ABSTRACT
Deep metagenomic shotgun sequencing has emerged as a powerful tool to interrogate composition and function of complex microbial communities. Computational approaches to assemble genome fragments have been demonstrated to be an effective tool for de novo reconstruction of genomes from these communities. However, the resultant “genomes” are typically fragmented and incomplete due to the limited ability of short-read sequence data to assemble complex or low-coverage regions. Here, we use single-molecule, real-time (SMRT) sequencing to reconstruct a high-quality, closed genome of a previously uncharacterized Corynebacterium simulans and its companion bacteriophage from a skin metagenomic sample. Considerable improvement in assembly quality occurs in hybrid approaches incorporating short-read data, with even relatively small amounts of long-read data being sufficient to improve metagenome reconstruction. Using short-read data to evaluate strain variation of this C. simulans in its skin community at single-nucleotide resolution, we observed a dominant C. simulans strain with moderate allelic heterozygosity throughout the population. We demonstrate the utility of SMRT sequencing and hybrid approaches in metagenome quantitation, reconstruction, and annotation.
IMPORTANCE
The species comprising a microbial community are often difficult to deconvolute due to technical limitations inherent to most short-read sequencing technologies. Here, we leverage new advances in sequencing technology, single-molecule sequencing, to significantly improve reconstruction of a complex human skin microbial community. With this long-read technology, we were able to reconstruct and annotate a closed, high-quality genome of a previously uncharacterized skin species. We demonstrate that hybrid approaches with short-read technology are sufficiently powerful to reconstruct even single-nucleotide polymorphism level variation of species in this a community.
INTRODUCTION
Microbial communities offer the potential for the discovery of a tremendous suite of previously unknown biological functions, for example, new bioactive compounds, antimicrobials, virulence factors, or metabolic pathways. Such discovery has relied on the ability to survey and deconvolute species from mixed microbial consortia. In particular, major value is derived from the ability to reconstruct genomes from metagenomic data. Metagenomics circumvents the requirement for cultivation-based discovery and its associated costs.
Short-read metagenomic shotgun sequencing, with typical median lengths of 100 to 150 bp for Illumina HiSeq technology, has been effectively applied to interrogate composition and function of microbial communities. However, annotation of metagenomic data sets often relies on sequenced reference genomes, which incompletely represent microbial diversity. Consequently, a significant fraction of metagenomic sequence data remains uncharacterized, ranging from 2 to 96% in the human skin, depending on sample origin (1). Hence, de novo, or reference-free approaches to analyze microbial communities are increasingly necessary. These in silico reconstructions are based primarily on de novo assembly of short reads into contigs, followed by abundance or compositional binning of contigs into species units (2–4). Such methods are highly dependent on constraints of sequencing depth, coverage, and community complexity, and correspondingly, often result in highly fragmented and incomplete reconstructions. Moreover, as species may also exist as a heterogeneous mix of subspecies, the presence of multiple similar genomes or repetitive elements can also result in poor-quality assemblies.
Single-molecule, real-time (SMRT) sequencing has proven highly effective in single-genome applications, where multikilobase sequences allow full coverage and closure of genome assemblies (5, 6). However, because this technology yields fewer reads than short-read technologies and is subject to uniformly distributed errors, primarily point insertions and deletions at a rate of ~15% (7), its utility in characterizing metagenomic populations is unknown. For example, the most common quantitation metrics, e.g., RPKM (reads per kilobase per million), rely on significant read numbers. However, the promise of long-read technology lies in its potential to reconstruct complex loci, similar genomes, and low-abundance species.
Here, we sought to use SMRT sequencing to address challenges in metagenomic characterization and assembly. Our goals were to evaluate the following: (i) the ability of SMRT sequence data to quantitatively characterize complex metagenomic communities, (ii) the value added and accuracy in using these long reads in metagenomic assemblies either alone or as a hybrid with matched short-read data, and (iii) the ability to deconvolute closely related strains in a population using a combination of short- and long-read data. Generating matched short- and long-read shotgun metagenomic data from skin samples, we report that even low-depth SMRT sequence data can accurately reconstruct taxonomic abundance in a complex community, and high-depth SMRT data can reconstruct a closed-genome sequence with high accuracy. Short-read Illumina data can then be leveraged for high-precision analyses, such as evaluating population-level heterozygosity of a strain. Our results demonstrate the first example of the utility of SMRT technology in shotgun characterizations of metagenomic communities, together with advances enabled by hybrid approaches incorporating both long- and short-read technologies.
RESULTS
Sample collection and sequencing.
To generate adequate biomass for sequencing, six skin samples were collected using a swab-scrape-swab procedure (1) and combined from the antecubital fossa, volar forearm, and hypothenar palm (representing an “arm” sample) and then the plantar heel, toe web space, and toenail (representing a “foot” sample) of a healthy individual. These skin sites were chosen because of the high microbial diversity reported for these sites in previous studies, which poses increased challenges for metagenomic annotation and assembly. We generated metagenomic shotgun sequence data using two platforms: linear PacBio RSII TdT (terminal deoxynucleotidyl transferase) sequencing and Illumina HiSeq. Host human-derived DNA was removed by mapping to the CHM1 human genome reference (8), which resulted in, on average, removal of an additional 5% of reads over previous hg19 references, likely due to the improved inclusion of low-complexity regions. Total sequence yield varied between sites, with the low-biomass arm site yielding relatively little nonhuman sequence data (27 and 805 Mbp for SMRT and HiSeq) compared to the foot (623 Mbp and 3.0 Gbp). The mean read lengths were 1,689 bp (range, 50 to 14,690 bp) for SMRT sequencing and 98 bp (50 to 101 bp) for Illumina. Finally, matched 16S rRNA (22,138 reads) and ITS1 (29,427 reads) amplicons were also generated to validate taxonomic assignments. All read statistics are reported in Table S1 in the supplemental material.
Read-based compositional analysis of skin communities.
Because SMRT sequencing typically yields orders of magnitude fewer reads than short-read methods (here, 511× and 86× fewer reads for arm and foot samples), we investigated SMRT sequencing’s accuracy in quantitating species abundance of mixed communities compared to HiSeq data. First, we estimated community coverage by tracking k-mer accumulation as a function of the number of reads sequenced. If the sequence (and as a proxy, species) diversity is adequately represented in a community, we would expect decreasing k-mer accumulation as the number of reads increases. Arm samples had low k-mer coverage regardless of the sequencing method (Fig. 1A), which is likely the result of insufficient sequencing depth for the community diversity represented. HiSeq sequencing of the foot site showed adequate community k-mer coverage, with accumulation leveling with high numbers of reads. However, we saw little k-mer redundancy in the SMRT read data. This likely results from both high community complexity and the uniform nature of error rates present in SMRT read data. Indeed, HiSeq-corrected SMRT data show a marked improvement in k-mer coverage for the foot (Fig. 1A), although with a modest cost in k-mer accumulation depth.
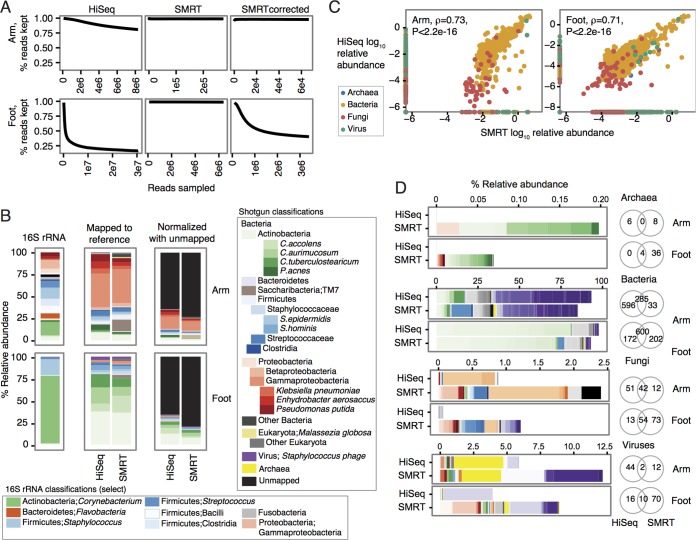
FIG 1 .
