ABSTRACT
The genomes of many photosynthetic and nonphotosynthetic bacteria encode numerous phytochrome superfamily photoreceptors whose functions and interactions are largely unknown. Cyanobacterial genomes encode particularly large numbers of phytochrome superfamily members called cyanobacteriochromes. These have diverse light color-sensing abilities, and their functions and interactions are just beginning to be understood. One of the best characterized of these functions is the regulation of photosynthetic light-harvesting antenna composition in the cyanobacterium Fremyella diplosiphon by the cyanobacteriochrome RcaE in response to red and green light, a process known as chromatic acclimation. We have identified a new cyanobacteriochrome named DpxA that maximally senses teal (absorption maximum, 494 nm) and yellow (absorption maximum, 568 nm) light and represses the accumulation of a key light-harvesting protein called phycoerythrin, which is also regulated by RcaE during chromatic acclimation. Like RcaE, DpxA is a two-component system kinase, although these two photoreceptors can influence phycoerythrin expression through different signaling pathways. The peak responsiveness of DpxA to teal and yellow light provides highly refined color discrimination in the green spectral region, which provides important wavelengths for photosynthetic light harvesting in cyanobacteria. These results redefine chromatic acclimation in cyanobacteria and demonstrate that cyanobacteriochromes can coordinately impart sophisticated light color sensing across the visible spectrum to regulate important photosynthetic acclimation processes.
IMPORTANCE
The large number of cyanobacteriochrome photoreceptors encoded by cyanobacterial genomes suggests that these organisms are capable of extremely complex light color sensing and responsiveness, yet little is known about their functions and interactions. Our work uncovers previously undescribed cooperation between two photoreceptors with very different light color-sensing capabilities that coregulate an important photosynthetic light-harvesting protein in response to teal, green, yellow, and red light. Other cyanobacteriochromes that have been shown to interact functionally sense wavelengths of light that are close to each other, which makes it difficult to clearly identify their physiological roles in the cell. Our finding of two photoreceptors with broad light color-sensing capabilities and clearly defined physiological roles provides new insights into complex light color sensing and its regulation.
INTRODUCTION
The phytochrome superfamily is an important group of photoreceptors whose members are widely distributed in photosynthetic and nonphotosynthetic prokaryotes and eukaryotes (1–3). These proteins were first identified and characterized in land plants, where they are known to control a number of light-dependent processes (1, 4–7). Phytochrome-class proteins are dual color light switches that contain one or more light-sensing bilin chromophores, each covalently attached to the protein at conserved cysteine residues. Every bilin chromophore found in phytochromes can exist in one of two forms, each of which maximally absorbs a different wavelength of light. The structural change from one form to the other is most effectively driven by the chromophore’s absorption of a photon of light. Switching between forms alters the structure of its attached protein in a unique way (8), leading to changes in how specific physiological processes are regulated.
Although all known phytochromes in plants and nonphotosynthetic bacteria isomerize between red light- and far-red light-absorbing forms (3, 4, 6, 7), phytochrome-like photoreceptors in large numbers of other species sense many additional wavelengths of light. The numbers, diversity, and range of light color-sensing capabilities of phytochrome superfamily members are the greatest in cyanobacteria and algae, with the filamentous cyanobacterium Fremyella diplosiphon containing as many as 27 different family members (9–11). In cyanobacteria, this variety is primarily due to the presence of a large number of cyanobacteriochromes, a subgroup within the phytochrome superfamily that uses only a GAF (cGMP phosphodiesterase/adenylate cyclase/FhlA) domain for light color sensing (12). A major unanswered question is how cyanobacteria actually use this capacity to respond to so many different light colors and how cross talk between photoreceptors is controlled.
A well-studied response that is regulated by a cyanobacteriochrome is type III chromatic acclimation (CA3), a process which involves changes in the composition of the photosynthetic light-harvesting antennae and is controlled by a cyanobacteriochrome called RcaE (13–15). In the CA3 model organism F. diplosiphon, RcaE directs a signal transduction pathway that in red light represses synthesis of the red-colored protein phycoerythrin and activates production of the blue-green-colored protein phycocyanin. In green light, RcaE does not repress phycoerythrin production or activate phycocyanin synthesis, resulting in an increased phycoerythrin-to-phycocyanin ratio. Consequently, because phycoerythrin and phycocyanin are major components of the photosynthetic light-harvesting antennae, which are called phycobilisomes, these changes result in a dramatic, reversible change in cell color phenotype, from blue-green in red light to brick-red in green light (16). Since phycoerythrin best absorbs green light and phycocyanin most effectively absorbs red light, CA3 provides a fitness advantage in changing light color environments by increasing the efficiency of photon capture for photosynthesis (17). RcaE is also a histidine kinase, switching between the kinase-active green-absorbing form (RcaEg) in red light and the kinase-inactive red-absorbing form (RcaEr) in green light (15). In red light, RcaEg phosphorylates a complex two-component system comprised of the single domain response regulator RcaF and complex response regulator/transcription factor RcaC. This represses transcription of cpeCDESTR (here cpeC) and cpeBA, operons required to produce phycoerythrin-containing phycobilisomes, and activates transcription of the phycocyanin-encoding operon cpcB2A2 (18, 19). There are only single copies of cpeC and cpeBA in the F. diplosiphon genome.
Here, we demonstrate that a previously uncharacterized cyanobacteriochrome named DpxA provides an additional level of photoreceptor-based regulation of phycoerythrin expression. DpxA maximally responds to teal and yellow light and regulates phycoerythrin abundance at least in part through a signal transduction pathway that is independent of the Rca system. Our data indicate that DpxA represses accumulation of phycoerythrin in yellow light and does not repress it in blue light. The absorption maxima of DpxA precisely flank the absorption maximum of RcaEg, allowing F. diplosiphon to use a second cyanobacteriochrome to fine-tune the RcaE regulation of phycoerythrin levels in the blue-to-yellow region of the light spectrum. These findings reveal how multiple cyanobacteriochromes are used by cyanobacteria to provide sophisticated light color sensing for the control of photosynthetic light-harvesting gene expression across the visible spectrum.
RESULTS
DpxA regulates phycoerythrin abundance.
