Abstract
Feline calicivirus (FCV) causes a variable syndrome of upper respiratory tract disease, mouth ulcers and lameness. A convenience-based prospective sample of oropharyngeal swabs (n=426) was obtained from five countries (France, Germany, Greece, Portugal and the UK). The prevalence of FCV by virus isolation was 22.2 per cent. Multivariable analysis found that animals presenting with lymphoplasmacytic gingivitis stomatitis complex were more likely to test positive for FCV infection. Furthermore, vaccinated cats up to 48 months of age were significantly less likely to be infected with FCV than unvaccinated animals of similar ages. Phylogenetic analysis based on consensus sequences for the immunodominant region of the capsid gene from 72 FCV isolates identified 46 strains. Thirteen of the 14 strains with more than one sequence were restricted to individual regions or sites in individual countries; the exception was a strain present in two sites close to each other in France. Four strains were present in more than one household. Five colonies, four of which were rescue shelters, had multiple strains within them. Polymerase sequence suggested possible rare recombination events. These locally, nationally and internationally diverse FCV populations maintain a continuous challenge to the control of FCV infection and disease.
Keywords: Feline calicivirus, Molecular epidemiology, Vesivirus, Epidemiology, Vaccines
Introduction
Feline calicivirus (FCV) is one of the most frequently reported pathogens causing upper respiratory tract and oral disease in cats (Radford and others 2009). Following FCV infection, most cats develop acute respiratory disease and oral ulceration (Pesavento and others 2008). Less frequently, more systemic signs including acute arthritis/limping syndrome (Pedersen and others 1983, Dawson and others 1994, TerWee and others 1997) and haemorrhagic-like fever may be seen (Pedersen and others 2000, Coyne and others 2006a). Most cats with a severe oral inflammatory syndrome called lymphoplasmacytic gingivitis stomatitis complex (LGSC) also test positive for FCV, although Koch's postulates have not been proven (Knowles and others 1991, Poulet and others 2000). Following infection with FCV, many cats shed virus beyond the resolution of clinical signs, and those shedding for more than 30 days are termed carriers (Wardley 1976, Wardley and Povey 1977). Most cats do seem to eventually clear infection, and it is thought the majority of ‘carriers’ are undergoing repeated reinfections, with only a minority being truly persistently infected (Coyne and others 2007b).
Vaccines against FCV have been available for over forty years. Several vaccine antigens are used including FCV-F9 in live vaccines, and in inactivated vaccines, either FCV-225 or a combination of FCV-431 and FCV-G1 (Radford and others 2009). Although pre-existing immunity, either from vaccination or previous infection, does ameliorate clinical signs, it does not induce sterilising immunity. As a result, the prevalence of FCV in the general cat population remains high (Wardley and others 1974), especially among cats living in colonies or shelters (Coutts and others 1994, Radford and others 2001b, Bannasch and Foley 2005, Helps and others 2005).
As an RNA virus, FCV evolves quickly (Radford and others 1999a, Coyne and others 2007b). Despite this, sufficient sequence and antigenic relatedness persists to allow FCV variants to be considered a single diverse serotype (Povey and Ingersoll 1975) and genotype (Glenn and others 1999). Phylogenetic analysis typically results in a ‘star-like’ phylogeny. Lack of statistical support for subspecies clusters means it has not proved possible to identify robust groupings of FCV either on spatial, temporal or clinical grounds.
Using this inherent genetic variability, the molecular epidemiology of FCV has been studied at various population levels. In individual households, one or a small number of strains can circulate often at high prevalence, with extensive genetic variation driven by positive selection (Radford and others 2003, Coyne and others 2007b). In rescue shelters, many strains can be present, and where biosecurity is good, these pass through the colony with little evidence of transmission (Radford and others 2001b, Coyne and others 2007a). At a regional or community level, a highly complex genetic landscape can exist with many 10 s of strains co-circulating, with only rare cases of transmission between practices at the national (UK) level (Coyne and others 2012).
The primary aim of this study was to see whether the population structure of viruses previously seen in the UK is mirrored when data from multiple countries is included.
Materials and methods
Sample collection
Samples were collected from five countries: France (five sites in the Paris area), UK (two sites: Bangor (Northern Ireland) and Northampton), Portugal (two sites: Mem Martins and Lisbon area), Greece (one site: Thessaloniki) and Germany (three sites: Lippstadt, Berlin and Achim). At each site, a private veterinary practitioner was recruited based on convenience by the project sponsor, and asked to sample two types of cats; healthy cats not vaccinated against FCV for at least 1 month (to reduce the probability of isolating vaccine strains), and those cats with clinical signs typical of FCV infection (for example coryza or LGSC).
For each cat whose owner gave consent, a plain cotton-tipped swab was used to collect an oropharyngeal saliva sample and placed in 2 ml viral transport medium (40 per cent L15/Mac Coy's (v/v) cell culture media, 60 per cent freezing excipient). These were stored locally at −18°C then shipped as a batch to the laboratory on dry ice. With each sample a short questionnaire was completed by the attending veterinary surgeon and owner, capturing demographic data, disease and vaccination history.
The non-invasive nature of this project and anonymised data collection meant that formal ethical committee approval was not necessary. However, informed owner consent was a stipulation before samples were collected.
Virus isolation
Diagnosis of FCV and feline herpesvirus type 1 (FeHV-1) was by demonstrating typical cytopathic effect on confluent monolayers of feline embryo A cells. Samples were only reported negative following a second negative passage. Positive isolates were stored at −80°C (Knowles and others 1990).
Statistical analysis
A parametrical bootstrap approach was used to test country and sites as random effects based on a hypothesis of the form H0: σ2=0. The estimate P value was 0.99 for country and 0.09 for sites. Thus, the level of between-country and between-site variability was not considered sufficient to incorporate random effects in a multivariable model.
A multivariable logistic regression analysis was conducted to model the probability of being infected with FCV. The model was fitted in R (V.3.01) using the ‘glm’ function from ‘aod’ package. Risk factors considered within the model included the country and site within the country where the samples were collected, sex, whether a cat lived outdoors, neutering status, FCV vaccination status, age and presence of the following clinical signs: ocular discharge, sneezing, gingivitis, nasal discharge, coughing, oral ulcers, conjunctivitis, stomatitis and LGSC. Visual inspection of the data indicated a curvilinear relationship between the proportion of FCV infected cats and age (Fig 1a), thus a quadratic term in age was included to fit the model. Age was centred to make the model interpretation easier and to reduce the correlation between the linear and quadratic terms. Two-way interactions between age and its quadratic term and the factors neutering and vaccination status were included in the initial multivariable model. A stepwise model selection by akaike information criterion (AIC) was conducted using the R command ‘step’ of the ‘stats’ package. A backward selection procedure was used to refit the model eliminating terms from the fitted model based on a likelihood ratio test (P>0.05)(Faraway 2006).