SMRT reads accurately reconstruct species abundances in metagenomic communities and recover rare species. (A) Estimation of sequencing coverage of the community. The number of reads subsampled for k-mer counting is shown as Reads sampled on the x axis. Reads are split into 20-mers, compared to a k-mer coverage table, and kept only if the median k-mer coverage is below 20× (percent reads kept shown on the y axis). If k-mer coverage is sufficiently deep for the community, one observes a decrease and leveling off in percent reads kept as the number of reads sampled increases. Whether the reads were generated with HiSeq, SMRT sequencing, or SMRT reads error corrected using HiSeq reads is indicated for each panel. (B) Relative abundance plots of the most abundant taxa per kingdom. “16S rRNA” classifications are to the genus level. “Mapped to reference” indicates relative abundances mapping to a multikingdom reference database containing Archaea, Bacteria, fungi, and viruses. “Normalized with unmapped” contextualizes the relative abundance of species generated by reference-based mapping to the fraction of reads from the sample that does not map to any reference. (C) Concordance of HiSeq and SMRT species classifications with Spearman correlation (ρ) calculated with the corresponding P value. (D) Differential detection of species with the two sequencing methods shown by kingdom. Venn diagrams show the shared number of species detected for the arm and foot samples. The colors indicate different taxonomic units for the Archaea, Bacteria, fungi, and viruses as follows. For Archaea, red colors indicate Crenarchaeota and green colors indicate Euryarchaeota. For Bacteria, red colors indicate Acidobacteria, Spirochaetes, Tenericutes, Thermotogae, and Verrucomicrobia; greens indicate Actinobacteria; blues indicate Bacteroidetes, Chlamydiae, Chloroflexi, and Cyanobacteria; oranges indicate Deinococcus-Thermus; grays indicate Firmicutes; yellows indicate Fusobacteria and Plantomycetes; purples indicate Proteobacteria. For fungi, reds, yellows, and purples indicate miscellaneous; greens indicate Apicomplexa; blues indicate Ascomycota; oranges indicate Basidiomycota; grays indicate Chlorophyta. For viruses, red and blue colors indicate miscellaneous and Fuselloviridae; greens indicate Herpesviridae; oranges indicate Myoviridae; grays indicate Papillomaviridae, Phycodnaviridae, Podoviridae, Polydnaviridae, and Polyomaviridae; yellows indicate Poxviridae; purples indicate Siphoviridae.
While limited sequencing depth may affect detection of low-abundance species in a community, read-based mapping to reference genomes can accurately reconstruct species abundances even at low coverages. We mapped the human-read-filtered shotgun reads to a multikingdom curated reference genome database containing bacterial, fungal, viral, and archaeal complete and draft genomes (1). Overall community composition was very similar at the species level by the two sequencing methods (Fig. 1B and C; see Table S2 in the supplemental material), showing a high diversity of Proteobacteria, Firmicutes, and to a lesser extent, Actinobacteria in the arm and predominantly Actinobacteria in the foot. Both approaches showed good concordance with genus-level designations generated by 16S rRNA sequencing for the foot (Fig. 1B). This suggests that even relatively few sequence reads can be effective at quantitating species abundances.
To investigate the discrepancy between the amplicon and metagenomic classifications in the arm sample, we examined the percentage of unclassifiable shotgun reads (Fig. 1B). More than 60% of reads remained unmapped, showing that reference-based analyses can underestimate the biodiversity of a sample. On the other hand, 16S rRNA sequences, while offering lower phylogenetic resolution than shotgun data, can place unclassifiable sequences on a larger phylogenetic tree to infer composition. As the major genera were present in both amplicon and shotgun-based methods, we believe that the significant amount of uncharacterizable reads is the primary factor behind compositional differences between methods.
Interestingly, we observed that a number of species were detected by only one of the two methods (Fig. 1D). Our initial assumption was that short-read sequencing possesses greater sensitivity in low-abundance detection because of greater achievable depth and coverage. We found that a large number of low-abundance species were detected with only long-read data (Fig. 1D; see Table S2 in the supplemental material), represented by additional archaeal, fungal, and viral species as well as bacterial phyla rarely isolated from skin, e.g., TM7. This recovery is consistent with recent reports supporting the increased ability of long reads to recover rare genomes over short reads (9). However, this difference was not explained by an increased ability to cover high-GC% genomes, as there was no significant difference in the GC content of the fungal, viral, or archaeal genomes that were recovered by only one sequencing method (P > 0.20 by Wilcoxon rank sum test). A more likely explanation for this increased detection of rare species could be amplification cycles during Nextera library construction, which could bias community composition toward more abundant species. Another possibility for the nonrecovery of archaeal genomes with short reads is an increased ability for long reads to discriminate high-homology regions that otherwise may be classified as bacterial. Finally, differences in sampling depth can cause discrepancies in organisms recovered, particularly those that are low abundance. While both short- and long-read methods similarly reconstruct abundant taxa in a community, additional microbial biodiversity can be uncovered in the skin with different technological approaches.
Metagenomic SMRT sequencing reconstructs a complete genome sequence of Corynebacterium simulans.
Because more than 80% of reads could not be assigned to a reference genome (Fig. 1B), we also pursued reference-independent approaches to reconstruct community composition. Our initial goal was first to define the ability of SMRT sequencing to reconstruct metagenomes in comparison to short-read data and second to investigate whether hybrid short- and long-read assemblies were improved with respect to length, quality, and coverage.
SMRT sequence reads were assembled using Pacific Biosciences’ Hierarchical Genome Assembly Process (HGAP), which assembles genomes using read overlap layouts, followed by a consensus algorithm to generate a final, high-quality genome sequence (10). Initial HGAP assembly of the foot metagenome resulted in 68 long contigs (see Table S3 in the supplemental material). However, the arm sample had insufficient coverage for the community’s complexity and did not produce any assemblies, a drawback of the HGAP approach, which requires significant read overlaps for preassembly and sequence coverage for consensus calling.
BLAST analysis of the four longest contigs in the foot assembly showed high similarity to a Corynebacterium aurimucosum genome (Fig. 2A). These contigs were connected manually into a single chromosome using overlapping contig ends and were polished using the Quiver consensus algorithm (10). The fidelity of the resulting assembly of 2.78 Mbp in length was validated by remapping SMRT reads to the assembled chromosome (Fig. 2A). We observed no obvious coverage breaks (mean coverage across the four contigs, 105.3 ± 13.8) and a mild coverage undulation with a peak at the origin of replication, which is likely due to genomic DNA being extracted from actively growing bacteria. We observed greater than 99.999% consensus concordance (quality value [QV] >50) between the sequencing data and the assembled chromosome, indicating high accuracy in reconstruction.
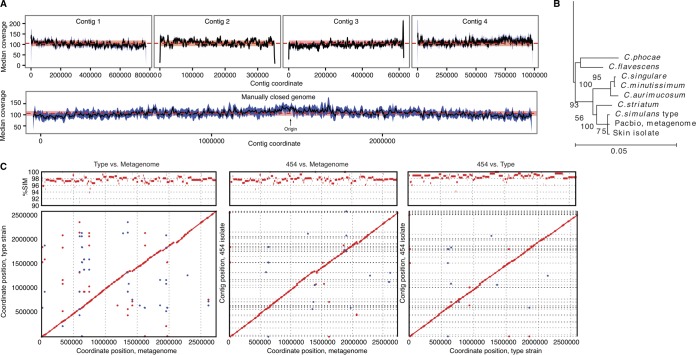
FIG 2 .