The F. diplosiphon genome is predicted to encode 27 phytochrome superfamily members (11), and most have no known function. To identify the functions of the uncharacterized cyanobacteriochromes in this organism, we created deletions of the genes encoding these putative proteins and compared the phenotypes of the mutants to that of wild-type cells. One of these deletions affected the color phenotype of cells when grown in natural-spectrum white light, with the mutant appearing black rather than the normal green color of the wild type (Fig. 1A). Whole-cell absorption spectra revealed that when normalized for chlorophyll absorbance, more green light (566 nm) was absorbed by this mutant than by wild-type cells, suggesting that phycoerythrin levels were higher in the mutant than in the wild type (Fig. 1A). Small but reproducible differences between mutant and wild-type cells in the absorption of long-wavelength light suggest that DpxA regulation of cell pigmentation may extend beyond control of phycoerythrin (Fig. 1A). Western blot analyses of total proteins extracted from mutant and wild-type cells using antibodies specific for the β subunits of phycoerythrin and phycocyanin revealed an approximately 2-fold-higher phycoerythrin level in the mutant than in the wild type but no difference in phycocyanin abundance (Fig. 1B). Therefore, the black pigmentation of the mutant cells, relative to the wild type, was due to elevated phycoerythrin levels. The disrupted gene was named dpxA because DpxA causes decreased phycoerythrin expression. DpxA has a calculated molecular mass of 54 kDa and is predicted to contain one GAF and one histidine kinase domain (see Fig. S1A in the supplemental material). Its location in the genome suggests that dpxA is part of an operon (see Fig. S1B). Two results indicate that the elevated phycoerythrin (PE) phenotype in the dpxA mutant is due to the loss of DpxA. First, wild-type dpxA provided in trans nearly completely complemented the mutant phenotype, demonstrating that the mutant phenotype was due to the loss of DpxA (see Fig. S2A and B). In addition, individual deletions of the genes downstream of dpxA had no effect on PE levels (see Fig. S2C). Thus, the dpxA mutant phenotype is due to the loss of DpxA.
FIG 1 .

Phenotype of a ΔdpxA mutant. (A) Whole-cell absorbance scans of wild-type (WT) and ΔdpxA cells after growth in natural-spectrum white light. Phycoerythrin (PE) and phycocyanin (PC) peaks are labeled. Spectra were normalized to the chlorophyll absorption peaks at both 440 nm and 680 nm, and each scan is an average from six independent experiments. Inset: photograph of wild-type and ΔdpxA cells after growth in natural-spectrum white light. (B) Western blot analysis using the soluble fraction of cell lysates of wild-type and ΔdpxA cells after growth in natural-spectrum white light. Mean values (below) and representative blots (above) are shown for PE (left) and PC (right). For each replicate, the same cell lysate was used for the antiphycoerythrin and antiphycocyanin assay blots. Values provided are normalized by the protein content value, quantified from the stained protein gel, from three independent protein extractions and Western blot replicates. ΔdpxA mutant values were set to 100%. Standard errors of the means are shown.
DpxA spectral qualities and kinase activity in vitro.
The DpxA GAF domain contains residues characteristic of bilin-binding photoreceptors, including a DXCF motif present in cyanobacteriochromes that sense the blue-to-orange region of the spectrum (20). The DpxA GAF domain sequence is similar to a DXCF-containing GAF domain from Nostoc punctiforme NpR5113g1 (see Fig. S1C in the supplemental material), which maximally absorbs teal and yellow-green light (10). Purified, full-length DpxA produced in phycocyanobilin-containing Escherichia coli fully photoconverted between the yellow-absorbing DpxAy form (absorbance maximum [λmax], 568 nm) in blue light and the teal-absorbing DpxAt form (λmax, 494 nm) in yellow light (Fig. 2A).
FIG 2 .

Light absorption and kinase activity of DpxA in response to yellow and blue light. (A) Difference spectrum of purified DpxA between protein exposed to yellow (λmax, 579 nm) minus blue light (λmax, 466 nm). Insets: photographs of purified protein exposed to yellow (top) and blue (bottom) light. (B) Autophosphorylation of purified DpxA preirradiated to the DpxAy (yellow line) or DpxAt (blue line) forms. The DpxAt 32P incorporation value at 20 min was set to 1.0. Each point is the average from three independent experiments, and standard errors of the means are shown. Inset: representative image of one experiment. Images shown are from the same gel.
Some cyanobacteriochromes are light-dependent kinases, and we predicted that the C-terminal domain of DpxA would show autokinase activity that would be regulated by yellow and teal light. Photoconversion of the full-length protein to DpxAt at the start of the kinase assay resulted in approximately 3-fold-higher in vitro phosphorylation than when converted into the DpxAy form (Fig. 2B). Thus, the kinase-active form of DpxA is DpxAt, which is produced by irradiation with yellow light. We then investigated if DpxA rapidly reverted to DpxAy or DpxAt in the dark after blue and yellow light treatments. When DpxA was converted into DpxAy, there was virtually no dark reversion to the DpxAt form over 22 h at 22°C, although some bleaching or degradation was apparent (see Fig. S3A in the supplemental material). There was slow and limited reversion to the DpxAy form over 22 h after DpxA was converted to DpxAt (see Fig. S3B). Therefore, DpxA does not undergo rapid dark reversion to either DpxAy or DpxAt, although the slow reversion from DpxAt to DpxAy suggests that DpxAy is the slightly more thermostable form of the protein.
Light-regulated effects of DpxA in vivo.
To determine the influence of the light-absorbing state of DpxA on phycoerythrin expression in F. diplosiphon cells, we grew cultures under light conditions which converted this photoreceptor to various ratios of DpxAy to DpxAt. Spectral scans of cells grown in blue light demonstrated that wild-type cells containing the DpxAy form accumulated phycoerythrin to the same level as did mutant cells lacking DpxA (Fig. 3A). In green light, the intermediate ratio of DpxAy to DpxAt resulted in a difference in phycoerythrin accumulation between the wild type and the mutant that was greater than that in blue light (Fig. 3B), while wild-type cells grown in yellow light and containing the DpxAt form accumulated far less phycoerythrin than did mutant cells lacking DpxA (Fig. 3C). Therefore, DpxAt must be the active form of this photoreceptor and increasingly repress phycoerythrin accumulation as the cells are shifted from blue to yellow light, while DpxAy is the inactive form and lacks the ability to repress phycoerythrin expression when cells are grown in blue light. The in vivo results are consistent with those of the autokinase assay (Fig. 2B), in which DpxAt was also the active form. Interestingly, DpxA most strongly repressed phycoerythrin accumulation in yellow light, a color in which the photoreceptor RcaE does not repress phycoerythrin accumulation. Also, DpxA had very little effect on phycoerythrin accumulation when the cells were grown in red light (Fig. 3D), although it directly or indirectly affected pigmentation in the 480- to 520-nm range.
