SUMMARY
Repair of DNA double-strand breaks (DSBs) by non-homologous end-joining is critical for neural development, and brain cells frequently contain somatic genomic variations that might involve DSB intermediates. We now use an unbiased, high-throughput approach to identify genomic regions harboring recurrent DSBs in primary neural stem/progenitor cells (NSPCs). We identify 27 recurrent DSB clusters (RDCs) and, remarkably, all occur within gene bodies. Most of these NSPC RDCs were detected only upon mild, aphidicolin-induced replication stress, providing a nucleotide-resolution view of replication-associated genomic fragile sites. The vast majority of RDCs occur in long, transcribed, and late-replicating genes. Moreover, almost 90% of identified RDC-containing genes are involved in synapse function and/or neural cell adhesion, with a substantial fraction also implicated in tumor suppression and/or mental disorders. Our characterization of NSPC RDCs reveals a basis of gene fragility and suggests potential impacts of DNA breaks on neurodevelopment and neural functions.
INTRODUCTION
Evolutionarily conserved DSB repair pathways are required for maintenance of genome stability in mammalian cells (Lieber, 2010). Classical non-homologous end joining (C-NHEJ) is a critical somatic cell DSB repair pathway that is not dependent on sequence homology and which functions throughout the cell cycle (Alt et al., 2013). Evolutionarily conserved “core” C-NHEJ proteins include XRCC4 and DNA Ligase 4 (Lig4), which form an end-ligation complex (Alt et al., 2013; Boboila et al., 2012). C-NHEJ to a degree relies on DSB detection by the Ataxia telangiectasia-mutated (ATM) DNA damage response protein (Alt et al., 2013). Deficiency for C-NHEJ factors, or ATM and its downstream factors, leads to persistence of DSBs and their more frequent joining to other DSBs to generate chromosomal rearrangements, including translocations, deletions, inversions, and amplifications (Alt et al., 2013; Gapud and Sleckman, 2011). In the absence of C-NHEJ, such chromosomal rearrangements employ an alternative end-joining pathway (A-EJ) (Boboila et al., 2012).
C-NHEJ DSB repair is required for both immune and nervous system development (Gao et al., 1998). Inactivation of Xrcc4 or Lig4 in the mouse germline blocks lymphocyte development owing to the requirement for C-NHEJ to join antigen receptor variable region gene segments during V(D)J recombination (Alt et al., 2013). Xrcc4 or Lig4 inactivation also severely impairs neural development, leading to widespread apoptotic death of early post-mitotic neurons and associated late embryonic lethality (Barnes et al., 1998; Gao et al., 1998; Frank et al., 2000). Neuronal loss and embryonic lethality in C-NHEJ-deficient mice are rescued by p53 deficiency, indicating that both result from a p53-dependent checkpoint response to unrepaired DSBs (Frank et al., 2000; Gao et al., 2000). However, V(D)J recombination and, correspondingly, B cell development, is not rescued in C-NHEJ/p53 double-deficient mice, which routinely develop lethal progenitor B cell lymphomas with clonal translocations and amplifications involving fusion of V(D)J recombination-associated DSBs in the immunoglobulin heavy chain (IgH) and c-Myc oncogene loci via A-EJ (Difilippantonio et al., 2002; Hu et al., 2015; Zhu et al., 2002). Notably, C-NHEJ/p53 double-deficient mice also develop medulloblastomas (MBs) in situ (Lee and McKinnon, 2002; Zhu et al., 2002). Moreover, neural stem/progenitor cell (NSPC)-specific inactivation of Xrcc4 in p53-deficient mice leads to MBs that harbor recurrent clonal translocations, amplifications and deletions (Yan et al., 2006).
Brain cells frequently contain somatic genomic variations, including deletions, and rearrangements, which in some cases are linked to retrotransposition (Erwin et al., 2014; McConnell et al., 2013; Poduri et al., 2013). In this regard, single-cell sequencing of human frontal cortex neurons revealed that up to 41% had at least one megabase (Mb)-scale de novo copy number variation (CNV), most of which were deletions (McConnell et al., 2013). Due to technical limitations of such analyses, the actual frequency of these CNVs might be even higher (Erwin et al., 2014). Such somatic changes have been speculated to generate neuronal diversity and result in greater variance of cellular and organismal phenotypes (Erwin et al., 2014; Muotri and Gage, 2006). In theory, genomic aberrations in NSPCs might be transmitted to daughter cells and, thereby, contribute to genomic mosaicism in individual neurons or glial cells, where they could influence aspects of normal or abnormal brain function (Poduri et al., 2013). A better understanding of potential impacts of such genomic alterations in neural cells awaits elucidation of underlying mechanisms (Erwin et al., 2014; Poduri et al., 2013).
We have developed an unbiased high-throughput, genome-wide, translocation sequencing (HTGTS) approach to map, at nucleotide resolution, genome-wide DSBs based on their ability to translocate to endogenous or ectopic “bait” DSBs at a specific chromosomal location (Chiarle et al., 2011; Dong et al., 2015; Frock et al., 2015; Hu et al., 2015). HTGTS and a related approach revealed that off-target activities of lymphocyte-specific antigen receptor gene diversification enzymes generate recurrent DSBs or DSB clusters across the genome of B lineage cells (Chiarle et al., 2011; Hu et al., 2015; Klein et al., 2011; Meng et al., 2014; Zhang et al., 2012). For both mouse and human cells, recurrent DSBs or classes of DSBs are evident in genome-wide translocation landscapes, regardless of chromosomal location. The ability of such clusters of DSBs across the genome to be revealed by HTGTS results from cellular heterogeneity in 3-D genome organization (Alt et al., 2013; Frock et al., 2015; Zhang et al., 2012), a phenomenon that allows recurrent DSBs to be reliably identified by HTGTS baits on a different chromosome (Frock et al., 2015). In the absence of recurrent DSBs, proximity causes DSBs in cis along a given chromosome to preferentially join (Dong et al., 2015; Frock et al., 2015; Zhang et al., 2012). Within a cis chromosome, translocation frequency is further enhanced between sequences within topological domains or loops due to increased interaction or other processes (Alt et al., 2013; Hu et al., 2015; Zhang et al., 2012). Together, these properties of chromosomal translocations allow use of HTGTS as a remarkably sensitive DSB detection method.
We now apply an enhanced linear amplification-mediated HTGTS approach (Frock et al., 2015) to map DSBs in NSPCs. These studies reveal a large set of recurrently broken genes and suggest potential mechanisms underlying their origin.
RESULTS
High-throughput Mapping of DSBs and Translocations in NSPCs
For initial studies, we performed HTGTS on NSPCs isolated from mice deficient for XRCC4 and p53 (Xrcc4−/−p53−/− mice); since based on our prior studies we expected this background to be a rich source of NSPC DSBs (Gao et al., 1998; Yan et al., 2006). We used a Cas9:sgRNA approach to generate an initial HTGTS bait DSB as we described for other studies (Dong et al., 2015; Frock et al., 2015; Hu et al., 2015). Specifically, we designed an sgRNA (“Chr12-sgRNA-1”) that targets a Cas9:sgRNA-generated bait DSB to an intergenic region approximately 52 kb telomeric of N-myc on chromosome (Chr) 12 (Figure 1A, top). The Chr12-sgRNA-1 was introduced into cultured NSPCs, which were then maintained for 4.5 days and harvested for HTGTS. We used a primer that allowed us to identify endogenous “prey” DSBs genome-wide that joined to centromeric broken ends of a Chr12-sgRNA-1 generated “bait” DSB (Figure 1A, top). In four separate experiments, we identified 32,144 independent HTGTS junctions. We visualized overall junction patterns along each individual chromosome via modified Circos plots (Frock et al., 2015) of the mouse genome separated into 2.5-Mb bins. These studies revealed that 61.4% (19,734) of HTGTS junctions mapped within 500 kb of the Chr12-sgRNA-1 target site, with the majority of these not representing translocations but rather representing rejoining of a given bait DSB following resection (Figure 1A; Figure S1A,B and Table S1) (Chiarle et al., 2011; Frock et al., 2015).