Reconstruction of a closed C. simulans metagenome from HGAP assemblies. (A) (Top) Mean coverage from remapping SMRT reads to the four longest contigs that share a lowest common ancestor of C. aurimucosum. (Bottom) These four contigs were manually linked to form a single chromosome using contig overlap information. The predicted origin of replication is indicated. (B) Taxonomic assignment of the reconstructed genome. 16S rRNA gene sequences were predicted from the chromosome and placed on a phylogenetic tree of full-length Corynebacterium rRNA gene sequences. A portion of the tree is shown with bootstrap values (1,000 iterations), showing placement of the reconstructed genome (“Pacbio, metagenome”) with C. simulans (“type”). For comparisons, we also included a previously sequenced 454 skin isolate typed as C. simulans (“Skin isolate”). (C) Synteny plots compare similarity of the de novo C. simulans metagenome, the sequenced C. simulans type strain, and the 454-sequenced skin isolate, generated by NUCmer. The top panels show the percent similarity across the genomes, ordered by nucleotide position. In the dot plots (bottom panels), aligned segments up to 3 kb in length are represented as dots or lines and the orientation of contigs is shown (forward [red] and reverse [blue]). Scale bar represents an estimate of relative times of divergence between nodes.
Phylogenetic placement (Fig. 2B, 1,000 bootstrap iterations, 100% confidence) of the 16S rRNA extracted from the de novo genome placed it not with C. aurimucosum but with a type strain of Corynebacterium simulans, a facultative, nonlipophilic species isolated from a human axillar lymph node (11). For genome comparisons and to assess the quality of the de novo C. simulans genome (termed “metagenome” in the comparison below), we also SMRT sequenced and assembled this C. simulans type strain with HGAP. For comparison, we also compared both genomes to a draft genome assembly of another C. simulans skin isolate previously sequenced by 454 technology (12). NUCmer alignment showed high similarity between these genomes (Fig. 2C) with average 96.8%, 97.1%, and 96.2% identity over aligned regions for type versus metagenome, type versus 454 isolate, and metagenome versus 454 isolate, respectively. While the three strains were predominantly collinear, we observed a modest number of inversions and insertions. The total sizes for the metagenome, type strain, and 454 isolate were 2.74 Mb, 2.60 Mb, and 2.65 Mbp (estimated), respectively, with 59% GC content for all three strains.
Comparison of long-read sequence assemblies with short and hybrid approaches.
We next evaluated short-read assemblies, in particular those using HiSeq sequencing, where coverage depth markedly exceeds SMRT sequencing capacity. We also examined whether hybrid assemblies were an effective alternative to improve assembly statistics, an important consideration given the relatively higher cost of SMRT sequencing. We created hybrid assemblies using different numbers of long reads to assess the relationship between SMRT sequencing depth and assembly improvement.
Short reads were assembled independently using different k-mer ranges and several different assemblers, including Velvet, Megahit, and SPAdes. Although SPAdes was not originally designed for metagenomic assembly, its short-read assembly yielded the highest combination of metrics of assembly quality for both the arm assemblies and the foot assemblies, including cumulative contig length (Fig. 3A and B; see Table S3 in the supplemental material), contig size (Fig. 3C and D), number of bases incorporated into the assembly, and concordance of reads mapping back to contigs (Table S2). Long reads were assembled by using HGAP, and hybrid assemblies were assembled by using SPAdes. Because HGAP assembly did not produce contigs for the arm, we used the unassembled SMRT read data as a proxy for contigs.
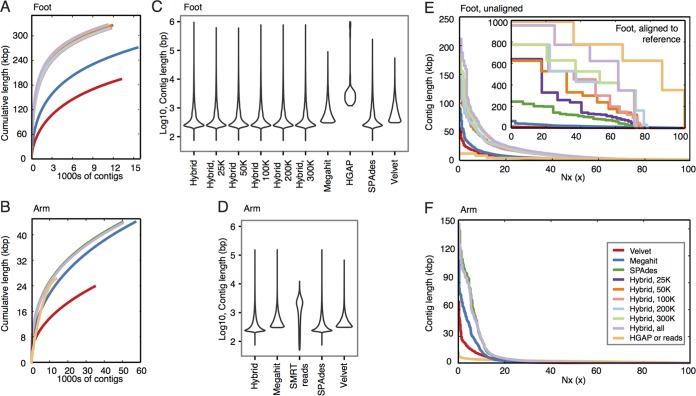
FIG 3 .
Metagenomic assembly comparisons between long-read, short-read, and hybrid approaches. (A and B) Line plots show the cumulative length of contigs generated by each of the assembly methods for the foot and arm samples, respectively. The assembly methods are indicated by the colors shown in the legend in panel F. (C and D) Violin plots are boxplots whose shapes show the density distribution of contig lengths (log10) for each of the assembly methods. (E and F) Modified Nx plots show the length for which the contigs of that length or longer covers x percentage of the assembly. For the foot, plots are separated by what aligned to the C. simulans metagenome as a reference (inset) or for contigs that did not align (unaligned). For the arm, all contigs are shown.
In foot samples, short reads, hybrid assemblies, and HGAP assemblies performed equally well in cumulative contig length, a proxy for reconstruction of genetic diversity of the community (Fig. 3A). The advantage of long reads lay predominantly in contig scaffolding, with all hybrid assemblies producing markedly longer contigs than short-read-only assemblies (Fig. 3C). Nx plots, which show the length for which the contigs of that length or longer covers x percentage of the assembly, showed very significant assembly improvements with the incorporation of long reads (Fig. 3E, inset; see Table S2 in the supplemental material). Surprisingly, even 25,000 reads (about half an SMRT cell yield) were sufficient to significantly improve contig scaffolding, although we expect this estimated depth to be dependent on community complexity. Finally, a limitation of HGAP assembly lies in the generation of smaller or less-abundant contigs, which are less informative for gene calling but can better reconstruct low-abundance organisms. Short-read or hybrid methods generated markedly shorter contigs than HGAP did (Fig. 3E, unaligned contigs), again reflecting its requirement for significant overlap for preassembly.
HGAP assembly of arm reads was ineffective. As the arm is a low-biomass skin site, sequencing depth for both SMRT and HiSeq methods was lower than for the foot. SMRT reads reconstructed a much lower fraction of the community than other assembly methods (Fig. 3B), likely due to the relatively fewer reads generated. Low depth compounded with the high community complexity typical of arm skin sites resulted in assemblies comprised primarily of a large number of small contigs (Fig. 3D). While the median length of SMRT reads exceeds that of short-read-based contigs, hybrids showed no improvement over short-read-only assemblies (Fig. 3F).
Finally, to assess the quality of short-read reconstructions of C. simulans, we extracted contigs with sequence similarity to the C. simulans metagenome and reassembled those reads. Because de novo assembly often results in chimeric assemblies, we also investigated the rate of misassemblies compared to the closed genome (see Materials and Methods for a description of criteria). We observed few misassemblies and high genome coverage, regardless of the assembler used, although the number of contigs differed significantly depending on the assembler used (Fig. 4A). This suggests a convergent but fragmented reconstruction using short reads and further supports the value of incorporating long-read data into shotgun metagenomic analyses.
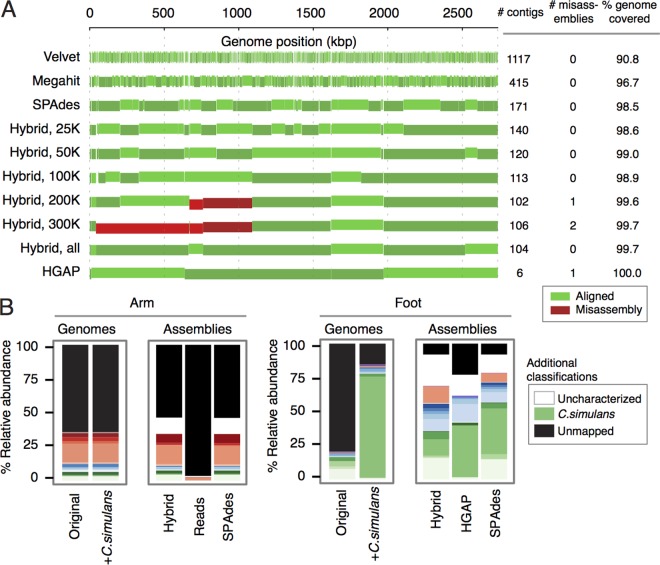
FIG 4 .