FIG 3 .
Effect of DpxA on phycoerythrin levels across the visible spectrum. Whole-cell absorbance scans of wild-type and ΔdpxA cells grown in blue (λmax, 466 nm) (A), green (λmax, 520 nm) (B), yellow (λmax, 579 nm) (C), or red (λmax, 645 nm) (D) light at 12 µmol photons m−2 s−1. Cell cultures were scanned in late logarithmic growth phase, and each scan is an average from six independent experiments. Spectra were normalized to the chlorophyll absorption peaks at both 440 nm and 680 nm. PC, phycocyanin.
Integration of DpxA and RcaE sensing.
Together, the photobiological properties of DpxA and RcaE provide color-sensing capability across the visible spectrum (Fig. 4). In blue light (466 nm), purified photoreceptors were in the DpxAy and RcaEr forms. Both of these are the kinase-inactive forms of the proteins. Exposing the photoreceptors to the longer wavelengths of green (520 nm) and yellow (579 nm) light gradually shifted DpxA from DpxAy to DpxAt while RcaE remained in its RcaEr form. Treatment with the longer wavelengths of amber (594 nm) and orange (610 nm) light changed RcaEr to RcaEg while not affecting DpxAt. In red light (645 nm), RcaE was completely converted to RcaEg, whereas DpxAt and DpxAy were present at approximately equivalent levels. This was observed after a 4-min red light treatment, regardless of the state of DpxA prior to the treatment. The DpxA equilibrium that existed after red light treatment was similar to the equilibrium observed for green light-treated DpxA. One possible explanation for this is that both red and green light-emitting diodes (LEDs) emit wavelengths at DpxA’s isosbestic point (Fig. 2A). Interestingly, when the light absorption profiles of all of the forms of DpxA and RcaE were plotted on the same graph (Fig. 5), the separation of the absorption maxima of DpxAt and DpxAy was determined to be only 72 nm, and these maxima evenly straddled the relatively broad absorption maximum of RcaEg (Fig. 5) (15). In addition, the absorption maxima for RcaEg and RcaEr were separated by 129 nm, nearly double that of the two forms of DpxA (Fig. 5).
FIG 4 .

DpxA and RcaE photostationary states after light treatments across the visible spectrum. Absorbance scans of purified full-length DpxA (dashed lines) and the RcaE GAF domain (solid lines) after treatment with light of the wavelength indicated. The colored vertical lines indicate the peak wavelengths of light to which the proteins were exposed. Insets: photographs of purified DpxA (left) and RcaE (right) in the light color specified.
FIG 5 .
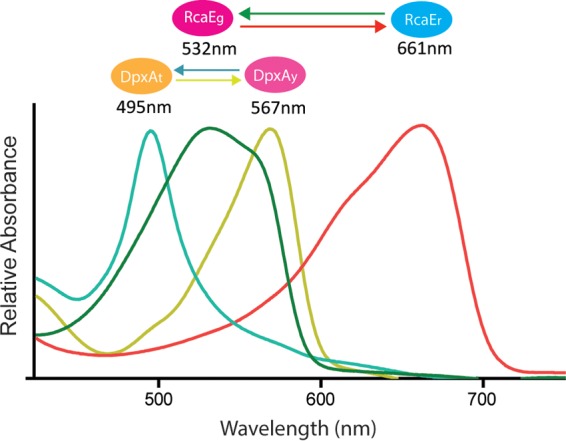
Absorbance spectra of DpxAt (teal line), DpxAy (yellow line), RcaEg (green line), and RcaEr (red line). Each form is indicated by a colored oval above its peak absorption wavelength, which is provided below the oval. The regions of the spectrum maximally absorbed by DpxAy and RcaEr are the wavelengths where phycoerythrin is repressed by DpxA and RcaE, respectively.
We investigated whether the effect of DpxA on phycoerythrin accumulation was through a signal transduction pathway that was independent of the Rca system. Although dpxA is adjacent to a gene that encodes a putative response regulator of a two-component system in the F. diplosiphon genome (see Fig. S1 in the supplemental material), it remained possible that DpxA’s influence on phycoerythrin abundance was exclusively through RcaF and/or RcaC, the response regulators that are necessary for repression of phycoerythrin during CA3. Because RcaC is the final component within the Rca pathway (21), we compared phycoerythrin levels in ΔrcaC and ΔrcaC ΔdpxA mutants grown in natural-spectrum white light and found that DpxA influence on phycoerythrin accumulation was not dependent on RcaC (see Fig. S4). This demonstrates that there must be a signal transduction pathway through which DpxA regulates phycoerythrin expression that is independent of the Rca system.
DISCUSSION
The capacity of DpxA and RcaE to respond to different regions of the light spectrum allows F. diplosiphon to actively regulate the abundance of phycoerythrin in almost all light colors and provides a new, previously unrecognized degree of sophistication in the regulation of photosynthetic gene expression by light color (Fig. 5). The influence of the DpxA and RcaE systems is evident across the visible spectrum. In red light, RcaE is in its active form and the influence of the Rca system is strong, activating phycocyanin expression and repressing phycoerythrin expression. The effect of DpxA in red light is minor (Fig. 3D), perhaps because DpxA is an approximately equimolar mixture of active and inactive forms in this light color (Fig. 4). Shifting cells to yellow light, the DpxA system is the predominant means of repressing phycoerythrin accumulation, as RcaE has largely converted into its inactive form (Fig. 4). In the blue region of the spectrum, both RcaE and DpxA are in their inactive states and the repression of phycoerythrin abundance by these two photoreceptors is at its minimum (Fig. 4 and 5). This may be a mechanism through which the cells fully commit to phycoerythrin accumulation only when the wavelengths of light between red and green are no longer available for photosynthesis. The possible existence of a color-sensing mechanism capable of fine-tuning light-harvesting biogenesis in the green region of the spectrum such as the one that we describe here has been previously suggested but not identified until now (10).
DpxA responds to a narrower range of wavelengths than the well-characterized cyanobacteriochrome RcaE (15) (Fig. 5), and although both repress phycoerythrin, DpxA does not activate phycocyanin in wild-type cells (Fig. 1B). Thus, DpxA functions as a fine-tuning modifier of phycobilisome biogenesis, affecting phycoerythrin accumulation in response to a subset of light colors within the green range, a spectral region in which RcaEg absorbs broadly and thus is not useful for fine color discrimination and where phycocyanin regulation is less relevant. Cyanobacteriochrome family members in other cyanobacterial species vary in the range of wavelengths to which they respond (10). Thus, the integration of the DpxA and RcaE color-sensing systems in the control of chromatic acclimation may be an indication of similar systems yet to be discovered in other organisms.