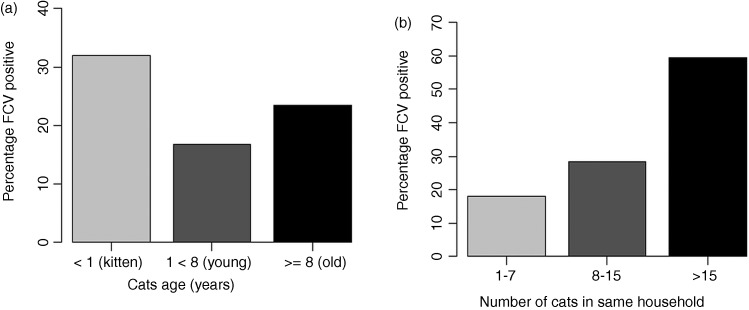
FIG 1:
Percentage of cats testing positive for feline calicivirus (FCV) by age−years (a) and by number of cats living in the same household (b)
Detailed information about the number of cats living in the same household (NCH) was only available for 66 per cent of the observations in the multivariable analysis. Therefore, to avoid reducing the numbers included in the multivariable model, the impact of NCH on FCV infection was only assessed by univariable logistic regression.
RNA extraction, reverse transcription-PCR
RNAs were extracted from positive samples (second passage or less) (QIAmp viral RNA Mini Kit; Qiagen). For every three samples, one negative control was included from uninfected cell cultures. Reverse transcription was performed using 200 ng random hexamers (Superscript III, Invitrogen), according to manufacturer's guidelines.
Capsid amplification: A 529-nucleotide region of the capsid gene, equivalent to residues 6406-6934 of FCV strain F9 (Carter and others 1992) and incorporating immunodominant regions C and E (Seal and others 1993, Radford and others 1999b), was amplified as previously described (Coyne and others 2007b, 2012). Briefly, each 50 µl reaction contained 2 µl cDNA, 45 µl 1.1×Reddy mix (Thermo Scientific), 1 µl nuclease-free water and 3.2 ng each of forward and reverse primers (Table 1). In early experiments, primers M13cap2F/T7cap2R were most cross-reactive and so all PCRs were initially conducted with these: only samples testing negative were subsequently attempted with the additional primers.
TABLE 1:
Primers used for partial capsid and polymerase gene amplification
| Primer | Sequence (5′→3′) | Binding site (nt)* |
|---|---|---|
| Capsid primers | ||
| M13cap2F | CAGGAAACAGCTATGACCCCTTTGTCTTCCARGCHAAYCG | 6406–6428 |
| T7cap2R | TAATACGACTCTCAATAGGGCCTCACCAATTCCNGTRTANCC | 6934–6913 |
| A1 forward | CCCTTCGTCTTTCAGGCCAACCG | 6406–6428 |
| A2 reverse | CCTCGCCAATCCCAGTGTAGCC | 6934–6913 |
| P1 forward | CCGTTTGTGTTTCAAGCAAACCG | 6406–6428 |
| p2 reverse | CCTCACCTATACCAGTGTAACC | 6934–6913 |
| M13cap1F | CAGGAAACAGCTATGACCCVTTTGTNTTYCARGCHAAYCG | 6406–6428 |
| T7cap1R | TAATACGACTCTCAATAGGGSYTCDCCRATNCCNGTRTANCC | 6934–6913 |
| Polymerase primers | ||
| M13_53D | CAGGAAACAGCTATGACGAYATGATGACYTAYGGKGAYGAYGG | 4766–4791 |
| T7_33D | TAATACGACTCTCAATAGGGCCGCGCYTCCACRCCRTTRAAYTG | 5251–5228 |
Underlined regions illustrate the M13 forward and T7 reverse primer binding site sequences used for sequencing amplicons produced with these otherwise degenerate primers. Nucleotide sequences are shown using the single-letter IUB codes for degeneracy: R=A/G purine; Y=T/C pyrimidine; K=T/G
*Nucleotide numbers of the binding sites relate to feline calicivirus (FCV) strain F9 (GenBank accession no. M86379) (Carter and others 1992)
Thermal cycling consisted of DNA denaturation (95°C, 2 min), followed by 40 cycles of denaturation (95°C, 30 s), primer annealing (45–55°C, 30 s) and primer extension (72°C, 90 s). A final extension was performed at 72°C (5 min).
Polymerase amplification: A 486-nucleotide region corresponding to the 3′ end of the FCV POL region was amplified from the same cDNA template using Reddy-Mix (ABgene) according to the manufacturers’ instructions, in 50 μl reactions containing 100 ng each of the primers M13-53D and T7-33D (Coyne and others 2006b) (Table 1). Thermal cycling consisted of denaturation (95°C, 4 minutes), followed by 40 cycles of denaturation (95°C, 60 s), primer annealing (55°C, 60 s) and primer extension (72°C, 3 minutes). A final extension was performed at 72°C (5 minutes).
Nucleotide sequence and phylogenetic analysis
Amplicons were purified (QIAquick PCR purification, Qiagen), quantified (Genequant) and sequenced bidirectionally (Source Bioscience). The authors have previously found this method to be >99 per cent reproducible (Coyne and others 2007b). Forward and reverse sequences were aligned and manually corrected (Chromas Pro, Technelysium). Pairwise p-distances between sequences, and Kimura 2-parameter Neighbour-joining trees with 1000 bootstrap replicates were calculated using MEGA V.5.2.2 (Tamura and others 2011). A pairwise distance approach was taken to avoid excluding a high percentage of aligned columns associated with sporadic nucleotide ambiguities in individual sequences. A 20 per cent uncorrected nucleotide distance threshold between capsid sequences was used to define distinct strains (Radford and others 2001b, Prikhodko and others 2014).
Results
Study sample
A total of 426 samples were collected from 13 sites in five countries. For 17 samples, the viral status could not be assessed due to bacterial overgrowth. For the remaining 409, FCV and FeHV-1 was isolated from 91 (22.2 per cent) and 18 (4.4 per cent), respectively (Table 2). For FCV, 16.2 per cent and 34.2 per cent of healthy and sick (at least one clinical sign) cats tested positive for FCV, respectively. For FeHV-1, the figures were 2.6 per cent and 8.0 per cent.
