Abstract
Irritable bowel syndrome (IBS) is a common condition with a complex and largely unknown etiology. There is no cure, and treatment options are mainly directed to the amelioration of symptoms. IBS causes reduced quality of life and poses considerable repercussions on health and socioeconomic systems. There is a heritable component in IBS, and genetic research is a valuable tool for the identification of causative pathways, which will provide important insight into the pathophysiology. However, although some gene-hunting efforts have been conducted and a few risk genes proposed, IBS genetic research is lagging behind compared to other complex diseases. In this mini-review, we briefly summarize existing genetic studies, discuss the main challenges in IBS genetic research, and propose strategies to overcome these challenges for IBS gene discovery.
Keywords: Genetics, Genome-wide association study, Irritable bowel syndrome
Irritable bowel syndrome prevalence
Irritable bowel syndrome (IBS) is the most commonly diagnosed functional gastrointestinal disorder (FGID) with a worldwide prevalence of 10–20 %, predominantly among women [1]. Its clinical appearance varies but is usually characterized by recurrent abdominal pain or discomfort accompanied by changes in bowel habits and diarrhea (IBS-D), constipation (IBS-C), or both (mixed, IBS-M). In the absence of reliable biomarkers or specific laboratory tests, consensus criteria towards a positive diagnosis have been developed: the symptom-based Rome III criteria [2]. Due to a complex and not fully elucidated pathophysiology, there is no cure, and available treatment options can only be directed to amelioration of symptoms in a trial-and-error approach based on patients’ individual symptomatology. Comorbidity with other FGIDs as well as psychological conditions such as depression and anxiety is observed, as well as certain non-gastrointestinal non-psychological comorbidities including fibromyalgia, chronic fatigue syndrome, and chronic pelvic pain [3]. IBS negatively affects quality of life, which reflects in increased work and school absenteeism, decreased work productivity, and higher utilization of the health care system, with considerable repercussions on health and socioeconomic systems [3, 4].
IBS pathophysiology
Structural abnormalities, tissue damage, or other organic explanations are typically absent in IBS. Therefore, this condition is traditionally classified as a functional gastrointestinal disorder. Due to significant associations with anxiety, depression, and other psychiatric conditions, IBS has often been considered to be a psychosomatic disorder. However, more recent research has contributed to the elucidation of several mechanisms that may play important roles in IBS [5–7], see Fig. 1. We now understand that mucosal immune activation, inflammatory cells, and elevated inflammatory markers may be present in IBS, at least in a subset of patients. Prior gastroenteritis (post-infectious IBS; PI-IBS), described effects of antibiotics and probiotics on IBS symptoms, and the observation of different gut microbiota profiles in patients and controls all speak for an important role of gut flora in IBS [7]. In addition, diet is recognized in some patients as an important trigger of gut symptoms, and dietary therapeutic approaches have been recently developed, such as the low-FODMAP diet where poorly absorbed and rapidly fermentable carbohydrates are avoided [8]. Visceral hypersensitivity and altered pain perception also appear to be important features of IBS, as demonstrated in rectal sensitivity experiments. Together with the observation of abnormal gut motility and alterations in enteroendocrine and neuropeptide systems in relation to the enteric and central nervous system as well as gut microbiota, perturbations in the gut-brain axis and its bidirectional communication have been proposed among the central mechanisms in IBS pathophysiology [5–7]. Other specific factors may instead be important only for specific subgroups of IBS patients, such as altered bile acid synthesis in IBS-D [5, 9].
Fig. 1.
