ABSTRACT
The methicillin resistance factor encoded by fmtA is a core member of the Staphylococcus aureus cell wall stimulon, but its function has remained elusive for the past two decades. First identified as a factor that affects methicillin resistance in S. aureus strains, FmtA was later shown to interact with teichoic acids and to localize to the cell division septum. We have made a breakthrough in understanding FmtA function. We show that FmtA hydrolyzes the ester bond between d-Ala and the backbone of teichoic acids, which are polyglycerol-phosphate or polyribitol-phosphate polymers found in the S. aureus cell envelope. FmtA contains two conserved motifs found in serine active-site penicillin-binding proteins (PBPs) and β-lactamases. The conserved SXXK motif was found to be important for the d-amino esterase activity of FmtA. Moreover, we show that deletion of fmtA (ΔfmtA) led to higher levels of d-Ala in teichoic acids, and this effect was reversed by complementation of ΔfmtA with fmtA. The positive charge on d-Ala partially masks the negative charge of the polyol-phosphate backbone of teichoic acids; hence, a change in the d-Ala content will result in modulation of their charge. Cell division, biofilm formation, autolysis, and colonization are among the many processes in S. aureus affected by the d-Ala content and overall charge of the cell surface teichoic acids. The esterase activity of FmtA and the regulation of fmtA suggest that FmtA functions as a modulator of teichoic acid charge, thus FmtA may be involved in S. aureus cell division, biofilm formation, autolysis, and colonization.
IMPORTANCE
Teichoic acids are involved in cell division, cell wall synthesis, biofilm formation, attachment of bacteria to artificial surfaces, and colonization. However, the function of teichoic acids is not fully understood. Modification by glycosylation and/or d-alanylation of the polyol-phosphate backbone of teichoic acids is important in the above cell processes. The intrinsic negative charge of teichoic acid backbone plays a role in the charge and/or pH of the bacterial surface, and d-alanylation represents a means through which bacteria modulate the charge or the pH of their surfaces. We discovered that FmtA removes d-Ala from teichoic acids. We propose FmtA may provide a temporal and spatial regulation of the bacterial cell surface charge in two ways, by removing the d-Ala from LTA to make it available to wall teichoic acid (WTA) in response to certain conditions and by removing it from WTA to allow the cell to reset its surface charge to a previous condition.
INTRODUCTION
Staphylococcus aureus is the leading cause of hospital- and community-acquired infections (1). S. aureus was once inherently susceptible to most antibiotics. However, it is now a pathogen of great concern due to its intrinsic virulence and its remarkable ability to rapidly adapt to different environmental conditions by mutation and DNA transfer (2–4). Multiple-drug-resistant S. aureus strains, such as methicillin-resistant S. aureus (MRSA), have become notoriously difficult to treat, with 20 to 40% of cases causing mortality (1). Emergence of MRSA strains resistant to vancomycin (5), an antibiotic reserved for the treatment of severe MRSA infections, has led to limited treatment options for S. aureus infections (6–9). To make matters worse, infections caused by multiple MRSA strains have reached epidemic proportions (8, 10).
The need for novel antibiotics for the treatment of S. aureus-related infections is as urgent today as it was in 1940, when penicillin was introduced to treat S. aureus infections (11, 12). Antibiotics that target cell wall biosynthesis (referred to as cell wall inhibitors), such as β-lactams and glycopeptides, are among the most efficient antibacterial agents for treating S. aureus infections; however, their biological activities have been compromised by the emergence of resistance mechanisms (11, 12). Recent reports have shown that MRSA can be resensitized to β-lactams and vancomycin by inhibiting nonessential genes involved in the biosynthesis of cell envelope components, such as peptidoglycan and teichoic acids, and these reports have rekindled interest in targeting the cell wall for drug discovery and provide evidence that antibiotic potency can be rescued (13–19).
Komatsuzawa et al. reported that fmtA was a factor in the methicillin resistance of MRSA strains (20) and that deletion of fmtA reduced the methicillin MIC for S. aureus Col (an MRSA strain) from 1,024 µg/ml to 128 µg/ml (20). In addition, deletion of fmtA was shown to disrupt the homogeneity of methicillin resistance (20). Further, fmtA was identified as a core member of the cell wall stimulon; fmtA expression increased in the presence of cell wall inhibitors and when genes involved in cell wall biosynthesis were deleted (21–24). The primary structure of FmtA shares similarities with d,d-carboxypeptidase from Streptomyces R61 (d,d-carboxypeptidase R61) and class C β-lactamases (25). FmtA harbors two of the three conserved motifs, SXXK and SND, found in the serine active site of penicillin-binding proteins (PBPs) and β-lactamases. The third conserved motif, KTG, has not been identified in FmtA. Our previous studies on FmtA revealed that it interacts covalently with β-lactams via a serine residue located in the conserved SXXK motif. We also showed that FmtA has very weak d,d-carboxypeptidase activity and interacts with teichoic acids (25). These observations led to the proposal that FmtA may be a PBP (25, 26). S. aureus has four native PBPs. PBP1 and PBP2 are essential enzymes (27). PBP1 and PBP4 are transpeptidases, and PBP2 is a bifunctional enzyme. The function of PBP3 remains elusive. PBP2 and PBP4 are involved in the synthesis of highly cross-linked peptidoglycan (28).
The interaction of FmtA with teichoic acids (26) is peculiar, considering that teichoic acids have been found to play roles in the temporal and spatial regulation of AtlA and PBP4 (29, 30). There are two types of teichoic acids in S. aureus. Teichoic acids that are bound to the outer leaflet of the cytoplasmic membrane are referred to as lipoteichoic acids (LTAs), whereas teichoic acids that are bound to peptidoglycan are referred to as wall teichoic acids (WTAs) (31, 32). LTAs are phosphate-rich polyglycerol polymers, and WTAs are phosphate-rich polyribitol polymers (Fig. 1) (33, 34). Their biosynthetic pathways differ. LTA synthesis takes place in the periplasm, and WTA synthesis takes place in the cytoplasm (35–37). Both polymers are postsynthetically glycosylated with N-acetylglucosamine and/or esterified with d-alanine, as shown in Fig. 1 (32, 38, 39). Modification of WTA with N-acetylglucosamine takes place in the cytoplasm, whereas modification with d-Ala takes place in the extracellular milieu (38, 40).
FIG 1 .
A schematic view of the cell surface of S. aureus. Highlighted are the cytoplasmic membrane, peptidoglycan (PG), lipoteichoic acid (LTA), and wall teichoic acid (WTA). The general chemical structures of LTA and WTA are shown.
The biological function of LTAs in S. aureus is not well understood, but they have been shown to interact with cell division proteins (41). It has also been suggested that LTAs are involved in osmoprotection of the cell (32). WTAs have been shown to act as scaffolds for many proteins, such as exogenous proteins required for phage infection (42) and endogenous proteins required for cell growth and division, such as AtlA (29, 43), PBP4 (30), and FmtA (26). Decoration of teichoic acids with d-Ala and α- or β-O-N-acetyl-d-glucosamine (GlcNAc) has been implicated in many processes. The d-Ala content of teichoic acids affects the bacterial surface charge by adding a positive charge to an otherwise negatively charged polyol-phosphate backbone of teichoic acids (44) and has been implicated in S. aureus virulence (45–47), attachment to artificial surfaces, initiation of biofilm formation (48, 49), cell autolysis (29, 34, 50), and susceptibility to cationic peptide antibiotics and vancomycin (47, 51). Glycosylation of WTAs with an α- or β-anomer of GlcNAc has also been reported to be important for methicillin resistance and horizontal gene transfer between bacterial pathogens (37, 52).