Reannotation of skin communities with metagenomes. (A) Reconstruction fidelity of the C. simulans genome for each of the assembly methods. The number of contigs, number of misassemblies (by Plantagora’s definition), and percentage of the genome covered are shown. A correct alignment of the contig is indicated in green, with each vertical bar representing a contig (the contigs or bars are staggered to distinguish contigs). A misassembly in the contig is indicated in red. (B) Improvement in the fraction of sequences that can be assigned to a taxonomical unit using reference-free methods. The community composition using the original reference genome database (Original) and with the addition of C. simulans genome to the database (+C.simulans) are indicated. Hybrid, SPAdes short-read, and HGAP assemblies were annotated using the NCBI nr database. Colors are as shown in the keys in Fig. 1B with additional colors indicated.
Reannotation of skin communities with metagenomes.
Metagenomic studies often focus on identification of taxa within samples but neglect to discuss results in the context of unmapped reads. For most microbial communities, the amount of this “dark matter,” or uncharacterized sequence space is significant (e.g., Fig. 4B). Analyses excluding this space likely underrepresent the biodiversity of the sample. To assess the extent to which metagenomic annotations improve with the addition of new genomes or de novo assemblies, we mapped HiSeq reads to databases incorporating these new references.
Adding the new C. simulans genome to the reference genome database very significantly increased the number of sequence reads accounted for in the foot (Fig. 4B) from an alignment rate of 20.3% to 84.9%. For assembly-based mapping, reads were mapped to contigs of >300 bp from either hybrid, HGAP, or SPAdes short-read-only assemblies that had been assigned a species by homology to the NCBI nonredundant (nr) database. While providing more-fragmented assemblies, SPAdes and hybrid assemblies accounted for the greatest improvement, each to 92.0%, and 77.2% for HGAP-only assemblies (Fig. 4B). However, we observed some differences in abundances in the foot, depending on whether reference genomes or contigs were used for taxonomic assignments. This likely arises from (i) lack of annotation in nr, (ii) homology between conserved regions (as our genome-mapping pipeline reassigns reads mapping to multiple loci to a “most likely” genome), or (iii) reads not represented in one or the other sequencing method (e.g., rare species in Fig. 1).
Finally, despite this significant increase in mapped space, a large fraction, up to 23.0% of contigs, remained uncharacterizable by BLAST search. This further underscores the value of reconstructing complete genomes from metagenomic analysis. Linking these uncharacterized genes to a larger genome context will allow more-accurate taxonomic assignment and enable use of different prediction models to reconstruct function. More generally, improved methods for functional annotation remain in great need.
Conversely, relatively little improvement was seen in arm assemblies, where relative abundances of Corynebacterium are typically low (Fig. 4B). As expected, proxying arm SMRT reads for contigs resulted in low-read mapping efficacy (3.7% alignment rate) due to SMRT read error rates. An approximately 46% read alignment was observed for either hybrid or SPAdes short-read assemblies, with annotated reads mapping primarily to Proteobacteria, consistent with genome-based annotations. We hypothesize that in the arm samples, high community complexity and inadequate sequencing depth greatly hinder reconstruction of low-abundance genomes and that the biodiversity of arm skin is poorly understood.
Population-level heterozygosity of C. simulans.
Recent studies have shown that many species contain a significant pan-genome, defined as the aggregate coding potential of substrains within that clade. Strain variation is an important consideration, as flexible regions of the genome can encode different properties of transmissibility, virulence, antibiotic resistance, or other functions. Previously, we showed that strain-level populations of common skin commensals are heterogeneous and comprised of multiple subspecies clades (1), and so we explored the ability of hybrid data to identify strain-level variation in the C. simulans populations of the foot.
We explored the sequence space around the C. simulans “metagenome” by mapping the 15.7 million HiSeq read pairs to the reconstructed reference, of which 76% mapped. Initial analyses showed that most variants were present at allele frequencies that were too low for standard variant callers to reliably detect, suggesting a dominant strain in the population. To identify low-frequency alleles, we used the LoFreq variant caller, which is able to confidently call variants at allele frequencies near the error rate for the sequencing platform (13). A total of 411 variants were called with a median allele frequency of 0.05 and median read depth of 445× (Fig. 5; see Table S4 in the supplemental material). Variants occurred primarily in protein-coding regions with 386 variants found in 55 protein-coding genes, most of which were part of highly conserved protein complexes (Table S4). For example, 16 of the 55 genes encode ribosomal components. However, some of these variants may be derived from non-Corynebacterium genomes, as it is difficult to separate intergenus variants from intragenus variants for these highly conserved genes.
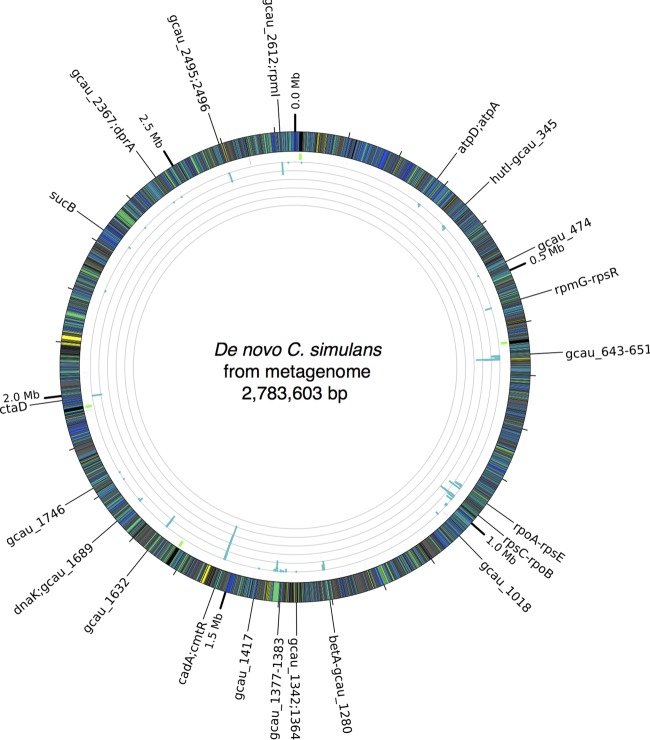
FIG 5 .
Population-wide heterozygosity of C. simulans strains in the metagenome. Low-frequency variant calls mapped to the C. simulans de novo metagenome. The outer ring is colored by TIGR (the Institute for Genomic Research; now the J. Craig Venter Institute) roles for the protein-coding genes in shades of green and blue. Mobile elements (e.g., transposases, integrases) are yellow. Genes with hypothetical functions are gray. RNA genes are indicated on the next ring, with rRNA genes in green and tRNA genes in magenta. The innermost ring shows a histogram of variant calls per 1,000-nucleotide window. The scale for each gray circle is 10 variants. Genes and gene clusters with one or more variants are annotated along the outer edge.
Other genes with variant calls appear to be specific to the Corynebacterium genus. For example, the cadA gene, encoding a P-type ATPase cadmium transporter, has 60 variants (the most of any gene) and is found predominantly in corynebacteria and close relatives. Of these 60 variants, 49 were synonymous and 11 were nonsynonymous; no nonsense mutations were detected. The dltA gene, encoding a d-alanine–poly(phosphoribitol) ligase, has 21 variants (10 synonymous and 11 nonsynonymous) and is also restricted to Corynebacterium. Finally, a putative septicolysin toxin gene was found to have three variants, two of them nonsynonymous, and appears to be a novel protein sharing only 93% protein identity with the closest homologs belonging to other corynebacteria. No paralogs were detected for any of these genes in the reconstructed reference. This highlights an important feature of this complementary approach: long-read genome assembly followed by deep short-read variant calling. Short-read assemblies can merge paralogs, which could subsequently confound variant calling. Conversely, variant calling is more difficult with the higher error rate of individual SMRT reads, and in a heterogeneous metagenomic population, it would be very difficult to separate read error from a true variant.
Functional characterization of C. simulans and phage.
