ABSTRACT
Much of the morbidity and mortality associated with influenza virus respiratory infection is due to bacterial coinfection with pathogens that colonize the upper respiratory tract such as methicillin-resistant Staphylococcus aureus (MRSA) and Streptococcus pneumoniae. A major component of the immune response to influenza virus is the production of type I and III interferons. Here we show that the immune response to infection with influenza virus causes an increase and restructuring of the upper respiratory microbiota in wild-type (WT) mice but not in Il28r−/− mutant mice lacking the receptor for type III interferon. Mice lacking the IL-28 receptor fail to induce STAT1 phosphorylation and expression of its regulator, SOCS1. Il28r−/− mutant mice have increased expression of interleukin-22 (IL-22), as well as Ngal and RegIIIγ, in the nasal cavity, the source of organisms that would be aspirated to cause pneumonia. Proteomic analysis reveals changes in several cytoskeletal proteins that contribute to barrier function in the nasal epithelium that may contribute to the effects of IL-28 signaling on the microbiota. The importance of the effects of IL-28 signaling in the pathogenesis of MRSA pneumonia after influenza virus infection was confirmed by showing that WT mice nasally colonized before or after influenza virus infection had significantly higher levels of infection in the upper airways, as well as significantly greater susceptibility to MRSA pneumonia than Il28r−/− mutant mice did. Our results suggest that activation of the type III interferon in response to influenza virus infection has a major effect in expanding the upper airway microbiome and increasing susceptibility to lower respiratory tract infection.
IMPORTANCE
S. aureus and influenza virus are important respiratory pathogens, and coinfection with these organisms is associated with significant morbidity and mortality. The ability of influenza virus to increase susceptibility to S. aureus infection is less well understood. We show here that influenza virus leads to a change in the upper airway microbiome in a type III interferon-dependent manner. Mice lacking the type III interferon receptor have altered STAT1 and IL-22 signaling. In coinfection studies, mice without the type III interferon receptor had significantly less nasal S. aureus colonization and subsequent pneumonia than infected WT mice did. This work demonstrates that type III interferons induced by influenza virus contribute to nasal colonization and pneumonia due to S. aureus superinfection.
INTRODUCTION
Bacterial superinfection is a major cause of the morbidity and mortality associated with influenza (1). Streptococcus pneumoniae, Haemophilus influenzae, and Staphylococcus aureus have been the most prevalent respiratory pathogens; however, highly virulent methicillin-resistant S. aureus (MRSA) strains common in the United States are now increasingly frequent (2, 3). These organisms are often part of the commensal flora colonizing the nasal cavity, which provides a source for subsequent aspiration into the lung. While the normal host readily clears the small numbers of routinely aspirated flora, in the setting of influenza virus infection, there is significantly increased susceptibility to severe bacterial pneumonia (4–6). Substantial epidemiological data have documented that nasal colonization with S. aureus is a major predisposing factor for subsequent infection (7–10). These data have spurred screening for nasal carriage of MRSA and universal decolonization of intensive care unit patients (11) to prevent infection, since the carriage of these organisms is so common (12). Several investigators have demonstrated that the bacterial inoculum required to cause S. pneumoniae (4) or S. aureus pneumonia (13, 14) in a murine model is significantly decreased in the setting of antecedent influenza virus infection, and the multiple immune mechanisms responsible have been recently reviewed (15). The density of pathogen colonization in the upper respiratory tract is likely a contributing factor in susceptibility to subsequent pneumonia. Influenza vaccination with live attenuated virus increases the amounts of both pneumococci and staphylococci in the upper airway (16), suggesting that the immune response activated by influenza virus affects the factors that normally regulate the upper airway microbiome.
A major component of the initial immune response to influenza virus infection is activation of type I and III interferons (IFNs), resulting in the induction of over 300 genes that comprise the interferome (17–19). Type I IFN signaling is mediated through IFNAR, a receptor that responds to 13 IFN-α subtypes, IFN-β, IFN-ε, and IFN-ω through JAK-STAT signaling and activation of an autocrine loop (20). The type III IFNs, IFN-λ, or IL-28A/B and IL-29 are sensed through a compound receptor composed of the IL-28 receptor (IL-28R) and IL-10RB (21), which is not ubiquitous like IFNAR and is expressed predominantly on mucosal surfaces (22) and on neutrophils (23). Type I and III IFN signaling contributes to the pathology elicited by S. aureus pulmonary infection (24, 25) and influenza virus-induced type I IFN contributes to greater S. aureus-induced lung pathology and poorer prognosis (6). Thus, once these pathogens reach the lower airways, they are likely to cause more severe disease in the setting of influenza. However, it is not clear whether influenza and IFNs enable pathogens such as MRSA to colonize and proliferate in the nasal cavity, which is a major source of subsequent pulmonary infection.
We postulated that bacterial superinfection after influenza virus infection requires two factors: first, the accumulation of an adequate inoculum of potential pathogens in the upper respiratory tract, and second, impaired innate immune defenses within the lung itself, attributed to the type I/III IFNs and other factors (5, 24, 26). The combination of both events results in pneumonia instead of bacterial clearance. In the studies presented here, we demonstrate that type III IFN-STAT1 signaling elicited by influenza alters the composition of the nasal microbiome and proteome, increases susceptibility to nasal colonization, and impairs the clearance of S. aureus from the lower airways.
RESULTS
Influenza virus infection results in expansion and restructuring of the nasal microbiome.
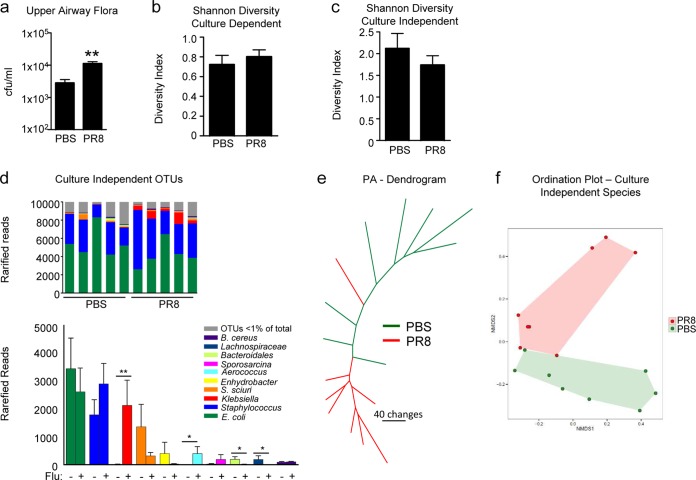
We investigated the influence of mouse-adapted influenza virus strain PR8 on the upper respiratory microbiome of wild-type (WT) C57BL/6J mice (Fig. 1). There was a significant increase in the total numbers of bacteria recovered from the upper airway following 3 days of infection (P = 0.002) (Fig. 1A). Although the overall diversity of the microbial community was unchanged, as measured by culture-dependent and -independent techniques (Fig. 1B and C), we observed specific changes in the relative abundance of certain bacterial species (Table 1; Fig. 1D), including murine commensal staphylococci. In addition, culture-independent microbial community profiles clustered on the basis of the presence of influenza virus, suggesting reproducible shifts in the structure of the commensal microbiome in response to infection (Fig. 1E and F).
FIG 1 .
Changes in the upper respiratory microbiome in response to influenza virus infection. WT mice were infected with PR8 or treated with PBS for 3 days. (a) Total bacterial load (CFU) recovered from the upper airway. n = 11. (b, c) Shannon-Weiner diversity of bacteria cultured (b) or identified with culture-independent techniques (c) from the upper airway. n = 11 (b) or 8 (c). (d) Culture-independent analysis of community composition in the upper airways of WT mice infected with PR8 or treated with PBS as a control. Relative abundance plots show differences between PR8-infected and PBS-treated mice. The bar plot shows differences in rarefied reads for the top 10 OTUs (ranked by relative abundance), as indicated by color coding. The top 10 OTUs constitute >99% of the rarefied reads. n = 8. (e) Maximum-parsimony tree based on OTU presence in or absence from the upper airways of mice infected with PR8 or treated with PBS. Branch length is proportional to the number of gain/loss changes in OTUs (scale bar represents 40 changes). Branches representing influenza virus-infected samples are red, and PBS-inoculated control samples are green. (f) Ordination plot of culture-independent analysis demonstrating segregation of PR8-infected mice (red) and PBS-treated controls (green); shaded areas are the minimum convex polygons enclosing the data points in each group. P = 0.004. Data are representative of at least two independent experiments. Graphs display mean values and standard errors. **, P < 0.01; *, P < 0.05.