TABLE 2:
Summary of samples, isolates, amplification and strains identified in each country and site
| Country | France |
UK |
Portugal |
Greece | Germany |
Total | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Site | FR1 | FR2 | FR3 | FR4 | FR5 | Total | UK1 | UK2 | Total | PT1 | PT2 | Total | GR1 | GE1 | GE2 | GE3 | Total | |
| Samples (N) | 41 | 7 | 33 | 7 | 14 | 102 | 49 | 34 | 83 | 48 | 32 | 80 | 42 | 40 | 28 | 34 | 102 | 409 |
| FCV Positive, N (%) | 17 (41) | 1 (14) | 1 (3) | 2 (29) | 6 (43) | 27 (26) | 9 (18) | 4 (12) | 13 (16) | 14 (29) | 4 (13) | 18 (23) | 10 (24) | 10 (25) | 8 (29) | 5 (15) | 23 (23) | 91 (22) |
| FCV Capsid PCR+ | 16 | 1 | 1 | 2 | 5 | 25 | 6 | 2 | 8 | 14 | 3 | 17 | 9 | 8 | 4 | 3 | 15 | 74 |
| FCV Capsid sequence | 14 | 1 | 1 | 2 | 5 | 23 | 6 | 2 | 8 | 14 | 3 | 17 | 9 | 8 | 4 | 3 | 15 | 72 |
| Strains | 5 | 1 | 1 | 2 | 4 | 12 | 3 | 2 | 5 | 9 | 3 | 12 | 8 | 4 | 3 | 2 | 9 | 46 |
FCV, feline calicivirus
Risk factors for FCV infection
Complete information about all the variables considered within the multivariable model was only available for 299 samples. The final model included the continuous variables age and a quadratic term for age (age2), as well as the categorical variables vaccination status against FCV (FCV-V) and whether or not the animal was presenting with LGSC, and the two-way interactions FCV-V by age and FCV-V by age2 (Table 3). Other individual clinical signs were not found to be significant in this population.
TABLE 3:
Final multivariable logistic regression model of factors associated with FCV infection
| Variable | Beta | OR | 95% CI |
P value | |
|---|---|---|---|---|---|
| Lower | Upper | ||||
| LGSC | |||||
| No | Reference | ||||
| Yes | 2.23 | 9.33 | 3.18 | 29.45 | <0.001 |
| Vaccinated | |||||
| No | Reference | ||||
| Yes | 0.35 | 1.42 | 0.54 | 4.07 | 0.5 |
| Age centered* (months) | −0.014 | 0.99 | 0.98 | 0.99 | 0.003 |
| Age centred2 (months) | 0.0002 | 1.0002 | 1.0001 | 1.0003 | <0.001 |
| Vaccinated×age centred | |||||
| Not vaccinated, age centred | Reference | ||||
| Vaccinated, age centred | 0.03 | 1.03 | 1.016 | 1.05 | <0.001 |
| Vaccinated×age centred2 | |||||
| Not vaccinated, age centred2 | Reference | ||||
| Vaccinated, age centred2 | −0.0004 | 0.9996 | 0.9993 | 0.9998 | <0.001 |
*Age centred: mean age was 71.7 months
FCV, feline calicivirus; LGSC, lymphoplasmacytic gingivitis stomatitis complex
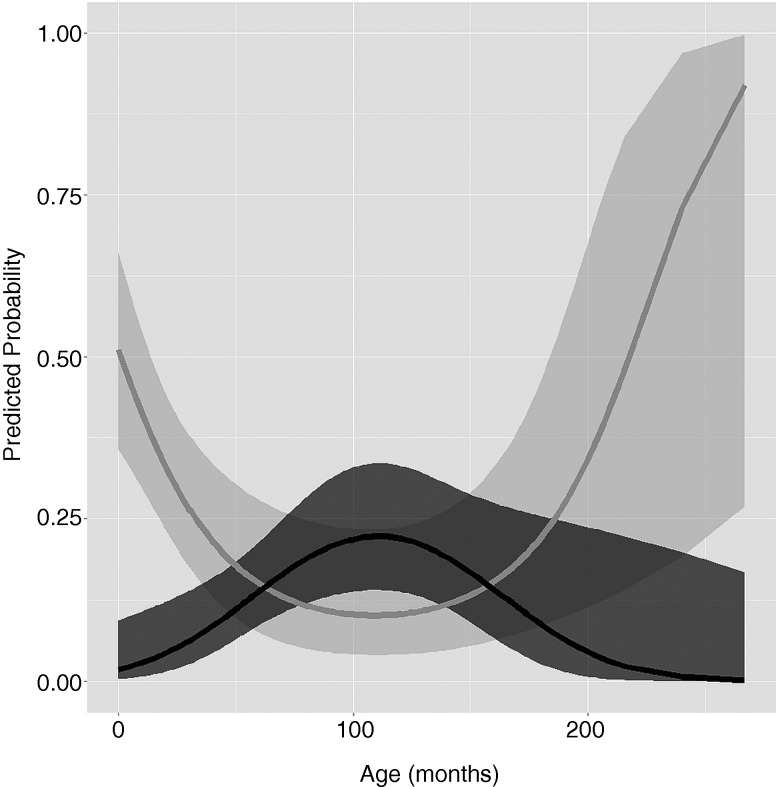
Cats presenting with LGSC were 9.33 (95% CI 3.18 to 29.45) times more likely to present with FCV infection than cats without LGSC. The relationship between the probability of FCV infection and age and vaccination status was more complex due to an interaction between these risk factors. In short, in unvaccinated animals (n=119) there was a reduction in risk each month for the first 8 years, followed by a plateau for the next 2.5 years and an increase again every month for the next 10 years (Fig 2). In vaccinated animals (n=218), there was an increase in risk each month for the first 8.5 years, followed by a plateau for the next 1.5 years and a decrease every month for the next 12.5 years (Fig 2). In vaccinated animals up to 48 months of age, the probability of being FCV infected was lower than in unvaccinated animals of similar ages. From 48 months of age, 95% CIs of the probability for FCV infection in vaccinated and unvaccinated animals overlapped, indicating a similar probability between both groups of animals during that age (Fig 2). The main effect of vaccination on the odds of FCV infection was not significant for animals with a centred age of 0 (i.e. a mean age of 71.7 months, Table 3 and Fig 2). One limitation of the study was the small sample size, especially for older animals, reflected by wide CIs in the predicted probability of FCV infection for those animals (Fig 3).