Other regulatory systems that influence phycoerythrin abundance exclusively have been described in F. diplosiphon and related species. The Cgi pathway has been shown to repress phycoerythrin accumulation in red light through a posttranscription initiation mechanism (22, 23), and a translation initiation factor 3 named IF3α has been identified in the light color-mediated repression of phycoerythrin expression in F. diplosiphon (24). Interactions between DpxA, the Cgi system, and IF3α have not yet been discovered. During type II chromatic acclimation (CA2) in Nostoc punctiforme, phycoerythrin is also repressed in red light, using the green-red-sensing cyanobacteriochrome CcaS (25, 26). It was postulated that CcaS may be similar to the sensor for the Cgi system. However, DpxA controls a teal-yellow-sensing system, distinguishing it from the green-red-sensing CcaS-controlled pathway. Furthermore, despite the existence of DpxA homologs with 67% or greater sequence identity in 15 other cyanobacterial species (see Fig. S5 in the supplemental material), DpxA has only 28% sequence identity with full-length CcaS. The DpxA system therefore appears to be independent of the RcaE-controlled portion of the CA3 system, the CA2 system, and perhaps the Cgi system, and is a previously unrecognized mechanism for acclimation of phycoerythrin in the green light range.
The identification of DpxA as a repressor of phycoerythrin abundance makes it the first yellow-light-sensing cyanobacteriochrome with a known physiological role. DpxA-like proteins must be broadly important for light sensing in cyanobacteria, since a large number of predicted photoreceptors with strong sequence relatedness (greater than or equal to 67% identity) to the entire length of DpxA exist in many cyanobacteria (see Fig. S5 in the supplemental material). The large number of phytochrome-class photoreceptors in F. diplosiphon also raises the possibility that another cyanobacteriochrome plays a role for RcaEr that is similar to the role DpxA plays for RcaEg, fine-tuning phycocyanin abundance in the red light region of the spectrum. If so, we would expect the absorption maxima for such a photoreceptor to approximately evenly flank the 661-nm absorption maximum of RcaEr.
The independent color-sensing capabilities of DpxA and RcaE, combined with the overlapping roles of these two photoreceptors, provide an important new perspective on how cyanobacteriochromes operate in the natural environment, where light is not monochromatic. Chromatic acclimation was first described in Oscillatoria sancta as a dramatic switch of cell color from red when grown in green light to blue-green when grown in red light (27). Since then, other cyanobacteria have been shown to change pigmentation in response to two light colors (16, 22, 28–30). Our discovery that CA3 in F. diplosiphon is controlled by two sensing systems that track four different light colors demonstrates that this process is simultaneously responding to many different ratios of light color in the environment. The presence of these two acclimation systems in one organism also underscores the value of fine-tuning the composition of the photosynthetic light-harvesting pigments to match subtle changes that occur in the spectral distribution of ambient light color.
Recent studies have shown that multiple cyanobacteriochromes can coordinately regulate phototaxis and aggregation (31, 32). However, the absorption characteristics of the photoreceptors involved in these processes are similar (UV/blue for phototaxis and blue/green for aggregation). Therefore, the physiological role of using more than one photoreceptor in these systems does not appear to be to significantly expand the range of light color sensing. It is possible that the photoreceptors in these systems serve intensity-sensing roles, which could be conferred by different reversion kinetics and/or output functions for each of the photoreceptors.
The results presented here provide a novel example of how cyanobacteriochromes interact by demonstrating that DpxA and RcaE have very different light absorption properties yet share overlapping functional roles in the regulation of phycoerythrin abundance. The gradual, coordinated release of the tandem repressing functions of these two photoreceptors as cells transition from red to blue light, especially through the yellow/teal region of the spectrum, demonstrates a much higher level of sophistication in the light color regulation of phycobilisome biogenesis than initially believed (16, 27–29, 33). In fact, it is not entirely surprising that such precise regulation exists, since efficient light harvesting is critical for cyanobacterial fitness under changing light conditions (17) and the production of these antennae, which may comprise up to 60% of the total soluble protein in a cyanobacterial cell (33), represents a major investment of resources for these organisms.
MATERIALS AND METHODS
Strains and growth conditions.
The wild-type strain of F. diplosiphon UTEX 481 (also called Tolypothrix sp. strain PCC 7601) used was the shortened-filament mutant strain SF33 (34). Cultures were grown in BG-11 medium from an absorbance at 750 nm of 0.01, as described previously (35), in light irradiances of 12 µmol photons m−2 s−1. Custom-built light-emitting diode (LED) panels provided red light (Digi-Key; catalog no. 160-1415-2-ND), green light (Digi-Key; catalog no. 754-1099-2-ND), blue light (Digi-Key; catalog no. 516-2672-1-ND), yellow light (Digi-Key; catalog no. 516-2673-1-ND), amber light (Novelty Lights, Inc.; catalog no. CGWA50/2.5-W-YE), and orange light (Novelty Lights, Inc.; catalog no. CGWG50/2.5-W-AM). Solux 4,700 K halogen lamps (Eiko Ltd.; catalog no. Q50MR16/CG/47/36) provided white light that closely matched the natural visible spectrum (36). The emission spectra of all LEDs are provided in Fig. S6 in the supplemental material, and those for the white light were as previously described (36).
Construction and complementation of the dpxA and rcaC deletion mutants.
To create dpxA, response regulator gene, hybrid histidine kinase gene, and rcaC single and double null mutant strains, two fragments of each gene were PCR amplified from F. diplosiphon genomic DNA. Primers are listed in Table S1 in the supplemental material. Primers 1 and 2 in each set were used to amplify the N-terminal region of the target gene. Primers 3 and 4 of each set were used to amplify the C-terminal region of the target gene. The fragments were fused by fusion PCR as previously described (24, 37). The resulting fusion fragment was cut with NcoI, which cleaved within the primer sequences, and ligated into pJCF276 (38), which had also been cut with NcoI. The sequences of the deletion constructs of dpxA and rcaC and double mutants were confirmed by sequencing. F. diplosiphon SF33 was transformed by triparental mating, and mutants were obtained by two selection procedures: first with neomycin and then by counterselection with sucrose as previously described (38). Three independent colonies of each null mutant were selected and tested by PCR amplification, sequencing, and Southern blot analyses. dpxA mutants were complemented by cloning dpxA and its native promoter into the shuttle vector pPL2.7 (34) using the primers listed in Table S1 in the supplemental material. Three independent growth experiments were conducted for each cell type and light condition.