TABLE 1 .
Microbiota changes based on culture with influenza and rIL-28a
| Species | No. of mice with bacteriab |
P value | Mean no. of CFU/50 µl of BALF |
P value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Influenza− | Influenza+ | PBS | rIL28 | Influenza− | Influenza+ | PBS | rIL28 | |||
| Gram positive | ||||||||||
| Staphylococcus lentus | 9 | 10 | 1.0000 | 268 | 483 | 0.3910 | ||||
| Staphylococcus xylosus | 9 | 9 | 1.0000 | 19 | 192 | 0.0264c | ||||
| Staphylococcus nepalensis | 8 | 7 | 1.0000 | 24 | 123 | 0.4698 | ||||
| Enterococcus faecalis | 3 | 4 | 1.0000 | 21 | 21.25 | 0.3545 | ||||
| Bacillus thuringiensis | 4 | 1 | 0.3108 | 8 | 1 | 0.2556 | ||||
| Enterococcus gallinarum | 2 | 1 | 1.0000 | 3 | 4 | >0.9999 | ||||
| Staphylococcus cohnii | 2 | 1 | 1.0000 | 11 | 106 | >0.9999 | ||||
| Aerococcus urinaeequi | 0 | 2 | 0.4762 | 0 | 13 | 0.4762 | ||||
| Jeotgalicoccus halotolerans | 0 | 2 | 0.4762 | 0 | 58 | 0.4762 | ||||
| Gram negative | ||||||||||
| Klebsiella oxytoca | 3 | 5 | 0.6594 | 2 | 43 | 0.1883 | ||||
| Enterobacter hormaechei | 4 | 2 | 0.6351 | 32 | 1,224 | 0.6351 | ||||
| Enterobacter absurie | 1 | 4 | 0.3108 | 7 | 464 | 0.1454 | ||||
| Gram positive | ||||||||||
| Staphylococcus xylosus | 10 | 10 | 1.0000 | 59 | 284 | 0.1639 | ||||
| Staphylococcus lentus | 7 | 9 | 0.5820 | 3 | 230 | 0.0034c | ||||
| Staphylococcus nepalensis | 3 | 9 | 0.0198c | 4 | 120 | 0.0014c | ||||
| Staphylococcus cohnii | 5 | 6 | 1.0000 | 59 | 283 | 0.1948 | ||||
| Enterococcus faecalis | 5 | 5 | 1.0000 | 24 | 244 | 0.5076 | ||||
| Aerococcus urinaeequi | 0 | 5 | 0.0325c | 0 | 289 | 0.0325c | ||||
| Gram negative | ||||||||||
| Enterobacter hormaechei | 4 | 4 | 1.0000 | 57 | 190 | 0.8114 | ||||
| Enterobacter absuriae | 1 | 0 | 1.0000 | 194 | 0 | >0.9999 | ||||
Culture-dependent identification of bacteria recovered from the upper airways of WT mice infected with PR8 or treated with PBS for 3 days or with BSA or rIL28B after 18 h is shown. Data are representative of at least three independent experiments. For presence data, the Fisher exact test was used. For mean CFU counts, the Mann-Whitney nonparametric test was used. For the median values and interquartile ranges, see Tables S1 to S4 in the supplemental material.
Total n = 20.
Significantly different.
IL-28R signaling has a major effect on the density of the upper airway flora.
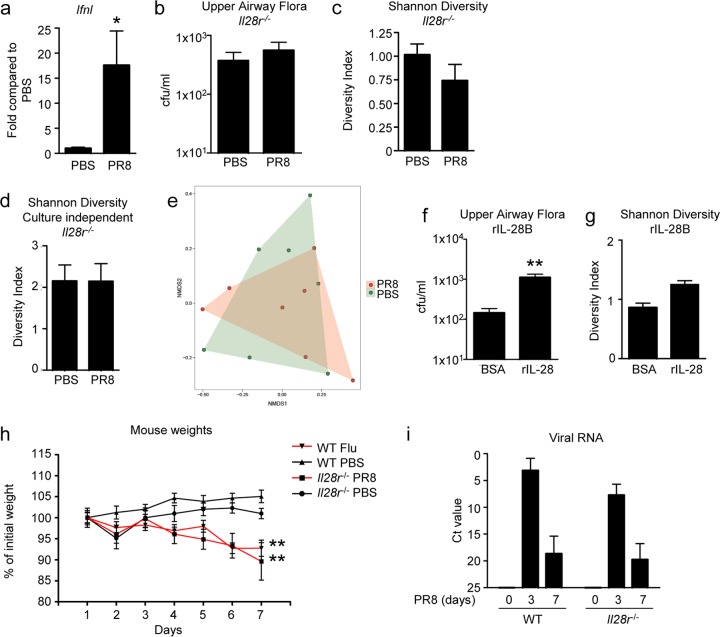
We next addressed the role of type III IFN signaling in the control of the nasal microbiome. Il28 gene expression in the upper airway was documented to be significantly upregulated in response to PR8 (Fig. 2A). There was no increase in the abundance or change in diversity of the nasal flora observed in the Il28r−/− mutant mice after influenza virus infection (Fig. 2B and C), in contrast to the significant increase in upper airway flora in WT mice after influenza virus infection. Culture-independent experiments further confirmed a lack of change in diversity after influenza virus infection (Fig. 2D) and also showed no major changes in the structure or composition of the microbiome (Fig. 2E).
FIG 2 .
Effects of type III IFN signaling on upper airway microbiome. (a) WT mice were infected for 3 days with PR8. Nasal lavage fluid was assessed by qRT-PCR for expression of IFN-λ (Il28). n = 5 (PBS) or 4 (PR8). Il28r−/− mutant mice were infected with PR8 or treated with PBS for 3 days. (b, c) Bacterial load (n = 8) (b) and diversity of culture-dependent organisms isolated from upper airways (c). n = 8 (PBS) or 7 (PR8). (d) Diversity analysis from culture-independent data. n = 7. (e) Ordination plot of culture-independent analysis demonstrating lack of segregation of PR8-infected (red) and PBS-treated control (green) Il28r−/− mutant mice. Shaded areas are the minimum convex polygons enclosing the data points in each group. WT mice were treated with 1 µg of recombinant murine IL-28 or with BSA as a control, and the total bacterial load (f) (n = 10) and diversity of culture-dependent organisms (g) were determined the following day. n = 10. (h) Weights of WT and Il28r−/− mutant mice infected with PR8 or treated with PBS. n = 7. (i) qRT-PCR analysis of viral RNA in the lungs of WT and Il28r−/− mutant mice infected with PR8 for 3 or 7 days. n = 6. Data are representative of at least two independent experiments. Graphs display mean values and standard errors. **, P < 0.01.
To confirm that type III IFN signaling was responsible for alterations in flora, recombinant IL-28B (IL-28A and -B are 96% identical [27]) was instilled into the airways of WT mice, which resulted in significant increases in the number of bacteria recovered from the upper airway (P = 0.0011) (Fig. 2F), to levels comparable to those observed with influenza virus infection. There were no significant changes in the overall diversity of the microbiota, but as with influenza virus infection, some species increased disproportionately (Fig. 2G; Table 1; see Tables S1 to S4 in the supplemental material). These results suggested that IL-28R–IFN-λ signaling is specifically involved in the restructuring of the microbial ecology of the upper airway and the increase in specific local flora in response to influenza virus infection. The differences in flora observed were not due to the severity of influenza virus infection in the different mouse strains. Weight loss over the course of infection (Fig. 2I) and levels of viral RNA found in the lungs of WT and Il28r−/− mutant mice were similar (Fig. 2I).
IL-28R signals through STAT1.