Figure 1. Elucidation of DSBs in Xrcc4−/−p53−/− NSPCs.
(A) Illustration of N-myc locus and sgRNA target site (vertical black arrowhead) and location and orientation of HTGTS primer (green arrowhead). Cen, centromere; Tel, telomere. E, exon. (B) Circos plot of the mouse genome divided into individual chromosomes showing the genome-wide HTGTS junction pattern of Chr12-sgRNA-1-mediated bait DSBs in Xrcc4−/−p53−/− NSPCs binned into 2.5-Mb regions (black bars); bar height indicates number of translocations per bin on a log scale. 20,000 junctions from four independent experiments are plotted. Red line indicates recurrent translocations between Chr12 bait DSBs (red arrowhead) and an RDC within Lsamp on Chr16; an RDC within Npas3 on Chr12 is denoted by green line. Blue star denotes translocations to sgRNA off-target site. See also Figure S1.
After excluding the break-site resections, a substantial fraction (~8%) of the remaining Chr12 junctions involved prey DSBs spread over Chr12 (Figure 1A), a phenomenon resulting from joining of bait DSBs to wide-spread low level DSBs in cis due to 3-D spatial proximity (Alt et al., 2013; Zhang et al., 2012). We estimate that the frequency of prey DSBs participating in such break-site chromosome translocations in XRCC4-deficient NSPCs is, at a minimum, about 8 per cell (Table S2). Indeed, the actual DSB frequency likely is much higher since most DSBs are rejoined locally and do not translocate (Alt et al., 2013). Beyond the break-site junctions, the remainder of the 9,966 (31%) HTGTS junctions distributed broadly throughout the genome (Table S1 and Figure 1A; for convenience, bins with less than 5 junctions are not illustrated on Circos plots but examples are shown in Figure S1C).
We used the SICER (Spatial clustering approach for the Identification of ChIP-Enriched Regions) algorithm (Zang et al., 2009) to perform an unbiased assay of the HTGTS library data with the goal of identifying significantly enriched junction clusters across the XRCC4-deficient NSPC genome (see Supplemental Experimental Procedures). This analysis revealed three recurrent translocation clusters; notably, two of these clusters were located specifically within the limbic system-associated membrane protein (Lsamp) gene on Chr16 and the neuronal PAS domain protein 3 (Npas3) gene on Chr12, while the other represented a Chr12-sgRNA-1 off-target (OT) site on Chr12 (Figure 1). As the prey DSBs participating in recurrent translocations to Lsamp and Npas3 were spread broadly across these long genes (see below), we refer to them as “recurrent DSB clusters” (RDCs). Finally, we also found the same three enriched junction clusters by an independent custom MACS-based pipeline (See Supplemental Experimental Procedures).
The Lsamp and Npas3 Genes are Prone to DSBs and Translocations in NSPCs
To elucidate potential underlying mechanisms, we examined HTGTS junctions between Chr12-sgRNA-1 bait DSBs and prey DSBs across the 2.2 Mb-long Lsamp gene in Xrcc4−/−p53−/− NSPCs. By convention, prey HTGTS junctions are denoted “+” if the prey is read from the junction in a centromere-to-telomere direction and “−“ if in the opposite direction (Figure 2A, top; Chiarle et al., 2011). Lsamp translocations occurred at similar levels to prey DSBs in both the plus (+) and minus (−) orientations, indicating that Chr12-sgRNA-1 bait DSBs can join to either end of a prey DSB (Figure 2A), similar to what is found for translocation of bait DSBs to prey DSBs genome-wide in B cells (Chiarle et al., 2011). Translocation junctions distributed broadly across Lsamp, but were most enriched over an approximately 600-kb internal region (Figure 2A). About 0.5% (51/9,966) of total inter-chromosomal translocations involved Lsamp (Table S1). To independently confirm accumulation of recurrent DSBs in Lsamp in Xrcc4−/−p53−/− NSPCs, we used a Cas9:sgRNA (Chr16-sgRNA-1) to introduce bait DSBs in an intergenic region approximately 8 Mb upstream of Lsamp (Figure 2B). We found that Chr16-sgRNA-1 bait junctions were again substantially enriched across Lsamp in both + and − orientations, with Lsamp translocations occurring at a level of about 2% (151/7,965) of total inter-chromosomal translocations (Table S1), consistent with anticipated proximity effects (Alt et al., 2013; Zhang et al., 2012). For comparison, when normalized as described above for wide-spread DSBs, we estimate that 60% of NSPCs have one Lsamp DSB that translocates to a bait DSB (Table S2); again, the number of Lsamp DSBs could be much higher, because we only include in our estimate the small fraction of total DSBs that translocate (See Discussion for details)
Figure 2. Identification and Characterization of Lsamp RDC.
(A) Translocation cluster between Chr12-sgRNA-1-mediated bait DSBs and prey DSBs on Chr16 in Xrcc4−/−p53−/− NSPCs. Upper; diagram of translocation outcomes (see text for details). Green arrowhead denotes HTGTS primer. Middle; graph of Chr16 prey junctions (normalized to 7,070 inter-chromosomal junctions from four independent experiments). Junctions in centromere-to-telomere orientation (+) are in blue, and junctions in telomere-to- centromere orientation (−) are in red. Bin size, 1 Mb. Lower; enlarged view of region around Lsamp showing HTGTS junctions (related to panel above as indicated by dashed lines; genomic coordinates are below). Junction enrichment within Lsamp (highlighted in yellow) was significant (P=3.33×10−7; see HTGTS Junction Enrichment Analysis in Supplemental Experimental Procedures). (B) Upper; illustration of intra-chromosomal translocations formed between Lsamp-proximal Chr16-sgRNA-1-mediated bait DSBs and prey DSB cluster (highlighted in yellow). Middle; prey junctions captured by Lsamp-proximal bait DSBs over a 16-Mb Chr16 region, combined from three independent Xrcc4−/−p53−/− experiments. Bin size, 100 kb. Details as in panel A. Lower; enlarged view of region around Lsamp showing HTGTS junctions (related to panel above with dashed lines with genomic coordinates indicated at the bottom). RefGene and GRO-seq data are shown (ordinate indicates normalized GRO-seq counts; reads are shown in plus (blue) and minus (red) orientation). Junction enrichment within Lsamp was highly significant (P=1.54×10−13), as described in (A). 5,917 junctions (945 intra-chromosomal translocations on Chr16 more than 10 kb from the bait-DSB site and 4,972 inter-chromosomal translocations) are plotted. (C) Upper; illustration of translocation outcomes between c-Myc25xI−SceI cassette (yellow box) bait DSBs and prey DSBs on Chr16 with details as in panel A. Middle; Chr16 prey junctions from four independent experiments in ATM−/−ROSAI−SceI−GRc-Myc25xI−SceI NSPCs with Lsamp RDC in yellow. A purple rectangle and star indicates region corresponding to Igλ, and a green rectangle and star indicates regions corresponding to Bcl-6 and Lpp. Lower; enlarged view of indicated RDC-containing region, as described for panel (B). RefGene and GRO-seq reads from ATM−/−ROSAI−SceI−GRc-myc25xI−SceI NSPCs are shown as for panel (B). 7,070 inter-chromosomal junctions are plotted. Junctions within Lsamp were significantly enriched (P=5.43×10−6), as described in (A). (D) HTGTS analysis of activated ATM−/−ROSAI−SceI−GRc-myc25xI−SceI B cells and GRO-seq analyses of activated B cells (Meng et al., 2014), displayed as described for panel B. See also Table S1.