FmtA exhibits very weak d,d-carboxypeptidase activity. It was proposed that FmtA might require binding to a ligand for activation (26). In light of our previous finding that FmtA interacts with teichoic acids, we aimed to evaluate the role of teichoic acids on the d,d-carboxypeptidase activity of FmtA. We found that FmtA acts as an esterase and removes d-Ala groups from WTAs and LTAs. Deletion of fmtA led to a 7-fold increase in the d-Ala content of WTAs, and complementation of the fmtA deletion mutant (ΔfmtA) with wild-type fmtA restored the d-Ala content to the level in WTAs from wild-type S. aureus. These findings have important implications for two aspects of d-Ala modification of teichoic acids. First, it has been proposed that the d-Ala of LTAs serves as the d-Ala source for d-alanylation of WTAs (33), but no study has shown how d-Ala is removed from LTA. Although it has been reported that d-Ala can spontaneously be removed from LTA, these processes are very slow (53). Second, studies have shown that the d-Ala content in teichoic acids is affected by a number of environmental factors, such as salt concentration, pH, and temperature (54–56), which suggests that, under certain environmental conditions, d-Ala from teichoic acids must be removed (54). Our results suggest that FmtA modulates d-alanylation of teichoic acids in S. aureus.
RESULTS
FmtA has d-amino esterase activity toward teichoic acids.
d-Ala is a small molecule that lacks optical properties to enable its monitoring by classical spectrophotometric methods, such as the use of a UV-visible (UV-Vis) spectrophotometer or fluorimeter. For this purpose, we used a fluorescence-based coupled enzyme assay to assess the d,d-carboxypeptidase activity of FmtA, measured as release of free d-Ala, in the absence and presence of WTA. The amount of free d-Ala measured in the reaction mixtures, which contained the tripeptide Nα,Nε-diacetyl-Lys-d-Ala-d-Ala (6 mM), WTA (0.1 µg/µl to 0.3 µg/µL), and FmtA, was 3- to 4-fold higher than in the reaction mixtures lacking WTA (Fig. 2A). This increase in free d-Ala was also observed in the absence of the tripeptide, indicating that WTA was the source of the free d-Ala in the assay.
FIG 2 .

Analysis of d-Ala removal from the tripeptide Nα,Nε-diacetyl-Lys-d-Ala-d-Ala (TP, 6 mM) by FmtA (10 µM) in the absence or presence of WTA (0.1 µg/µl, 0.2 µg/µl, 0.3 µg/µl) (A) or in the presence of Δdlt WTA (B). Free d-Ala was measured using a fluorescence-based coupled enzymatic assay. Error bars represent standard deviations from three independent experiments.
WTAs are d-alanylated at the C-2 position of ribitol (Fig. 1). To exclude the possibility that WTAs are an adventitious source of free d-Ala in our assays, we performed the assay using WTAs isolated from an S. aureus strain in which the dltABCD operon was deleted (Δdlt WTAs). This operon is responsible for incorporating d-Ala into WTAs (51). d-Ala release was not observed in the presence of Δdlt WTAs (Fig. 2B) suggesting that WTAs are the source of d-Ala and that FmtA may remove d-Ala from WTAs. PBP4 and PBP2a activities toward WTA was also investigated, but no hydrolysis activity was detected (see Fig. S1A and B in the supplemental material).
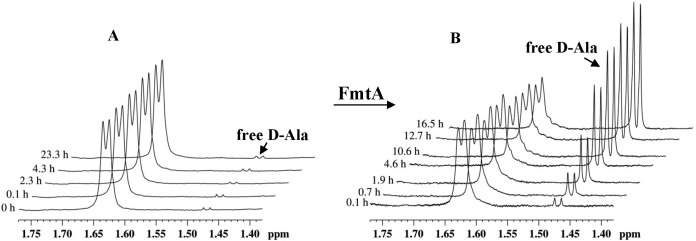
We used proton nuclear magnetic resonance spectroscopy (1H-NMR) to further investigate the activity of FmtA on WTAs. The release of free d-Ala was identified by the appearance of a new resonance signal at 1.47 ppm in the 1H-NMR spectrum of WTA and the disappearance of the resonance peak at 1.63 ppm that corresponds to bound d-Ala (Fig. 3A and B). In the absence of FmtA, free d-Ala was not detected after incubation for 2 days (Fig. 3C). To investigate the specificity of this FmtA activity, we incubated WTAs with heat-denatured FmtA, and no significant d-Ala release was detected (data not shown).
FIG 3 .
(A) 1H-NMR spectra of WTA isolated from S. aureus RN4220. (B) 1H-NMR spectra centered at 1.60 ppm in the absence of FmtA. (C) 1H-NMR spectra centered at 1.60 ppm in the presence of FmtA. Experiments were carried out in 10 mM sodium phosphate (pH 7.2) at 25°C with 5 µg/µl of WTA.
We also investigated the activity of FmtA on LTAs by 1H-NMR. LTA modification with d-Ala occurs on C2 of glycerol (57). Removal of the d-Ala from LTA is identified by the disappearance of the resonance peak at 1.63 ppm (bound d-Ala) and the appearance of the free d-Ala resonance peak at 1.47 ppm. In the absence of FmtA, there was no significant removal of d-Ala from LTA (Fig. 4).
FIG 4 .
1H-NMR spectra of LTA from S. aureus (Sigma). 1H-NMR spectra were centered at 1.80 ppm in the absence (A) and presence (B) of 10 µM FmtA. Experiments were carried out in 10 mM sodium phosphate (pH 7.2) at 25°C with 5 µg/µl of WTA.
To investigate whether the esterase activity of FmtA is related to the presence of the conserved PBP motifs in FmtA, we investigated the activity of two PBPs, PBP2a (a nonnative PBP from S. aureus) and PBP4 (a native PBP from S. aureus) toward WTAs. Both PBP2a and PBP4 have been shown to have transpeptidase activity (58–60) and to interact with WTAs, although the interaction of PBP4 with WTA is weaker than with FmtA (26) (see Fig. S2 in the supplemental material). We found that PBP2a and PBP4 did not have significant activity toward WTAs (Fig. 5); these data agree with our fluorescence data (see Fig. S1 in the supplemental material).
FIG 5 .

1H-NMR spectra of WTA isolated from S. aureus RN4220. 1H-NMR spectra were centered at 1.60 ppm in the presence of 10 µM PBP2 and 10 µM PBP4 at various incubation times. Experiments were carried out in 10 mM sodium phosphate buffer (pH 7.2) at 25°C with 5 µg/µl of WTA.
Kinetics of FmtA esterase activity.
The NMR studies described above provided the necessary means of monitoring of free d-Ala, which otherwise lacks any special optical properties, and the much-needed sensitivity for our experimental settings, where we are limited by the amount of WTA isolated from S. aureus, about 1 mg per liter of cell culture. In addition, the use of the 700 MHz NMR enabled us to use reaction volumes as low as 250 µl and employ pseudo-first-order rate kinetics in characterization of the rate constant of d-Ala removal from WTA by FmtA.