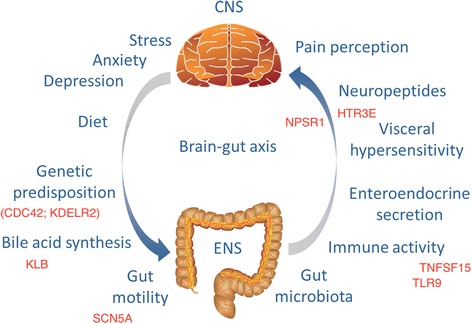
Factors proposed to be involved in the complex pathophysiology of irritable bowel syndrome. CNS central nervous system, ENS enteric nervous system. Prominent, proposed IBS-risk genes from Table 1 are also reported, by positioning them based on the (most likely) mechanistic involvement in the mentioned pathways
The genetic architecture of IBS
A heritable component of IBS has been demonstrated in twin and family studies, although heritability estimates have varied between 0 and 57 % [10]. Recently, however, a Swedish nationwide survey including more than 50,000 cases showed increased IBS risk among first-, second- and third-degree relatives, clearly indicating that a genetic component exists [11]. Evidence is now accumulating that genetic risk in IBS spans from complex polygenic conditions with combinations of common variants, to cases with rare single gene abnormalities [12]. For the majority of IBS patients, the genetic background will be constituted by a large set of common genetic variants, each contributing a small risk effect. At the same time, there may be subsets of patients where highly penetrant genetic variability in individual genes accounts for most of the phenotype. A good example of this phenomenon recently came from a study at the Mayo Clinic in collaboration with our group [13]: sequencing of the SCN5A gene in 584 IBS patients and 1380 asymptomatic controls identified functionally deleterious mutations in 2.2 % of IBS cases but none among controls. The SCN5A gene encodes the NaV1.5 ion channel responsible for the pacemaker function of the heart but is also present on interstitial cells of cajal, the ‘pacemaker’ cells of the gut. Most of the mutations identified were loss-of-function and carriers most often constipation-predominant. Of note, a severe IBS-C case with a highly penetrating SCN5A loss-of-function mutation could be successfully treated with mexiletine, a drug known to restore NaV1.5 channel function. In addition, also common SCN5A single nucleotide polymorphisms (SNPs) were found to affect IBS risk in our pilot genome-wide association study (GWAS) of ~5000 subjects (described further down), as well as in four independent case-control cohorts from Sweden, Italy, Greece, and the USA. These findings hence strengthen the hypothesis that both rare mutations and common variants may be implicated in IBS. However, overall, gene-hunting efforts in IBS have so far been scarce [12], and large-scale efforts remain to be carried out. Recognizing that IBS spans from complex polygenic conditions to rare single-gene forms, we need to adopt different strategies to identify these genetic factors.
The challenge in finding IBS-risk genes: past and future perspectives
More than 60 candidate genes have been studied for IBS, including genes involved in serotonin synthesis and reuptake, mucosal immune activation and inflammation, neuropeptide signaling, nociception, bile acid synthesis, and intestinal secretion. These genetic findings have been reviewed and discussed thoroughly before, and a selection of genes showing best evidence of association to IBS is reported in Table 1 [10, 12]. However, all of these studies are candidate-gene case-control studies conducted in relatively small sample sets. Many of the identified associations have not been successfully replicated in independent studies, and although p values have been nominally significant, none of the findings are close to sustain the correction for a GWA study. Therefore, IBS-risk genes thus far proposed mostly still represent non-validated hits rather than true predisposing factors, and we have very few convincing IBS-risk genes identified so far. One exception may be the TNFSF15 gene, which was convincingly associated in our original study [14] and then successfully replicated in independent cohorts [15–17]. In the original study, we hypothesized that genes involved in immune responses and host-microbe interactions may also contribute to IBS susceptibility, since these are mechanisms suggested to be involved in IBS pathophysiology [5, 7]. Therefore, we tested the association of polymorphisms (SNPs) from 30 known Crohn’s disease risk loci on IBS risk in two case-control studies from Sweden and the USA (total n = 1992). The G allele (risk allele for Crohn’s) of rs426839 in TNFSF15 was significantly associated with increased risk of IBS (p = 2.2e-05; OR 1.37) and even more in IBS-C (p = 8.7e-07; OR 1.79). This gene encodes the TL1A protein expressed in immune cells, which promotes inflammatory response in the gut mucosa. Carrying the risk allele is suggested to result in higher TL1A expression and thereby stronger T cell activation and immune response. Hence, the association with TNFS15 indicates that inflammatory response may be an important mechanism also in IBS.
Table 1.