To further assess potential mechanisms of Lsamp translocations, we employed I-SceI-mediated bait DSBs within c-Myc (“c-Myc25xI−SceI”) on Chr15 (Chiarle et al., 2011) for HTGTS analyses of ATM-deficient (ATM−/−) NSPCs. These studies revealed overall translocation patterns, including the presence of an Lsamp RDC, that were generally similar to those observed for Xrcc4−/−p53−/− NSPCs (Figure 2C and data not shown). Because we had previously generated HTGTS libraries from the same c-Myc25xI−SceI bait DSBs in B cells (Meng et al., 2014), we could directly compare HTGTS translocation junctions along Chr16 in B cells versus those in NSPCs (Table S1). In this regard, HTGTS libraries from primary IgH class switch recombination (CSR)-stimulated B lymphocytes did not reveal any junction enrichment in Lsamp (Figure 2D). On the other hand, activated B-cell HTGTS libraries exhibited two HTGTS junction peaks in Chr16 not present in NSPCs libraries (Figure 2D; compare with Figure 2C). One B-cell peak (purple star) contained junctions spread broadly over the Igλ light chain locus and the other (green star) contained two focal peaks of junctions in Bcl-6 and in a transcribed region near Lpp. Notably, the latter two are known off-targets of activation-induced cytidine deaminase (AID), the B-cell enzyme that induces DSB formation for IgH CSR (Meng et al., 2014). For comparison, in size-matched HTGTS libraries (normalized to 7,000 inter-chromosomal junctions), activated B-cell libraries contained 12 junctions targeted to a site of convergent transcription downstream of the TSS of Bcl-6 (the strongest Chr16 AID off-target gene; Meng et al., 2014), while NSPC libraries contained over 40 junctions spread across the body of Lsamp. By performing global run-on sequencing analyses (GRO-seq; Core et al., 2008), we found active transcription over the entire Lsamp gene in Xrcc4−/−p53−/− and ATM−/− NSPCs (lower panels in Figure 2B and C). In contrast, examination of GRO-seq analyses of activated B cells (Meng et al., 2014) revealed that Lsamp is not detectably transcribed (Figure 2D, lower panel).
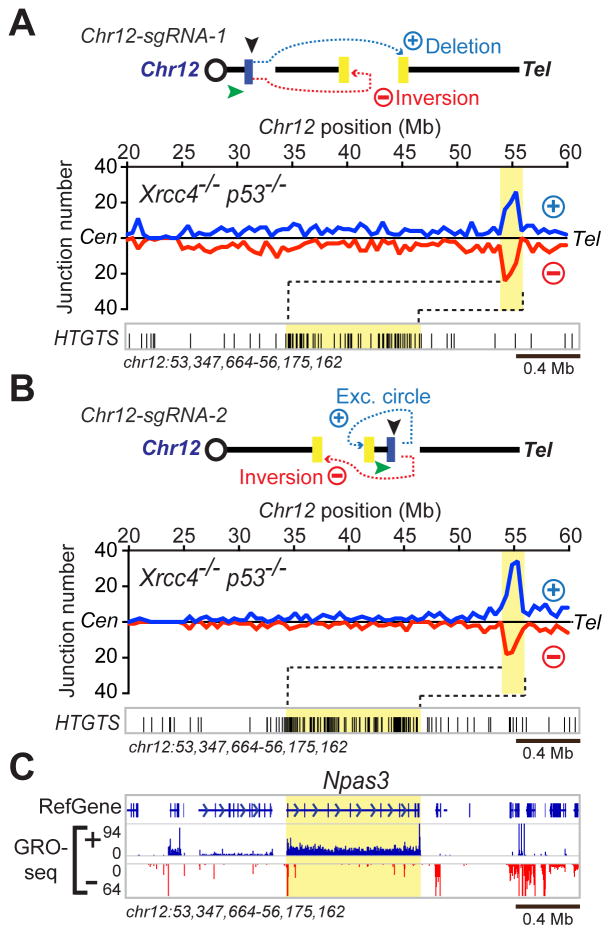
We also examined the Npas3 RDC in detail in Xrcc4−/−p53−/− NSPCs (Figure 3). Similar to junctions identified in Lsamp, junctions were detected in both orientations across Npas3 when cloned from the Chr12-sgRNA-1 bait DSB site located 40 Mb centromeric of the gene (Figure 3A). These intra-chromosomal junctions to the 825-kb Npas3 gene occurred at a frequency that corresponded to about 1% of all inter-chromosomal HTGTS junctions (Table S1). Junction enrichment in Npas3 again was further enhanced when a different sgRNA (Chr12-sgRNA-2) was used to move the bait DSB approximately 6 Mb telomeric to Npas3 (Figure 3B), with intra-chromosomal translocations to Npas3 DSBs occurring at a level corresponding to almost 3% of inter-chromosomal translocations captured (Table S1). GRO-seq analyses of Xrcc4−/−p53−/− NSPCs indicated active transcription over the entire Npas3 gene (Figure 3C).
Figure 3. Identification of Recurrent DSB Cluster in Npas3.
(A) Upper; illustration of intra-chromosomal translocation outcomes between Chr12-sgRNA-1-mediated bait DSBs and Chr12 prey DSBs in Xrcc4−/−p53−/− NSPCs. Lower; prey junctions identified from Chr12-sgRNA-1 bait DSBs over a 40-Mb Chr12 region containing the Npas3 RDC. Data are combined from four independent experiments; bin size, 500 kb. 13,455 junctions (3,489 junctions located more than 10 kb from either side of the bait DSB and 9,966 inter-chromosomal junctions) are plotted. Junction enrichment within Npas3 was highly significant (P=2.63×10−15; see HTGTS Junction Enrichment Analysis in Supplemental Experimental Procedures). Other details as in Figure 2A. (B) Upper, illustration of intra-chromosomal translocation outcomes between Chr12-sgRNA-2 bait DSBs and Chr12 prey DSBs, presented as in (A). Lower; prey junctions identified from Chr12-sgRNA-2 bait DSBs over a 40-Mb Chr12 region containing the Npas3 RDC. Data combined from three independent experiments are presented as in (A); bin size, 500 kb. 5,471 total junctions (1,366 Chr12 junctions located more than 10 kb from either side of the bait DSB and 4,105 inter-chromosomal junctions) are plotted. Junction enrichment within Npas3 region was significant (P =2.03×10−14), as described in (A). (C) GRO-seq and RefGene information (bottom) shown as described for Figure 2B. See also Table S1.
HTGTS studies with the c-Myc25xI−SceI bait DSBs revealed the Lsamp RDC in both wild-type (WT) as well as in ATM-deficient NSPCs, while the Chr12-sgRNA-1 revealed the Lsamp RDC in Xrcc4−/−p53−/−, but not wild-type NSPCs (Table S1). None of the bait DSBs used revealed Npas3 RDCs in WT HTGTS libraries and only the Chr12-sgRNAs revealed the Npas3 RDC in the Xrcc4−/−p53−/− NSPCs (Figures 1 and 3; data not shown). We suspect that the differential recovery of these two RDCs may be related to the frequency at which the different bait and prey DSBs are induced or persist in the different genotypes (Dong et al., 2015), as both the Lsamp and Npas3 RDCs were readily apparent in HTGTS studies employing bait DSBs on Chr12, -15, and -16, respectively, in Xrcc4−/−p53−/− NSPCs under conditions in which these prey DSBs are further enhanced; and Lsamp and Npas3 were also detected under such conditions by bait DSBs on Chr15 or Chr12 in WT NSPCs (Figures 4–6; see below).