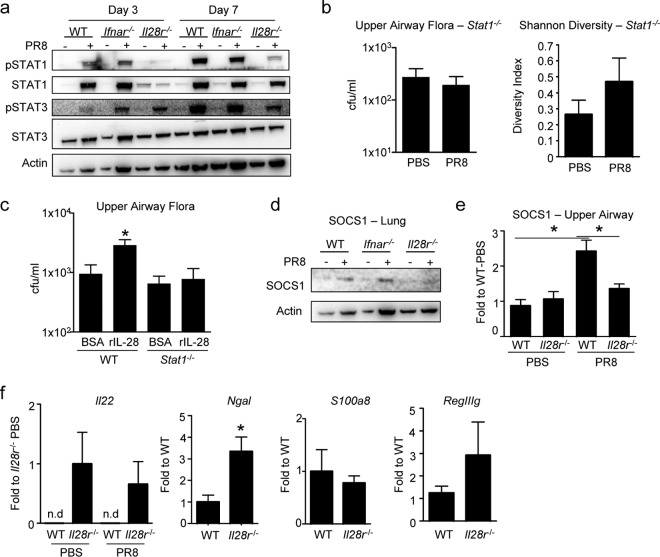
The type III IFNs are involved in multiple downstream signaling cascades that could affect the abundance of nasal flora through the action of antimicrobial peptides, cytokines that alter barrier function and immune cells recruited across the mucosal surface. The effects of type III IFNs are mediated by JAK/STAT signaling, initiated by phosphorylation of STAT1 (28, 29). The relative induction of STAT1 and STAT3 phosphorylation in WT and Il28r−/− mutant mice, exposed to influenza virus or phosphate-buffered saline (PBS), were compared, and Ifnar−/− mutant mice were included as a control for the type I IFNs (Fig. 3A). STAT1 and STAT3 phosphorylation was induced by influenza virus infection in WT and Ifnar−/− mutant mice at days 3 and 7 postinfection (Fig. 3A). However, minimal amounts of STAT1 or phosphorylated STAT1 were detected in Il28r−/− mutant mice. Even at 7 days postinfection, substantially less phosphorylated STAT1 was detected in Il28r−/− mutant mice than in WT or Ifnar−/− mutant controls. To determine if these differences in STAT1 activation were associated with changes in the upper airway microbiome after influenza virus infection, we quantified commensal flora in STAT1 null mice before and following influenza virus infection. As was observed in the Il28r−/− mutant mice, in the absence of STAT1, there was no increase in the abundance or diversity of nasal flora after influenza virus infection (Fig. 3B). To confirm that the effect of type III IFN on flora was through STAT1, we applied purified IL-28B to the airways of WT and Stat1−/− mutant mice. While addition of IL-28B to WT mice led to expansion of the resident flora and increases in specific species, Stat1−/− mutant mice had no change in the numbers of bacteria present (Fig. 3C; see Tables S5 to S8 in the supplemental material). Thus, IFN-λ-dependent regulation of the nasal microbiome is mediated by STAT1-dependent signals.
FIG 3 .
Mutant (Il28r−/−) mice lack STAT1-associated signaling. (a) Immunoblot assays of P-STAT1, STAT1, P-STAT3, STAT3, and actin in WT and Ifnar−/− and Il28r−/− mutant mice infected with PR8 for 3 or 7 days or treated with PBS as a control. Stat1−/− mutant mice were infected with PR8 or treated with PBS as a control for 3 days. (b) Bacterial load isolated from the upper airway and Shannon-Weiner analysis of the diversity of recovered bacteria. n = 4 (WT, PBS), 5 (WT, PR8), 8 (Il28r−/− mutant, PBS), or 7 (Il28r−/− mutant, PR8). (c) WT or Stat1−/− mutant mice were treated with 1 µg of recombinant murine IL-28 or with BSA as a control, and the culture-dependent bacterial load was measured. n = 6. (d) Immunoblot assays of SOCS1 and actin in WT and Ifnar−/− and Il28r−/− mutant mice infected with PR8 for 3 days or treated with PBS as a control. (e) qRT-PCR analysis of Socs1 expression in their upper airways. n = 4 (PBS) or 5 and 6 (left and right; PR8). (f) qRT-PCR analysis of Il22, Ngal, S100a8, and RegIIIγ in the upper airways of WT and Il28r−/− mutant mice. Il22 data are shown for both PBS-treated and PR8-infected mice. Il22, n = 8; Ngal, n = 5 and 9, respectively; S100a8, n = 9 and 8, respectively; RegIIIγ, n = 8 and 7, respectively. Data are representative of at least two independent experiments. Graphs display mean values and standard errors. n.d, not detectable. *, P < 0.05.
Effects of STAT1-SOCS1 interactions in the upper airway.
STAT1 signaling could affect the abundance of the upper airway flora through several mechanisms. STAT1 induces and is regulated by SOCS1, which interacts directly with JAK kinase (30, 31). SOCS1 regulates the expression of antimicrobial peptides that would control the abundance of commensal flora (32). We predicted that, in the absence of STAT1 signaling, there would be decreased SOCS1 expression, as was observed in the lung lysates of Il28r−/− mutant mice compared with those of WT or Ifnar−/− mutant mice, both of which express STAT1 (Fig. 3D). In the nasal cavities of WT mice, there was significant upregulation of Socs1 expression in response to influenza virus infection, but no increase was observed in the Il28r−/− mutant mice that lacked P-STAT1 (P = 0.0101) (Fig. 3E).
SOCS1 involvement in the suppression of NF-κB- and STAT1-dependent genes could impact the abundance of antimicrobial peptides in the upper airway. We compared the expression of selected antimicrobial peptides and cytokines in the nasal cavities of WT and Il28r−/− mutant mice (Fig. 3F). While Il22 expression was undetectable in WT mice, it was prominently expressed in Il28r−/− mutant mice. There was upregulation of the IL-22-dependent antimicrobial peptide RegIIIγ in Il28r−/− mutant mice but no significant differences in S100A8 (33, 34). NGAL (lipocalin), an NF-κB-dependent gene product, was also significantly upregulated in the absence of IFN-λ signaling.
Proteomic analysis of upper airway lavage fluids reflects consequences of IL-28 signaling for epithelial barrier function.
A global analysis of the effects of type III IFN signaling on the proteome of the upper airway was performed. Nasal lavage fluids were harvested from WT and Il28r−/− mutant mice with and without influenza virus infection (Fig. 4). At the baseline, we found significant amounts of the antimicrobial peptide NGAL in Il28r−/− mutant mice, which was likely to have an effect on the overall composition of the nasal microbiota (Fig. 4A). Classification of the families of proteins differentially expressed in WT and Il28r−/− mutant mice indicated major differences in proteins that control barrier function. Consistent with the observed upregulation of Il22 in Il28r−/− mutant mice and proposed effects of IL-22 on mucosal barrier function, we detected differential expression of proteins that were enriched for actin depolymerization and the cytoskeleton, including radixin and moesin, in Il28r−/− mutant mice (Fig. 4B). Fewer differences were seen in the nasal proteome of Il28r−/− mutant mice after influenza virus infection, consistent with the major role of IL-28R in mediation of the local responses to influenza virus infection (Fig. 4C). One protein differentially upregulated in Il28r−/− mutant mice in response to influenza virus infection was AGR2, which interacts with Muc-2 and has an anti-inflammatory role in the gut (35). Another protein with a similar expression profile, PP1A, is a phosphatase that plays a role in HIV-1 transcription (36). Many more differentially expressed proteins were observed in WT mice following influenza virus infection. These findings suggest a change in the proteome reflecting consequences of IL-28 signaling in response to viral influenza virus infection.
FIG 4 .
Upper airway proteomic changes in WT and Il28r−/− mutant mice. Two micrograms of protein from nasal lavage fluid was proteolytically cleaved by trypsin and analyzed by LC-MS/MS. (a) The upper airway proteomes of WT and Il28r−/− mutant mice in the resting state were analyzed by an MS-based approach. Relative protein abundance among biological samples is expressed by spectral counts on a log scale. The color scale bar indicates the range of protein expression levels. Each column represents an individual mouse. Significantly different proteins (P < 0.05) are presented and clustered by protein expression profiles among biological samples. (b) Functional annotation of proteins differentially expressed in WT and Il28r−/− mutant mice. Shown are functional categories enriched and statistically significant within the data set. Data were analyzed with David. (c) WT and Il28r−/− mutant mice were infected with PR8 or treated with PBS as a control, and upper airway nasal lavage fluid was analyzed by an MS-based approach. Each column represents an individual mouse. Representative data are shown.
Type III IFN signaling promotes MRSA nasal colonization and pulmonary infection.