A great benefit of finished genomes or high-quality, long metagenomes is the increased power for functional annotation and prediction models. For example, gene calling and comparative genomics are most effective in complete genomes. As another example, genome mining to predict biosynthetic pathways relies on clustering of genes that cooccur within a larger locus (14). Therefore, such analyses are contingent on long stretches of contiguous DNA sequence or complete genomes. Here, we present several characterizations of the C. simulans genome to highlight examples of analyses enabled by this metagenomic reconstruction.
A side-by-side gene comparison of the de novo C. simulans metagenome and type genomes showed significant variable gene content. Of 2,423 and 2,571 genes for the type strain and metagenome, 223 and 376 were not shared, respectively (Fig. 6A; see Table S5 in the supplemental material). The majority of this variable, or pangenomic content was comprised of uncharacterized genes by COG (clusters of orthologous groups of proteins) annotation (Fig. 6B). The type strain possessed more transport and metabolism genes, potentially a reflection on its specialization as a pathogen in the lymph node where it was isolated. The type strain also possessed more defense mechanisms, particularly components of a type I restriction modification system.
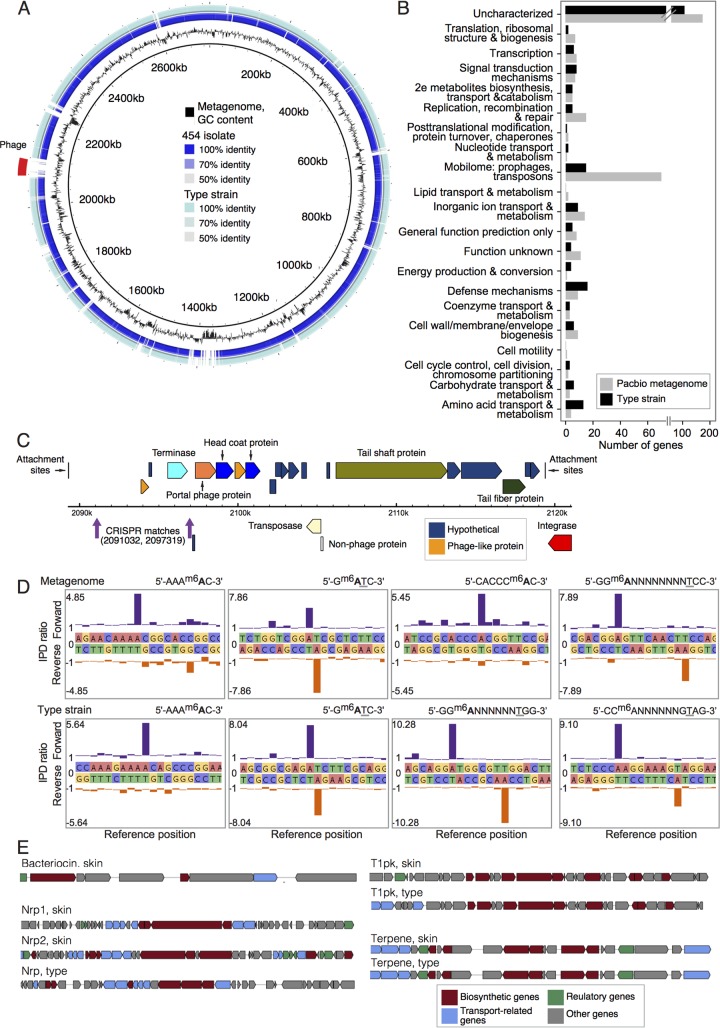
FIG 6 .
Select functional characterizations of C. simulans. (A) Whole-genome comparisons of the C. simulans genomes described in this study. The de novo C. simulans metagenome is used as a reference (innermost ring in black). The GC content of the metagenome is shown in the second ring in black. Ordered contigs from the 454-sequenced skin isolate are shown in the third ring. Alignment of the C. simulans type strain is shown in the fourth or outermost ring. Intensity of color shows the percent identity of the match. (B) COG categories of variable genes, those that are absent in either one of the two complete genomes. (C) Genome structure of the bacteriophage identified in panel A. The metagenome-derived C. simulans contains two CRISPR spacers that are a 100% match to this phage genome and are indicated in purple. (D) Epigenome analysis of C. simulans. Examples of kinetic modification detection signals of 6-methyladenine (m6A) in the C. simulans metagenome (top) and type strain (bottom). The x axis shows the template position and base calls, and the y axis shows the ratio of average interpulse durations (IPDs) for each DNA strand to the control. High deviations from the baseline level indicate a base modification, with the forward strand shown in purple and the reverse strand shown in orange. The methylated bases are indicated in bold type, and the underlined bases indicate methylation on the opposite DNA strand. (E) Examples of biosynthetic pathways predicted from the two complete genomes using antiSMASH 3.0.
The skin strain contained markedly more mobile elements with at least five clustered loci containing multiple elements (or remnants) of transposon and transposase machinery. Surprisingly, we also found a previously undescribed, integrated C. simulans phage that is also absent in the 454-sequenced skin strain (Fig. 6A). This phage, unlikely to result from misassembly due to the high continuity of SMRT reads at flanking regions, is 31.8 kb in length with well-defined attachment sites, a complete head, and tail proteins, integrase, and type VI secretion system (Fig. 6C). This phage also contains lytic elements that encode chitinase-like proteins, which are similar to lysozyme-like families of hydrolases that drive virulence.
Both genomes contained a single, well-defined clustered regularly interspaced short palindromic repeat (CRISPR) locus with similar numbers of spacers (29 and 30 for metagenome and type strain, respectively). Interestingly, only the skin strain had two spacers that were a 100% match to the C. simulans prophage. However, the remaining spacers had little to no homology to GenBank plasmids or phage databases. This likely reflects limited characterization of Corynebacterium plasmids and phages, or little exposure of these strains to currently described Corynebacterium phages—Corynebacterium phages are particularly poorly characterized with only two characterized genomes, neither of which occur in greater than trace amounts in larger skin populations (1).
Aside from the newly characterized phage, two additional spacers in the type strain mapped to a Corynebacterium plasmid and Mycobacterium phage, and two spacers in the skin strain mapped to Corynebacterium and Bacillus plasmids (see Table S5 in the supplemental material). Spacers between strains showed little similarity, as could be expected from random spacer acquisition even from exposure to common foreign hosts, which suggests no recent common ancestor for or horizontal transfer between the two C. simulans strains. Finally, homology to spacers was also not found elsewhere in the metagenomic assemblies, suggesting either a distant spacer acquisition or a near-complete elimination of the invasive species.
A unique feature of SMRT sequencing is the ability to detect epigenetic modifications by measuring fluorescent nucleotide incorporation kinetics (15), as base modifications alter typical rates of polymerase progression. Base methylation serves not only as a protective mechanism against restriction digestion of foreign DNA, but epigenetic diversity can correspondingly increase transcriptional diversity, even within otherwise clonal lineages. We detected four unique methyltransferase motifs containing N6-methyladenosine (m6A) in each of the genomes (Fig. 6D; see Table S5 in the supplemental material). Of the four motifs, two were common in the type and skin strains, and two in each strain were unique. It is likely that the two shared motifs, 5′-Gm6ATC-3′ and 5′-AAAm6AC-3′ (the methylated base indicated in bold type; the underlined base indicates methylation on the opposite DNA strand), are caused by the same methyltransferase genes. Further investigation of the methylome of both species and at the community level will likely provide new insights into gene regulation, virulence mechanisms, DNA damage and repair, and community dynamics.
Finally, secondary metabolites are sophisticated natural products that can modulate interspecies interactions and can reflect an organism’s interface with its larger society. However, these methods typically require complete genomes. A more general prediction of biosynthetic pathways using de novo prediction as well as homology to known gene clusters (14) showed nonribosomal peptide synthetases (Nrps), type I polyketide synthases (T1pks), and terpene synthesis common to both strains (Fig. 6C). Terpene metabolites are widespread in bacteria with poorly characterized function aside from production of odorant molecules (16), and the terpene biosynthesis loci were nearly identical in the two strains. Nrp and polyketide synthases are large enzyme complexes that produce complex secondary metabolites with a wide range of functions, most notably antimicrobial or immunosuppressive functions. Again, T1pks and Nrp clusters were structurally similar in both strains, although an additional Nrp was detected in the skin strain. Finally, the skin strain harbored a bacteriocin locus containing transport and biosynthetic genes with limited homology to lactococcins, which typically are bactericidal by forming pores in the cytoplasmic membranes of sensitive cells.