FIG 2:
Predicted probability of a cat being infected with feline calicivirus by age and vaccination status. Black and grey lines depict the predicted probability in a vaccinated and unvaccinated cat, respectively. Shaded boundaries represent 95% CIs
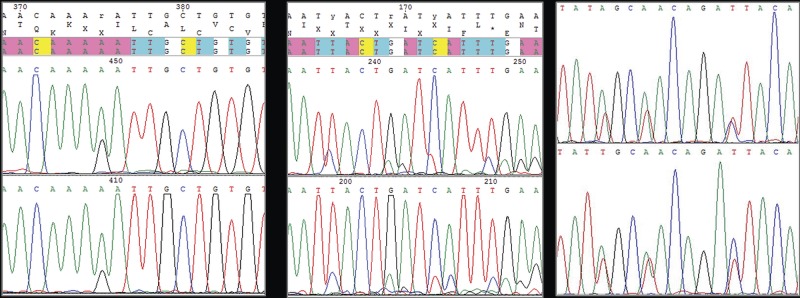
FIG 3:
Examples of types of consensus sequences obtained. (a) Clear consensus with rare ambiguity (<5 per cent) in 71 sequences. (b) Mixed sequence (>20 per cent ambiguous) but with clear major and minor sequences which can be manually resolved for further analyses (FR1_11). (c) Mixed sequences (>20 per cent ambiguous), with approximately equal peak heights making meaningful manual resolution impossible (FR1_37 and FR1_40)
Visual inspection of the data indicated a linear relationship between the percentage of cats testing positive for FCV and NCH (Fig 1b). The number of cats living in a household was found to be significantly associated with the probability of being FCV infected on the univariable analysis. Thus, for one-unit increase in NCH, the odds of being FCV infected increase 1.5% (OR=1.015, 95% CI 1.004 to 1.034, p=0.043).
Sequence analysis
In total, 72 consensus sequences of the major immunodominant region of the FCV capsid were obtained from the 91 FCV isolates (Table 2). This included 71 clear consensus sequences containing a small number (<5 per cent) of ambiguous nucleotide positions typical for a calicivirus (Fig 3a). In contrast, one sample (FR1_11), contained many ambiguities strongly suggesting a mixed infection (Fig 3b). One of the two sequences in this sample was present in a clear majority allowing FR1_11 sequence to be manually ‘split’ into its predicted major and minor sequences, as well as its consensus (average). Whilst the accuracy of these three individual sequences is likely to be lower, the authors included them to gain further insight into the diversity of viruses circulating in this population.
Two additional samples (FR1_37 and FR1_40) gave very clear sequence, and were also identified as mixed infections. However, unlike FR1_11, the two sequences were present in approximately equal proportions making it impossible to manually resolve them (Fig 3c). These sequences were not available for further analyses and would require a cloning approach to separate them.
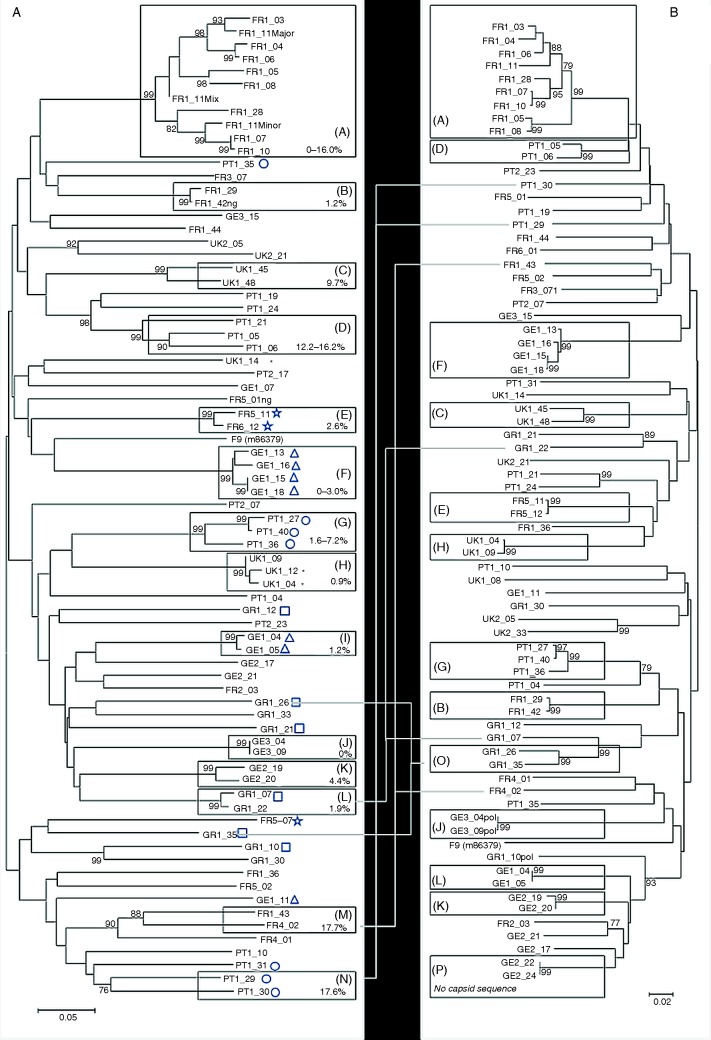
The final capsid phylogeny (Fig 4a) included 74 usable sequences (71 consensus, three for FR1_11). In total, 46 strains (pairwise genetic distance >20 per cent) were identified; the number of strains in each country and site ranged from 5 to 12 and 1 to 9, respectively (also Table 2). There was no evidence of wide-scale clustering at the geographical level; rather strains from each country were dispersed throughout the tree.
FIG 4:
Unrooted Kimura 2-parameter Neighbour joining tree of (a) 74 partial capsid sequences (including three sequences for FR1_11) and (b) 73 partial polymerase sequences obtained in this study. Each sequence has a unique ID made up of country code (FRance, GErmany GReece, PorTugal and UK) and site number (1–5) followed by sample number. Those strains represented by more than a single sequence (<20 per cent capsid divergence) are boxed, additionally labelled A–N, and the intrastrain capsid diversity indicated in the box. Where multiple sequences come from a single household they are indicated by an additional symbol (□, ○, Δ, *, ☆). Two new strain clusters O and P with high bootstrap support are also indicated. Where clustering varies between capsid and polymerase phylogenies, the sequences are linked in each phylogeny by a thin grey line. The feline calicivirus (FCV) vaccine strain F9 is also included (GenBank accession No. M86379). The percentage of replicate trees in which the associated taxa clustered together in bootstrap tests (1000 replicates) is shown next to the branches; only bootstrap values >75 per cent are shown. Distances are drawn to scale and relate to the distance bar
Of the 46 strains identified, 32 were represented by single sequences, the remaining 14 each being represented by more than 1 isolate and supported by bootstrap values of >75 per cent (A–N on Fig 4a). The most numerous of these was strain A (9 samples and 11 sequences, including 3 for FR1_11). Of these 14 strains, 13 were geographically restricted to single sites, the exception being strain M, found in two sites in France (FR1_43 and FR4_02). Eight of these strains had low levels of diversity (<5 per cent) suggestive of relatively acute or recent infection. The remaining six strains (A, C, D, G, M and N) showed greater diversity (5–20 per cent); these high levels of strain diversity have been found previously in endemically infected households (Coyne and others 2007b). Four of the strains (D, J, L and M) were present in more than one household/colony (data not presented). Five of the households had more than one strain of virus (range two to six; Fig 4a).