Expression and purification of His-tagged DpxA and RcaE.
For the expression of DpxA and the GAF domain of RcaE in E. coli, each insert was PCR amplified using F. diplosiphon genomic DNA and cloned into the pETDuet vector (Novagen) after cutting with BamHI and SacI. All primer sequences used are listed in Table S1 in the supplemental material. RcaE GAF expression primers were based upon previously published sequences (15). All junctions and PCR amplification products were confirmed by sequencing. These plasmids were transformed into E. coli BL21(DE3) cells containing pPcyA (39). E. coli BL21(DE3) colonies containing pPcyA and the pETDuet vector with dpxA or rcaE inserts were selected on LB medium plates with 30 µg ⋅ ml−1 chloramphenicol and 50 µg ⋅ ml−1 ampicillin. A 5-ml culture of cells grown in LB medium with chloramphenicol and ampicillin with shaking at 37°C overnight was added to 1 liter of LB medium containing 30 µg ⋅ ml−1 chloramphenicol and 50 µg ⋅ ml−1 ampicillin and grown with shaking at 25°C for 4 h. A final concentration of 0.2 mM isopropyl-β-d-thiogalactoside was added to induce expression of the genes encoding heme oxygenase 1 (ho1), phycocyanobilin ferredoxin-dependent oxidoreductase (pcyA), and six-histidine-tagged versions of either DpxA or the RcaE GAF domain. Cells were grown overnight with shaking at 25°C and then harvested by centrifugation at 8,000 × g for 10 min at 4°C. RcaE and DpxA proteins from E. coli were then isolated as previously described (14). Kinase assays were performed as previously described (40), with DpxA and RcaE fully converted to the desired state by 5-min treatments with either blue or yellow light before conducting the remainder of the experiment in darkness.
Measurement of phycoerythrin and phycocyanin abundance.
Total cellular proteins were isolated from F. diplosiphon cells (41), and Western blot analyses using antiphycoerythrin and antiphycocyanin polyclonal antibodies (a gift from Wendy Schluchter, University of New Orleans) and goat anti-rabbit IgG-horseradish peroxidase (HRP) antibody (Santa Cruz Biotechnology) were performed according to the manufacturer’s instructions. The same cell lysates were used for the two antibodies. Samples were taken at the same optical density at 750 nm of 1.0, and 10 µl of lysate was added to each lane. Phycoerythrin and phycocyanin proteins were detected using SuperSignal West Femto chemiluminescent substrate (Thermo Scientific), according to the manufacturer’s instructions, and the images were viewed using the Bio-Rad ChemiDoc MP system. Each final value was calculated by normalizing the antibody image value by the protein content value for that lane, which was quantified from the protein gel.
Spectral measurements.
A Beckman DU640B spectrophotometer was used to measure the spectra of F. diplosiphon cultures from 375 to 725 nm. Spectral scans were recorded at an absorbance at 750 nm of 1.0 to 1.2. For all analyses, six scans recorded from independent experiments were normalized to the average of the two peak chlorophyll absorption values at 440 and 680 nm. Each scan presented is the average of the six scans after normalization to the two chlorophyll values. A Synergy Mx plate reader (BioTek Instruments) was used to measure the absorbance and difference spectra of DpxA full-length protein and the RcaE GAF domain as previously described (36). The light sources used for each of these experiments were provided using the LEDs with emission spectra shown in Fig. S6 in the supplemental material and described under “Strains and growth conditions.” Light irradiances of each LED source were measured using a Jaz spectrometer (Ocean Optics) and an LI-250 photometer with a cosine-corrected sensor (Li-Cor). For the absorbance measurements, purified DpxA and RcaE GAF domain were irradiated for 4 min with 12 µmol photons m−2 s−1 of the appropriate light wavelength before each spectral measurement. Each whole-cell scan shown in the manuscript is the average of six independent experiments. Each protein scan is representative of three independent experiments, with two technical replicates for each.
Bioinformatics.
Protein sequences closely matching DpxA were identified by using F. diplosiphon DpxA sequence to search the National Center for Biotechnology Information (NCBI) genome database using BLASTP (42). Matches with an E value of 0.0 were downloaded in FASTA format and aligned using the multiple sequence alignment Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) (43). The identity threshold between pairs was determined using the percent Identity Matrix Feature, also available through the Clustal Omega program.