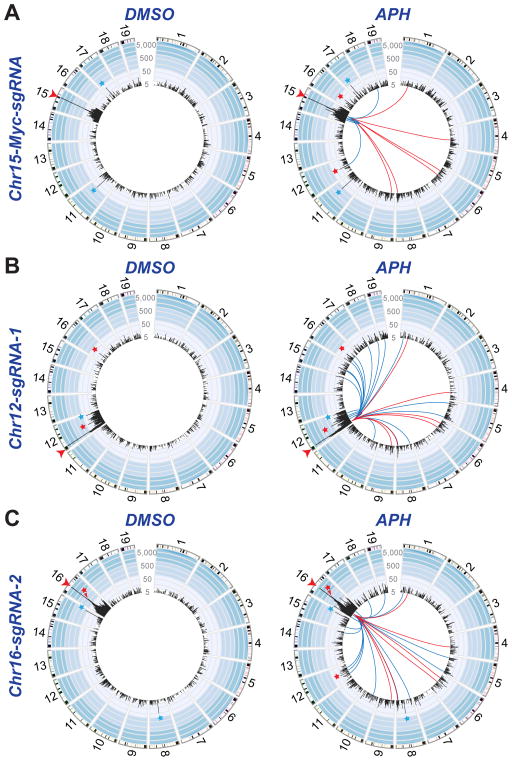
Figure 4. Genome-wide Identification of Replication Stress-induced RDCs in NSPCs.
(A) Circos plot showing HTGTS junctions from Cas9:sgRNA-mediated bait DSBs on Chr15 (Chr15-Myc-sgRNA) in DMSO- (left) or APH-treated (right) Xrcc4−/−p53−/− NSPCs. Junctions from three independent experiments per condition were combined and randomly down-sampled so that identical numbers of junctions for each condition (n=17,701 junctions) could be shown in each plot. (B) HTGTS junctions from bait DSBs on Chr12 (Chr12-sgRNA-1), as in (A). (C) HTGTS junctions identified in three (DMSO, left) or four (APH, right) experiments from Chr16-sgRNA-2-mediated bait DSBs; other details as in (A). For all panels, the bait DSB site (red arrowhead) and sgRNA off-target sites (blue stars) are denoted. Lines in the middle of the plot connect the break-site to the SICER-identified replication stress-induced RDCs that were identified for that particular break-site. Red lines indicate 6 RDCs detected by bait DSBs on all three tested chromosomes. Blue lines in each plot indicate RDCs detected by bait DSBs on two of the three tested break-sites, which numbered 5 for the Chr15-Myc-sgRNA break-site (panel A), 19 for Chr12-sgRNA-1 break site (panel B), and 16 for the Chr16-sgRNA-2 break site (panel C). Red stars indicate location of Lsamp and Npas3. See also Figure S3.
Figure 6. Replication Stress-induced RDCs in Repair-proficient NSPCs.
(A) Detection of RDCs on a different chromosome from the bait DSBs on Chr15 or Chr12. Three are shown, including Lsamp (B), Nrxn1 (C) and Ctnna2 (D); others are shown in Figure S5. Libraries were normalized as described in Figure 4 (Chr15 bait libraries, 14,525 junctions; Chr12 bait libraries, 10,088 junctions). Details are as in Figure 5. (E, F) Detection of RDCs in Csmd3 (E) or Nrxn3 (F) from two bait DSBs of which one lies on the RDC-containing chromosome. Libraries were normalized as described above. Other details as in Figure 5. See also Figure S5.
Elucidation of Replication Stress-induced DSBs and Translocations in NSPCs
Given that NSPCs undergo extensive cell division both in vivo and in vitro (McKinnon, 2013), we investigated potential effects of DNA replication stress on DSB generation. Treatment with low doses of aphidicolin (APH), a DNA polymerase inhibitor, induces replication stress and, thus, has been widely used for common fragile site (CFS) analyses (Durkin and Glover, 2007; Glover et al., 1984). To identify genomic regions subject to DNA replication stress-associated DSBs, we treated Xrcc4−/−p53−/− NSPCs with either APH or vehicle control (dimethyl sulfoxide; DMSO) and performed HTGTS with bait DSBs generated, respectively, on either Chr12 (Chr12-sgRNA-1), Chr16 (Chr16-sgRNA-2), or Chr15 (Chr15-Myc-sgRNA). For each of the three bait DSBs, we performed at least three independent HTGTS experiments on control- or APH-treated cells. These experiments all were analyzed separately to confirm reproducibility, and then pooled, normalized to the same number of total junctions, and plotted in modified Circos plots to facilitate comparison of APH-induced RDCs found in the different bait libraries (Figures 4 and S2).
For the unbiased identification of junction enrichment across the genome in APH-treated versus control samples, we again employed SICER, which also is a method of choice for comparing two identical samples with or without a specific treatment (Zang et al. 2009) (Figure S2; see Supplemental Experimental Procedures). This analysis revealed 282, 156, and 294 candidate replication stress-induced RDCs, respectively, in HTGTS libraries generated from Chr12-, Chr15-, and Chr16-bait DSBs. For further analysis, we only considered RDCs that showed a significantly higher translocation density in libraries from APH-treated versus vehicle control-treated cells (P<0.05, one-tailed t test; see Supplemental Experimental Procedures). This criterion reduced the number of cluster candidates that were significantly enriched across all biological replicates to 69, 158, and 133 in Chr15-Myc-sgRNA-, Chr12-sgRNA-1-, and Chr16-sgRNA-2-based libraries, respectively (Table S3). While many of these might be bona fide replication stress-induced RDCs, for more detailed analyses we only considered APH-induced RDCs that were independently detected by at least two HTGTS bait DSB locations on different chromosomes (Figure S2A). Based on this stringent criterion, 26 of the 360 candidate replication stress-induced RDCs were identified from at least two bait DSB locations (Figure 4); strikingly, all of these, like the majority of all candidate RDCs, were in gene bodies (Figures 5 and S2-4). Notably, we verified these 26 RDCs with the MACS-based, custom pipeline mentioned above (Table S4). Translocation junctions within these RDCs occurred similarly in + and − orientations, again indicating that the bait DSB end could join to one or the other end of a given prey DSB within the RDC (Figure S3). Six of the 26 RDC-containing genes (“RDC-genes”) were detected by bait DSBs located on three different chromosomes (Figures 5A–C and S4A). Finally, as expected based on proximity effects (Alt et al., 2013), we found higher junction densities in replication stress-induced RDCs that were on the same chromosome as the bait DSBs that detected them (Figures 5D–F and S4C).
Figure 5. Characterization of Replication Stress-induced RDCs in XRCC4/p53-deficient NSPCs.
(A) APH-induced RDCs in Xrcc4−/−p53−/− NSPCs identified from bait DSBs located on three different chromosomes. Six APH-induced inter-chromosomal translocation clusters were detected by all three HTGTS strategies; the Ctnna2 (B) and Cdh13 (C) RDCs are shown and the other four are shown in Figure S4A. (B, C) HTGTS junctions in either DMSO- or APH-treated libraries prepared from the indicated bait DSBs. Genomic regions corresponding to RDCs are highlighted in yellow. RefGene tracks are shown. Libraries were normalized as described in Figure 4. (D–F) APH-induced RDCs in Xrcc4−/−p53−/− NSPCs in Csmd3 (D), Nrxn3 (E), and Cadm2 (F) identified from bait DSBs located on two different chromosomes. The panels are organized as for panels A,B, and C. All panels show 2 Mb on either side of the indicated RDC. See Figure S4 for additional examples of proximity-facilitated RDC identification.
We performed an identical set of assays for replication stress-induced RDCs in wild-type NSPCs, except that we only employed HTGTS bait DSBs from Chr15 or Chr12. Although wild-type NSPC HTGTS experiments yielded somewhat lower total junction numbers than Xrcc4−/−p53−/− NSPC experiments, they revealed 13 of the 26 RDCs detected in Xrcc4−/−p53−/− NSPCs (Figures 6 and S5A-F). In addition, Lsamp appeared in WT cells as a replication stress-induced RDC. In total, six of the 14 wild-type RDCs (including Lsamp) were detected from both bait DSBs (Figures 6A–D and S5B). These studies show that replication stress-associated RDCs form in both WT and C-NHEJ (XRCC4)-deficient cells. As in repair-deficient NSPCs, location of the replication-stress induced RDC on the break-site chromosome in WT NSPCs resulted in higher junction densities (Figure 6E and F).