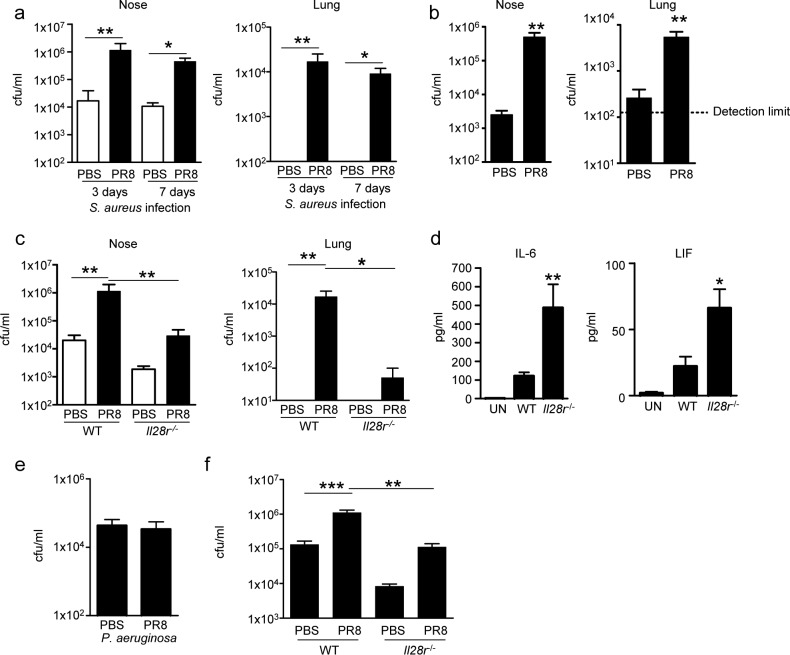
To determine if the density of the human commensal/pathogen MRSA USA300 is similarly affected by IFN-λ signaling, we examined the consequences of influenza virus infection on the nasal carriage of MRSA (Fig. 5A). Following influenza virus infection, mice were intranasally inoculated with MRSA in a small volume such that bacteria were not aspirated into the lungs, animals infected with influenza virus were found to retain significantly more S. aureus in their nasal tissue, i.e., 67-fold (at 3 days) and 41-fold (at 7 days) more than PBS-inoculated controls (Fig. 5A). Influenza virus infection also led to significant recovery of S. aureus from the lower airways compared to PBS-inoculated mice that did not aspirate S. aureus into their lungs (Fig. 5A). Similar results were obtained with mice precolonized with S. aureus prior to influenza virus, which had a 197-fold greater nasal bacterial burden (P < 0.01) and a 21-fold (P < 0.01) greater amount of bacteria recovered from the lungs than control mice (Fig. 5B).
FIG 5 .
Type III IFN signaling contributes to S. aureus colonization of the upper airway and development of bacterial pneumonia. (a) WT C57BL/6J mice were infected intranasally with 50 PFU of influenza virus PR8 or treated with PBS for 7 days prior to the local intranasal application of 108 CFU of S. aureus USA300. Mice were euthanized 3 and 7 days after S. aureus infection. Bacterial counts in mouse nasal and lung homogenates were assessed. n = 6 (3-day data), 4 (PBS, 7 days), or 3 (PR8, 7 days). (b) WT mice were intranasally inoculated locally with 108 CFU of S. aureus USA300 7 days before intranasal infection with 50 PFU of influenza virus PR8 for a further 7 days before euthanasia. Bacterial counts in mouse nasal and lung homogenates were assessed. n = 5. (c) WT and Il28r−/− mutant mice were infected with PR8 or treated with PBS in the lungs for 7 days prior to the local administration of S. aureus (108 CFU). Mice were euthanized 3 days later. Bacterial counts in nasal and lung homogenates are shown. n = 6 (WT and Il28r−/−, PBS) or 4 (Il28r−/−, PR8). (d) Cytokine differences in BALF from mice nasally infected with PR8 and S. aureus (c). n = 3 (uninfected [UN]), 6 (WT), or 4 (Il28r−/−). (e) WT mice were intranasally infected with 107 CFU of P. aeruginosa PAK in the lungs for 24 h after 7 days of PR8 infection or treatment with PBS as a control and 24 h after PAK inoculation. Bacterial counts in the lungs are shown. n = 7. (f) Counts of S. aureus bacteria in the lungs of WT and Il28r−/− mutant mice after 7 days of PR8 infection or treatment with PBS as a control and 24 h after intranasal inoculation with S. aureus USA300 (107 CFU) under anesthesia. n = 13 (WT, PBS), 11 (WT, PR8), or 7 (Il28r−/−). Data are representative of at least two independent experiments. Graphs display mean values and standard errors. ***, P < 0.001; **, P < 0.01; *, P < 0.05.
When we repeated the experiment by comparing WT and Il28r−/− mutant mice, influenza virus-infected Il28r−/− mutant mice had 40-fold fewer (P < 0.01) S. aureus bacteria in their nasal tissue than did infected WT mice (Fig. 5C). Likewise, in their lung tissue, Il28r−/− mutant mice had 99.7% fewer bacteria than similarly infected WT mice (Fig. 5C). Analysis of cytokine production in bronchoalveolar lavage fluid (BALF) from Il28r−/− mutant mice showed significant increases in IL-6 and leukemia inhibitory factor (Fig. 5D) that may have aided in bacterial clearance from the airway.
To determine if the effects of type III IFN signaling on infection are specific for the pathogens typically associated with human infection after influenza virus infection, we repeated the influenza studies by using the opportunistic pathogen Pseudomonas aeruginosa. No increase in subsequent pulmonary infection was observed (Fig. 5E), suggesting that the consequences of IL-28R signaling on the antimicrobial milieu of the upper airway are somewhat pathogen specific (Fig. 5E). To further confirm that IL-28 signaling is a major factor in the pathogenesis of pulmonary MRSA infection after influenza virus infection, PBS-treated or influenza virus-infected mice were inoculated with a standard intranasal inoculum of S. aureus USA300. As expected, there was significantly increased susceptibility of WT mice to MRSA pneumonia, with a bacterial burden in the lungs 8.3-fold greater than that of Il28r−/− mutant mice (Fig. 5F).
DISCUSSION
Influenza virus infection predisposes the host to severe bacterial pneumonia, which is associated with substantial morbidity and mortality (1, 3, 37, 38). Numerous mechanisms of immune impairment after influenza virus infection have been described that contribute to increased severity of infection, as well as increased susceptibility to smaller inocula of either S. pneumoniae (39, 40) or S. aureus (13) after influenza virus infection. Our findings suggest that IFN-λ production induced by influenza virus infection is a major factor contributing to subsequent bacterial pneumonia. Induction of IL-28R/STAT1/SOCS1 signaling alters the antimicrobial milieu of the upper airway, affecting the expression of antimicrobial peptides and cytoskeletal components that regulate the mucosal barrier function. IFN-λ signaling in response to influenza virus infection results in proliferation of the upper airway microbiota, increasing the likelihood that colonizing pathogens such as MRSA will be aspirated and cause pneumonia.
Both type I and III IFNs are induced in response to influenza virus infection, although the type III response, mediated by mucosal IL-28R, has been thought to predominate (41). STAT1 and STAT3 phosphorylation comprises the major signaling cascade activated by influenza virus infection contributing to induction of the expression of over 300 genes that comprise the interferome, responses that are critical for antiviral activity (17–19). We found that the induction of STAT1 phosphorylation by influenza virus was dependent upon IL-28R, consistent with the dominant role of the type III IFNs in this infection. Our results suggest that, in the lung during influenza virus infection, the type III IFN pathway predominantly regulates STAT1 activation. Therefore, while type I IFN is capable of activating this pathway, perhaps type III IFNs are needed in this model to drive production of the protein. In the absence of STAT1 signaling and the associated effects on SOCS1, Il28r−/− mutant mice had significant protection from both endogenous and exogenously acquired organisms, and specifically from MRSA pneumonia, whether acquired before or after influenza virus infection.