SUPPLEMENTAL MATERIAL
(A) Predicted domain organization of DpxA. DpxA is 474 amino acids in length. The GAF domain length is 140 amino acids, and the histidine kinase domain length is 216 amino acids. (B) Genomic context of the dpxA gene within the Fremyella diplosiphon genome. DpxA is the “GAF histidine kinase.” Protein accession numbers are as follows: hybrid histidine kinase, WP_045867494; response regulator, WP_045867495; DpxA, WP_045867496; CpeS-like, WP_045867497; PBS lyase, WP_045867498; RF1-like, WP_045867499; hypothetical, WP_045867500. The gene immediately 3′ of the hybrid histidine kinase gene is encoded by the opposite strand of DNA. (C) Clustal Omega alignment of the GAF domains of DpxA from F. diplosiphon and NpR5113g1 from Nostoc punctiforme. Residues filled in black are identical, and residues filled in gray are similar, between the GAF domains. Download
Complementation of the ΔdpxA mutant. (A and B) Photographs (A) and whole-cell absorption spectra (B) of whole-cell cultures of wild-type cells, ΔdpxA cells, and ΔdpxA cells transformed with pPL2.7-dpxA. Scans and pictures are representative of three independent experiments. Phycoerythrin (PE) and phycocyanin (PC) peaks are labeled. (C) Effect of the deletion of dpxA, dpxB, and dpxC in the wild-type background. In each case, cultures were grown to late log phase in yellow light and scans are representative of three independent experiments. Phycocyanin (PC) and phycoerythrin (PE) are indicated. Whole-cell absorbance scans of wild type (black line), ΔdpxA (blue line), response regulator gene deletion downstream of dpxA (orange line), and hybrid histidine kinase gene deletion downstream of dpxA (purple line). Download
Dark reversion kinetics of the two forms of DpxA. Analysis of DpxA absorption characteristics during a dark treatment after exposure to blue (A) or yellow (B) light. Line colors and numbers in the key indicate the length of time of the dark treatment after the initial light treatment. Blue and yellow lines are the absorption spectra obtained after irradiation with either yellow light (YL) or blue light (BL), which was given after the last dark treatment time point. Download
DpxA operates through a non-Rca pathway. Whole-cell absorption spectra of the ΔrcaC and ΔrcaC ΔdpxA mutants after growth in natural-spectrum white light. Download
Comparison of the sequence of DpxA from F. diplosiphon to proteins with similar sequences from 15 different species of cyanobacteria. Regions containing the GAF domains are indicated with a red line above the sequences, and stars indicate conserved cysteine residues involved in bilin binding. Residues filled in black are identical, and residues filled in gray are similar, between at least 60% of proteins analyzed. Amino acid sequence numbers are indicated on the right. Genus/species abbreviations are as follows: S_hofmanni, Scytonema hofmanni PCC 7110; C_thermalis, Chroococcidiopsis thermalis PCC 7203; Calothrix7103, Calothrix sp. PCC 7103; L_boryana, Leptolyngbya boryana PCC 6306; M_testarum, Mastigocoleus testarum; Pseudanabaena6802, Pseudanabaena PCC 6802; Calothrix6303, Calothrix sp. PCC 6303; Synechocystis7509, Synechocystis sp. PCC 7509; Cyanothece7425, Cyanothece sp. PCC 7425; C_minutus6605, Chamaesiphon minutus PCC 6605; Leptolyngba7375, Leptolyngbya sp. PCC 7375; S_tolypothrichoides_VB61278, Scytonema tolypothrichoides VB-61278; C_cyanobacterium_CENA595, Chroococcales cyanobacterium CENA595; S_millei, Scytonema millei VB511283. Download
Emission spectra for the LED lights used in this study. The maximum light emission values for each LED are as follows: blue, 466 nm; green, 520 nm; yellow, 579 nm; amber, 594 nm; orange, 610 nm; red, 645 nm; far red, 740 nm. Download
DNA sequences of primers used in this study.
ACKNOWLEDGMENTS
We thank Wendy Schluchter and Christina Kronfel for the gift of the antiphycoerythrin and antiphycocyanin antibodies. We also thank the members of the Kehoe laboratory for their thoughtful comments on the manuscript.
This research was entirely supported by National Science Foundation grant MCB-1029414 to David M. Kehoe. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Wiltbank LB, Kehoe DM. 2016. Two cyanobacterial photoreceptors regulate photosynthetic light harvesting by sensing teal, green, yellow, and red light. mBio 7(1):e02130-15. doi:10.1128/mBio.02130-15.
REFERENCES
- 1.Rockwell NC, Su YS, Lagarias JC. 2006. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol 57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rockwell NC, Lagarias JC. 2010. A brief history of phytochromes. Chemphyschem 11:1172–1180. doi: 10.1002/cphc.200900894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auldridge ME, Forest KT. 2011. Bacterial phytochromes: more than meets the light. Crit Rev Biochem Mol Biol 46:67–88. doi: 10.3109/10409238.2010.546389. [DOI] [PubMed] [Google Scholar]
- 4.Kendrick RE, Kronenberg GHM. 1994. Photomorphogenesis in plants, 2nd ed. Kluwer Academic Publishers, Dordrecht, Netherlands. [Google Scholar]
- 5.Mathews S. 2006. Phytochrome-mediated development in land plants: red light sensing evolves to meet the challenges of changing light environments. Mol Ecol 15:3483–3503. doi: 10.1111/j.1365-294X.2006.03051.x. [DOI] [PubMed] [Google Scholar]
- 6.Franklin KA, Quail PH. 2010. Phytochrome functions in Arabidopsis development. J Exp Bot 61:11–24. doi: 10.1093/jxb/erp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Possart A, Fleck C, Hiltbrunner A. 2014. Shedding (far-red) light on phytochrome mechanisms and responses in land plants. Plant Sci 217-218:36–46. doi: 10.1016/j.plantsci.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 8.Takala H, Björling A, Berntsson O, Lehtivuori H, Niebling S, Hoernke M, Kosheleva I, Henning R, Menzel A, Ihalainen JA, Westenhoff S. 2014. Signal amplification and transduction in phytochrome photosensors. Nature 509:245–248. doi: 10.1038/nature13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma Q, Hua HH, Chen Y, Liu BB, Krämer AL, Scheer H, Zhao KH, Zhou M. 2012. A rising tide of blue-absorbing biliprotein photoreceptors—characterization of seven such bilin-binding GAF domains in Nostoc sp. PCC7120. FEBS J 279:4095–4108. doi: 10.1111/febs.12003. [DOI] [PubMed] [Google Scholar]
- 10.Rockwell NC, Martin SS, Gulevich AG, Lagarias JC. 2012. Phycoviolobilin formation and spectral tuning in the DXCF cyanobacteriochrome subfamily. Biochemistry 51:1449–1463. doi: 10.1021/bi201783j. [DOI] [PubMed] [Google Scholar]