None of the sequences obtained clustered with live vaccine strain FCV-F9 (as represented by a published sequence M86379) or with the inactivated vaccine strain FCV_255 (data not presented). The median uncorrected nucleotide distances between F9 and sequences obtained in this study were 39.2 per cent (SD 3.1). This was similar to that obtained when the sequences obtained in this study were compared pairwise with themselves (median 40.1 per cent, SD 6.9).
Phylogenetic analysis of partial polymerase sequences was generally congruent with that based on the capsid (Fig 4b). However, the two sequences in polymerase clade O (GR1_26 and 35) were identified as distinct strains based on their capsid sequence suggesting possible recombination. No capsid sequence was available for those isolates in polymerase clade P (GE2_22 and 24). A similar finding was apparent for capsid clade L containing GR1_07 and 22; these sequences were closely related based on their capsid sequences but dispersed as distinct strains in the polymerase phylogeny. Finally, sequences clustering in clades M and N of the capsid phylogeny, albeit on the margins of the strain diversity threshold (20 per cent) of the present study, were also absent from the polymerase phylogeny.
Discussion
Previously the authors have been reconstructing the molecular epidemiology of FCV at a local and national level in the UK. These studies have pointed to both high FCV prevalence and strain diversity, produced by a combination of rapid evolution and occasional geographical dispersal. Here the authors report for the first time how FCVs are dispersed internationally, showing that, subject to limitations associated with the authors’ sampling methodology, FCV prevalence remains high across Europe, with little evidence for wide geographical dispersal of individual strains.
The isolation rates reported here are somewhat higher than previously reported, possibly reflecting different sampling methodologies. In two previous UK studies, in which cats were not selected based on clinical signs, isolation rates of 10 per cent (Porter and others 2008) and 9–11 per cent (Coyne and others 2012) for FCV and <1 per cent for FeHV-1 were achieved. In contrast, in the study reported here, some of the 426 cats were recruited specifically based on disease, notably the presence of LGSC (N=26; 6 per cent). This inclusion rate is much higher than would be expected in the general population where the prevalence of LGSC is estimated to be 0.7 per cent (Healey and others 2007). Since previous studies have shown this condition to be strongly associated with FCV (Belgard and others 2010), with 80 per cent or more of cats testing positive (Knowles and others 1989), over-representation of such cases in the study population might be expected to increase the FCV prevalence over that found when cats were more randomly recruited (Radford and others 2007).
Other risk factors associated with FCV shedding included age and vaccination status, confirming results of previous studies (Porter and others 2008, Coyne and others 2012). In this population, vaccinated cats up to 48 months of age were less likely to shed FCV than unvaccinated animals of similar ages, suggesting in this age group, vaccination may have reduced infection rates. Experimental studies have shown both vaccinated cats (Pedersen and Hawkins 1995, Poulet and others 2005) and cats recovered from acute FCV infection (Povey and Ingersoll 1975) remain susceptible to subsequent infection. However, such previous exposure is often associated with a reduction in the shedding of subsequent heterologous challenge (Povey and Ingersoll 1975, Poulet and others 2005), consistent with the reduced risk of testing FCV positive identified here. The loss of a demonstrable protective effect of vaccination on FCV infection over 48 weeks of age could represent the acquisition of immunity in the unvaccinated cats following field infection, a consequence of the high prevalence of FCV in the population. However, since the authors did not model here the time since last vaccine, the apparent loss of protective effect may also reflect increasing time since the last vaccination in older animals.
Sequence analyses identified 46 strains among the 72 isolates sequenced, confirming the high levels of strain diversity seen previously (Coyne and others 2012, Prikhodko and others 2014). Although the authors have not assessed the antigenicity of these viruses, it seems likely that because of the known localisation of neutralising epitopes in the regions sequenced, such genetic diversity would be associated with similar antigenic diversity. These levels of diversity pose a continual challenge to FCV control by acquired immune responses, something vaccine companies evaluate in relation to the antigens they choose (Hohdatsu and others 1999, Addie and others 2008, Porter and others 2008). In this context, it was interesting that, despite the time that has passed since its first isolation (Bittle and others 1960), FCV-F9 has not become a phylogenetic outlier, remaining within the genetic diversity of the more recent isolates sequenced here. This suggests the evolution of the antigenic domain of FCV may behave differently from other rapidly evolving viruses like influenza.
By comparing the phylogeny with the geographical origin of the isolates, the authors found no evidence of widespread international spread of FCV strains. Among the 46 strains described, 32 were only represented by a single isolate, and in only 13 and 14 cases were multiple variants of strains found in the polymerase and capsid phylogenies, respectively. These strain clusters were always confined to single countries, and generally to individual sites within a country. There was some limited support for geographical clustering beyond the existing strain definition in the present study in several countries best exemplified by a Portuguese cluster of five isolates including those in strain D (Fig 4a), suggesting occasionally more widespread and longer-term virus transmission as suggested previously in Japan (Sato and others 2002). However, the overall picture remains one of lack of international promiscuity, very distinct from human caliciviruses where some strains or variants such as human norovirus genotype ii.4 are widely distributed internationally (Ramani and others 2014). The authors’ current hypothesis is that the rate of FCV evolution is so high that evolutionary signals become saturated rapidly, leading to a loss of phylogenetic resolution over relatively short time periods (Coyne and others 2012). This will only be resolved when longer sequences are obtained.
Some of the strains the authors identified were associated with high levels of within-strain diversity, notably strains A, C, D and G. These levels of diversity are reminiscent of those seen in endemically infected, multicat households, where the authors hypothesised that high levels of population immunity lead to rapid evolution by positive selection (Coyne and others 2007b).