Prioritized IBS-risk genes based on existing published statistical evidence
| Gene | Chr location | Gene name | Gene function | Gene region | Phenotype | Key reference |
|---|---|---|---|---|---|---|
| TNFSF15 | 9q32 | Tumor necrosis factor (ligand) superfamily, member 15 | Codes for TNF-like ligand 1A (TL1A), which contributes to the modulation of inflammatory responses. | Intron | IBS, IBS-C | [14] |
| Intron and upstream | IBS-D | [15] | ||||
| Intron | IBS-A | [16] | ||||
| Intron | IBS, IBS-C | [17] | ||||
| TLR9 | 3p21.3 | Toll-like receptor 9 | A Toll-like receptor that activate the immune system through recognition of specific patterns on microorganisms. | Intron and upstream | PI-IBS | [26] |
| Upstream | IBS-D | [27] | ||||
| HTR3E | 3q27.1 | 5-hydroxytryptamine (serotonin) receptor 3E, ionotropic | A receptor for serotonin, a neurotransmitter in the central nervous system and gastrointestinal tract. | 3′UTR | IBS-D | [28] |
| NPSR1 | 7p14.3 | Neuropeptide S receptor 1 | Receptor for neuropeptide S, expressed in brain and enteroendocrine cells, involved in anxiety, inflammation, nociception, etc. | Intron and coding polymorphism | Colonic transit time, sensory ratings (pain, gas, urgency) | [19] |
| 5′ near gene and beginning of gene | RAP | [21] | ||||
| KLB | 4p14 | Klotho beta | Co-receptor of fibroblast growth factor 19 (FGFR4) on hepatocyte membrane, required for suppression of bile acid synthesis in liver. | Coding polymorphism | Colonic transit in IBS-D | [9] |
| SCN5A | 3p21 | Sodium channel, voltage gated, type V alpha subunit | Voltage-gated sodium channel (NaV1.5) in pacemaker cells of the heart and interstitial cells of cajal (ICC) cells of the gut important for smooth muscle contraction. | Rare coding mutations and signal of common SNPs in middle of gene | IBS, IBS-C | [13] |
| CDC42 | 1p36.1 | Cell division cycle 42 | A small GTPase of the Rho-subfamily involved in cell cycle regulation and possibly epithelial barrier function through intestinal stem cell differentiation and proliferation. | Intron | IBS-C | [16] |
| KDELR2 | 7p22.1 | KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 | Belongs to a family of KDEL motif binding receptors, mediating the retrograde transport of proteins to the endoplasmatic reticulum. | Intron | IBS | [25] |
Chr chromosomal location, IBS-D diarrhea-predominant IBS, IBS-C constipation-predominant IBS, IBS-A alternating IBS, PI-IBS post-infectious IBS, RAP recurrent abdominal pain
An alternative approach to the discovery of IBS relevant genes is to study the endophenotypes, otherwise called intermediate phenotypes of disease. These are usually quantitative traits related to IBS, such as colonic transit time and visceral sensitivity ratings. To focus on biological observations instead of clinical entities for establishing, diagnosis was proposed already 50 years ago, and this alternative strategy has been used successfully for genetic studies in the psychiatric field where illnesses often are, as IBS, complex and heterogeneous [18]. The concept of endophenotypes is to use disease-associated phenotypes in an attempt to reduce the complexity and to increase chances of finding genes important to biological processes underlying disease etiology. For the case of IBS, suitable traits to use for this approach include bowel movement frequency, Bristol stool form scale (a subjective measure of stool consistency, related to colonic transit time), pain and sensation ratings in response to visceral stimuli, and others. Previous work from our group has shown associations between polymorphisms in the neuropeptide S receptor gene (NPSR1) and colonic transit time, as well as gastrointestinal (GI) sensory ratings, such as gas, pain, and urgency [19]. The NPSR1 protein is a receptor for neuropeptide S, a neuropeptide involved in anxiety, response to stress and fear, inflammation, and nociception. Neuropeptides act in the brain-gut axis and have been implicated in IBS before [2, 6]. We also showed that the neuropeptide S (NPS)-NPSR1 system can induce the expression of other neuropeptides in vitro [19, 20]. Hence, this gene was a plausible candidate to investigate also for its potential genetic associations with abdominal pain. In the Swedish BAMSE birth cohort, we performed a candidate-gene study in relation to recurrent abdominal pain (RAP) [21], which occurs frequently among children and is one of the cardinal symptoms of FGIDs. The mechanisms of visceral pain and RAP are not fully understood, a heritable component has been demonstrated, and a few candidate genes proposed. We observed association with RAP at 7/24 tested SNPs, with the strongest signal in correspondence of a putative regulatory region upstream NPSR1 where they may exert their genetic effects through the modulation of gene expression.