The rate of FmtA esterase activity was monitored by 1H-NMR. The amount of free d-Ala released during the reaction was quantified and normalized to the GlcNAc resonance peak, as the GlcNAc content of WTA does not change over time. The normalized free d-Ala amounts measured at different time points were used to construct progress curves in which the data were fitted to pseudo-first-order rate kinetics. The observed pseudo-first-order rate constant (kobs) was 0.57 ± 0.01 h−1 with 10 µM FmtA and 5 mg/ml WTA. The reaction rates increased with increased enzyme concentrations as indicated from the calculated kobs values, which increased from 0.46 h−1 with 5 µM FmtA to 1.11 h−1 with 20 µM FmtA ([WTA] = 5 mg/ml) (Fig. 6A). In contrast, FmtA reaction rates increased with the decrease of WTA concentrations. The kobs value increased 3-fold, to 1.3 ± 0.2 h−1, when the WTA concentration was reduced from 5 mg/ml to 0.5 mg/ml (Fig. 6B). The kobs value measured at 0.5 mg/ml WTA is the highest pseudo-first-order rate constant that we could measure; as such, it can serve as an approximation for the apparent first-order rate constant of the esterase reaction catalyzed by FmtA, although it is an underestimation of the true first-order rate constant for this reaction. From this kobs value, we estimated the time required to hydrolyze 50% of the d-alanyl ester bonds in WTA (half time [t1/2]) to be 32 min.
FIG 6 .

Kinetics of d-Ala release from WTA by FmtA. (A) d-Ala release was monitored in the presence of different FmtA concentrations (5 µM [squares], 10 µM [circles], and 20 µM [triangles]). (B) d-Ala release was monitored in the presence of different WTA concentrations (0.5 µg/µl [asterisks], 1 µg/µl [triangles], 2.5 µg/µl [squares], and 5 µg/µl [circles]). (C) Effects of the FmtA-Ser127Ala and FmtA-Lys130Ala mutations on the removal of d-Ala from WTA (circles, wild-type FmtA; triangles, FmtA-Ser127Ala FmtA; squares, FmtA-Lys130Ala).
The reduction in FmtA esterase activity found with increased WTA concentrations could be a result of structural changes in FmtA upon WTA binding. Indeed, an earlier investigation of the interaction of FmtA with WTA by circular dichroism revealed that the secondary structural elements of FmtA were altered extensively in the presence of high WTA concentrations (26). We have observed that high concentrations of WTA (≥10 mg/ml) cause aggregation of FmtA. This may be due to a salting-out effect produced by the highly negatively charged WTAs.
To investigate the significance of the conserved PBP motif SXXK for the esterase activity of FmtA, we constructed two variants of FmtA by replacing Ser127 and Lys130 with Ala. By determining the ratios of the observed pseudo-first-order rate constants of the mutants and wild-type FmtA, we found that FmtA-Ser127Ala and FmtA-Lys130Ala had 16% of the activity of wild-type FmtA (Fig. 6C). The seemingly high remaining activity of FmtA mutant could result from relative instability of ester moieties in slight acidic or basic aqueous solutions and binding of the WTA to FmtA; i.e., the topology of the FmtA active site may introduce enough constraints to the carbonyl C of the d-Ala bound to WTA to lead to its increased electrophilicity and attack by weak nucleophiles such as water molecules in long incubation times.
To investigate whether FmtA can catalyze esterification of WTA by d-Ala, we set up several assays. We incubated d-Ala (6 mM) with ribitol (10 mM) or with Δdlt WTAs (2.5 mg/ml) and monitored the reaction by 1H-NMR in the absence and presence of FmtA (20 µM). In addition, LTA (0.5 mg/ml) and Δdlt WTA (2.5 mg/ml) were incubated together in the absence and presence of FmtA (20 µM) to investigate whether d-Ala released from LTA can be transferred to WTA by FmtA. Lastly, we incubated d-Ala with ATP (5 mM), MgCl2 (10 mM), dithiothreitol (DTT) (1 mM), and Δdlt WTA in the absence and presence of FmtA (20 µM). None of the above conditions led to the incorporation of d-Ala to WTA (data not shown).
Specificity of FmtA esterase activity.
To determine the substrate specificity of FmtA, we tested several carboxylesterase substrates, including p-nitrophenyl butyrate (p-NPB) and p-nitrophenyl acetate (p-NPA). FmtA showed very low esterase activity on p-NPB and p-NPA as assessed by continuous spectrophotometric assay (data not shown). We also used 1H-NMR to monitor hydrolysis of p-NPB and p-NPA and detected very little activity on p-NPB and p-NPA (data not shown). In addition, we investigated potential α-amino-acid esterase and aminopeptidase activities in FmtA with d-alanine methyl ester and l-alanine p-nitroanilide, respectively. However, we did not observe any catalytic activity against these substrates. Carboxylesterases have been reported to catalyze the hydrolysis of short-chain aliphatic and aromatic carboxylic ester compounds (61) suggesting that FmtA may be specific for the d-Ala ester attached at the C-2 of ribitol-5-phosphate and glycerol-3-phosphate, which are the repeating units of teichoic acids.
Assessment of FmtA esterase activity in cells.
To investigate the significance of our findings in S. aureus, we constructed three S. aureus RN4220 mutant strains (see Text S1 in the supplemental material). The fmtA deletion (ΔfmtA) strain was constructed using a pMAD vector (62). The complementation strain was created by cloning fmtA into a pMK4 vector (63) and introducing the construct into S. aureus RN4220. We also constructed an fmtA conditional mutant (CM) in which fmtA expression is under the control of an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter, Pspac (Pspac-fmtA), using a pMUTIN integration vector (Bacillus Genetic Stock Center, The Ohio University).
We isolated WTAs from S. aureus RN4220 and the ΔfmtA, complemented ΔfmtA, and Pspac-fmtA strains in the presence or absence of 0.5 mM IPTG. WTAs from each strain were analyzed by 1H-NMR. The amount of d-Ala attached to ribitol relative to the amount of N-acetylglucosamine attached to ribitol was determined by integrating the resonance peaks of N-acetylglucosamine at 2.08 ppm and d-Ala at 1.63 ppm. Deletion of fmtA resulted in a 7-fold increase in d-alanylation of WTAs (Table 1; Fig. 7). Interestingly, this increased d-alanylation was also seen in WTAs isolated from the Pspac-fmtA strain in the absence of IPTG (4.5-fold increase), but in the presence of IPTG, the WTAs exhibited wild-type levels of d-alanylation. WTAs isolated from the complemented ΔfmtA strain also exhibited wild-type levels of d-alanylation.
TABLE 1 .
Quantitative analysis of the relative amount of d-Ala on WTAs isolated from various S. aureus strainsa
| Strain or description | Relative integration (GlucNac/d-Ala) |
|---|---|
| RN4220 | 0.63 ± 0.03 |
| ΔfmtA | 4.0 ± 0.8 |
| Pspac-fmtA | 2.6 ± 0.5 |
| Pspac-fmtA + IPTG | 0.6 ± 0.1 |
| ΔfmtA::pMK4:fmtA | 0.5 ± 0.1 |
Values are the averages of three independent measurements and the errors represent the standard deviations.
FIG 7 .

1H-NMR spectra of WTA isolated from S. aureus RN4220 (WT) and the Pspac-fmtA mutant (CM), the ΔfmtA mutant, the Pspac-fmtA mutant (CM) with 0.5 mM IPTG, and the complemented ΔfmtA (ΔfmtA+) strain. 1H-NMR spectra were centered at 1.80 ppm.
DISCUSSION
We discovered that FmtA has esterase activity specific to teichoic acids. We determined that the pseudo-first-order rate constant of the FmtA esterase activity was 1.3 h−1 at 5 µM FmtA and 0.5 mg/ml WTA. The time that it would take FmtA to remove 50% of the d-Ala from teichoic acid was estimated using the above observed pseudo-first-order rate constant, and it was determined that t1/2 does not exceed 32 min. Taking into account that S. aureus doubles every 30 min, we can conclude that the esterase activity of FmtA is significant and relevant in vivo. Evidence for alanyl turnover in S. aureus LTAs has been reported by Haas et al., and their study determined that the t1/2 of alanyl turnover in S. aureus LTA was 37 min (64), which is in remarkable agreement with our estimated t1/2. By comparing the kinetics of alanyl turnover in S. aureus LTA to the kinetics of alanyl turnover in a base-catalyzed reaction (53), Haas et al. suggested that d-alanyl removal from LTA is likely to be an enzyme-catalyzed process (64). Our study suggests that FmtA is the enzyme that catalyzes the alanyl turnover of teichoic acids. The FmtA esterase activity that we observed in vitro strongly correlates with our in vivo data in which WTA isolated from S. aureus ΔfmtA had a higher d-Ala content than WTA isolated from wild-type S. aureus. Complementation of the ΔfmtA mutant with wild-type fmtA restored the d-Ala level to wild-type.