A likely mechanism of the resistance of Il28r−/− mutant mice to bacterial superinfection is the ability to suppress the nasal microbiota through increased production of antimicrobial peptides. IL-22 and IL-22-dependent antimicrobial peptides are important in the control of the gut microbiota and mucosal barrier integrity (42). IL-22 functions to ameliorate pulmonary pathology in influenza virus infection and decrease the severity of pneumonia due to S. pneumoniae superinfection (43). Il28r−/− mutant mice had markedly increased constitutive IL-22 expression, which likely contributed to their resistance to superinfection. Increased IL-22 expression may be a compensatory response, as IFN-λ and IL-22 have been shown to have synergistic effects in activating the phosphorylation of STAT1 (44). Our findings are consistent with this observation, as in the absence of IL-28R, IL-22 was not sufficient to activate STAT1. IL-22 itself does not have direct antiviral or antibacterial effects; thus, much of its beneficial effects are thought to be due to maintenance of epithelial barrier function (45). We observed some expected and possible consequences of IL-22 signaling on antimicrobial peptide expression and epithelial barrier function in the upper airway. There was a substantial increase in the expression of RegIIIγ, but not S100A8, both IL-22 dependent (33, 34), in Il28r−/− mutant mice, consistent with their ability to control microbial proliferation in the nasal cavity. Additional antimicrobial peptides and cytokines with antimicrobial activity were also found in the nasal secretions of Il28r−/− mutant mice. Ngal was highly induced in the absence of IFN-λ signaling, detected by both quantitative reverse transcription (qRT)-PCR and proteomic analysis. The suppressive effects of IL-28 signaling on the expression of these and likely additional antimicrobial peptides help to account for the differences in the microbiotas isolated from WT and influenza virus-infected mice, differences not observed in Il28r−/− mutant mice.
The upregulation of several classes of proteins that function in maintaining the dynamic responses and integrity of the mucosal barrier were the major changes found in the upper airways of Il28r−/− mutant mice. In the absence of IL-28R, the nasal mucosal secretions contained significantly increased amounts of the actin cytoskeleton and proteins involved in actin depolymerization. Also significantly increased were the ERM proteins radixin and moesin that mediate the interactions between the plasma membrane, where pathogens are sensed, and the cytoskeleton (46). These findings suggest involvement of IL-28R in the suppression of cytoskeletal dynamics that enable immune cell transmigration, even in the maintenance of the normal microbiota. Whether these changes in cytoskeletal proteins are directly due to IL-28R, IL-22, or some other effectors remains to be established.
The accumulating literature provides a clearer understanding of the pathogenesis of bacterial superinfection after influenza virus infection and suggests several targets to prevent this potentially lethal infection. Our data indicate that induction of IFN-λ signaling in response to influenza virus infection and the activation of both STAT1 and its regulator SOCS1, while critical in regulating the inflammatory response associated with antiviral activities, are major factors in enhancing the proliferation of the nasal microbiome, which often includes potential pathogens. Nasal IL-22 expression is limited under resting conditions and not induced by influenza virus infection. Its protective effects in inducing the expression of antimicrobial peptides and in activating dynamic responses of the mucosal barrier to facilitate signaling and immune cell recruitment could be exploited therapeutically. Strategies already in place, such as universal MRSA decolonization (11), could be more widely implemented in the setting of early influenza virus infection and, combined with the development of local immunomodulators of STAT1 or therapeutic delivery of IL-22, may be useful in preventing bacterial superinfection in influenza.
MATERIALS AND METHODS
Ethics statement.
Animal work in this study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, the Animal Welfare Act, and U.S. federal law. The protocol was approved by the Institutional Animal Care and Use Committee of Columbia University (protocol AAAF4851).
Viral and bacterial growth.
Mouse-adapted influenza virus A/Puerto Rico/8/34 (PR8; H1N1) was grown on Madin-Darby canine kidney cells and stored as previously described (13). S. aureus (LAC USA300) and P. aeruginosa (PAK) were grown to stationary phase overnight at 37°C in Luria-Bertani broth, diluted 1:100 in the morning, and grown to an optical density at 600 nm of 1.0 (USA300) or 0.50 (PAK) for infection (25).
Animal models.
Seven-week-old C57BL/6J WT mice were bred in house as previously described (24). Il28r−/− mutant mice were provided by Bristol Myers Squibb and bred in house, and Ifnar−/− mutant mice have been described previously (25, 47). Both Ifnar−/− and Il28r−/− mutant mice are bred on the C57BL/6J background. Stat1−/− mutant mice were generously provided by Christian Schindler, Columbia University (C57BL/6J background), and from Taconic (129S6/SvEv background). Mice were anesthetized with ketamine and xylazine and intranasally inoculated with influenza virus (50 PFU/mouse) or PBS with 0.1% bovine serum albumin (BSA) as a control. To determine the effect of IFN-λ on the nasal flora, mice were intranasally treated with 1 µg of recombinant mIL-28B (PBL Assay Science; <1 EU/µg) in 50 µl or with PBS-BSA as a control, resulting in delivery to both the upper and lower respiratory tracts. Nasal flora, upper airway RNA, and lung tissue were harvested 3 days following infection. Nasal lavage was performed by inserting a sterile angiocatheter intratracheally, flushing with sterile UV-treated PBS, and collecting lavage fluid from the nose. Selected animals were reinfected 7 days following initial infection with exponentially growing S. aureus USA300 (4 × 107 CFU/mouse). These mice were sacrificed 24 h following bacterial infection, and the numbers of bacteria present in their lungs were determined as previously described (24). Colonization studies were performed by intranasal inoculation of 108 CFU of S. aureus in a small volume (10 µl into the nares without anesthesia) to limit infection to the nasopharynx. Colonization studies were performed for 3 or 7 days postinfection after a 1-week influenza virus infection. S. aureus was applied to the nares of precolonized mice 7 days prior to influenza virus infection. Bacteria were isolated from homogenized nasal septum tissue. Infection of mice while they were under anesthesia was conducted with 1 × 107 CFU of exponentially growing P. aeruginosa PAK in 50 µl.
Culture-dependent analysis of flora.
For each sample, 2 ml of mouse nasal lavage fluid was centrifuged at 1,400 rpm for 5 min. A 1.7-ml volume of the supernatant was removed, and the pellet was resuspended in the remaining 300 µl. Fifty microliters of each sample was plated on CHROMagar Orientation and CHROMagar S. aureus plates and incubated overnight at 37°C. For colony PCR from CHROMagar plates, the 16S rRNA gene sequence was amplified with primers Bact-8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and Bact-1391R (5′-GACGGGCGGTGTGTRCA-3′). At least two colonies per unique color/morphology per plate were picked. The PCR used Taq 5× master mix (New England Biolabs) and 0.5 µM of forward and reverse primers. The cycling conditions were 95°C for 1 min; 30 cycles of 95°C for 30 s, 60°C for 1 min, and 68°C for 1 min; 68°C for 5 min; and holding at 4°C. The amplified 16S rRNA gene sequences were sequenced by GeneWiz, Inc., with the Bact-8F primer. The resulting sequences were identified by 16S rRNA gene sequence (Bacteria and Archaea) nucleotide BLAST (basic local alignment search tool) searching at http://blast.ncbi.nlm.nih.gov/Blast.cgi. The resulting alignments were viewed, and the highest-scoring alignment with an E (error) value of 0.0 was taken as the species identification. For each sample, the identifying information recorded for each 16S rRNA gene PCR template colony was used to identify the assumed species of all colonies of that unique color on both CHROMagar plates. Using the calculated number of CFU per species for each nasal lavage fluid sample, the mean total CFU count, taxonomic richness, Shannon-Weiner diversity index, and species evenness were calculated for each experimental condition, as were the mean CFU count for each species and the species prevalence.
Culture-independent analysis.
Nasal lavage fluid microbial DNA was isolated with the Powersoil DNA Isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA) according to the manufacturer’s instructions with modifications. For each sample, 200 µl of mouse nasal lavage fluid was used. Subsequently, the V4 region of the 16S rRNA gene was PCR amplified in triplicate as described previously (48) with primer pair 515f/806r. PCR amplicons were sequenced on an Illumina MiSeq at the University of Colorado Next Generation Sequencing Facility. The raw, paired-end reads were merged, quality filtered, and clustered into operational taxonomic units (OTUs) at the ≥97% identity level with the UPARSE pipeline (49). A taxonomic identity was assigned to each OTU with the QIIME (50) implementation of the Ribosomal Database Project classifier (51) and the August 2013 release of the GreenGenes database (52). To account for variability in sequence depth, communities were rarefied by randomly selecting 10,000 sequences from each sample. Bray-Curtis dissimilarities in community structure across our sample set were calculated in the vegan package in R v. 3.0.0 after log transformation of relative OTU abundances (53). The resulting DNA sequences and OTU table are available for download at http://dx.doi.org/10.5061/dryad.v1ff0.