- 11.Yerrapragada S, Shukla A, Hallsworth-Pepin K, Choi K, Wollam A, Clifton S, Qin X, Muzny D, Raghuraman S, Ashki H, Uzman A, Highlander SK, Fryszczyn BG, Fox GE, Tirumalai MR, Liu Y, Kim S, Kehoe DM, Weinstock GM. 2015. Extreme sensory complexity encoded in the 10-megabase draft genome sequence of the chromatically acclimating cyanobacterium Tolypothrix sp. PCC 7601. Genome Announc 3:e00355-15. doi: 10.1128/genomeA.00355-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikeuchi M, Ishizuka T. 2008. Cyanobacteriochromes: a new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem Photobiol Sci 7:1159–1167. doi: 10.1039/b802660m. [DOI] [PubMed] [Google Scholar]
- 13.Kehoe DM, Grossman AR. 1996. Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science 273:1409–1412. doi: 10.1126/science.273.5280.1409. [DOI] [PubMed] [Google Scholar]
- 14.Terauchi K, Montgomery BL, Grossman AR, Lagarias JC, Kehoe DM. 2004. RcaE is a complementary chromatic adaptation photoreceptor required for green and red light responsiveness. Mol Microbiol 51:567–577. doi: 10.1046/j.1365-2958.2003.03853.x. [DOI] [PubMed] [Google Scholar]
- 15.Hirose Y, Rockwell NC, Nishiyama K, Narikawa R, Ukaji Y, Inomata K, Lagarias JC, Ikeuchi M. 2013. Green/red cyanobacteriochromes regulate complementary chromatic acclimation via a protochromic photocycle. Proc Natl Acad Sci U S A 110:4974–4979. doi: 10.1073/pnas.1302909110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kehoe DM, Gutu A. 2006. Responding to color: the regulation of complementary chromatic adaptation. Annu Rev Plant Biol 57:127–150. doi: 10.1146/annurev.arplant.57.032905.105215. [DOI] [PubMed] [Google Scholar]
- 17.Stomp M, van Dijk MA, van Overzee HM, Wortel MT, Sigon CA, Egas M, Hoogveld H, Gons HJ, Huisman J. 2008. The timescale of phenotypic plasticity and its impact on competition in fluctuating environments. Am Nat 172:169–185. doi: 10.1086/591680. [DOI] [PubMed] [Google Scholar]
- 18.Chiang GG, Schaefer MR, Grossman AR. 1992. Complementation of a red-light-indifferent cyanobacterial mutant. Proc Natl Acad Sci U S A 89:9415–9419. doi: 10.1073/pnas.89.20.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kehoe DM, Grossman AR. 1997. New classes of mutants in complementary chromatic adaptation provide evidence for a novel four-step phosphorelay system. J Bacteriol 179:3914–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rockwell NC, Martin SS, Lagarias JC. 2015. Identification of DXCF cyanobacteriochrome lineages with predictable photocycles. Photochem Photobiol Sci 14:929–941. doi: 10.1039/c4pp00486h. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Alvey RM, Bezy RP, Kehoe DM. 2008. Inverse transcriptional activities during complementary chromatic adaptation are controlled by the response regulator RcaC binding to red and green light-responsive promoters. Mol Microbiol 68:286–297. doi: 10.1111/j.1365-2958.2008.06151.x. [DOI] [PubMed] [Google Scholar]
- 22.Gutu A, Kehoe DM. 2012. Emerging perspectives on the mechanisms, regulation, and distribution of light color acclimation in cyanobacteria. Mol Plant 5:1–13. doi: 10.1093/mp/ssr054. [DOI] [PubMed] [Google Scholar]
- 23.Bezy RP, Wiltbank L, Kehoe DM. 2011. Light-dependent attenuation of phycoerythrin gene expression reveals convergent evolution of green light sensing in cyanobacteria. Proc Natl Acad Sci U S A 108:18542–18547. doi: 10.1073/pnas.1107427108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gutu A, Nesbit AD, Alverson AJ, Palmer JD, Kehoe DM. 2013. Unique role for translation initiation factor 3 in the light color regulation of photosynthetic gene expression. Proc Natl Acad Sci U S A 110:16253–16258. doi: 10.1073/pnas.1306332110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirose Y, Narikawa R, Katayama M, Ikeuchi M. 2010. Cyanobacteriochrome CcaS regulates phycoerythrin accumulation in Nostoc punctiforme, a group II chromatic adapter. Proc Natl Acad Sci U S A 107:8854–8859. doi: 10.1073/pnas.1000177107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirose Y, Shimada T, Narikawa R, Katayama M, Ikeuchi M. 2008. Cyanobacteriochrome CcaS is the green light receptor that induces the expression of phycobilisome linker protein. Proc Natl Acad Sci U S A 105:9528–9533. doi: 10.1073/pnas.0801826105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaidukov N. 1903. Die Farbervanderung bei den Prozessen der Komplementaren chromatischen Adaptation. Ber Dtsch Bot Ges 21:517–522. [Google Scholar]
- 28.Grossman AR. 2003. A molecular understanding of complementary chromatic adaptation. Photosynth Res 76:207–215. doi: 10.1023/A:1024907330878. [DOI] [PubMed] [Google Scholar]
- 29.Tandeau de Marsac N. 2003. Phycobiliproteins and phycobilisomes: the early observations. Photosynth Res 76:193–205. doi: 10.1023/A:1024954911473. [DOI] [PubMed] [Google Scholar]
- 30.Gan F, Zhang S, Rockwell NC, Martin SS, Lagarias JC, Bryant DA. 2014. Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light. Science 345:1312–1317. doi: 10.1126/science.1256963. [DOI] [PubMed] [Google Scholar]
- 31.Song JY, Cho HS, Cho JI, Jeon JS, Lagarias JC, Park YI. 2011. Near-UV cyanobacteriochrome signaling system elicits negative phototaxis in the cyanobacterium Synechocystis sp. PCC 6803. Proc Natl Acad Sci U S A 108:10780–10785. doi: 10.1073/pnas.1104242108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Enomoto G, Win NN, Narikawa R, Ikeuchi M. 2015. Three cyanobacteriochromes work together to form a light color-sensitive input system for c-di-GMP signaling of cell aggregation. Proc Natl Acad Sci U S A. 112:8082–8087. doi: 10.1073/pnas.1504228112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bogorad L. 1975. Phycobiliproteins and complementary chromatic adaptation. Annu Rev Plant Physiol Plant Mol Biol 26:369–401. doi: 10.1146/annurev.pp.26.060175.002101. [DOI] [Google Scholar]
- 34.Cobley JG, Zerweck E, Reyes R, Mody A, Seludo-Unson JR, Jaeger H, Weerasuriya S, Navankasattusas S. 1993. Construction of shuttle plasmids which can be efficiently mobilized from Escherichia coli into the chromatically adapting cyanobacterium, Fremyella diplosiphon. Plasmid 30:90–105. doi: 10.1006/plas.1993.1037. [DOI] [PubMed] [Google Scholar]