Analysis of translocation junctions between bait DSBs and replication stress-mediated RDCs revealed, strikingly, that approximately 60% of junctions in WT NSPCs were microhomology (MH)-mediated, while more than 90% of junctions in Xrcc4−/−p53−/− NSPCs were MH-mediated (Figure S5G; Table S5). Genome-wide translocation junctions showed a similar shift in MH usage between WT and Xrcc4−/−p53−/− NSPCs (Figure S5G). Together, these findings show that both the C-NHEJ DSB repair pathway and A-EJ pathways (that are biased towards longer MH usage) can mediate translocations of replication stress-associated DSBs and translocations to DSBs genome-wide in NSPCs.
Replication Stress-associated DSBs and Translocations Target Long, Actively Transcribed, Neural Genes
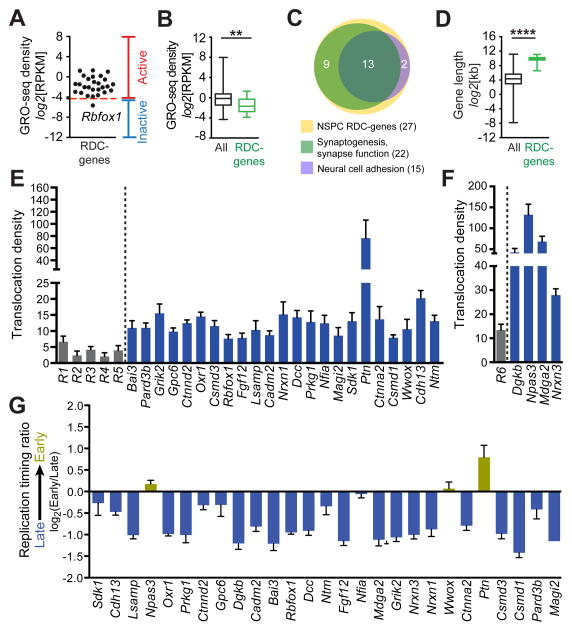
All 27 (including Lsamp in WT NSPCs) replication stress-induced RDCs identified by HTGTS and our unbiased, genome-wide enrichment analysis were located within genes (Figures 5, 6, S4, and S5), with all but one clearly being actively transcribed, albeit on average at slightly lower levels than other active genes in NSPCs (Figure 7A and B). Strikingly, detailed analysis of these RDC-genes revealed that 15/27 (55.6%) are involved in neural cell adhesion and 22/27 (81.5%) have roles in synaptogenesis and synaptic function (Figure 7C; Table S6). Moreover, the vast majority of these genes have been linked to neural disorders in mice and/or in humans (Table S6). We note, however, that expression of some of these genes is not restricted to neural cells. For example, Lsamp is expressed in fibroblasts where it is also fragile (Le Tallec et al., 2011); and Wwox, Pard3b, Oxr1, and Nfia are all expressed in B cells (Meng et al, 2014), with Wwox also being fragile in lymphocytes (Le Tallec et al. 2013). Likewise, Dcc is expressed in most normal tissues and is deleted in colon cancer (Fearon et al., 1990) (See also Discussion).
Figure 7. Replication Stress-induced RDCs in Long, Actively Transcribed, Neural Genes.
(A) Transcriptional activity (GRO-Seq) of the identified 27 genes containing replication stress-induced RDCs. Transcriptional activity cut-off value (RPKM = 0.05) is indicated by dashed red line. (B) Transcription rate of all active (RPKM ≥0.05) NSPC genes (black) and active replication stress-induced RDC-genes (green). Whiskers show minimum and maximum values; top and bottom edge of box plots correspond to 25th and 75th percentile, respectively; horizontal lines indicate the median; **P < 0.005, Kolmogorov-Smirnov (K–S) test. (C) Venn diagram of the indicated molecular functions among the 27 identified RDC-genes (yellow circle); 22 of 27 genes (81.5%; light green circle) have roles in synaptogenesis and synapse function. 15 of the 27 genes (55.6%; purple circle) have roles in neural cell adhesion, with the majority (13 of 15 genes, 86.7%) also having roles in synaptogenesis and synapse function. See Table S6 for a detailed description. (D) Gene length comparison of all active NSPC genes (black) and NSPC RDC-genes (green). Box-and-whisker plots show the binary logarithm of kb gene length; graph details as in (A); ****P < 0.0001, K-S test. (E) Five groups (R1-5) of 50 actively transcribed 15–20 kb genes each were randomly selected from three independent Xrcc4−/−p53−/− Chr12-sgRNA-1 bait DSB libraries and junction numbers within the concatenated regions determined (gray bars). Junction numbers within the indicated inter-chromosomal RDCs were determined in the same libraries (blue bars). Translocation density is indicated as junctions per Mb. (F) Translocation densities of concatenated average-size (15–25 kb) active genes on Chr12 (R6, n=62, gray bar) or intra-chromosomal Chr12 RDCs (blue bars). Data represent mean and SEM of libraries from three independent Chr12-sgRNA-1 bait DSB experiments. (G) Replication timing analysis of RDC-genes (see Experimental Procedures for details). Average and SEM are shown. See also Figures S6, S7, and Table S6.
With the exception of Ptn, all genes harboring replication stress-induced RDCs in NSPCs were longer than 100 kb, which is significantly above the average gene length in the mouse genome (Figure 7D). To test whether these long genes incur more translocations and, thus, form RDCs simply because of their larger target size, we computationally sampled and concatenated randomly selected, active genes of average size (15–25 kb) from HTGTS libraries into regions of approximately 1 Mb, and compared size-normalized junction density in these regions to that of the 27 RDC genes (Figure 7E and F). Even when normalized by size, the large genes harboring RDCs in NSPCs showed higher junction density than predicted by size alone (Figure 7E and F, Figure S6A and B). Moreover, the large genes harboring RDCs represented only a small fraction (1.5%) of the 1,761 actively transcribed NSPC genes larger than 100 kb, which further indicates that the observed accumulation of DSBs in these genes in response to replication stress is not just due to size per se. These findings indicate that this subset of long genes in NSPCs is disproportionately susceptible to DSB-induced genomic instability.
To gain further insight into potential underlying mechanisms, we investigated the replication timing of the 27 identified RDC-genes in NSPCs by examining existing murine neural progenitor replication timing data (Hiratani et al., 2008; Pope et al., 2014). Whereas a few of these genes show relatively neutral or early replication timing (Npas3, Nfia, Wwox, and Ptn), the majority replicate late (Figures 7G and S6C). Notably, the 27 RDCs on average replicate significantly later in NSPCs than other genes larger than 100 kb (P.W., A.N.C., J.K., Z.D., R.M.M., F.W.A., B.S., unpublished data). Because the 27 genes we identify as being prone to genomic instability in NSPCs are highly conserved between mouse and man, we also examined existing replication timing data of their human orthologs in neural progenitors (Ryba et al., 2010; Figure S6D). Nearly 90% of these human orthologs showed conserved replication timing with their mouse counterparts (Figure S6D), suggesting that the majority of genes we identify as sensitive to replication stress-induced genomic instability in murine NSPCs could potentially be prone to replication stress-induced fragility in humans.
DISCUSSION
Detection of Recurrent Classes of DSBs in NSPCs
Development of NSPCs into post-mitotic neurons in vivo is dependent on repair of DSBs by C-NHEJ (Gao et al., 1998), suggesting critical roles for DSBs and/or their repair in neural cells. We now have employed HTGTS to identify tens of thousands of endogenous DSBs across the genomes of XRCC4/p53-deficient and WT NSPCs, based on their translocation to bait DSBs on several different chromosomes. Our findings reveal multiple different sources of recurrent DSBs in NSPCs, of which a large fraction corresponds to general classes of DSBs observed in other cell types (e.g., Chiarle et al., 2011; Frock et al., 2015; see below). Beyond these, our unbiased approach revealed 27 clear RDC sites in NSPCs, as they were recurrently detected from HTGTS bait DSBs located on different chromosomes. Strikingly, all 27 RDCs occurred in gene bodies. Moreover, they mainly occur in large genes encoding proteins involved in neural development or function, with a significant subset having been implicated as rearranged in neural and other cancers. Based on detection from a single HTGTS bait-site, we identified 333 additional, likely lower level, RDC candidates. As spatial proximity of bait and prey DSBs on the same chromosome clearly enhances detection of replication stress-induced RDCs in NSPCs (Figures 5, 6, S4C and S5D), HTGTS with additional bait DSB locations may eventually allow confirmation of many of these additional apparent RDCs. Due to the high sensitivity of HTGTS as a DSB identification approach, we expect that, with appropriate means of delivering bait DSBs, our approach could be extended to other neural lineage cells, including mature neurons.