Presence/absence analysis.
For the PA dendrogram analysis, a presence/absence matrix was constructed where any number of rarefied reads for a particular OTU was designated present. Each sample was treated as a taxon, and each OTU was treated as a binary character. The dendrogram was then constructed on the basis of a maximum-parsimony algorithm as implemented in the program PAUP (54). All characters and character state transformations were given equal weight. We used Mesquite (55) to define specific OTUs that were exclusively present in or absent from the cluster of samples defined by influenza virus. All but one virus positive-sample clustered on the basis of the parsimony analysis. To further explore characteristic attributes of the virus-positive samples, we performed the same analysis by forcing all virus-positive samples into the same group.
ELISA and immunoblotting.
BALF was analyzed for cytokine and chemokine content by enzyme-linked immunosorbent assay (ELISA) (R&D Biosystems, PBL Assay Science, or eBioscience) and multiplex analysis (Eve Technologies). Lung homogenates were lysed in radioimmunoprecipitation assay buffer (20 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100) with Halt protease and phosphatase inhibitor (Pierce). Protein separation, transfer, and immunoblotting were performed as described previously (26). Anti-phospho-STAT1 (Abcam), -STAT1, -phospho-STAT3, -STAT3, -SOCS1 (Cell Signaling), and -β-actin (Sigma) antibodies were used to measure expression.
qRT-PCR.
RNA was recovered with the PureLink RNA minikit (Life Technologies). Briefly, the upper airways were washed with RNA lysis buffer by inserting a catheter into the trachea and pushing the lysis buffer through the nares. RNA was purified according to the manufacturer’s instructions. cDNA was synthesized with the High Capacity cDNA RT kit (Applied Biosystems). qRT-PCR was performed with Power SYBR green PCR master mix in a StepOne Plus thermal cycler (Applied Biosystems). β-Actin was used to normalize samples. Primers for actin, Ifnb, and Ifnl have been described previously (24). The following primers were used: HA, 5′-ATGCAGACAATATGTATAGGC-3′ (sense) and 5′-GATACTGAGCTCAATTGCTC (antisense); mReg3β, 5′-ACTCCCTGAAGAATATACCCTCC-3′ (sense) and 5′-CGCTATTGAGCACAGATACGAG-3′ (antisense); Lipocalin-2 (Ngal), 5′-TGCAAGTGGCCACCACGGAG-3′ (sense) and 5′-GCATTGGTCGGTGGGGACAGAGA-3′ (antisense); S100a8, 5′-AAATCACCATGCCCTCTACAAG-3′ (sense) and 5′-CCCACTTTTATCACCATCGCAA-3′ (antisense); SOCS1, 5′-CTGCGGCTTCTATTGGGGAC-3′ (sense) and 5′-AAAAGGCAGTCGAAGGTCTCG-3′ (antisense); Il22, 5′-ATGAGTTTTTCCCTTATGGGGAC-3′ (sense) and 5′-GCTGGAAGTTGGACACCTCAA-3′ (antisense).
Shotgun proteomic analysis. (i) Materials.
High-performance liquid chromatography (HPLC) grade LC buffers, dithiothreitol, acetonitrile (ACN), ammonium bicarbonate, trifluoroacetic acid, and iodoacetamide were purchased from Thermo, Fisher Scientific (Waltham, MA). Trypsin Gold, Mass Spectrometry Grade, was purchased from Promega (Madison, WI). Nanopure water was prepared with a Milli-Q water purification system (Millipore, Billerica, MA).
(ii) Sample preparation.
Proteins were precipitated from 500 µl of mouse nasal lavage fluids with methanol-chloroform and resuspended in 20 µl of 4 M urea in 50 mM ammonium bicarbonate. The protein concentration in the mouse lavage fluid was determined by the EZQ Protein Quantification Assay (Life Technology Corp.). Two micrograms of protein from mouse lavage fluid was digested with 150 ng of trypsin (1:40) along with 2 mM CaCl2 and incubated at 37°C for 16 h. Samples were centrifuged for 30 min at 14,000 rpm, and the cleared supernatants were transferred to fresh tubes to be acidified with 90% formic acid (FA) (2% final) to stop proteolysis. The soluble peptide mixtures were collected for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
(iii) LC-MS/MS analysis.
The concentrated peptide mixture was reconstituted in a solution of 2% ACN–2% FA for MS analysis. Peptides were loaded with the autosampler directly onto a 2-cm C18 PepMap precolumn and eluted from the ID PepMap RSLC C18 2-µm column (50 cm by75 µm) with a Thermo Dionex 3000 and a 98-min gradient ranging from 2% buffer B to 30% buffer B (100% acetonitrile, 0.1% FA). The gradient was switched from 30% to 85% buffer B over 5 min and held constant for 1 min. Finally, the gradient was changed from 85% buffer B to 98% buffer A (100% water, 0.1% FA) over 2 min and then held constant at 98% buffer A for 25 min. The application of a 2.0-kV distal voltage electrosprayed the eluting peptides directly into the mass spectrometer equipped with an Easy-Spray Source (Thermo Finnigan, San Jose, CA). The full mass spectra of the peptides were recorded over an m/z range of 400 to 1,500 at a 120,000 resolution, followed by MS/MS collision-induced dissociation events for a total of a 3-s cycle. Charge state-dependent screening was turned off, and peptides with charge states of 2 to 6 were analyzed. Mass spectrometer scanning functions and HPLC gradients were controlled by the Xcalibur data system (Thermo Finnigan, San Jose, CA). Three technical replicates of each sample were run, and MS/MS data from technical replicates were merged for a subsequent database search.
(iv) Database search and interpretation of MS/MS data.
Tandem mass spectra from .raw files were searched against a human protein database with the Proteome discoverer 1.4 (Thermo Finnigan, San Jose, CA). The Proteome Discoverer application extracts relevant MS/MS spectra from the .raw file and determines the precursor charge state and the quality of the fragmentation spectrum. The Proteome Discoverer probability-based scoring system rates the relevance of the best matches found by the SEQUEST algorithm (56). The mouse database was downloaded as FASTA-formatted sequences from the UniProt protein database (released in December 2014) (57). The peptide mass search tolerance was set to 10 ppm. A minimum sequence length of 7 amino acid residues was required. Only fully tryptic peptides were considered. To calculate confidence levels and false-positivity rates (FDR), Proteome Discoverer generates a decoy database containing reverse sequences of the nondecoy protein database and performs the search against this concatenated database (nondecoy plus decoy) (58). The discriminant score was set at a 1% FDR determined on the basis of the number of accepted decoy database peptides to generate protein lists for this study. Spectral counts used to identify each protein were used for expression profiling analysis. Qlucore Omics Explorer (Qlucore AB, Sweden) was used to perform statistical analysis of quantifiable proteins among biological replicates (t test, P < 0.05). Differentially expressed proteins were analyzed with David (59).
Statistics.
Significance of data was determined by a nonparametric Mann-Whitney test. Multiple comparisons were performed by analysis of variance (ANOVA) with a Bonferroni comparison posttest. These tests were conducted with GraphPad Prism software, and significance was defined as P < 0.05.
For culture-independent community analyses, principal-coordinate analysis was used to visualize pairwise similarities in composition. Permutational multivariate ANOVA (PERMANOVA, with 999 permutations) was used to test whether influenza virus-infected mice harbored bacterial communities that were significantly different in composition from those found in controls. The ordination and PERMANOVA analyses were conducted in R v. 3.0.0.
SUPPLEMENTAL MATERIAL
Prevalence and median colony counts by species—WT mice, influenza virus infected versus PBS-treated control.
Interquartile range by species—WT mice, influenza virus infected versus PBS-treated control.
Prevalence and median colony counts by species—WT mice, IFN-λ versus PBS-treated control.
Interquartile range by species—WT mice, IFN-λ versus PBS-treated control.
Prevalence and median colony counts by species—WT 129S6/SvEv mice, IFN-λ or BSA-treated control.
Interquartile range by species—WT 129S6/SvEv mice, IFN-λ or BSA-treated control.
Prevalence and median colony counts by species—STAT1 mice, IFN-λ or BSA-treated control.