Overall, IBS poses some major challenges, and it appears that in order to overcome these and be able to identify true unequivocal risk genes and variants, we need to implement larger scale analyses. Genetic-risk genes and variants have been successfully identified and replicated in a plethora of complex diseases using the powerful and hypothesis-free approach of GWAS studies and their meta-analyses (see the NHGRI-EBI GWAS Catalog; www.ebi.ac.uk/gwas) [22]. For instance in the GI field, a recent study reported the identification of 38 additional susceptibility loci for inflammatory bowel disease (IBD), bringing the total tally of confirmed IBD risk loci to 200 [23]. On the contrary, IBS genetics is very much lagging behind, and no similar GWAS effort had been attempted before our Screening-Across Life Span Twin (SALT) study (see below).
GWA studies are powerful approaches to discovering genetic risk loci in complex diseases [24], though this has never even been attempted in IBS. In order to reach the statistical power necessary to detect meaningful association, very large sample size is required. Unfortunately, this is currently unfeasible in the IBS community, as the number and size of well-characterized cohorts around the world is still surprisingly slim. In fact, only a portion of individuals suffering from IBS according to the Rome III criteria seek medical attention for their symptoms, with numbers reported ranging from 10 to 70 % [1]. In order to reach more individuals, we propose to shift approach and instead make use of general population samples for discovery purposes in IBS genetics. Large biobanks and general population cohorts offer a great opportunity to perform genetic epidemiological association studies with considerably larger sample sizes. When available, Rome questionnaire data may be used to identify IBS cases and asymptomatic controls in these large study populations. In addition, genotype data may also be linked to phenotype in these studies using the International Classification of Diseases (ICD codes) and electronic medical records (EMR). By shifting to a general population approach for the discovery phase, we gain considerable sample size and improve in the definition of controls, as they are co-sampled with the cases and classified with the very same investigative tools (for example questionnaires). However, once IBS-risk genes and variants have been discovered, validation of results should be performed by replication and targeted analyses in case-control cohorts from gastro clinics with well-characterized IBS patients and healthy controls. Furthermore, investigating rare variants/mutations in IBS, next-generation sequencing in selected cases and controls will also be necessary.
Our group recently conducted the very first pilot GWAS of IBS [25]. In this study, Rome II criteria from questionnaires were used to identify 534 IBS cases and 4932 asymptomatic controls from the SALT study of the Swedish Twin Registry. A GWAS was performed and top regions replicated in six independent case-control studies with patients and controls recruited from European and US clinics. Confirmatory, one of the loci (harboring the KDELR2 gene) was successfully replicated in all these samples (total sample size of 8977) with consistent effect of association. Even though sample size was still too small to reach genome-wide significance, this study corroborated our hypothesis that general population-based cohorts with associated genetic and epidemiological/healthcare data provide excellent opportunities to study the genetic architecture of IBS and related GI symptoms.
Overall, the ultimate value of genetic research in IBS is to identify key physiological mechanisms, which will help us understand its pathophysiology. These findings will help improve IBS diagnosis and classification and therefore impact therapeutic strategies and personalized treatments.
Acknowledgements
Mauro D’Amato’s research in IBS is supported by grants from the Swedish Vetenskapsrådet (VR).
Abbreviations
- FGID
functional gastrointestinal disorder
- GI
gastrointestinal
- GWAS
genome-wide association study
- IBS
irritable bowel syndrome
- IBS-C
constipation-predominant IBS
- IBS-D
diarrhea-predominant IBS
- IBS-M
mixed IBS phenotyped
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
MH and MD both wrote and approved the final manuscript.
Contributor Information
Maria Henström, Email: maria.henstrom@ki.se.
Mauro D’Amato, Phone: +34658025711, Email: mauro.damato@osakidetza.eus.