In five cases, individual households/communities of cats were infected with more than one strain (Fig 4a). The most extreme example was a Greek rescue shelter where all six isolates sequenced were distinct strains. This observation suggests lots of viruses coming together into a single population but with little transmission, a pattern either attributable to very good internal biosecurity (Radford and others 2001b, Coyne and others 2007a), or suggesting that the cats were sampled on arrival in the shelter before transmission had a chance to occur. A more detailed understanding of the population demographics and husbandry of these populations may shed more light on the behaviours that underlie these phylogenetic patterns.
In the present study, the authors found no FCV F9-like sequences, a common strain used in live vaccines. This is in contrast to previous studies which have reported albeit rare occurrences of F9-like viruses in the general cat population (Radford and others 2001a, Coyne and others 2007b, 2012). The origins of these F9-like viruses are unknown but because of their reported close sequence similarity to the original F9 sequence, it seems reasonable they would have originated from regular use of live vaccines containing this strain. The authors’ failure to find them in this study may reflect the fact that cats within 1 month of receiving a live vaccination were excluded from the study population. Together, this suggests that on the rare occasions cats do shed vaccine-derived virus following vaccination, the duration is short, and the potential for onward transmission limited, consistent with reversion to virulence assays and dissemination experiments conducted for registration of live vaccines. This allows these vaccine-derived viruses to sporadically appear at a low level in the population in a state of evolutionary stasis.
The close co-circulation of distinct strains, and lack of sterilising immunity in individual cats, provides an ideal opportunity for mixed infections (Coyne and others 2006b). Sequence data suggested such mixed infections both at the quasispecies (within-strain) level and cats infected with more than one strain (FR1_11, 37 and 40). These diverse populations within an individual host provide an ideal opportunity for recombination (Lai 1992), and the identification of several incongruences between capsid and polymerase phylogenies presented here provide some evidence of recombination in these populations. Recombination is a feature of the evolution of many RNA viruses (Lai 1992), and has been described previously for caliciviruses in general and FCV in particular (Coyne and others 2006b); there is some evidence that a hot spot for the necessary template switch exists at the junction between ORF1 and ORF2, in between the polymerase and capsid sequences generated in this study (Oliver and others 2004, Bull and others 2005), driven by predicted RNA secondary structure (Jiang and others 1993, Porter 2004, Coyne and others 2006b, Prikhodko and others 2014). Formal conformation of recombination requires sequencing across the putative recombination site, but was beyond the scope of this project.
For 19 (21 per cent) of the 91 samples that showed FCV-like cytopathic effect in cell culture the authors were unable to generate PCR products, despite using several primer combinations (see methods and table 1). This is a well recognised phenomenon for RNA viruses, especially when trying to amplify the most variable regions of the virus. In a previous study this ‘failure rate’ was much higher (30–45 per cent) (Coyne and others 2012), possibly reflecting an improvement in primer design and PCR technology in the present study. These samples refractory to sequencing represent an intriguing resource for future studies using new technologies such as next-generation sequencing (Radford and others 2012).
Study limitations: This study is the largest of its kind to prospectively study FCV evolution and epidemiology in Europe. However, in order to make sample collection feasible, a convenience-based study design was required. This may impact some of the epidemiological and phylogenetic conclusions, limiting their generalisability. Further analyses with a bigger sample size would increase the power of the study. Ultimately, longer sequences would be needed to confirm recombination and to increase the resolution of the phylogenies in space and time.
The authors have described for the first time the molecular epidemiology of a prospective sample of FCV isolates from a sample of European countries, confirming both the strain diversity of FCV, and the lack of dominant strains within the population.
Footnotes
Funding: VO'H was on a Nuffield Sponsored Summer research placement. The work was funded by Virbac.
Competing interests: DM, CL and TA work for Virbac, a company that makes FCV vaccines based on FCV strain F9. ADR also receives support from other vaccine manufacturers including MSD Animal Health.
References
- ADDIE D., POULET H., GOLDER M. C., MCDONALD M., BRUNET S., THIBAULT J. C. & HOSIE M. J. (2008) Ability of antibodies to two new caliciviral vaccine strains to neutralise feline calicivirus isolates from the UK. Veterinary Record 163, 355–357 10.1136/vr.163.12.355 [DOI] [PubMed] [Google Scholar]
- BANNASCH M. J. & FOLEY J. E. (2005) Epidemiologic evaluation of multiple respiratory pathogens in cats in animal shelters. Journal of Feline Medicine & Surgery 7, 109–119 10.1016/j.jfms.2004.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- BELGARD S., TRUYEN U., THIBAULT J. C., SAUTER-LOUIS C. & HARTMANN K. (2010) Relevance of feline calicivirus, feline immunodeficiency virus, feline leukemia virus, feline herpesvirus and Bartonella henselae in cats with chronic gingivostomatitis. Berliner und Munchener tierarztliche Wochenschrift 123, 369–376 [PubMed] [Google Scholar]
- BITTLE J. L., YORK C. J., NEWBERNE J. W. & MARTIN M. (1960) Serologic relationship of new feline cytopathogenic viruses. American Journal of Veterinary Research 21, 547–550 [Google Scholar]