Potential Sources of General Classes of Endogenous DSBs in NSPCs
In XRCC4/p53-deficient NSPCs, a large proportion of bait DSB junctions involve rejoining of the two bait DSB ends subsequent to resection (e.g., Figure S1), similar to what occurs in other cell types (Chiarle et al., 2011; Frock et al., 2015). Beyond the immediate break site, junctions were enriched along each tested XRCC4/p53-deficient NSPC break-site chromosome (i.e., Chr12, -15 and -16) relative to other chromosomes, consistent with spatial proximity influencing preferential joining of bait DSBs to the subset of widespread, low-level chromosomal DSBs that occur in cis (Frock et al., 2015; Zhang et al., 2012). Previously, this phenomenon was most prominently observed in cells harboring widespread DSBs generated by ionizing radiation or by non-specific activities of certain nucleases (Frock et al., 2015; Zhang et al., 2012). While we have not elucidated the source of widespread low-level DSBs in NSPCs, such DSBs might arise from various endogenous sources, including replicative, transcriptional, or oxidative stresses (e.g., Aguilera and Garcia-Muse, 2013; Erwin et al., 2014; Kim and Jinks-Robertson, 2012). In this regard, ATM deficiency, which increases oxidative stress (Paull, 2015), led to the greatest levels of this class of DSBs in NSPCs (Table S1). Notably, low-level widespread DSBs and overall RDC DSBs appear to similarly contribute as major DSB sources detectable in NSPCs. Finally, DSBs captured by HTGTS baits also are enriched near the TSSs of active genes in NSPCs; but they are not frequent enough to be considered recurrent in any given gene; for example, they occur at negligible frequency in RDC gene TSSs compared to the frequency of DSBs across the gene body (P.W., A.N.C., J.K., Z.D., R.M.M., F.W.A., B.S., unpublished data).
Mechanisms Promoting Replication Stress-induced Genomic Instability of Neural Genes in NSPCs
Of the 27 genes harboring robust RDCs in NSPCs, 25 were evident only in response to APH-induced replication stress; moreover, APH-treatment increased the DSB frequency in the two genes, Npas3 and Lsamp, that had RDCs in the absence of treatment (Figures 4–6, S4C, and S5B). APH is well known to induce CFS instability (Durkin and Glover, 2007). Consistent with characteristics often associated with CFSs, most replication stress-induced RDCs in NSPCs are within actively transcribed, large, and late-replicating genes (Figure 7). Thus, as proposed for CFSs, these characteristics, and potentially others, may contribute to the DSBs that generate NSPC RDCs by increasing the frequency of collisions between transcription and replication factors and/or mitotic entry with incomplete replication (Gao and Smith, 2014; Helmrich et al., 2011; Le Tallec et al., 2014). In this regard, Lsamp is the largest, actively transcribed NSPC gene and it replicates late, potentially predisposing it to frequent DSBs and RDC formation in the absence of APH treatment. The mechanism(s) of Npas3 fragility may be distinct, as this gene has neutral to early replication timing. In this context, we also identify an RDC in Ptn, which is not an exceptionally large gene (95.7 kb), replicates early, and is highly transcribed relative to surrounding regions, reminiscent of the ERFSs identified in B lymphocytes (Barlow et al., 2013). Notably, DSBs in ERFSs have also been linked to collisions between transcription and replication, but ERFSs are not induced by APH treatment (Barlow et al., 2013).
Mapping of suspected CFSs generally has been achieved mostly through experimental approaches involving cytogenetic studies of metaphase chromosomes from a limited number of cells (Durkin and Glover, 2007). Thus, the majority of CFSs have been characterized at low resolution (Savelyeva and Brueckner, 2014). In the mouse, only 8 CFSs have been molecularly mapped and only in lymphocytes (Helmrich et al., 2006); one of these (Wwox, FRA8E1; Krummel et al., 2002) was identified as an RDC in our study of NSPCs. The orthologous human gene (WWOX) is also located within a CFS (FRA16D; Krummel et al., 2002). In human cells, only 9 CFSs have been fine-mapped to a resolution of about 150 kb; although others have been implicated at lower resolution (several megabases), mostly in transformed cell lines (Savelyeva and Brueckner, 2014). Remarkably, of these implicated human CFSs, 6 span genes (Bosco et al., 2010; Le Tallec et al., 2011; Le Tallec et al., 2013) that correspond to RDCs that we identified at high resolution in NSPCs (Pard3b, Fgf12, Prkg1, Gpc6, Lsamp, Sdk1; Table S6). Thus, HTGTS elucidates CFSs, and other types of genomic fragility, at nucleotide resolution. Such resolution is critical for understanding underlying mechanisms. For example, based on analysis of large numbers of HTGTS junctions, we find that both RDC translocation junctions and genome-wide translocation junctions in XRCC4-deficient NSPCs have a markedly increased frequency and extent of MH-usage as compared to their counterparts in WT NSPCs (Figure S5G). Thus, in contrast to earlier conclusions based on more limited approaches studying mouse ES cells (Arlt et al., 2012), our studies indicate that both C-NHEJ and A-EJ pathways can mediate the various types of translocations we observe in NSPCs.
RDC-Genes in NSPCs are Implicated in Neural Processes, Neural Disorders, and Cancer
The great majority (24 of 27) of RDC-genes in NSPCs have roles in neural cell adhesion and/or regulation of synapse formation and function (Figure 7; also see Table S6). These include the cadherin-associated proteins Ctnna2 and Ctnnd2; cadherin Cdh13; synaptic cell adhesion molecule Cadm2; neural cell adhesion molecules Bai3, Csmd1, Csmd3, Dcc, Lsamp, Mdga2, Magi2, Ntm and Sdk1; excitatory neurotransmitter receptor Grik2; and two members of the neurexin family of synaptic cell surface proteins (Nrxn1, Nrxn3) (See Table S6). In addition, nearly all NSPC RDC-containing genes have been linked, either in mice, humans or both, to neurodevelopmental and neuropsychiatric disorders, including autism-spectrum disorder (44%; 12/27), schizophrenia (37%; 10/27), bipolar disorder (29.6%; 8/27), and intellectual disability (22.2%; 6/27) (Table S6). In the above contexts, recurrent DSB-mediated genomic alterations in NSPC RDC-genes might generate neuronal diversity and, thereby, affect neural physiology and/or predispose to neurodevelopmental disorders.
It is perhaps notable that the human orthologs of 9 of the RDCs identified in our study are found in relatively focal (5.8–15.4 Mb) CNVs detected by single-cell sequencing of human frontal cortex neurons (McConnell et al., 2013; Figure S7). While the relevance of this finding awaits further studies, it is tempting to speculate that the human orthologs of RDCs we define in NSPCs may give rise to at least some of these neuronal CNVs. In this regard, NSPCs harboring RDCs may be positively selected; and/or DSBs leading to RDC formation may occur at high frequency. Consistent with the latter possibility, we estimate that, when considered in aggregate, 12 DSBs per cell translocate to the 27 RDCs in XRCC4/p53-deficient NSPCs (Table S2). However, the actual DSB frequency in these cells is likely much higher. In this regard, we have used the XRCC4/p53-deficient NSPCs to enhance ability to find recurrent endogenous DSB clusters via HTGTS. Thus, while XRCC4 deficiency has no known impact on DSB generation, it enhances DSB persistence, thereby, enhancing their translocation and facilitating their detection by HTGTS (Alt et al. 2013). Notably, however, even in XRCC4-deficient NSPCs, most DSBs are still joined locally near the break site by A-EJ, resulting in our HTGTS results estimating only the minimal DSB frequency in any given RDC (e.g., Table S2 and data not shown). Finally, our finding of RDCs in WT NSPCs, where an even greater fraction of DSBs are joined locally by C-NHEJ (Table S2 and data not shown), emphasizes that actual DSB frequency in RDC genes is much greater than minimal numbers revealed by HTGTS.