References
- 1.Canavan C, West J, Card T. The epidemiology of irritable bowel syndrome. Clin Epidemiol. 2014;6:71–80. doi: 10.2147/CLEP.S40245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drossman DA. The functional gastrointestinal disorders and the Rome III process. Gastroenterology. 2006;130:1377–90. doi: 10.1053/j.gastro.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Whitehead WE, Palsson O, Jones KR. Systematic review of the comorbidity of irritable bowel syndrome with other disorders: what are the causes and implications? Gastroenterology. 2002;122:1140–56. doi: 10.1053/gast.2002.32392. [DOI] [PubMed] [Google Scholar]
- 4.Camilleri M, Williams DE. Economic burden of irritable bowel syndrome. Proposed strategies to control expenditures. Pharmacoeconomics. 2000;17:331–8. doi: 10.2165/00019053-200017040-00003. [DOI] [PubMed] [Google Scholar]
- 5.Camilleri M. Peripheral mechanisms in irritable bowel syndrome. N Engl J Med. 2012;367:1626–35. doi: 10.1056/NEJMra1207068. [DOI] [PubMed] [Google Scholar]
- 6.Lee YJ, Park KS. Irritable bowel syndrome: emerging paradigm in pathophysiology. World J Gastroenterol. 2014;20:2456–69. doi: 10.3748/wjg.v20.i10.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kennedy PJ, Cryan JF, Dinan TG, Clarke G. Irritable bowel syndrome: a microbiome-gut-brain axis disorder? World J Gastroenterol. 2014;20:14105–25. doi: 10.3748/wjg.v20.i39.14105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibson PR, Varney J, Malakar S, Muir JG. Food components and irritable bowel syndrome. Gastroenterology. 2015;148:1158–74. doi: 10.1053/j.gastro.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Wong BS, Camilleri M, Carlson PJ, Guicciardi ME, Burton D, McKinzie S, Rao AS, Zinsmeister AR, Gores GJ. A Klothoβ variant mediates protein stability and associates with colon transit in irritable bowel syndrome with diarrhea. Gastroenterology. 2011;140:1934–42. doi: 10.1053/j.gastro.2011.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saito YA. The role of genetics in IBS. Gastroenterol Clin North Am. 2011;40:45–67. doi: 10.1016/j.gtc.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waehrens R, Ohlsson H, Sundquist J, Sundquist K, Zöller B. Risk of irritable bowel syndrome in first-degree, second-degree and third-degree relatives of affected individuals: a nationwide family study in Sweden. Gut. 2015;64:215–21. doi: 10.1136/gutjnl-2013-305705. [DOI] [PubMed] [Google Scholar]
- 12.D’Amato M. Genes and functional GI disorders: from casual to causal relationship. Neurogastroenterol Motil. 2013;25:638–49. doi: 10.1111/nmo.12173. [DOI] [PubMed] [Google Scholar]
- 13.Beyder A, Mazzone A, Strege PR, Tester DJ, Saito YA, Bernard CE, Enders FT, Ek WE, Schmidt PT, Dlugosz A, Lindberg G, Karling P, Ohlsson B, Gazouli M, Nardone G, Cuomo R, Usai-Satta P, Galeazzi F, Neri M, Portincasa P, Bellini M, Barbara G, Camilleri M, Locke GR, 3rd, Talley NJ, D'Amato M, Ackerman MJ, Farrugia G. Loss-of-function of the voltage-gated sodium channel NaV1.5 (channelopathies) in patients with irritable bowel syndrome. Gastroenterol. 2014;146:1659–68. doi: 10.1053/j.gastro.2014.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zucchelli M, Camilleri M, Andreasson AN, Bresso F, Dlugosz A, Halfvarson J, Törkvist L, Schmidt PT, Karling P, Ohlsson B, Duerr RH, Simren M, Lindberg G, Agreus L, Carlson P, Zinsmeister AR, D'Amato M. Association of TNFSF15 polymorphism with irritable bowel syndrome. Gut. 2011;60:1671–7. doi: 10.1136/gut.2011.241877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swan C, Duroudier NP, Campbell E, Zaitoun A, Hastings M, Dukes GE, Cox J, Kelly FM, Wilde J, Lennon MG, Neal KR, Whorwell PJ, Hall IP, Spiller RC. Identifying and testing candidate genetic polymorphisms in the irritable bowel syndrome (IBS): association with TNFSF15 and TNFα. Gut. 2013;62:985–94. doi: 10.1136/gutjnl-2011-301213. [DOI] [PubMed] [Google Scholar]