The presence of FmtA esterase activity toward teichoic acids and the absence of FmtA esterase activity toward small-molecule substrates of amidases, peptidases, and carboxylesterases suggest that FmtA has d-amino esterase activity, is specific for d-amino acids and may recognize the positive charge on the amino group of d-Ala; this d-amino esterase nomenclature is in line with the nomenclature of d,d-carboxypeptidases and d-aminopeptidases. Microbial esterases, commonly referred to as carboxylesterases, constitute a large group of enzymes that hydrolyze short-chain aliphatic and aromatic carboxylic ester compounds. They have been classified into eight families based on the their conserved sequence motifs and biological properties (65), and their esterase activity has primarily been attributed to the GXSXG motif. However, in family VIII esterases, represented by EstB from Burkholderia gladioli, the esterase activity is associated with a conserved SXXK motif, which is found in the serine active site of PBPs and β-lactamases (66), despite the presence of the GXSXG motif. Notably, these enzymes show sequence and structural similarities to class C β-lactamases and d,d-carboxypeptidase R61. EstB contains two of the three conserved motifs found in the serine active site of PBPs and β-lactamases, SXXK and SYN, whereas the third conserved KTG motif is replaced with WGG. However, EstB does not have β-lactamase or d,d-carboxypeptidase activity (66). EstB is not an exception in this respect; there are a large number of enzymes that exhibit a wide range of activities, such as d,d-endopeptidases (67), d,l-endopeptidases (68), d-amino acid amidases (69–71), d-aminopeptidases (72, 73), and d-esterases (66, 74), that also share the conserved motifs and structural folds found in the serine active sites of PBPs and β-lactamases but do not function as PBPs and β-lactamases (75). These enzymes are referred to as penicillin-recognizing enzymes (PRE) (75, 76). Many structural studies on PREs have shown that the divergence in their activities may be attributed to the presence of a core enzyme surrounded by specific structural modulators (69, 71, 77).
Our previous studies showed that FmtA has primary structure similarities with class C β-lactamases and d,d-carboxypeptidase R61 and has an overall structural fold similar to that of d,d-carboxypeptidase R61 (25); thus, FmtA can be considered a member of family VIII of esterases. However, FmtA does not have typical carboxylesterase activity. This lack of activity toward short-chain aliphatic carboxylic ester compounds indicates that FmtA recognizes the positive charge on the amino group of d-alanine. Recognition of the charge on an amino acid is a common structural feature of d-amino acid amidases (DAA), d-aminopeptidases (DAP), and d,d-carboxypeptidases. DAA and DAP recognize the positive charge on the amino group of d-Ala, whereas d,d-carboxypeptidases recognize the negative charge on the carboxylic group of d-Ala. Moreover, structural and mutagenesis studies on DAP from Ochrobactrum anthropi have revealed that this enzyme can be converted to a d,d-carboxypeptidase by removing two structural elements that interact with the amino group of d-Ala and introducing a structural element from d,d-carboxypeptidases that is known to interact with the carboxylic group of d-Ala (69). In the case of FmtA, the lack of activity toward a typical carboxylesterase substrate suggests that FmtA may have binding specificity for the glycerol-3-phosphate and ribitol-5-phosphate repeating units of teichoic acids.
The function of FmtA as a d-amino esterase of teichoic acids requires FmtA to be located in the extracellular milieu where the teichoic acids are located. Moreover, the d-Ala content, and as a result, the charge state of teichoic acids is expected to be dynamic and dependent on the environment, the growth state, and lifestyle (i.e., growth, division, planktonic or biofilm state) of the bacteria (31, 54). As such, the dynamic modulation of the charge state of teichoic acids requires temporal regulation of the d-Ala removal. The requirement for localization of FmtA in the extracellular milieu is met by FmtA, as earlier findings show that FmtA is an extracellular protein (20) and that green fluorescent protein (GFP)-labeled FmtA, which lacked the first 27 N-terminal amino acid residues, considered to constitute the signal peptide (25), localized to the cell division septum of S. aureus (26). The requirement for temporal regulation of d-Ala content of teichoic acids is also met by FmtA, as the expression of fmtA has been shown to be tightly regulated (24, 78), and we recently demonstrated that fmtA is controlled by the globular regulatory protein SarA (79). Of note, the involvement of SarA in the regulation of fmtA is in agreement with the esterase function of FmtA. Both SarA and FmtA have independently been shown to be involved in S. aureus biofilm formation (80–84). Further, S. aureus biofilm formation is dependent on the major autolysin AtlA (85, 86), activity of which is closely associated with the d-Ala content of WTA (29, 43).
Haas et al. showed that d-Ala removed from LTA can be incorporated into WTA (64). In addition, Reichmann et al. showed that WTA isolated from S. aureus lacking LTA contained a low level of d-Ala (87). The inference from these studies is that LTA may serve as the donor of d-Ala for d-alanylation of WTA. Because FmtA has esterase activity, FmtA may function to remove d-Ala from LTA so that it is available for uptake by WTA (31). The question that subsequently arises is that of how d-Ala is incorporated into WTA. It is generally accepted that the dltABCD operon is involved in the de novo esterification of LTA, but not WTA, with d-Ala (87). The esterase activity of FmtA raises the question of whether FmtA may function as a transesterase. As a transesterase, FmtA would remove d-Ala from LTA and transfer it to WTA. The major concern with this proposal comes from recent findings that show little spatial overlap between mature LTA and WTA polymers in the extracellular milieu. S. aureus LTAs are composed of approximately 25 glycerol-3-phosphate repeating units and are anchored to the membrane, and they do not stretch beyond the peptidoglycan layer (32, 41). In contrast, WTAs are composed of approximately 40 ribitol-5-phosphate repeating units, are anchored to peptidoglycan, and stretch well beyond the peptidoglycan (31). It could be argued that d-alanylation of WTA may occur prior to the attachment of WTA to peptidoglycan while it is anchored in the outer leaflet of the lipid bilayer and positioned close to LTA at the cell division septum. However, studies indicate that d-alanylation of WTA is not a one-time event in the life cycle of the cell. We did not observe any FmtA transesterase activity in this study. Further studies are needed to elucidate the mechanism of d-Ala incorporation into WTA.
Teichoic acids are involved in S. aureus cell division, cell wall synthesis, biofilm formation, and attachment to artificial surfaces and colonization. Although the role of teichoic acids in many of these processes remains elusive, glycosylation and/or esterification of the polyol-phosphate backbone of teichoic acids is seen as a factor in establishing interactions with bacterial extracellular proteins, host receptors, and phage receptors involved in the above-described processes (29, 30, 37, 44, 45, 52, 88). However, d-alanylation goes beyond transforming teichoic acids into recognition modules on the cell surface. It has been suggested that the intrinsic negative charge of the teichoic acid backbone functions as a bacterial cell surface charge and/or pH determinant (89); hence, d-alanylation of teichoic acids may be a mechanism through which bacteria modulate their surface charge and/or pH. In light of the FmtA enzymatic activity that we identified, we propose that FmtA may provide the cell with the ability to temporally and spatially regulate their cell surface charge by removing d-Ala from LTA so that it is available to WTA in response to certain conditions and by removing d-Ala from WTA to allow the cell to reset its surface charge to a previous condition.