Given that HTGTS does not reveal the precise frequency of DSBs at a given RDC, we compared the approximate frequency of spontaneous translocations to Lsamp in NSPCs to those occurring to Bcl-6 in activated B cells, in which Bcl-6 is a major AID off-target. This comparison is possible because we have done HTGTS on both NSPCs and on activated B cells from the same c-Myc bait DSBs in the same ATM-deficient background (Figure 2). We found that translocations to Lsamp in NSPCs occurred five times more frequently than translocations to Bcl-6 in B cells (Table S2). As Bcl-6 translocations occur at about 3% the level of translocations to an IgH class switch recombination region that breaks in at least 40–50% of activated B cells over a 4-day activation period (which is the same period over which we assayed NSPCs), this comparison suggests that DSBs occur frequently in Lsamp and, by extension, in other RDCs in the context of replication stress. An intriguing, unanswered question raised by our current findings is how the bulk of RDCs DSBs are repaired locally, in particular, whether they might frequently join to other DSBs within the same RDC. In this context, most of the 27 RDC genes fall within a single replication domain (Figure S6), which very often appear to correspond to topologically-associating domains (“TADs”) (Pope et al., 2014). The frequent joining of recurrent DSBs within a given TAD or chromosomal loop domain is exploited by lymphoid cells to promote frequent joining of DSBs within antigen receptor loci (Zarrin et al., 2007; Alt et al., 2013; Dong et al., 2015; Hu et al., 2015) and may also contribute to recurrent deletions found in certain cancers (Alt et al., 2013; Hu et al., 2015). In analogy to our recent HTGTS studies in which endogenous IgH switch region breaks were used as bait DSBs (Dong et al., 2015), we could begin to address such questions by using RDC regions with the highest DSB density as endogenous baits.
We have previously found that DSB repair by C-NHEJ suppresses development of medulloblastomas (MBs) with recurrent deletions, translocations, and amplification of N-myc and other genes (Yan et al., 2006). Notably, Cdh13, an NSPC RDC gene, has frequently been found to have copy number loss in human group III MBs (Northcott et al., 2012), as well as in other cancers, including ovarian, lung, liver, and breast cancers (See Table S6). In addition, NRXN3 amplification in double minutes has been detected in human MBs (Rausch et al., 2012). Several preliminary candidate RDCs lie within the centromeric portion of Chr12 where mouse N-myc is located. In this regard, RDC-gene fragility in NSPCs might be relevant to the speculation that frequent generation of endogenous DSBs during normal neuroblast differentiation contributes to N-myc amplification in human neuroblastomas (Kohl et al., 1983). Indeed, numerous NSPC RDC-genes are frequently deleted, rearranged, or amplified in various human cancers (Table S6). Thus, LSAMP is among the most frequently deleted genes in human cancers and NPAS3 is deleted in high-grade astrocytomas and glioblastomas (See Table S6). Likewise, three RDCs are recurrently deleted and rearranged (CADM2), rearranged and amplified (CSMD3), or involved in inter-chromosomal gene fusions (DGKB) in prostate cancer (see Table S6). These latter observations may well reflect fragility of some NSPC RDC-genes in other tissues and cell types in which they are expressed. HTGTS analyses of additional cell types for spontaneous or replication stress-induced RDCs could test this hypothesis and also identify RDCs specific to those other cell types.
EXPERIMENTAL PROCEDURES
NSPC Culture and DSB Induction
NSPCs from frontal brains of postnatal day (P) 8–14 mice were prepared and cultured as described in Supplemental Experimental Procedures. All related animal work was performed under protocol 14-10-2790R approved by the Institutional Animal Care and Use Committee of Boston Children’s Hospital. Bait DSB induction was achieved either via a Cas9:sgRNA approach (Frock et al., 2015) or via a TA-inducible I-SceI approach (Chiarle et al., 2011). Replication stress was induced by treatment with aphidicolin (APH, Sigma) for 96 hrs. See Supplemental Experimental Procedures for details.
Global Run-on Sequencing
GRO-seq libraries were prepared as previously described (Meng et al., 2014) from 5 – 8 × 106 NSPC nuclei. Three biological replicates per genotype (ATM−/−R26I−SceI−GRc-myc25xI−SceI or Xrcc4−/−p53−/−) were performed. GRO-seq data were aligned to mouse genome build mm9/NCBI37 by Bowtie2 and non-redundant, uniquely mapped sequence reads were retained. De novo transcripts were identified and gene expression levels were estimated as previously described (Meng et al., 2014).
HTGTS and Related Bioinformatic Analyses
EM-PCR-mediated HTGTS and LAM-HTGTS were performed and analyzed as described (Chiarle et al., 2011; Frock et al., 2015). Primers used and junction yield per experiment, as well as descriptions of bioinformatic methods used for HTGTS junction analyses, RDC identification, repair junction signature analysis (e.g., direct versus MH-mediated), and Cas9:sgRNA off-target site identification are described in Table S7 and Supplemental Experimental Procedures.
Replication Timing Analysis
Custom Python scripts were used to calculate median replication timing ratios of genomic regions based on Repli-chip data (Weddington et al., 2008). Replication timing data sets analyzed were mouse NPC 46C, TT2, and D3 (Hiratani et al., 2008) and two replicates of human NPC BG01 (Ryba et al., 2010). Replication timing ratios were displayed by IGV (Robinson et al., 2011).
Supplementary Material
Acknowledgments
We thank Drs. R. Axel, C. Boboila, and members of the Alt laboratory for helpful comments and stimulating discussions, Drs. Chunguang Guo, Monica Gostissa, Jiazhi Hu for experimental advice; and Drs. Yi Zhang, Li Shen, and Fei-Long Meng for DNA sequencing assistance. This work in the Alt lab was supported by the Porter Anderson Fund from Boston Children’s Hospital and the Howard Hughes Medical Institute. B.S. is a Martin D. Abeloff Scholar of The V Foundation for Cancer Research and is supported by NIA/NIH grant K01AG043630. P.W. is supported by a National Cancer Center postdoctoral fellowship.
Footnotes
ACCESSION NUMBERS
Sequencing data have been deposited to the Gene Expression Omnibus (GEO) under accession number GSE74356.