DISCUSSION
Here, we present a proof-of-principle study demonstrating the utility of long-read SMRT sequencing in the taxonomic and functional characterization of complex microbial skin communities. We also made a side-by-side comparison using short-read HiSeq data and demonstrated the value achievable by hybrid methods, e.g., in evaluating strain heterozygosity. At this time, we believe that short- and long-read technologies can provide complementary strengths in metagenomic community analysis. Both approaches provide similar but distinct assessments of biodiversity, with a large number of species detected by only one of the two methods. We further observed that long reads provide superior contiguity of genome information, and they provide epigenetic information. Deep short-read sequences allowed assessment of polymorphism and genetic heterogeneity in a sample and were better able to reconstruct short contigs from low-abundance genomes from a sample. Our results highlight the striking functional diversity that remains to be discovered in skin communities and the value of high-quality, long assemblies versus fragmented metagenomic reconstructions in which homology of a single gene is the standard unit of measurement for deciphering function.
We demonstrated that long-read data, while lower in coverage and depth, can reconstruct community composition very similarly to short-read data. However, consistent with a recent report using synthetic long-read Illumina Moleculo technology for metagenomic analysis (9), we uncovered a greater number of low-abundance species with long-read data. Despite significantly lower read counts, the SMRT data set yielded higher numbers of eukaryotic DNA viruses, fungi, and even substantial relative abundances of bacteria such as TM7. Unlike this previous report, however, our first assessment of taxonomic composition was based on reference genomes and not assemblies, so the ability of the long reads to reconstruct high-complexity regions is unlikely to underlie this differentiated sensitivity. One reason is that even the few amplification cycles during the Nextera-based HiSeq library preparation may bias the community against low-abundance and/or very high/low-complexity genomes, in contrast to amplification-free SMRT sequencing. A second possibility is that the longer reads may provide additional resolving power for low-abundance genomes, where short reads might be unassignable or assigned to a phylogenetically near neighbor. We speculate that significant phylogenetic diversity, especially in nonbacterial kingdoms, may yet be uncovered with complementary metagenomic sequencing approaches.
We show the first example of the utility of single-molecule sequencing technology in shotgun characterizations of metagenomic communities. We observed a considerable improvement in solo and hybrid assembly quality using even a modest number of SMRT reads. The improvement was such that we recovered and annotated a closed complete genome sequence of a previously uncharacterized species designated Corynebacterium simulans together with a previously uncharacterized lysogenic Corynebacterium bacteriophage. While the nature of such consensus “metagenomes” derived from mixed populations is such that highly conserved regions from other species may also be incorporated into the assembly, our dual approach using both long and short reads showed that the reconstructed C. simulans was a high-quality, closed genome. We were then able to define epigenetic markers for this metagenome using polymerase kinetics to detect base modifications.
We previously observed that populations of other common skin species such as Staphylococcus epidermidis and Propionibacterium acnes are typically heterogeneous with extensive polymorphism (1). Interestingly, the C. simulans strain population in this foot sample had low variability, which enabled assembly of a high-quality closed genome. While our reconstruction produced a dominant consensus strain, the addition of short reads allowed us to identify single-nucleotide variation at multiple loci across the reconstructed genome. Interestingly, genes with the highest single-nucleotide polymorphism (SNP) heterogeneity variability were those that may reflect a unique specialization to interspecies interactions or host interface, like cell surface components and antibiotic resistance (dltA), environmental adaptation (toxic heavy metals by cadA), and toxins (septicolysin). This heterogeneity suggests the coexistence of multiple closely related variants, which could be the result of a species’ longer-term evolutionary trajectory.
With continuing cost reductions and performance advances in SMRT technology, long-read approaches are poised to contribute substantially to microbial community analysis. While SMRT reads reconstruct compositional abundance similarly to deep short-read sequencing, its unique advantage lies in its ability to assemble long stretches of contiguous sequence de novo. For functional annotation or genome assembly of little-characterized microbiomes, this may prove a key advantage. Low-input library preparations for low-biomass samples like skin and circular consensus sequencing (CCS) approaches to produce low-error sequences will further enhance the generalizability of SMRT technology to complex microbiomes. In particular, CCS may obviate the benefits of hybrid approaches derived from a relatively shallow long-read sequencing effort complemented by deep short-read sequencing, although feasibility of scale for complex communities should be further investigated. Finally, single-molecule sequencing also holds great potential in deciphering community-wide epigenetic modifications. Epigenetic signatures could likely be used to bin contigs or SMRT reads into strain-level taxonomic groups. However, meta-epigenomes will also likely provide new insights into interspecies and intercellular dynamics within a community, as both inter- and intraspecies epigenetic heterogeneity likely drive microscale differences in transcriptional modulation. We already appreciate the contribution of species and subspecies variation to functional differences, but new technologies will allow communities to be examined with increasing resolution, such that even compositionally identical communities may prove phenotypically divergent.
MATERIALS AND METHODS
Sample collection and processing.
An aggregate foot sample, representing the left and right symmetric sites of the plantar heel, toe web space, and toenail, and an aggregate arm sample, representing the left and right symmetric sites of the hypothenar palm, volar forearm, and antecubital fossa were each collected from a healthy female in her 20s recruited from the Washington, DC, metropolitan region. This sample collection was approved by the Institutional Review Board of the National Human Genome Research Institute (http://www.clinicaltrials.gov/ct2/show/NCT00605878). Written informed consent was provided prior to participation, and the subject provided medical and medication history and underwent a physical examination. As previously described, samples from each skin site were collected using a swab-scrape-swab procedure (1), in which the defined anatomical skin area was separately swabbed with a swab (Catch-All sample collection swabs; Epicentre) premoistened with yeast cell lysis buffer (MasterPure yeast DNA purification kit; Epicentre), scraped via a sterile disposable surgical blade, and swabbed with the same swab again. Residual material from the scalpel and swab was collected and placed in lysis buffer. Toenail samples were cut with sterilized nail clippers and placed in lysis buffer. All samples were stored at −80°C until extraction. Samples were then incubated in yeast cell lysis buffer (MasterPure yeast DNA purification kit; Epicentre) and Readylyse (Epicentre) for 30 min at 37°C and then mechanically disrupted using 5-mm stainless steel beads (Qiagen) in a Tissuelyser (Qiagen) set at 30 Hz for 2 min. Samples were incubated for 30 min at 65°C and placed on ice for 5 min, and debris was spun down after treatment with MasterPure Complete (MPC) protein precipitation reagent. Samples were combined with 350 µl of 100% ethanol and column purified using the Invitrogen PureLink genomic DNA. Finally, samples were eluted in 30 µl of water (MoBio) and combined according to the body site, foot or arm.
Sample sequencing.
Each sample was then split for three separate sequencing protocols. For Pacific Biosciences terminal deoxynucleotidyl transferase (TdT) sequencing library preparation and SMRT sequencing, 60 ng (arm) and 100 ng (foot) of DNA from each sample was randomly sheared to 10-kb target size using a G-tube device and the standard procedure recommended by the manufacturer (Covaris Inc.). This reduced input is enabled by an experimental low-DNA-input protocol under development at Pacific Biosciences (see Text S1 in the supplemental material), which relies on TdT to add poly(dA) tails to the 3′ ends of DNA fragments for priming and magnetic bead loading. The poly(dA)-tailed DNA fragment library was then annealed with a poly(dT) sequencing primer and sequenced using DNA/polymerase binding kit 2.0 with Magbead loading kit and 120-min sequencing time on a Pacific Biosciences RS II instrument. For quality filtration, reads with a length of <50 bp and a quality score of <75 were excluded using the SMRT Analysis 2.0 software (https://github.com/PacificBiosciences/SMRT-Analysis.git). Read mapping to the Pacbio CHM1 human genome reference (8) using blasr (17) were then filtered. Total reads passing quality control were 16,388 (26.6 Mbp) for the arm sample and 355,610 (622.9 Mbp) for the foot sample. The data were deposited in SRA, and all sequences can be accessed under BioProject accession no. 46333.