- BULL R. A., HANSMAN G. S., CLANCY L. E., TANAKA M. M., RAWLINSON W. D. & WHITE P. A. (2005) Norovirus recombination in ORF1/ORF2 overlap. Emerging Infectious Diseases Dis 11, 1079–1085 10.3201/eid1107.041273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CARTER M. J., MILTON I. D., MEANGER J., BENNETT M., GASKELL R. M. & TURNER P. C. (1992) The complete nucleotide sequence of a feline calicivirus. Virology 190, 443–448 10.1016/0042-6822(92)91231-I [DOI] [PubMed] [Google Scholar]
- COUTTS A. J., DAWSON S., WILLOUGHBY K. & GASKELL R. M. (1994) Isolation of feline respiratory viruses from clinically healthy cats at UK cat shows. Veterinary Record 135, 555–556 [PubMed] [Google Scholar]
- COYNE K. P., CHRISTLEY R. M., PYBUS O. G., DAWSON S., GASKELL R. M. & RADFORD A. D. (2012) Large-scale spatial and temporal genetic diversity of feline calicivirus. Journal of Virology 86, 11356–11367 10.1128/JVI.00701-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- COYNE K. P., EDWARDS D., RADFORD A. D., CRIPPS P., JONES D., WOOD J. L., GASKELL R. M. & DAWSON S. (2007a) Longitudinal molecular epidemiological analysis of feline calicivirus infection in an animal shelter: a model for investigating calicivirus transmission within high-density, high-turnover populations. Journal of Clinical Microbiology 45, 3239–3244 10.1128/JCM.01226-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- COYNE K. P., GASKELL R. M., DAWSON S., PORTER C. J. & RADFORD A. D. (2007b) Evolutionary mechanisms of persistence and diversification of a calicivirus within endemically infected natural host populations. Journal of Virology 81, 1961–1971 10.1128/JVI.01981-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- COYNE K. P., JONES B. R., KIPAR A., CHANTREY J., PORTER C. J., BARBER P. J., DAWSON S., GASKELL R. M. & RADFORD A. D. (2006a) Lethal outbreak of disease associated with feline calicivirus infection in cats. Veterinary Record 158, 544–550 10.1136/vr.158.16.544 [DOI] [PubMed] [Google Scholar]
- COYNE K. P., REED F. C., PORTER C. J., DAWSON S., GASKELL R. M. & RADFORD A. D. (2006b) Recombination of Feline calicivirus within an endemically infected cat colony. Journal of General Virology 87, 921–926 10.1099/vir.0.81537-0 [DOI] [PubMed] [Google Scholar]
- DAWSON S., BENNETT D., CARTER S. D., BENNETT M., MEANGER J., TURNER P. C., CARTER M. J., MILTON I. & GASKELL R. M. (1994) Acute arthritis of cats associated with feline calicivirus infection. Research in Veterinary Science 56, 133–143 10.1016/0034-5288(94)90095-7 [DOI] [PubMed] [Google Scholar]
- FARAWAY J. J. (2006) Extending the linear model with R: generalized linear, mixed effects and nonparametric regression models. Boca Raton, FL: Chapman & Hall/CRC [Google Scholar]
- GLENN M., RADFORD A. D., TURNER P. C., CARTER M., LOWERY D., DESILVER D. A., MEANGER J., BAULCH-BROWN C., BENNETT M. & GASKELL R. M. (1999) Nucleotide sequence of UK and Australian isolates of feline calicivirus (FCV) and phylogenetic analysis of FCVs. Veterinary Microbiology 67, 175–193 10.1016/S0378-1135(99)00043-7 [DOI] [PubMed] [Google Scholar]
- HEALEY K. A., DAWSON S., BURROW R., CRIPPS P., GASKELL C. J., HART C. A., PINCHBECK G. L., RADFORD A. D. & GASKELL R. M. (2007) Prevalence of feline chronic gingivo-stomatitis in first opinion veterinary practice. Journal of Feline Medicine & Surgery 9, 373–381 10.1016/j.jfms.2007.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- HELPS C. R., LAIT P., DAMHUIS A., BJORNEHAMMAR U., BOLTA D., BROVIDA C., CHABANNE L., EGBERINK H., FERRAND G., FONTBONNE A., PENNISI M. G., GRUFFYDD-JONES T., GUNN-MOORE D., HARTMANN K., LUTZ H., MALANDAIN E., MOSTL K., STENGEL C., HARBOUR D. A. & GRAAT E. A. (2005) Factors associated with upper respiratory tract disease caused by feline herpesvirus, feline calicivirus, Chlamydophila felis and Bordetella bronchiseptica in cats: experience from 218 European catteries. Veterinary Record 156, 669–673 10.1136/vr.156.21.669 [DOI] [PubMed] [Google Scholar]
- HOHDATSU T., SATO K., TAJIMA T. & KOYAMA H. (1999) Neutralizing feature of commercially available feline calicivirus (FCV) vaccine immune sera against FCV field isolates. Journal of Veterinary Medical Science 61, 299–301 10.1292/jvms.61.299 [DOI] [PubMed] [Google Scholar]
- JIANG X., WANG M., WANG K. & ESTES M. K. (1993) Sequence and genomic organization of Norwalk virus. Virology 195, 51–61 10.1006/viro.1993.1345 [DOI] [PubMed] [Google Scholar]
- KNOWLES J. O., DAWSON S., GASKELL R. M., GASKELL C. J. & HARVEY C. E. (1990) Neutralisation patterns among recent British and North American feline calicivirus isolates from different clinical origins. Veterinary Record 127, 125–127 [PubMed] [Google Scholar]
- KNOWLES J. O., GASKELL R. M., GASKELL C. J., HARVEY C. E. & LUTZ H. (1989) Prevalence of feline calicivirus, feline leukaemia virus and antibodies to FIV in cats with chronic stomatitis. Veterinary Record 124, 336–338 10.1136/vr.124.13.336 [DOI] [PubMed] [Google Scholar]
- KNOWLES J. O., MCARDLE F., DAWSON S., CARTER S. D., GASKELL C. J. & GASKELL R. M. (1991) Studies on the role of feline calicivirus in chronic stomatitis in cats. Veterinary Microbiology 27, 205–219 10.1016/0378-1135(91)90148-9 [DOI] [PubMed] [Google Scholar]
- LAI M. M. (1992) RNA recombination in animal and plant viruses. Microbiology Reviews 56, 61–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- OLIVER S. L., BROWN D. W., GREEN J. & BRIDGER J. C. (2004) A chimeric bovine enteric calicivirus: evidence for genomic recombination in genogroup III of the Norovirus genus of the Caliciviridae. Virology 326, 231–239 10.1016/j.virol.2004.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEDERSEN N. C., ELLIOTT J. B., GLASGOW A., POLAND A. & KEEL K. (2000) An isolated epizootic of hemorrhagic-like fever in cats caused by a novel and highly virulent strain of feline calicivirus. Veterinary Microbiology 73, 281–300 10.1016/S0378-1135(00)00183-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEDERSEN N. C. & HAWKINS K. F. (1995) Mechanisms for persistence of acute and chronic feline calicivirus infections in the face of vaccination. Veterinary Microbiology 47, 141–156 [DOI] [PubMed] [Google Scholar]
- PEDERSEN N. C., LALIBERTE L. & EKMAN S. (1983) A transient febrile “limping” syndrome of kittens caused by two different strains of feline calicivirus. Feline Practice 13, 26–35 [Google Scholar]