MATERIALS AND METHODS
Materials and reagents.
Growth media were purchased from EMD Bioscience. Enzymes (trypsin, DNase, RNase, horseradish peroxidase, and d-amino acid oxidase) and chemicals (Tris, cytochrome c, and LTA) were purchased from Sigma (Oakville, Canada) and Thermo-Fisher (Whitby, Canada) unless otherwise stated. Amplex red (AR) was purchased from Molecular Probes, Inc. Oligonucleotides were acquired from Sigma.
Isolation and purification of FmtA, FmtA-Ser127Ala, and FmtA-Lys130Ala.
The fmtA gene from S. aureus strain Mu50 was cloned into pET24a(+) using the NdeI and HindIII restriction sites (25). The cloned fmtA lacked the first 81 nucleotides, corresponding to the 27-amino-acid signal peptide. The predicted conserved active-site residues in the SXXK motif in FmtA, Ser127 (encoded by TCA) and Lys130 (encoded by AAA), were mutated to alanine with the Quik-Change site-directed mutagenesis kit (Agilent). Specifically, pET24a(+)::fmtA was used as the template, and amplification was carried out using Pfu Turbo DNA polymerase and a pair of mutagenic primers for each substitution. The primer sets used were DirS127A (5′ CGATGTTTTTAATAGGTGCAGCTCAAAAATTTTC 3′)-RevS127A (5′ GAAAATTTTTGAGCTGCACCTATTAAAAACATCG 3′) and DirK130A (5′ GGTTCAGCTCAAGCATTTTCAACAGGGTTAC 3′)-RevK130A (5′ GTAACCCTGTTGAAAATGCTTGAGCTGAACC 3′) (mutations are in italics). The nucleotide sequences of the variants were verified by DNA sequencing (Core Facility, York University). E. coli BL21(DE3) cells were then transformed with the mutagenized vectors. Protein synthesis, isolation, and purification were performed as described previously (25).
Enzyme assays.
The d,d-carboxypeptidase activity of FmtA was assessed using a fluorescent enzyme-coupled assay as described previously (90). The esterase activity of FmtA was investigated by a continuous spectrophotometric assay using p-NPB or p-NPA as the substrate. The standard reaction mixture contained p-NPB or p-NPA (1 mM), FmtA, PBP4 or PBP2a (5 µM), and 50-mM sodium phosphate (pH 7.0) in a final volume of 0.5 ml and was incubated at room temperature. The reaction was initiated by the addition of substrate. Hydrolysis of p-NPB or p-NPA was monitored spectrophotometrically for formation of p-nitrophenol, which is detected at 405 nm (ε = 0.2 mM−1 cm−1).
Investigation of the enzymatic activity of FmtA by NMR.
WTAs were isolated from various strains (RN4220, ΔfmtA, complemented ΔfmtA, and CM) using trichloroacetic acid as described by Fridman et al. (91). LTA (S. aureus) was purchased from Sigma. Teichoic acids were lyophilized and resuspended in 100% D2O to a concentration of 5 to 10 mg/ml and were analyzed by NMR. One-dimensional (1D) 1H NMR spectra were collected at 25°C on a Bruker AV III 700 MHz spectrometer (operating frequencies of 700.28 MHz for 1H NMR and 176.096 MHz for 13C NMR). The spectrometer was controlled with TOPSPIN version 3.2 software and equipped with a 5-mm 1H/13C/15N cryoprobe. For quantitative 1D 1H spectra, 1D 1H T1 analysis was completed using the inversion recovery experiment. The enzymatic activity of FmtA toward teichoic acids was investigated using reaction mixtures containing 10 µM FmtA unless stated otherwise, various concentrations of WTA, 5 mg/ml LTA, and 10 mM sodium phosphate (pH 7.0). Residual solvent suppression was completed by excitation sculpting using the standard library pulse program. The progress of the reactions was monitored by recording the 1H-NMR spectra of the teichoic acids at various time intervals at 25°C.
To investigate the substrate specificity of FmtA by NMR, several esterase substrates, including p-NPB, p-NPA, and d-Ala-methyl ester, were dissolved in 300 µl of 10 mM sodium phosphate buffer (pH 7.0) (at a final concentration of 1 mM) and were incubated with FmtA (10 µM) and 300 µl of deuterium oxide. The p-NPA and p-NPB reactions were monitored by NMR for the appearance of peaks at 8.15 ppm and for the disappearance of peaks at 8.35 ppm at various time intervals at 25°C. The d-Ala methyl ester reactions were monitored for the appearance of peaks at 1.46 ppm and for the disappearance of peaks at 1.53 ppm.
SUPPLEMENTAL MATERIAL
Construction of S. aureus fmtA mutant strains. Download
Analysis of d-Ala removal from the tripeptide Nα,Nε-diacetyl-Lys-d-Ala-d-Ala (TP; 6 mM) by PBP2a (10 µM) (top) and PBP4 (10 µM) (bottom) in the absence or presence of WTA (0.1 µg/µl, 0.2 µg/µl, and 0.3 µg/µl). Free d-Ala was measured using a fluorescence-based coupled enzymatic assay. Error bars represent standard deviations obtained from three independent experiments. Download
Assessment of interactions of WTAs with FmtA (top) and PBP4 (bottom). FmtA (10 µM) or PBP4 (10 µM) was incubated with WTAs at concentrations varying from 0.5 to 20 µg/µl in 50 mM sodium phosphate buffer, pH 7.2. The samples were loaded onto a 15% native polyacrylamide gel and resolved by electrophoresis. Download
ACKNOWLEDGMENTS
We thank Chantal Bizet (Institut Pasteur, France) for providing the pMAD vector and Alexander Tomasz for providing the pMGP2 vector.
This research was supported by a grant from the Canadian Institutes of Health Research (Operating Grant 93530) and an Early Researcher Award from the Ministry of Economic Development and Innovation (Ontario, Canada) (ER09-06-134).
Footnotes
Citation Rahman MM, Hunter HN, Prova S, Verma V, Qamar A, Golemi-Kotra D. 2016. The Staphylococcus aureus methicillin resistance factor fmta is a d-amino esterase that acts on teichoic acids. mBio 7(1):e02070-15. doi:10.1128/mBio.02070-15.