Illumina libraries were created using Nextera library preparation with 50 ng of DNA used as input into the transposon fragmentation step. The manufacturer’s protocol was followed with the exception of using 10 cycles of PCR. Illumina libraries were then sequenced with 100-bp paired-end reads on an Illumina HiSeq system at the NIH Intramural Sequencing Center with a target of 100 million clusters. A total of 176,833,930 and 146,424,342 sequences were generated for the arm and foot, respectively. Sequencing data were processed to remove low-quality reads and any read pairs in which at least one read mapped to the hg19 human reference. Nextera adapter sequences were trimmed, if necessary, using crossmatch 1.090518 (http://www.phrap.org) and custom scripts. A second round of filtering for human reads using the CHM1 human reference improved the removal of human reads by 4.8% and 0.8%, respectively. Bases with a quality score below 20 were trimmed, and reads of <50 bp long were removed. Total reads passing quality filters were 8,369,943 and 30,554,290, respectively, representing 805.5 Mbp and 3.0 Gbp of sequence. Sequencing depth was evaluated using k-mer accumulation. SMRT reads were split into 100-bp fragments using pyfasta 0.5.2. Reads were then split into k-mers, compared to a k-mer coverage table using khmer v0.7.1 (18), and kept only if the median k-mer coverage was below a 5× cutoff. The resulting curves estimate the coverage of k-mer space as a function of sequencing effort.
Matched 16S rRNA and ITS1 amplicon libraries were created as previously described (19). Briefly, the V1-V3 region of the 16S rRNA gene was amplified using the barcoded 27F (F stands for forward) and 534R (R stands for reverse) primers, and the ITS1 region was amplified with 18SF and 5.8S-1R primers. Amplicon libraries were sequenced on a 454 GS FLX (Roche) instrument using titanium chemistry. Amplicon sequence data were then processed as previously described using mothur v1.34 (20). Briefly, 454 flow gram data were denoised and error trimmed, and chimeric sequences were removed prior to taxonomic classifications using the Ribosomal Database Project (RDP) classifier training set v10 (21).
Taxonomic characterization.
Quality-filtered, human-filtered SMRT sequencing reads were aligned using blasr to the reference genome collection developed by Oh et al. (1) with the database version containing 2,342 bacterial genomes, 389 fungal genomes, 1,375 viral genomes, and 67 archaeal genomes as described. Illumina short reads were mapped to the same database using the very sensitive bowtie2 version 2.2.3 (22). The top 10 hits were retrieved, and multiply mapping reads were reassigned to a “most likely” genome using a modified method based on Pathoscope v1.0 (23), which uses a Bayesian framework to examine each read’s sequence and mapping quality within the context of a global reassignment. Read hit counts were then normalized by genome length. Quality-filtered 16S rRNA sequences were classified using RDP training set 9 (21), and ITS1 sequences were classified using a custom ITS1 database (24).
De novo assembly. (i) Pacbio-only assembly.
Human-read-filtered SMRT sequencing reads were assembled using the Hierarchical Genome Assembly Process (HGAP) software package developed at Pacific Biosciences (10). Because of the limited number of reads from the arm sample, HGAP assembly was not effective on this sample; however, these data were used for hybrid assemblies. Where error-corrected reads were evaluated, SMRT sequence reads were error corrected using LSC (25) with default parameters.
(ii) Illumina-only and hybrid assemblies.
Human-read-filtered Illumina sequence data were assembled using SPAdes-3.5.0 (26), Megahit (27), and Velvet (28) stepping over a k-mer range of 25 to 71. Error-corrected reads were also used in hybrid assemblies with no improvement over using raw reads with SPAdes. Overall de novo assembly statistics were evaluated as a combination of percent paired or singleton reads realigning to the assembly, the number of bases incorporated into the assembly, and number of contigs of >300 bp. QUAST v2.3 was used to evaluate assembly quality, using the metagenome-derived C. simulans as a reference. In QUAST, a misassembly is defined under the following circumstances: (i) if the left flanking sequence aligns more than 1 kb away from the right flanking sequence, (ii) if overlap between contigs is greater than 1 kb, or (iii) if flanking sequences align on opposite strands (29). Genome coverage is calculated as the total number of aligned bases/genome size, where a contig must have at least one alignment to a base in the reference.
Construction and characterization of the C. simulans genome.
The four longest contigs produced by the HGAP assembly showed significant similarity to a Corynebacterium aurimucosum reference genome. These contigs were connected manually into a single chromosome using the overlapping contig ends and were polished using the Quiver consensus algorithm included in the SMRT Analysis software package. Remapping analysis with blasr was used for quality control of the final genome assembly. No read coverage gap was observed in the remapping result. To make a taxonomic assignment to the reconstructed uncharacterized genome, 16S rRNA sequences were obtained from RDP (30) using criteria to select for Corynebacterium type strains, isolates, and sequences of >1,200 bp for a total of 86 sequences. The archaeal Methanocaldococcus jannaschii was used as an outgroup. 16S rRNA sequences were predicted from full genomes using RNAmmer (31) and aligned using RDP’s rRNA models (21) and the Infernal aligner (32). A bootstrap consensus tree using neighbor joining was generated from 1,000 tests using MEGA (33). NUCmer (34) was used for genome comparisons and plot generation.
Evaluation of strain variation.
Variants were called by aligning the trimmed HiSeq read pairs to the reference using bowtie2 v. 2.2.3 with the very sensitive (end-to-end) alignment presets. Indels were realigned using the IndelRealigner in the Genome Analysis Toolkit (GATK) v. 3.2-2 (35). PCR and optical duplicates were removed using the MarkDuplicates tool in Picard tools v. 1.77 (http://broadinstitute.github.io/picard/). Variants were called using the low-frequency variant caller LoFreq v. 2.1.2 (13) with default parameters.
454 sequencing of C. simulans isolate.
The commensal skin isolate used for select genome comparisons was obtained from a swab from a healthy volunteer as described previously (24), classified by 16S rRNA gene sequencing, and verified with ribosomal protein signature provided by matrix-assisted laser desorption ionization−time of flight (MALDI-TOF) mass spectrometry. Isolates were grown on sheep blood agar and passaged three times to confirm the homogeneity of the strain. Genomic DNA was prepared using MoBio Laboratories UltraClean microbial DNA kit (MoBio Laboratories, Inc.) according to the manufacturer’s instructions. DNA was quantified prior to sequencing using the Quant-iT double-stranded DNA (dsDNA) broad-range (BR) assay (Life Technologies). Genomic libraries were constructed with the Roche 454 titanium kit (Roche Diagnostics GmbH, Mannheim, Germany) by the NIH Intramural Sequencing Center (NISC) using unidirectional fragment reads. Contig assembly was executed with the gsAssembler v.2.3 and exceeded the provisional assembly metrics set forth by the Human Microbiome Project (HMP) (http://www.hmpdacc.org). For comparative analyses, contigs were ordered against a C. aurimucosum reference.
C. simulans genome annotation.
Structural and functional annotation of the metagenome-derived C. simulans, the Pacbio-sequenced type strain, and the 454-sequenced skin isolate was performed using the Institute for Genome Sciences (IGS) Analysis Engine (36) at http://ae.igs.umaryland.edu/cgi/index.cgi. Manatee (http://manatee.sourceforge.net/) was used to view annotations. Manatee annotations are available at NCBI under BioProject accession no. 46333. PGAP (37) was used to verify nonoverlapping genes between the C. simulans type and metagenome strains. PHAST (38) was used to annotate the C. simulans integrated phage. AntiSMASH 3.0 (14) was used for biosynthetic pathway prediction. Brig (39) and CIRCOS (40) were used to generate circular genome plots. Epigenome annotation was performed through the SMRT Analysis 2.0 software. Briefly, kinetics of base addition are quantitated fluorescently during the SMRT sequencing process. Kinetic characteristics, such as time between base incorporations, are altered by DNA modifications and are quantitated as increases in interpulse durations (IPD). IPD ratios, as in Fig. 6, can then be reconstructed using references trained on kinetics of known characterized models, which can include N6-methyladenine, 5-methylcytosine, and 5-hydroxymethylcytosine, and others. Thus, polymerase kinetics can be reconstructed into base modifications in the DNA template.