- 35.Seib LO, Kehoe DM. 2002. A turquoise mutant genetically separates expression of genes encoding phycoerythrin and its associated linker peptides. J Bacteriol 184:962–970. doi: 10.1128/jb.184.4.962-970.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bussell AN, Kehoe DM. 2013. Control of a four-color sensing photoreceptor by a two-color sensing photoreceptor reveals complex light regulation in cyanobacteria. Proc Natl Acad Sci U S A 110:12834–12839. doi: 10.1073/pnas.1303371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noubir S, Luque I, Ochoa de Alda JA, Perewoska I, Tandeau de Marsac N, Cobley JG, Houmard J. 2002. Co-ordinated expression of phycobiliprotein operons in the chromatically adapting cyanobacterium Calothrix PCC 7601: a role for RcaD and RcaG. Mol Microbiol 43:749–762. [DOI] [PubMed] [Google Scholar]
- 38.Cobley JG, Clark AC, Weerasurya S, Queseda FA, Xiao JY, Bandrapali N, D’Silva I, Thounaojam M, Oda JF, Sumiyoshi T, Chu MH. 2002. CpeR is an activator required for expression of the phycoerythrin operon (cpeBA) in the cyanobacterium Fremyella diplosiphon and is encoded in the phycoerythrin linker-polypeptide operon (cpeCDESTR). Mol Microbiol 44:1517–1531. doi: 10.1046/j.1365-2958.2002.02966.x. [DOI] [PubMed] [Google Scholar]
- 39.Biswas A, Vasquez YM, Dragomani TM, Kronfel ML, Williams SR, Alvey RM, Bryant DA, Schluchter WM. 2010. Biosynthesis of cyanobacterial phycobiliproteins in Escherichia coli: chromophorylation efficiency and specificity of all bilin lyases from Synechococcus sp. strain PCC 7002. Appl Environ Microbiol 76:2729–2739. doi: 10.1128/AEM.03100-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirose Y, Shimada T, Narikawa R, Katayama M, Ikeuchi M. 2008. Cyanobacteriochrome CcaS is the green light receptor that induces the expression of phycobilisome linker protein. Proc Natl Acad Sci U S A 105:9528–9533. doi: 10.1073/pnas.0801826105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li L, Kehoe DM. 2005. In vivo analysis of the roles of conserved aspartate and histidine residues within a complex response regulator. Mol Microbiol 55:1538–1552. doi: 10.1111/j.1365-2958.2005.04491.x. [DOI] [PubMed] [Google Scholar]
- 42.Acland A, Agarwala R, Barrett T, Beck J, Benson DA, Bollin C, Bolton E, Bryant SH, Canese K, Church DM, Clark K, DiCuccio M, Dondoshansky I, Federhen S, Feolo M, Geer LY, Gorelenkov V, Hoeppner M, Johnson M, Kelly C, Khotomlianski V, Kimchi A, Kimelman M, Kitts P, Krasnov S, Kuznetsov A, Landsman D, Lipman DJ, Lu ZY, Madden TL, Madej T, Maglott DR, Marchler-Bauer A, Karsch-Mizrachi I, Murphy T, Ostell J, O’Sullivan C, Panchenko A, Phan L, Pruitt DPKD, Rubinstein W, Sayers EW, Schneider V, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Siyan K, Slotta D. 2013. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 41:D8–D20. doi: 10.1093/nar/gks1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Predicted domain organization of DpxA. DpxA is 474 amino acids in length. The GAF domain length is 140 amino acids, and the histidine kinase domain length is 216 amino acids. (B) Genomic context of the dpxA gene within the Fremyella diplosiphon genome. DpxA is the “GAF histidine kinase.” Protein accession numbers are as follows: hybrid histidine kinase, WP_045867494; response regulator, WP_045867495; DpxA, WP_045867496; CpeS-like, WP_045867497; PBS lyase, WP_045867498; RF1-like, WP_045867499; hypothetical, WP_045867500. The gene immediately 3′ of the hybrid histidine kinase gene is encoded by the opposite strand of DNA. (C) Clustal Omega alignment of the GAF domains of DpxA from F. diplosiphon and NpR5113g1 from Nostoc punctiforme. Residues filled in black are identical, and residues filled in gray are similar, between the GAF domains. Download
Complementation of the ΔdpxA mutant. (A and B) Photographs (A) and whole-cell absorption spectra (B) of whole-cell cultures of wild-type cells, ΔdpxA cells, and ΔdpxA cells transformed with pPL2.7-dpxA. Scans and pictures are representative of three independent experiments. Phycoerythrin (PE) and phycocyanin (PC) peaks are labeled. (C) Effect of the deletion of dpxA, dpxB, and dpxC in the wild-type background. In each case, cultures were grown to late log phase in yellow light and scans are representative of three independent experiments. Phycocyanin (PC) and phycoerythrin (PE) are indicated. Whole-cell absorbance scans of wild type (black line), ΔdpxA (blue line), response regulator gene deletion downstream of dpxA (orange line), and hybrid histidine kinase gene deletion downstream of dpxA (purple line). Download
Dark reversion kinetics of the two forms of DpxA. Analysis of DpxA absorption characteristics during a dark treatment after exposure to blue (A) or yellow (B) light. Line colors and numbers in the key indicate the length of time of the dark treatment after the initial light treatment. Blue and yellow lines are the absorption spectra obtained after irradiation with either yellow light (YL) or blue light (BL), which was given after the last dark treatment time point. Download
DpxA operates through a non-Rca pathway. Whole-cell absorption spectra of the ΔrcaC and ΔrcaC ΔdpxA mutants after growth in natural-spectrum white light. Download
Comparison of the sequence of DpxA from F. diplosiphon to proteins with similar sequences from 15 different species of cyanobacteria. Regions containing the GAF domains are indicated with a red line above the sequences, and stars indicate conserved cysteine residues involved in bilin binding. Residues filled in black are identical, and residues filled in gray are similar, between at least 60% of proteins analyzed. Amino acid sequence numbers are indicated on the right. Genus/species abbreviations are as follows: S_hofmanni, Scytonema hofmanni PCC 7110; C_thermalis, Chroococcidiopsis thermalis PCC 7203; Calothrix7103, Calothrix sp. PCC 7103; L_boryana, Leptolyngbya boryana PCC 6306; M_testarum, Mastigocoleus testarum; Pseudanabaena6802, Pseudanabaena PCC 6802; Calothrix6303, Calothrix sp. PCC 6303; Synechocystis7509, Synechocystis sp. PCC 7509; Cyanothece7425, Cyanothece sp. PCC 7425; C_minutus6605, Chamaesiphon minutus PCC 6605; Leptolyngba7375, Leptolyngbya sp. PCC 7375; S_tolypothrichoides_VB61278, Scytonema tolypothrichoides VB-61278; C_cyanobacterium_CENA595, Chroococcales cyanobacterium CENA595; S_millei, Scytonema millei VB511283. Download
Emission spectra for the LED lights used in this study. The maximum light emission values for each LED are as follows: blue, 466 nm; green, 520 nm; yellow, 579 nm; amber, 594 nm; orange, 610 nm; red, 645 nm; far red, 740 nm. Download
DNA sequences of primers used in this study.