- 16.Wouters MM, Lambrechts D, Knapp M, Cleynen I, Whorwell P, Agréus L, Dlugosz A, Schmidt PT, Halfvarson J, Simrén M, Ohlsson B, Karling P, Van Wanrooy S, Mondelaers S, Vermeire S, Lindberg G, Spiller R, Dukes G, D'Amato M, Boeckxstaens G. Genetic variants in CDC42 and NXPH1 as susceptibility factors for constipation and diarrhoea predominant irritable bowel syndrome. Gut. 2014;63:1103–11. doi: 10.1136/gutjnl-2013-304570. [DOI] [PubMed] [Google Scholar]
- 17.Czogalla B, Schmitteckert S, Houghton LA, Sayuk GS, Camilleri M, Olivo-Diaz A, Spiller R, Wouters MM, Boeckxstaens G, Bermejo JL, Niesler B. A meta-analysis of immunogenetic case-control association studies in irritable bowel syndrome. Neurogastroenterol Motil. 2015;27:717–27. doi: 10.1111/nmo.12548. [DOI] [PubMed] [Google Scholar]
- 18.Braff DL, Freedman R, Schork NJ, Gottesman II. Deconstructing schizophrenia: an overview of the use of endophenotypes in order to understand a complex disorder. Schizophr Bull. 2007;33:21–32. doi: 10.1093/schbul/sbl049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camilleri M, Carlson P, Zinsmeister AR, McKinzie S, Busciglio I, Burton D, Zucchelli M, D'Amato M. Neuropeptide S receptor induces neuropeptide expression and associates with intermediate phenotypes of functional gastrointestinal disorders. Gastroenterology. 2010;138:98–107. doi: 10.1053/j.gastro.2009.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sundman L, Saarialho-Kere U, Vendelin J, Lindfors K, Assadi G, Kaukinen K, Westerholm-Ormio M, Savilahti E, Mäki M, Alenius H, D'Amato M, Pulkkinen V, Kere J, Saavalainen P. Neuropeptide S receptor 1 expression in the intestine and skin—putative role in peptide hormone secretion. Neurogastroenterol Motil. 2010;22:79–87. doi: 10.1111/j.1365-2982.2009.01366.x. [DOI] [PubMed] [Google Scholar]
- 21.Henström M, Zucchelli M, Söderhäll C, Bergström A, Kere J, Melén E, Olén O, D'Amato M. NPSR1 polymorphisms influence recurrent abdominal pain in children: a population-based study. Neurogastroenterol Motil. 2014;26:1417–25. doi: 10.1111/nmo.12401. [DOI] [PubMed] [Google Scholar]
- 22.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–6. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, Abedian S, Cheon JH, Cho J, Daryani NE, Franke L, Fuyuno Y, Hart A, Juyal RC, Juyal G, Kim WH, Morris AP, Poustchi H, Newman WG, Midha V, Orchard TR, Vahedi H, Sood A, Sung JJ, Malekzadeh R, Westra HJ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–86. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ek WE, Reznichenko A, Ripke S, Niesler B, Zucchelli M, Rivera NV, Schmidt PT, Pedersen NL, Magnusson P, Talley NJ, Holliday EG, Houghton L, Gazouli M, Karamanolis G, Rappold G, Burwinkel B, Surowy H, Rafter J, Assadi G, Li L, Papadaki E, Gambaccini D, Marchi S, Colucci R, Blandizzi C, Barbaro R, Karling P, Walter S, Ohlsson B, Törnblom H, et al. Exploring the genetics of irritable bowel syndrome: a GWA study in the general population and replication in multinational case-control cohorts. Gut. 2015;64:1774–82. doi: 10.1136/gutjnl-2014-307997. [DOI] [PubMed] [Google Scholar]
- 26.Villani AC, Lemire M, Thabane M, Belisle A, Geneau G, Garg AX, Clark WF, Moayyedi P, Collins SM, Franchimont D, Marshall JK. Genetic risk factors for post-infectious irritable bowel syndrome following a waterborne outbreak of gastroenteritis. Gastroenterology. 2010;138:1502–13. doi: 10.1053/j.gastro.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 27.Camilleri M, Carlson P, McKinzie S, Zucchelli M, D'Amato M, Busciglio I, Burton D, Zinsmeister AR. Genetic susceptibility to inflammation and colonic transit in lower functional gastrointestinal disorders: preliminary analysis. Neurogastroenterol Motil. 2011;23:935–e398. doi: 10.1111/j.1365-2982.2011.01749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kapeller J, Houghton LA, Mönnikes H, Walstab J, Möller D, Bönisch H, Burwinkel B, Autschbach F, Funke B, Lasitschka F, Gassler N, Fischer C, Whorwell PJ, Atkinson W, Fell C, Büchner KJ, Schmidtmann M, van der Voort I, Wisser AS, Berg T, Rappold G, Niesler B. First evidence for an association of a functional variant in the microRNA-510 target site of the serotonin receptor-type 3E gene with diarrhea predominant irritable bowel syndrome. Hum Mol Genet. 2008;17:2967–77. doi: 10.1093/hmg/ddn195. [DOI] [PubMed] [Google Scholar]