SUPPLEMENTAL MATERIAL
Reduced-input TdT Pacbio library preparation protocol. Download
Sequence characteristics of samples generated in this study.
Taxonomic classifications of samples generated in this study.
Assembly statistics for HGAP and short-read assemblies.
Population-level SNP diversity in C. simulans metagenome.
Functional characterizations of C. simulans
ACKNOWLEDGMENTS
We thank the Institute for Genome Sciences Analysis Engine service at the University of Maryland School of Medicine for providing structural and functional annotation of the sequences. We also thank the IGS Analysis Engine team for their assistance in submission of the annotated sequences to GenBank. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, Md. (http://hpc.nih.gov).
This work was supported in part by the National Institutes of Health (NIH) NHGRI and NCI Intramural Research Programs.
J.K. and Y-C.T. are full-time employees at Pacific Biosciences, a company commercializing single-molecule sequencing technologies.
J.O. designed the study. C.D. prepared libraries for Illumina sequencing, which was carried out at the NISC Comparative Sequencing Program in Rockville, MD. J.A.S. contributed reagents for the study. H.H.K. provided the patient samples. J.O., S.C., and Y-C.T. analyzed the data. J.K. provided reagents for Pacbio sequencing and advice. J.O. wrote the manuscript.
Footnotes
Citation Tsai Y-C, Conlan S, Deming C, NISC Comparative Sequencing Program, Segre JA, Kong HH, Korlach J, Oh J. 2016. Resolving the complexity of human skin metagenomes using single-molecule sequencing. mBio 7(1):e01948-15. doi:10.1128/mBio.01948-15.
REFERENCES
- 1.Oh J, Byrd AL, Deming C, Conlan S, NISC Comparative Sequencing Program, Kong HH, Segre JA. 2014. Biogeography and individuality shape function in the human skin metagenome. Nature 514:59–64. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto J-M, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K, Wang J. 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 3.Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. 2013. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31:533–538. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- 4.Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ, Lahti L, Loman NJ, Andersson AF, Quince C. 2014. Binning metagenomic contigs by coverage and composition. Nat Methods 11:1144–1146. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- 5.Koren S, Harhay GP, Smith TP, Bono JL, Harhay DM, Mcvey SD, Radune D, Bergman NH, Phillippy AM. 2013. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol 14:R101. doi: 10.1186/gb-2013-14-9-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conlan S, Thomas PJ, Deming C, Park M, Lau AF, Dekker JP, Snitkin ES, Clark TA, Luong K, Song Y, Tsai Y-C, Boitano M, Dayal J, Brooks SY, Schmidt B, Young AC, Thomas JW, Bouffard GG, Blakesley RW, NISC Comparative Sequencing Program, Mullikin JC, Korlach J, Henderson DK, Frank KM, Palmore TN, Segre JA. 2014. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med 6:254ra126. doi: 10.1126/scitranslmed.3009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, Paxinos EE, Sebra R, Chin C-S, Iliopoulos D, Klammer A, Peluso P, Lee L, Kislyuk AO, Bullard J, Kasarskis A, Wang S, Eid J, Rank D, Redman JC, Steyert SR, Frimodt-Møller J, Struve C, Petersen AM, Krogfelt KA, Nataro JP, Schadt EE, Waldor MK. 2011. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med 365:709–717. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaisson MJP, Huddleston J, Dennis MY, Sudmant PH, Malig M, Hormozdiari F, Antonacci F, Surti U, Sandstrom R, Boitano M, Landolin JM, Stamatoyannopoulos JA, Hunkapiller MW, Korlach J, Eichler EE. 2015. Resolving the complexity of the human genome using single-molecule sequencing. Nature 517:608–611. doi: 10.1038/nature13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharon I, Kertesz M, Hug LA, Pushkarev D, Blauwkamp TA, Castelle CJ, Amirebrahimi M, Thomas BC, Burstein D, Tringe SG, Williams KH, Banfield JF. 2015. Accurate, multi-kb reads resolve complex populations and detect rare microorganisms. Genome Res 25:534–543. doi: 10.1101/gr.183012.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 11.Wattiau P, Janssens M, Wauters G. 2000. Corynebacterium simulans sp. nov., a non-lipophilic, fermentative Corynebacterium. Int J Syst Evol Microbiol 50:347–353. doi: 10.1099/00207713-50-1-347. [DOI] [PubMed] [Google Scholar]
- 12.Oh J, Conlan S, Polley EC, Segre JA, Kong HH. 2012. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med 4:77. doi: 10.1186/gm378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilm A, Aw PPK, Bertrand D, Yeo GHT, Ong SH, Wong CH, Khor CC, Petric R, Hibberd ML, Nagarajan N. 2012. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res 40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Müller R, Wohlleben W, Breitling R, Takano E, Medema MH. 2015. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. 2010. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods 7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamada Y, Kuzuyama T, Komatsu M, Shin-ya K, Omura S, Cane DE, Ikeda H. 2015. Terpene synthases are widely distributed in bacteria. Proc Natl Acad Sci U S A 112:857–862. doi: 10.1073/pnas.1422108112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaisson MJ, Tesler G. 2012. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13:238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howe AC, Jansson JK, Malfatti SA, Tringe SG, Tiedje JM, Brown CT. 2014. Tackling soil diversity with the assembly of large, complex metagenomes. Proc Natl Acad Sci U S A 111:4904–4909. doi: 10.1073/pnas.1402564111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh J, Freeman AF, Park M, Sokolic R, Candotti F, Holland SM, Segre JA, Kong HH. 2013. The altered landscape of the human skin microbiome in patients with primary immunodeficiencies. Genome Res 23:2103–2114. doi: 10.1101/gr.159467.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Horn DJV, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francis OE, Bendall M, Manimaran S, Hong C, Clement NL, Castro-Nallar E, Snell Q, Schaalje GB, Clement MJ, Crandall KA, Johnson WE. 2013. Pathoscope: species identification and strain attribution with unassembled sequencing data. Genome Res 23:1721–1729. doi: 10.1101/gr.150151.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, NIH Intramural Sequencing Center Comparative Sequencing Program, Kong HH, Segre JA. 2013. Topographic diversity of fungal and bacterial communities in human skin. Nature 498:367–370. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Au KF, Underwood JG, Lee L, Wong WH. 2012. Improving PacBio long read accuracy by short read alignment. PLoS One 7:e46679. doi: 10.1371/journal.pone.0046679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 28.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. 2014. Ribosomal Database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagesen K, Hallin P, Rødland EA, Staerfeldt H-H, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nawrocki EP, Kolbe DL, Eddy SR. 2009. Infernal 1.0: inference of RNA alignments. Bioinformatics 25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delcher AL, Phillippy A, Carlton J, Salzberg SL. 2002. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res 30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galens K, Orvis J, Daugherty S, Creasy HH, Angiuoli S, White O, Wortman J, Mahurkar A, Giglio MG. 2011. The IGS standard operating procedure for automated prokaryotic annotation. Stand Genomic Sci 4:244–251. doi: 10.4056/sigs.1223234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J. 2012. PGAP: pan-genomes analysis pipeline. Bioinformatics 28:416–418. doi: 10.1093/bioinformatics/btr655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (Brig): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krzywinski MI, Schein JE, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reduced-input TdT Pacbio library preparation protocol. Download
Sequence characteristics of samples generated in this study.
Taxonomic classifications of samples generated in this study.
Assembly statistics for HGAP and short-read assemblies.
Population-level SNP diversity in C. simulans metagenome.
Functional characterizations of C. simulans