REFERENCES
- 1.Lowy FD. 2003. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest 111:1265–1273. doi: 10.1172/JCI18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, Enright MC, Foster TJ, Moore CE, Hurst L, Atkin R, Barron A, Bason N, Bentley SD, Chillingworth C, Chillingworth T, Churcher C, Clark L, Corton C, Cronin A, Doggett J. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci U S A 101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fitzgerald JR, Sturdevant DE, Mackie SM, Gill SR, Musser JM. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc Natl Acad Sci U S A 98:8821–8826. doi: 10.1073/pnas.161098098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiramatsu K, Cui L, Kuroda M, Ito T. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol 9:486–493. doi: 10.1016/S0966-842X(01)02175-8. [DOI] [PubMed] [Google Scholar]
- 5.Bartley J. 2002. First case of VRSA identified in Michigan. Infect Control Hosp Epidemiol 23:480. [DOI] [PubMed] [Google Scholar]
- 6.Brown DFJ, Reynolds PE. 1980. Intrinsic resistance to beta-lactam antibiotics in Staphylococcus aureus. FEBS Lett 122:275–278. doi: 10.1016/0014-5793(80)80455-8. [DOI] [PubMed] [Google Scholar]
- 7.Bancroft EA. 2007. Antimicrobial resistance—it’s not just for hospitals. JAMA 298:1803–1804. [DOI] [PubMed] [Google Scholar]
- 8.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK, Active Bacterial Core surveillance (ABCs) MRSA Investigators . 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 9.Silver LL. 2003. Novel inhibitors of bacterial cell wall synthesis. Curr Opin Microbiol 6:431–438. doi: 10.1016/j.mib.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 10.David MZ, Daum RS. 2010. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh C. 2003. Where will new antibiotics come from? Nat Rev Microbiol 1:65–70. doi: 10.1038/nrmicro727. [DOI] [PubMed] [Google Scholar]
- 12.Bassetti M, Merelli M, Temperoni C, Astilean A. 2013. New antibiotics for bad bugs: where are we? Ann Clin Microbiol Antimicrob 12:22. doi: 10.1186/1476-0711-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swoboda JG, Meredith TC, Campbell J, Brown S, Suzuki T, Bollenbach T, Malhowski AJ, Kishony R, Gilmore MS, Walker S. 2009. Discovery of a small molecule that blocks wall teichoic acid biosynthesis in Staphylococcus aureus. ACS Chem Biol 4:875–883. doi: 10.1021/cb900151k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farha MA, Leung A, Sewell EW, D’Elia MA, Allison SE, Ejim L, Pereira PM, Pinho MG, Wright GD, Brown ED. 2013. Inhibition of WTA synthesis blocks the cooperative action of PBPs and sensitizes MRSA to beta-lactams. ACS Chem Biol 8:226–233. doi: 10.1021/cb300413m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell J, Singh AK, Santa Maria JP, Kim Y, Brown S, Swoboda JG, Mylonakis E, Wilkinson BJ, Walker S. 2011. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem Biol 6:106–116. doi: 10.1021/cb100269f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blake KL, O’Neill AJ, Mengin-Lecreulx D, Henderson PJ, Bostock JM, Dunsmore CJ, Simmons KJ, Fishwick CW, Leeds JA, Chopra I. 2009. The nature of Staphylococcus aureus MurA and MurZ and approaches for detection of peptidoglycan biosynthesis inhibitors. Mol Microbiol 72:335–343. doi: 10.1111/j.1365-2958.2009.06648.x. [DOI] [PubMed] [Google Scholar]
- 17.Gardete S, Ludovice AM, Sobral RG, Filipe SR, de Lencastre H, Tomasz A. 2004. Role of murE in the expression of beta-lactam antibiotic resistance in Staphylococcus aureus. J Bacteriol 186:1705–1713. doi: 10.1128/JB.186.6.1705-1713.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sobral RG, Ludovice AM, de Lencastre H, Tomasz A. 2006. Role of murF in cell wall biosynthesis: isolation and characterization of a murF conditional mutant of Staphylococcus aureus. J Bacteriol 188:2543–2553. doi: 10.1128/JB.188.7.2543-2553.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balibar CJ, Shen X, Tao J. 2009. The mevalonate pathway of Staphylococcus aureus. J Bacteriol 191:851–861. doi: 10.1128/JB.01357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komatsuzawa H, Sugai M, Ohta K, Fujiwara T, Nakashima S, Suzuki J, Lee CY, Suginaka H. 1997. Cloning and characterization of the fmt gene which affects the methicillin resistance level and autolysis in the presence of Triton X-100 in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 41:2355–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernal P, Lemaire S, Pinho MG, Mobashery S, Hinds J, Taylor PW. 2010. Insertion of epicatechin gallate into the cytoplasmic membrane of methicillin-resistant Staphylococcus aureus disrupts penicillin-binding protein (PBP) 2a-mediated beta-lactam resistance by delocalizing PBP2. J Biol Chem 285:24055–24065. doi: 10.1074/jbc.M110.114793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McAleese F, Wu SW, Sieradzki K, Dunman P, Murphy E, Projan S, Tomasz A. 2006. Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate-S. aureus-type resistance to vancomycin. J Bacteriol 188:1120–1133. doi: 10.1128/JB.188.3.1120-1133.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCallum N, Spehar G, Bischoff M, Berger-Bächi B. 2006. Strain dependence of the cell wall-damage induced stimulon in Staphylococcus aureus. Biochim Biophys Acta 1760:1475–1481. doi: 10.1016/j.bbagen.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 24.Utaida S, Dunman PM, Macapagal D, Murphy E, Projan SJ, Singh VK, Jayaswal RK, Wilkinson BJ. 2003. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149:2719–2732. doi: 10.1099/mic.0.26426-0. [DOI] [PubMed] [Google Scholar]
- 25.Fan X, Liu Y, Smith D, Konermann L, Siu KW, Golemi-Kotra D. 2007. Diversity of penicillin-binding proteins. Resistance factor FmtA of Staphylococcus aureus. J Biol Chem 282:35143–35152. doi: 10.1074/jbc.M706296200. [DOI] [PubMed] [Google Scholar]
- 26.Qamar A, Golemi-Kotra D. 2012. Dual roles of FmtA in Staphylococcus aureus cell wall biosynthesis and autolysis. Antimicrob Agents Chemother 56:3797–3805. doi: 10.1128/AAC.00187-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wada A, Watanabe H. 1998. Penicillin-binding protein 1 of Staphylococcus aureus is essential for growth. J Bacteriol 180:2759–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Łeski TA, Tomasz A. 2005. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J Bacteriol 187:1815–1824. doi: 10.1128/JB.187.5.1815-1824.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlag M, Biswas R, Krismer B, Kohler T, Zoll S, Yu W, Schwarz H, Peschel A, Götz F. 2010. Role of Staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol Microbiol 75:864–873. [DOI] [PubMed] [Google Scholar]
- 30.Atilano ML, Pereira PM, Yates J, Reed P, Veiga H, Pinho MG, Filipe SR. 2010. Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus. Proc Natl Acad Sci U S A 107:18991–18996. doi: 10.1073/pnas.1004304107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown S, Santa Maria JP Jr., Walker S. 2013. Wall teichoic acids of Gram-positive bacteria. Annu Rev Microbiol 67:313–336. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Percy MG, Gründling A. 2014. Lipoteichoic acid synthesis and function in Gram-positive bacteria. Annu Rev Microbiol 68:81–100. doi: 10.1146/annurev-micro-091213-112949. [DOI] [PubMed] [Google Scholar]
- 33.Reichmann NT, Gründling A. 2011. Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes. FEMS Microbiol Lett 319:97–105. doi: 10.1111/j.1574-6968.2011.02260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swoboda JG, Campbell J, Meredith TC, Walker S. 2010. Wall teichoic acid function, biosynthesis, and inhibition. Chembiochem 11:35–45. doi: 10.1002/cbic.200900557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koch HU, Haas R, Fischer W. 1984. The role of lipoteichoic acid biosynthesis in membrane lipid metabolism of growing Staphylococcus aureus. Eur J Biochem 138:357–363. doi: 10.1111/j.1432-1033.1984.tb07923.x. [DOI] [PubMed] [Google Scholar]