Interquartile range by species—STAT1 mice, IFN-λ versus BSA-treated control.
ACKNOWLEDGMENTS
P.J.P., D.P., T.S.C., and A.S.P. designed the experiments. P.J.P., H.S., T.H., N.F., C.R., and D.O. analyzed nasal flora. D.P., T.S.C., and P.J.P performed in vivo experiments and analyzed host signaling pathways. E.I.C. conducted proteomic analysis. P.J.P., D.P., T.S.C., and A.S.P. wrote the manuscript.
Funding Statement
This work was supported by funding from the NIH (K08AI101005) and the John M. Driscoll Jr. MD Childrens Fund Scholarship to PJP, NIH (R56HL125653) to DP, Parker B. Francis Fellowship to TC, NIH (P30CA013696-39S3) to EIC, and NIH R01HL079395 to AP.
Footnotes
Citation Planet PJ, Parker D, Cohen TS, Smith H, Leon JD, Ryan C, Hammer TJ, Fierer N, Chen EI, Prince AS. 2016. Lambda interferon restructures the nasal microbiome and increases susceptibility to Staphylococcus aureus superinfection. mBio 7(1):e01939-15. doi:10.1128/mBio.01939-15.
REFERENCES
- 1.Morens DM, Taubenberger JK, Fauci AS. 2008. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis 198:962–970. doi: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Randolph AG, Vaughn F, Sullivan R, Rubinson L, Thompson BT, Yoon G, Smoot E, Rice TW, Loftis LL, Helfaer M, Doctor A, Paden M, Flori H, Babbitt C, Graciano AL, Gedeit R, Sanders RC, Giuliano JS, Zimmerman J, Uyeki TM, Pediatric Acute Lung Injury and Sepsis Investigator’s Network, National Heart Lung and Blood Institute ARDS Clinical Trials Network . 2011. Critically ill children during the 2009–2010 influenza pandemic in the United States. Pediatrics 128:e1450-1458. doi: 10.1542/peds.2011-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chertow DS, Memoli MJ. 2013. Bacterial coinfection in influenza: a grand rounds review. JAMA 309:275–282. doi: 10.1001/jama.2012.194139. [DOI] [PubMed] [Google Scholar]
- 4.Sun K, Metzger DW. 2008. Inhibition of pulmonary antibacterial defense by interferon-gamma during recovery from influenza infection. Nat Med 14:558–564. doi: 10.1038/nm1765. [DOI] [PubMed] [Google Scholar]
- 5.Shahangian A, Chow EK, Tian X, Kang JR, Ghaffari A, Liu SY, Belperio JA, Cheng G, Deng JC. 2009. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J Clin Invest 119:1910–1920. doi: 10.1172/JCI35412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kudva A, Scheller EV, Robinson KM, Crowe CR, Choi SM, Slight SR, Khader SA, Dubin PJ, Enelow RI, Kolls JK, Alcorn JF. 2011. Influenza A inhibits Th17-mediated host defense against bacterial pneumonia in mice. J Immunol 186:1666–1674. doi: 10.4049/jimmunol.1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wertheim HF, Vos MC, Ott A, van Belkum A, Voss A, Kluytmans JA, van Keulen PH, Vandenbroucke-Grauls CM, Meester MH, Verbrugh HA. 2004. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 364:703–705. doi: 10.1016/S0140-6736(04)16897-9. [DOI] [PubMed] [Google Scholar]
- 8.Stevens AM, Hennessy T, Baggett HC, Bruden D, Parks D, Klejka J. 2010. Methicillin-resistant Staphylococcus aureus carriage and risk factors for skin infections, southwestern Alaska, USA. Emerg Infect Dis 16:797–803. doi: 10.3201/eid1605.091851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corne P, Marchandin H, Jonquet O, Campos J, Bañuls AL. 2005. Molecular evidence that nasal carriage of Staphylococcus aureus plays a role in respiratory tract infections of critically ill patients. J Clin Microbiol 43:3491–3493. doi: 10.1128/JCM.43.7.3491-3493.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tilahun B, Faust AC, McCorstin P, Ortegon A. 2015. Nasal colonization and lower respiratory tract infections with methicillin-resistant Staphylococcus aureus. Am J Crit Care 24:8–12. doi: 10.4037/ajcc2015102. [DOI] [PubMed] [Google Scholar]
- 11.Huang SS, Septimus E, Kleinman K, Moody J, Hickok J, Avery TR, Lankiewicz J, Gombosev A, Terpstra L, Hartford F, Hayden MK, Jernigan JA, Weinstein RA, Fraser VJ, Haffenreffer K, Cui E, Kaganov RE, Lolans K, Perlin JB, Platt R, CDC Prevention Epicenters Program, AHRQ DECIDE Network, Healthcare-Associated Infections Program . 2013. Targeted versus universal decolonization to prevent ICU infection. N Engl J Med 368:2255–2265. doi: 10.1056/NEJMoa1207290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorwitz RJ, Kruszon-Moran D, McAllister SK, McQuillan G, McDougal LK, Fosheim GE, Jensen BJ, Killgore G, Tenover FC, Kuehnert MJ. 2008. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004. J Infect Dis 197:1226–1234. doi: 10.1086/533494. [DOI] [PubMed] [Google Scholar]
- 13.Lee MH, Arrecubieta C, Martin FJ, Prince A, Borczuk AC, Lowy FD. 2010. A postinfluenza model of Staphylococcus aureus pneumonia. J Infect Dis 201:508–515. doi: 10.1086/650204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iverson AR, Boyd KL, McAuley JL, Plano LR, Hart ME, McCullers JA. 2011. Influenza virus primes mice for pneumonia from Staphylococcus aureus. J Infect Dis 203:880–888. doi: 10.1093/infdis/jiq113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCullers JA. 2014. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat Rev Microbiol 12:252–262. doi: 10.1038/nrmicro3231. [DOI] [PubMed] [Google Scholar]
- 16.Mina MJ, McCullers JA, Klugman KP. 2014. Live attenuated influenza vaccine enhances colonization of Streptococcus pneumoniae and Staphylococcus aureus in mice. mBio 5:e01040-13. doi: 10.1128/mBio.01040-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Der SD, Zhou A, Williams BR, Silverman RH. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A 95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta S, Yan H, Wong LH, Ralph S, Krolewski J, Schindler C. 1996. The SH2 domains of Stat1 and Stat2 mediate multiple interactions in the transduction of IFN-alpha signals. EMBO J 15:1075–1084. [PMC free article] [PubMed] [Google Scholar]
- 19.Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, Chapman R, Hertzog PJ. 2013. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res 41:D1040–D1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker D, Prince A. 2011. Type I interferon response to extracellular bacteria in the airway epithelium. Trends Immunol 32:582–588. doi: 10.1016/j.it.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levy DE, Marié IJ, Durbin JE. 2011. Induction and function of type I and III interferon in response to viral infection. Curr Opin Virol 1:476–486. doi: 10.1016/j.coviro.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sommereyns C, Paul S, Staeheli P, Michiels T. 2008. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog 4:e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blazek K, Eames HL, Weiss M, Byrne AJ, Perocheau D, Pease JE, Doyle S, McCann F, Williams RO, Udalova IA. 2015. IFN-λ resolves inflammation via suppression of neutrophil infiltration and IL-1β production. J Exp Med 212:845–853. doi: 10.1084/jem.20140995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen TS, Prince AS. 2013. Bacterial pathogens activate a common inflammatory pathway through IFNλ regulation of PDCD4. PLoS Pathog 9:e1003682. doi: 10.1371/journal.ppat.1003682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker D, Planet PJ, Soong G, Narechania A, Prince A. 2014. Induction of type I interferon signaling determines the relative pathogenicity of Staphylococcus aureus strains. PLoS Pathog 10:e1003951. doi: 10.1371/journal.ppat.1003951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin FJ, Gomez MI, Wetzel DM, Memmi G, O’Seaghdha M, Soong G, Schindler C, Prince A. 2009. Staphylococcus aureus activates type I IFN signaling in mice and humans through the Xr repeated sequences of protein A. J Clin Invest 119:1931–1939. doi: 10.1172/JCI35879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, Ostrander C, Dong D, Shin J, Presnell S, Fox B, Haldeman B, Cooper E, Taft D, Gilbert T, Grant FJ, Tackett M, Krivan W, McKnight G, Clegg C, Foster D, Klucher KM. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol 4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 28.Leitner NR, Lassnig C, Rom R, Heider S, Bago-Horvath Z, Eferl R, Müller S, Kolbe T, Kenner L, Rülicke T, Strobl B, Müller M. 2014. Inducible, dose-adjustable and time-restricted reconstitution of STAT1 deficiency in vivo. PLoS One 9:e86608. doi: 10.1371/journal.pone.0086608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Odendall C, Dixit E, Stavru F, Bierne H, Franz KM, Durbin AF, Boulant S, Gehrke L, Cossart P, Kagan JC. 2014. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat Immunol 15:717–726. doi: 10.1038/ni.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prêle CM, Woodward EA, Bisley J, Keith-Magee A, Nicholson SE, Hart PH. 2008. SOCS1 regulates the IFN but not NFkappaB pathway in TLR-stimulated human monocytes and macrophages. J Immunol 181:8018–8026. doi: 10.4049/jimmunol.181.11.8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piganis RA, De Weerd NA, Gould JA, Schindler CW, Mansell A, Nicholson SE, Hertzog PJ. 2011. Suppressor of cytokine signaling (SOCS) 1 inhibits type I interferon (IFN) signaling via the interferon alpha receptor (IFNAR1)-associated tyrosine kinase Tyk2. J Biol Chem 286:33811–33818. doi: 10.1074/jbc.M111.270207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tokumaru S, Sayama K, Shirakata Y, Komatsuzawa H, Ouhara K, Hanakawa Y, Yahata Y, Dai X, Tohyama M, Nagai H, Yang L, Higashiyama S, Yoshimura A, Sugai M, Hashimoto K. 2005. Induction of keratinocyte migration via transactivation of the epidermal growth factor receptor by the antimicrobial peptide LL-37. J Immunol 175:4662–4668. doi: 10.4049/jimmunol.175.7.4662. [DOI] [PubMed] [Google Scholar]
- 33.Kinnebrew MA, Ubeda C, Zenewicz LA, Smith N, Flavell RA, Pamer EG. 2010. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J Infect Dis 201:534–543. doi: 10.1086/650203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zindl CL, Lai JF, Lee YK, Maynard CL, Harbour SN, Ouyang W, Chaplin DD, Weaver CT. 2013. IL-22-producing neutrophils contribute to antimicrobial defense and restitution of colonic epithelial integrity during colitis. Proc Natl Acad Sci U S A 110:12768–12773. doi: 10.1073/pnas.1300318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park SW, Zhen G, Verhaeghe C, Nakagami Y, Nguyenvu LT, Barczak AJ, Killeen N, Erle DJ. 2009. The protein disulfide isomerase AGR2 is essential for production of intestinal mucus. Proc Natl Acad Sci U S A 106:6950–6955. doi: 10.1073/pnas.0808722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ammosova T, Jerebtsova M, Beullens M, Voloshin Y, Ray PE, Kumar A, Bollen M, Nekhai S. 2003. Nuclear protein phosphatase-1 regulates HIV-1 transcription. J Biol Chem 278:32189–32194. doi: 10.1074/jbc.M300521200. [DOI] [PubMed] [Google Scholar]
- 37.Hall MW, Geyer SM, Guo CY, Panoskaltsis-Mortari A, Jouvet P, Ferdinands J, Shay DK, Nateri J, Greathouse K, Sullivan R, Tran T, Keisling S, Randolph AG, Pediatric Acute Lung Injury and Sepsis Investigators (PALISI) Network, PICFlu Study Investigators . 2013. Innate immune function and mortality in critically ill children with influenza: a multicenter study. Crit Care Med 41:224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ziakas PD, Anagnostou T, Mylonakis E. 2014. The prevalence and significance of methicillin-resistant Staphylococcus aureus colonization at admission in the general ICU setting: a meta-analysis of published studies. Crit Care Med 42:433–444. doi: 10.1097/CCM.0b013e3182a66bb8. [DOI] [PubMed] [Google Scholar]
- 39.Siegel SJ, Roche AM, Weiser JN. 2014. Influenza promotes pneumococcal growth during coinfection by providing host sialylated substrates as a nutrient source. Cell Host Microbe 16:55–67. doi: 10.1016/j.chom.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwasaki A, Pillai PS. 2014. Innate immunity to influenza virus infection. Nat Rev Immunol 14:315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jewell NA, Cline T, Mertz SE, Smirnov SV, Flaño E, Schindler C, Grieves JL, Durbin RK, Kotenko SV, Durbin JE. 2010. Lambda interferon is the predominant interferon induced by influenza A virus infection in vivo. J Virol 84:11515–11522. doi: 10.1128/JVI.01703-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shih VF, Cox J, Kljavin NM, Dengler HS, Reichelt M, Kumar P, Rangell L, Kolls JK, Diehl L, Ouyang W, Ghilardi N. 2014. Homeostatic IL-23 receptor signaling limits Th17 response through IL-22-mediated containment of commensal microbiota. Proc Natl Acad Sci U S A 111:13942–13947. doi: 10.1073/pnas.1323852111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ivanov S, Renneson J, Fontaine J, Barthelemy A, Paget C, Fernandez EM, Blanc F, De Trez C, Van Maele L, Dumoutier L, Huerre MR, Eberl G, Si-Tahar M, Gosset P, Renauld JC, Sirard JC, Faveeuw C, Trottein F. 2013. Interleukin-22 reduces lung inflammation during influenza A virus infection and protects against secondary bacterial infection. J Virol 87:6911–6924. doi: 10.1128/JVI.02943-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hernández PP, Mahlakõiv T, Yang I, Schwierzeck V, Nguyen N, Guendel F, Gronke K, Ryffel B, Hölscher C, Dumoutier L, Renauld JC, Suerbaum S, Staeheli P, Diefenbach A. 2015. Interferon-lambda and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat Immunol 16:698–707. doi: 10.1038/ni.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sonnenberg GF, Fouser LA, Artis D. 2010. Functional biology of the IL-22-IL-22R pathway in regulating immunity and inflammation at barrier surfaces. Adv Immunol 107:1–29. doi: 10.1016/B978-0-12-381300-8.00001-0. [DOI] [PubMed] [Google Scholar]
- 46.Arpin M, Chirivino D, Naba A, Zwaenepoel I. 2011. Emerging role for ERM proteins in cell adhesion and migration. Cell Adh Migr 5:199–206. doi: 10.4161/cam.5.2.15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parker D, Prince A. 2012. Staphylococcus aureus induces type I IFN signaling in dendritic cells via TLR9. J Immunol 189:4040–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 50.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2013. Vegan: community ecology package. R package version 2.0-7. R Foundation for Statistical Computing, Vienna, Austria: http://CRAN.R-project.org/package=vegan. [Google Scholar]
- 54.Wilgenbusch JC, Swofford D. 2003. Inferring evolutionary trees with PAUP*. Curr Protoc Bioinformatics Chapter 6:Unit 6.4. doi: 10.1002/0471250953.bi0604s00. [DOI] [PubMed] [Google Scholar]
- 55.Maddison WP, Maddison DR. 2015. Mesquite: a modular system for evolutionary analysis, version 2.7. Tangient LLC, San; Francisco, CA: http://mesquiteproject.org. [Google Scholar]
- 56.Yates JR III, Eng JK, McCormack AL, Schieltz D. 1995. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem 67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 57.UniProt Consortium 2014. Activities at the universal protein resource (UniProt). Nucleic Acids Res 42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elias JE, Gygi SP. 2007. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 59.Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using David bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Prevalence and median colony counts by species—WT mice, influenza virus infected versus PBS-treated control.
Interquartile range by species—WT mice, influenza virus infected versus PBS-treated control.
Prevalence and median colony counts by species—WT mice, IFN-λ versus PBS-treated control.
Interquartile range by species—WT mice, IFN-λ versus PBS-treated control.
Prevalence and median colony counts by species—WT 129S6/SvEv mice, IFN-λ or BSA-treated control.
Interquartile range by species—WT 129S6/SvEv mice, IFN-λ or BSA-treated control.
Prevalence and median colony counts by species—STAT1 mice, IFN-λ or BSA-treated control.
Interquartile range by species—STAT1 mice, IFN-λ versus BSA-treated control.