- PESAVENTO P. A., CHANG K. O. & PARKER J. S. (2008) Molecular virology of feline calicivirus. Veterinary Clinics of North America: Small Animal Practice 38, 775–786, vii 10.1016/j.cvsm.2008.03.002 [DOI] [PubMed] [Google Scholar]
- PORTER C. J. (2004) Studies on the molecular biology of feline calicivirus replication. University of Liverpool [Google Scholar]
- PORTER C. J., RADFORD A. D., GASKELL R. M., RYVAR R., COYNE K. P., PINCHBECK G. L. & DAWSON S. (2008) Comparison of the ability of feline calicivirus (FCV) vaccines to neutralise a panel of current UK FCV isolates. Journal of Feline Medicine & Surgery 10, 32–40 10.1016/j.jfms.2007.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- POULET H., BRUNET S., LEROY V. & CHAPPUIS G. (2005) Immunisation with a combination of two complementary feline calicivirus strains induces a broad cross-protection against heterologous challenges. Veterinary Microbiology 106, 17–31 10.1016/j.vetmic.2004.12.010 [DOI] [PubMed] [Google Scholar]
- POULET H., BRUNET S., SOULIER M., LEROY V., GOUTEBROZE S. & CHAPPUIS G. (2000) Comparison between acute oral/respiratory and chronic stomatitis/gingivitis isolates of feline calicivirus: pathogenicity, antigenic profile and cross-neutralisation studies. Archives of Virology 145, 243–261 10.1007/s007050050021 [DOI] [PubMed] [Google Scholar]
- POVEY C. & INGERSOLL J. (1975) Cross-protection among feline caliciviruses. Infection and Immunity 11, 877–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRIKHODKO V. G., SANDOVAL-JAIME C., ABENTE E. J., BOK K., PARRA G. I., ROGOZIN I. B., OSTLUND E. N., GREEN K. Y. & SOSNOVTSEV S. V. (2014) Genetic characterization of feline calicivirus strains associated with varying disease manifestations during an outbreak season in Missouri (1995–1996). Virus Genes 48, 96–110 10.1007/s11262-013-1005-0 [DOI] [PubMed] [Google Scholar]
- RADFORD A. D., ADDIE D., BELÁK S., BOUCRAUT-BARALON C., EGBERINK H., FRYMUS T., GRUFFYDD-JONES T., HARTMANN K., HOSIE M. J., LLORET A., LUTZ H., MARSILIO F., PENNISI M. G., THIRY E., TRUYEN U. & HORZINEK M. C. (2009) Feline calicivirus infection. ABCD guidelines on prevention and management. Journal of Feline Medicine & Surgery 11, 556–564 10.1016/j.jfms.2009.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RADFORD A. D., BENNETT M., MCARDLE F., DAWSON S., TURNER P. C., WILLIAMS R. A., GLENN M. A. & GASKELL R. M. (1999a) Quasispecies evolution of a hypervariable region of the feline calicivirus capsid gene in cell culture and persistently infected cats. Veterinary Microbiology 69, 67–68 10.1016/S0378-1135(99)00090-5 [DOI] [PubMed] [Google Scholar]
- RADFORD A. D., CHAPMAN D., DIXON L., CHANTREY J., DARBY A. C. & HALL N. (2012) Application of next-generation sequencing technologies in virology. Journal of General Virology 93, 1853–1868 10.1099/vir.0.043182-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RADFORD A. D., COYNE K. P., DAWSON S., PORTER C. J. & GASKELL R. M. (2007) Feline calicivirus. Veterinary Research 38, 319–335 10.1051/vetres:2006056 [DOI] [PubMed] [Google Scholar]
- RADFORD A. D., DAWSON S., RYVAR R., COYNE K., JOHNSON D. R., COX M. B., ACKE E. F., ADDIE D. D. & GASKELL R. M. (2003) High genetic diversity of the immunodominant region of the feline calicivirus capsid gene in endemically infected cat colonies. Virus Genes 27, 145–155 10.1023/A:1025772409539 [DOI] [PubMed] [Google Scholar]
- RADFORD A. D., SOMMERVILLE L., RYVAR R., COX M. B., JOHNSON D. R., DAWSON S. & GASKELL R. M. (2001a) Endemic infection of a cat colony with a feline calicivirus closely related to an isolate used in live attenuated vaccines. Vaccine 19, 4358–4362 10.1016/S0264-410X(01)00191-8 [DOI] [PubMed] [Google Scholar]
- RADFORD A. D., SOMMERVILLE L. M., DAWSON S., KERINS A. M., RYVAR R. & GASKELL R. M. (2001b) Molecular analysis of isolates of feline calicivirus from a population of cats in a rescue shelter. Veterinary Record 149, 477–481 10.1136/vr.149.16.477 [DOI] [PubMed] [Google Scholar]
- RADFORD A. D., WILLOUGHBY K., DAWSON S., MCCRACKEN C. & GASKELL R. M. (1999b) The capsid gene of feline calicivirus contains linear B-cell epitopes in both variable and conserved regions. Journal of Virology 73, 8496–8502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAMANI S., ATMAR R. L. & ESTES M. K. (2014) Epidemiology of human noroviruses and updates on vaccine development. Current Opinion in Gastroenterology 30, 25–33 10.1097/MOG.0000000000000022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SATO Y., OHE K., MURAKAMI M., FUKUYAMA M., FURUHATA K., KISHIKAWA S., SUZUKI Y., KIUCHI A., HARA M., ISHIKAWA Y. & TANENO A. (2002) Phylogenetic analysis of field isolates of feline calcivirus (FCV) in Japan by sequencing part of its capsid gene. Veterinary Research Communications 26, 205–219 10.1023/A:1015253621079 [DOI] [PubMed] [Google Scholar]
- SEAL B. S., RIDPATH J. F. & MENGELING W. L. (1993) Analysis of feline calicivirus capsid protein genes: identification of variable antigenic determinant regions of the protein. Journal of General Virology 74(Pt 11), 2519–2524 10.1099/0022-1317-74-11-2519 [DOI] [PubMed] [Google Scholar]
- TAMURA K., PETERSON D., PETERSON N., STECHER G., NEI M. & KUMAR S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- TERWEE J., LAURITZEN A. Y., SABARA M., DREIER K. J. & KOKJOHN K. (1997) Comparison of the primary signs induced by experimental exposure to either a pneumotrophic or a ‘limping’ strain of feline calicivirus. Veterinary Microbiology 56, 33–45 10.1016/S0378-1135(96)01344-2 [DOI] [PubMed] [Google Scholar]
- WARDLEY R. C. (1976) Feline calicivirus carrier state. A study of the host/virus relationship. Archives of Virology 52, 243–249 10.1007/BF01348021 [DOI] [PubMed] [Google Scholar]
- WARDLEY R. C., GASKELL R. M. & POVEY R. C. (1974) Feline respiratory viruses--their prevalence in clinically healthy cats. Journal of Small Animal Practice 15, 579–586 10.1111/j.1748-5827.1974.tb06538.x [DOI] [PubMed] [Google Scholar]
- WARDLEY R. C. & POVEY R. C. (1977) The clinical disease and patterns of excretion associated with three different strains of feline caliciviruses. Research in Veterinary Science 23, 7–14 [PubMed] [Google Scholar]