- 36.Allison SE, D’Elia MA, Arar S, Monteiro MA, Brown ED. 2011. Studies of the genetics, function, and kinetic mechanism of TagE, the wall teichoic acid glycosyltransferase in Bacillus subtilis 168. J Biol Chem 286:23708–23716. doi: 10.1074/jbc.M111.241265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown S, Xia G, Luhachack LG, Campbell J, Meredith TC, Chen C, Winstel V, Gekeler C, Irazoqui JE, Peschel A, Walker S. 2012. Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc Natl Acad Sci U S A 109:18909–18914. doi: 10.1073/pnas.1209126109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perego M, Glaser P, Minutello A, Strauch MA, Leopold K, Fischer W. 1995. Incorporation of d-alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis. Identification of genes and regulation. J Biol Chem 270:15598–15606. doi: 10.1074/jbc.270.26.15598. [DOI] [PubMed] [Google Scholar]
- 39.Neuhaus FC, Baddiley J. 2003. A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in Gram-positive bacteria. Microbiol Mol Biol Rev 67:686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kovács M, Halfmann A, Fedtke I, Heintz M, Peschel A, Vollmer W, Hakenbeck R, Brückner R. 2006. A functional dlt operon, encoding proteins required for incorporation of d-alanine in teichoic acids in gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J Bacteriol 188:5797–5805. doi: 10.1128/JB.00336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reichmann NT, Piçarra Cassona C, Monteiro JM, Bottomley AL, Corrigan RM, Foster SJ, Pinho MG, Gründling A. 2014. Differential localization of LTA synthesis proteins and their interaction with the cell division machinery in Staphylococcus aureus. Mol Microbiol 92:273–286. doi: 10.1111/mmi.12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chatterjee AN. 1969. Use of bacteriophage-resistant mutants to study the nature of the bacteriophage receptor site of Staphylococcus aureus. J Bacteriol 98:519–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peschel A, Vuong C, Otto M, Götz F. 2000. The d-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob Agents Chemother 44:2845–2847. doi: 10.1128/AAC.44.10.2845-2847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collins LV, Kristian SA, Weidenmaier C, Faigle M, Van Kessel KP, Van Strijp JA, Götz F, Neumeister B, Peschel A. 2002. Staphylococcus aureus strains lacking d-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J Infect Dis 186:214–219. doi: 10.1086/341454. [DOI] [PubMed] [Google Scholar]
- 45.Weidenmaier C, Kokai-Kun JF, Kristian SA, Chanturiya T, Kalbacher H, Gross M, Nicholson G, Neumeister B, Mond JJ, Peschel A. 2004. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat Med 10:243–245. doi: 10.1038/nm991. [DOI] [PubMed] [Google Scholar]
- 46.Park KH, Kurokawa K, Zheng L, Jung DJ, Tateishi K, Jin JO, Ha NC, Kang HJ, Matsushita M, Kwak JY, Takahashi K, Lee BL. 2010. Human serum mannose-binding lectin senses wall teichoic acid glycopolymer of Staphylococcus aureus, which is restricted in infancy. J Biol Chem 285:27167–27175. doi: 10.1074/jbc.M110.141309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abachin E, Poyart C, Pellegrini E, Milohanic E, Fiedler F, Berche P, Trieu-Cuot P. 2002. Formation of d-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol Microbiol 43:1–14. doi: 10.1046/j.1365-2958.2002.02723.x. [DOI] [PubMed] [Google Scholar]
- 48.Gross M, Cramton SE, Götz F, Peschel A. 2001. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect Immun 69:3423–3426. doi: 10.1128/IAI.69.5.3423-3426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fabretti F, Theilacker C, Baldassarri L, Kaczynski Z, Kropec A, Holst O, Huebner J. 2006. Alanine esters of enterococcal lipoteichoic acid play a role in biofilm formation and resistance to antimicrobial peptides. Infect Immun 74:4164–4171. doi: 10.1128/IAI.00111-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steen A, Palumbo E, Deghorain M, Cocconcelli PS, Delcour J, Kuipers OP, Kok J, Buist G, Hols P. 2005. Autolysis of Lactococcus lactis is increased upon d-alanine depletion of peptidoglycan and lipoteichoic acids. J Bacteriol 187:114–124. doi: 10.1128/JB.187.1.114-124.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Götz F. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem 274:8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 52.Winstel V, Liang C, Sanchez-Carballo P, Steglich M, Munar M, Bröker BM, Penadés JR, Nübel U, Holst O, Dandekar T, Peschel A, Xia G. 2013. Wall teichoic acid structure governs horizontal gene transfer between major bacterial pathogens. Nat Commun 4:2345. doi: 10.1038/ncomms3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Childs WC III, Taron DJ, Neuhaus FC. 1985. Biosynthesis of d-alanyl-lipoteichoic acid by Lactobacillus casei: interchain transacylation of d-alanyl ester residues. J Bacteriol 162:1191–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koprivnjak T, Mlakar V, Swanson L, Fournier B, Peschel A, Weiss JP. 2006. Cation-induced transcriptional regulation of the dlt operon of Staphylococcus aureus. J Bacteriol 188:3622–3630. doi: 10.1128/JB.188.10.3622-3630.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koch HU, Döker R, Fischer W. 1985. Maintenance of d-alanine ester substitution of lipoteichoic acid by reesterification in Staphylococcus aureus. J Bacteriol 164:1211–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.MacArthur AE, Archibald AR. 1984. Effect of culture pH on the d-alanine ester content of lipoteichoic acid in Staphylococcus aureus. J Bacteriol 160:792–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fischer W. 1988. Physiology of lipoteichoic acids in bacteria. Adv Microb Physiol 29:233–302. doi: 10.1016/S0065-2911(08)60349-5. [DOI] [PubMed] [Google Scholar]
- 58.Pinho MG, de Lencastre H, Tomasz A. 2001. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc Natl Acad Sci U S A 98:10886–10891. doi: 10.1073/pnas.191260798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pinho MG, Filipe SR, de Lencastre H, Tomasz A. 2001. Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP2) by the drug resistance protein PBP2A in Staphylococcus aureus. J Bacteriol 183:6525–6531. doi: 10.1128/JB.183.22.6525-6531.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qiao Y, Lebar MD, Schirner K, Schaefer K, Tsukamoto H, Kahne D, Walker S. 2014. Detection of lipid-linked peptidoglycan precursors by exploiting an unexpected transpeptidase reaction. J Am Chem Soc 136:14678–14681. doi: 10.1021/ja508147s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chahinian H, Sarda L. 2009. Distinction between esterases and lipases: comparative biochemical properties of sequence-related carboxylesterases. Protein Pept Lett 16:1149–1161. doi: 10.2174/092986609789071333. [DOI] [PubMed] [Google Scholar]
- 62.Arnaud M, Chastanet A, Débarbouillé M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, Gram-positive bacteria. Appl Environ Microbiol 70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sullivan MA, Yasbin RE, Young FE. 1984. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene 29:21–26. doi: 10.1016/0378-1119(84)90161-6. [DOI] [PubMed] [Google Scholar]
- 64.Haas R, Koch HU, Fischer W. 1984. Alanyl turnover from lipoteichoic acid to teichoic-acid in Staphylococcus aureus. FEMS Microbiol Lett 21:27–31. doi: 10.1111/j.1574-6968.1984.tb00180.x. [DOI] [Google Scholar]
