 (2010). 10.1073/pnas.0909636107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cardenas A. E., Jas G. S., DeLeon K. Y., Hegefeld W. A., Kuczera K., and Elber R., J. Phys. Chem. B 116(9), 2739–2750 (2012). 10.1021/jp2102447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Becker N. B., Allen R. J., and ten Wolde P. R., J. Chem. Phys. 136(17), 174118 (2012). 10.1063/1.4704810 [DOI] [PubMed] [Google Scholar]
- 92.Swenson D. W. H. and Bolhuis P. G., J. Chem. Phys. 141(4), 044101 (2014). 10.1063/1.4890037 [DOI] [PubMed] [Google Scholar]
- 93.Kirmizialtin S., Nguyen V., Johnson K. A., and Elber R., Structure 20(4), 618–627 (2012). 10.1016/j.str.2012.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Juraszek J. and Bolhuis P. G., Biophys. J. 95(9), 4246–4257 (2008). 10.1529/biophysj.108.136267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kreuzer S. M., Moon T. J., and Elber R., “Catch bond-like kinetics of helix cracking: Network analysis by molecular dynamics and Milestoning,” J. Chem Phys. 139(12), 121902 (2013). 10.1063/1.4811366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sarich M., Noe F., and Schutte C., Multiscale Model. Simul. 8(4), 1154–1177 (2010). 10.1137/090764049 [DOI] [Google Scholar]
- 97.Noe F., Schutte C., Vanden-Eijnden E., Reich L., and Weikl T. R., Proc. Natl. Acad. Sci. U. S. A. 106(45), 19011–19016 (2009). 10.1073/pnas.0905466106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chodera J. D., Singhal N., Pande V. S., Dill K. A., and Swope W. C., J. Chem. Phys. 126(15), 155101 (2007). 10.1063/1.2714538 [DOI] [PubMed] [Google Scholar]
- 99.Ulitsky A. and Shalloway D., J. Chem. Phys. 109(5), 1670–1686 (1998). 10.1063/1.476882 [DOI] [Google Scholar]
- 100.Shalloway D., J. Chem. Phys. 105(22), 9986–10007 (1996). 10.1063/1.472830 [DOI] [Google Scholar]
- 101.Schutte C., Noe F., Lu J. F., Sarich M., and Vanden-Eijnden E., J. Chem. Phys. 134(20), 204105 (2011). 10.1063/1.3590108 [DOI] [PubMed] [Google Scholar]
- 102.Mori H., Fujisaka H., and Shigematsu H., “A new expansion of the master equation,” Prog. Theor. Phys. 51, 109 (1974). 10.1143/PTP.51.109 [DOI] [Google Scholar]
- 103.Chodera J. D., Swope W. C., Pitera J. W., and Dill K. A., Multiscale Model. Simul. 5(4), 1214–1226 (2006). 10.1137/06065146X [DOI] [Google Scholar]
- 104.Noe F., Horenko I., Schutte C., and Smith J. C., J. Chem. Phys. 126(15), 155102 (2007). 10.1063/1.2714539 [DOI] [PubMed] [Google Scholar]
- 105.Hummer G. and Szabo A., J. Phys. Chem. B 119(29), 9029–9037 (2015). 10.1021/jp508375q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Buchete N. V. and Hummer G., J. Phys. Chem. B 112(19), 6057–6069 (2008). 10.1021/jp0761665 [DOI] [PubMed] [Google Scholar]
- 107.Shalloway D. and Faradjian A. K., J. Chem. Phys. 124(5), 054112 (2006). 10.1063/1.2161211 [DOI] [PubMed] [Google Scholar]
- 108.Valeriani C., Allen R. J., Morelli M. J., Frenkel D., and ten Wolde P. R., J. Chem. Phys. 127(11), 114109 (2007). 10.1063/1.2767625 [DOI] [PubMed] [Google Scholar]
- 109.Hinrichs N. S. and Pande V. S., J. Chem. Phys. 126(24), 244101 (2007). 10.1063/1.2740261 [DOI] [PubMed] [Google Scholar]
- 110.Pronk S., Pouya I., Lundborg M., Rotskoff G., Wesen B., Kasson P. M., and Lindahl E., J. Chem. Theory Comput. 11, 2600–2608 (2015). 10.1021/acs.jctc.5b00234 [DOI] [PubMed] [Google Scholar]
- 111.Ciccotti G. and Jacucci G., Phys. Rev. Lett. 35(12), 789–792 (1975). 10.1103/PhysRevLett.35.789 [DOI] [Google Scholar]