- 65.Arpigny JL, Jaeger KE. 1999. Bacterial lipolytic enzymes: classification and properties. Biochem J 343:177–183. doi: 10.1042/bj3430177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wagner UG, Petersen EI, Schwab H, Kratky C. 2002. EstB from Burkholderia gladioli: a novel esterase with a beta-lactamase fold reveals steric factors to discriminate between esterolytic and beta-lactam cleaving activity. Protein Sci 11:467–478. doi: 10.1110/ps.33002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Asano Y, Ito H, Dairi T, Kato Y. 1996. An alkaline d-stereospecific endopeptidase with beta-lactamase activity from Bacillus cereus. J Biol Chem 271:30256–30262. doi: 10.1074/jbc.271.47.30256. [DOI] [PubMed] [Google Scholar]
- 68.Bourne DG, Riddles P, Jones GJ, Smith W, Blakeley RL. 2001. Characterisation of a gene cluster involved in bacterial degradation of the cyanobacterial toxin microcystin LR. Environ Toxicol 16:523–534. doi: 10.1002/tox.10013. [DOI] [PubMed] [Google Scholar]
- 69.Delmarcelle M, Boursoit MC, Filée P, Baurin SL, Frère JM, Joris B. 2005. Specificity inversion of Ochrobactrum anthropi d-aminopeptidase to a d,d-carboxypeptidase with new penicillin binding activity by directed mutagenesis. Protein Sci 14:2296–2303. doi: 10.1110/ps.051475305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Komeda H, Asano Y. 2000. Gene cloning, nucleotide sequencing, and purification and characterization of the d-stereospecific amino-acid amidase from Ochrobactrum anthropi SV3. Eur J Biochem 267:2028–2035. doi: 10.1046/j.1432-1327.2000.01208.x. [DOI] [PubMed] [Google Scholar]
- 71.Okazaki S, Suzuki A, Komeda H, Yamaguchi S, Asano Y, Yamane T. 2007. Crystal structure and functional characterization of a d-stereospecific amino acid amidase from Ochrobactrum anthropi SV3, a new member of the penicillin-recognizing proteins. J Mol Biol 368:79–91. doi: 10.1016/j.jmb.2006.10.070. [DOI] [PubMed] [Google Scholar]
- 72.Asano Y, Nakazawa A, Kato Y, Kondo K. 1989. Properties of a novel d-stereospecific aminopeptidase from Ochrobactrum anthropi. J Biol Chem 264:14233–14239. [PubMed] [Google Scholar]
- 73.Fanuel L, Thamm I, Kostanjevecki V, Samyn B, Joris B, Goffin C, Brannigan J, Van Beeumen J, Frère JM. 1999. Two new aminopeptidases from Ochrobactrum anthropi active on d-alanyl-p-nitroanilide. Cell Mol Life Sci 55:812–818. doi: 10.1007/s000180050334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Petersen EI, Valinger G, Sölkner B, Stubenrauch G, Schwab H. 2001. A novel esterase from Burkholderia gladioli which shows high deacetylation activity on cephalosporins is related to beta-lactamases and DD-peptidases. J Biotechnol 89:11–25. doi: 10.1016/S0168-1656(01)00284-X. [DOI] [PubMed] [Google Scholar]
- 75.Joris B, Ghuysen JM, Dive G, Renard A, Dideberg O, Charlier P, Frère JM, Kelly JA, Boyington JC, Moews PC. 1988. The active-site-serine penicillin-recognizing enzymes as members of the streptomyces R61 DD-peptidase family. Biochem J 250:313–324. doi: 10.1042/bj2500313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frère J, Joris B, Dideberg O, Charlier P, Ghuysen J. 1988. Penicillin-recognizing enzymes. Biochem Soc Trans 16:934–938. doi: 10.1042/bst0160934. [DOI] [PubMed] [Google Scholar]
- 77.Cougnoux A, Gibold L, Robin F, Dubois D, Pradel N, Darfeuille-Michaud A, Dalmasso G, Delmas J, Bonnet R. 2012. Analysis of structure-function relationships in the colibactin-maturating enzyme ClbP. J Mol Biol 424:203–214. doi: 10.1016/j.jmb.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 78.Komatsuzawa H, Ohta K, Labischinski H, Sugai M, Suginaka H. 1999. Characterization of fmtA, a gene that modulates the expression of methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 43:2121–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao Y, Verma V, Belcheva A, Singh A, Fridman M, Golemi-Kotra D. 2012. Staphylococcus aureus methicillin-resistance factor fmtA is regulated by the global regulator SarA. PLoS One 7:e43998. doi: 10.1371/journal.pone.0043998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wolz C, Pöhlmann-Dietze P, Steinhuber A, Chien YT, Manna A, van Wamel W, Cheung A. 2000. Agr-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in Staphylococcus aureus. Mol Microbiol 36:230–243. doi: 10.1046/j.1365-2958.2000.01853.x. [DOI] [PubMed] [Google Scholar]
- 81.Cheung AL, Bayer AS, Zhang G, Gresham H, Xiong YQ. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol Med Microbiol 40:1–9. doi: 10.1016/S0928-8244(03)00309-2. [DOI] [PubMed] [Google Scholar]
- 82.Trotonda MP, Xiong YQ, Memmi G, Bayer AS, Cheung AL. 2009. Role of mgrA and sarA in methicillin-resistant Staphylococcus aureus autolysis and resistance to cell wall-active antibiotics. J Infect Dis 199:209–218. doi: 10.1086/595740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boles BR, Thoendel M, Roth AJ, Horswill AR. 2010. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One 5:e10146. doi: 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tu Quoc PH, Genevaux P, Pajunen M, Savilahti H, Georgopoulos C, Schrenzel J, Kelley WL. 2007. Isolation and characterization of biofilm formation-defective mutants of Staphylococcus aureus Infect Immun 75:1079–1088. doi: 10.1128/IAI.01143-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bayles KW. 2007. The biological role of death and lysis in biofilm development. Nat Rev Microbiol 5:721–726. doi: 10.1038/nrmicro1743. [DOI] [PubMed] [Google Scholar]
- 86.Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. 2009. Modulation of eDNA release and degradation affects Staphylococcus biofilm maturation. PLoS One 4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reichmann NT, Cassona CP, Gründling A. 2013. Revised mechanism of d-alanine incorporation into cell wall polymers in Gram-positive bacteria. Microbioly 159:1868–1877. doi: 10.1099/mic.0.069898-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xia G, Kohler T, Peschel A. 2010. The wall teichoic acid and lipoteichoic caid polymers of Staphylococcus aureus Int J Med Microbiol 300:148–154. doi: 10.1016/j.ijmm.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 89.Biswas R, Martinez RE, Göhring N, Schlag M, Josten M, Xia G, Hegler F, Gekeler C, Gleske AK, Götz F, Sahl HG, Kappler A, Peschel A. 2012. Proton-binding capacity of Staphylococcus aureus wall teichoic acid and its role in controlling autolysin activity. PLoS One 7:e41415. doi: 10.1371/journal.pone.0041415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gutheil WG, Stefanova ME, Nicholas RA. 2000. Fluorescent coupled enzyme assays for d-alanine: application to penicillin-binding protein and vancomycin activity assays. Anal Biochem 287:196–202. doi: 10.1006/abio.2000.4835. [DOI] [PubMed] [Google Scholar]
- 91.Fridman M, Williams GD, Muzamal U, Hunter H, Siu KW, Golemi-Kotra D. 2013. Two unique phosphorylation-driven signaling pathways crosstalk in Staphylococcus aureus to modulate the cell-wall charge: Stk1/Stp1 meets GraSR. Biochemistry 52:7975–7986. doi: 10.1021/bi401177n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Construction of S. aureus fmtA mutant strains. Download
Analysis of d-Ala removal from the tripeptide Nα,Nε-diacetyl-Lys-d-Ala-d-Ala (TP; 6 mM) by PBP2a (10 µM) (top) and PBP4 (10 µM) (bottom) in the absence or presence of WTA (0.1 µg/µl, 0.2 µg/µl, and 0.3 µg/µl). Free d-Ala was measured using a fluorescence-based coupled enzymatic assay. Error bars represent standard deviations obtained from three independent experiments. Download
Assessment of interactions of WTAs with FmtA (top) and PBP4 (bottom). FmtA (10 µM) or PBP4 (10 µM) was incubated with WTAs at concentrations varying from 0.5 to 20 µg/µl in 50 mM sodium phosphate buffer, pH 7.2. The samples were loaded onto a 15% native polyacrylamide gel and resolved by electrophoresis. Download