AUTHOR CONTRIBUTIONS
F.W.A. and B.S. conceived of and planned the study. B.S., P.W., A.N.C., Z.D., R.M.M., and F.W.A. designed experiments. P.W., A.N.C., J.K, and B.S. performed research. B.S., P.W., A.N.C., J.K., Z.D., R.M.M., and F.W.A. analyzed and interpreted data. B.S., P.W., and F.W.A. designed figures and wrote the manuscript. Other authors helped polish the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguilera A, Garcia-Muse T. Causes of genome instability. Annu Rev Genet. 2013;47:1–32. doi: 10.1146/annurev-genet-111212-133232. [DOI] [PubMed] [Google Scholar]
- Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152:417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt MF, Rajendran S, Birkeland SR, Wilson TE, Glover TW. De novo CNV formation in mouse embryonic stem cells occurs in the absence of Xrcc4-dependent nonhomologous end joining. PLoS Genet. 2012;8:e1002981. doi: 10.1371/journal.pgen.1002981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JH, Faryabi RB, Callen E, Wong N, Malhowski A, Chen HT, Gutierrez-Cruz G, Sun HW, McKinnon P, Wright G, et al. Identification of early replicating fragile sites that contribute to genome instability. Cell. 2013;152:620–632. doi: 10.1016/j.cell.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes DE, Stamp G, Rosewell I, Denzel A, Lindahl T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr Biol. 1998;8:1395–1398. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv Immunol. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- Bosco N, Pelliccia F, Rocchi A. Characterization of FRA7B, a human common fragile site mapped at the 7p chromosome terminal region. Cancer Genet Cytogenet. 2010;202:47–52. doi: 10.1016/j.cancergencyto.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, Myers DR, Choi VW, Compagno M, Malkin DJ, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio MJ, Petersen S, Chen HT, Johnson R, Jasin M, Kanaar R, Ried T, Nussenzweig A. Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J Exp Med. 2002;196:469–480. doi: 10.1084/jem.20020851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Panchakshari RA, Zhang T, Zhang Y, Hu J, Volpi SA, Meyers RM, Ho YJ, Du Z, Robbiani DF, et al. Orientation-specific joining of AID-initiated DNA breaks promotes antibody class switching. Nature. 2015;525:134–139. doi: 10.1038/nature14970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- Erwin JA, Marchetto MC, Gage FH. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat Rev Neurosci. 2014;15:497–506. doi: 10.1038/nrn3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon ER, Cho KR, Nigro JM, Kern SE, Simons JW, Ruppert JM, Hamilton SR, Preisinger AC, Thomas G, Kinzler KW, et al. Identification of a chromosome 18q gene that is altered in colorectal cancers. Science. 1990;247:49–56. doi: 10.1126/science.2294591. [DOI] [PubMed] [Google Scholar]
- Frank KM, Sharpless NE, Gao Y, Sekiguchi JM, Ferguson DO, Zhu C, Manis JP, Horner J, DePinho RA, Alt FW. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol Cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- Frock RL, Hu J, Meyers RM, Ho YJ, Kii E, Alt FW. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Smith DI. Very large common fragile site genes and their potential role in cancer development. Cell Mol Life Sci. 2014;71:4601–4615. doi: 10.1007/s00018-014-1753-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Ferguson DO, Xie W, Manis JP, Sekiguchi J, Frank KM, Chaudhuri J, Horner J, DePinho RA, Alt FW. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature. 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- Gao Y, Sun Y, Frank KM, Dikkes P, Fujiwara Y, Seidl KJ, Sekiguchi JM, Rathbun GA, Swat W, Wang J, et al. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- Gapud EJ, Sleckman BP. Unique and redundant functions of ATM and DNA-PKcs during V(D)J recombination. Cell Cycle. 2011;10:1928–1935. doi: 10.4161/cc.10.12.16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover TW, Berger C, Coyle J, Echo B. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum Genet. 1984;67:136–142. doi: 10.1007/BF00272988. [DOI] [PubMed] [Google Scholar]
- Helmrich A, Ballarino M, Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell. 2011;44:966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Helmrich A, Stout-Weider K, Hermann K, Schrock E, Heiden T. Common fragile sites are conserved features of human and mouse chromosomes and relate to large active genes. Genome Res. 2006;16:1222–1230. doi: 10.1101/gr.5335506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schubeler D, Gilbert DM. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Zhang Y, Zhao L, Frock RL, Du Z, Meyers RM, Meng FL, Schatz DG, Alt FW. Chromosomal Loop Domains Direct the Recombination of Antigen Receptor Genes. Cell. 2015;163:947–959. doi: 10.1016/j.cell.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- Kim N, Jinks-Robertson S. Transcription as a source of genome instability. Nature reviews Genetics. 2012;13:204–214. doi: 10.1038/nrg3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, Di Virgilio M, Bothmer A, Nussenzweig A, Robbiani DF, et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147:95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl NE, Kanda N, Schreck RR, Bruns G, Latt SA, Gilbert F, Alt FW. Transposition and amplification of oncogene-related sequences in human neuroblastomas. Cell. 1983;35:359–367. doi: 10.1016/0092-8674(83)90169-1. [DOI] [PubMed] [Google Scholar]
- Krummel KA, Denison SR, Calhoun E, Phillips LA, Smith DI. The common fragile site FRA16D and its associated gene WWOX are highly conserved in the mouse at Fra8E1. Genes Chromosomes Cancer. 2002;34:154–167. doi: 10.1002/gcc.10047. [DOI] [PubMed] [Google Scholar]
- Le Tallec B, Dutrillaux B, Lachages AM, Millot GA, Brison O, Debatisse M. Molecular profiling of common fragile sites in human fibroblasts. Nat Struct Mol Biol. 2011;18:1421–1423. doi: 10.1038/nsmb.2155. [DOI] [PubMed] [Google Scholar]
- Le Tallec B, Koundrioukoff S, Wilhelm T, Letessier A, Brison O, Debatisse M. Updating the mechanisms of common fragile site instability: how to reconcile the different views? Cell Mol Life Sci. 2014;71:4489–4494. doi: 10.1007/s00018-014-1720-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Tallec B, Millot GA, Blin ME, Brison O, Dutrillaux B, Debatisse M. Common fragile site profiling in epithelial and erythroid cells reveals that most recurrent cancer deletions lie in fragile sites hosting large genes. Cell reports. 2013;4:420–428. doi: 10.1016/j.celrep.2013.07.003. [DOI] [PubMed] [Google Scholar]
- Lee Y, McKinnon PJ. DNA ligase IV suppresses medulloblastoma formation. Cancer Res. 2002;62:6395–6399. [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madabhushi R, Gao F, Pfenning AR, Pan L, Yamakawa S, Seo J, Rueda R, Phan TX, Yamakawa H, Pao PC, et al. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell. 2015;161:1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell MJ, Lindberg MR, Brennand KJ, Piper JC, Voet T, Cowing-Zitron C, Shumilina S, Lasken RS, Vermeesch JR, Hall IM, et al. Mosaic copy number variation in human neurons. Science. 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon PJ. Maintaining genome stability in the nervous system. Nat Neurosci. 2013;16:1523–1529. doi: 10.1038/nn.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng FL, Du Z, Federation A, Hu J, Wang Q, Kieffer-Kwon KR, Meyers RM, Amor C, Wasserman CR, Neuberg D, et al. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell. 2014;159:1538–1548. doi: 10.1016/j.cell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri AR, Gage FH. Generation of neuronal variability and complexity. Nature. 2006;441:1087–1093. doi: 10.1038/nature04959. [DOI] [PubMed] [Google Scholar]
- Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, Stutz AM, Korshunov A, Reimand J, Schumacher SE, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488:49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT. Mechanisms of ATM Activation. Annu Rev Biochem. 2015;84:711–738. doi: 10.1146/annurev-biochem-060614-034335. [DOI] [PubMed] [Google Scholar]
- Poduri A, Evrony GD, Cai X, Walsh CA. Somatic mutation, genomic variation, and neurological disease. Science. 2013;341:1237758. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515:402–405. doi: 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, Jager N, Remke M, Shih D, Northcott PA, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20:761–770. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelyeva L, Brueckner LM. Molecular characterization of common fragile sites as a strategy to discover cancer susceptibility genes. Cell Mol Life Sci. 2014;71:4561–4575. doi: 10.1007/s00018-014-1723-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weddington N, Stuy A, Hiratani I, Ryba T, Yokochi T, Gilbert DM. ReplicationDomain: a visualization tool and comparative database for genome-wide replication timing data. BMC Bioinformatics. 2008;9:530. doi: 10.1186/1471-2105-9-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan CT, Kaushal D, Murphy M, Zhang Y, Datta A, Chen C, Monroe B, Mostoslavsky G, Coakley K, Gao Y, et al. XRCC4 suppresses medulloblastomas with recurrent translocations in p53-deficient mice. Proc Natl Acad Sci U S A. 2006;103:7378–7383. doi: 10.1073/pnas.0601938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrin AA, Del Vecchio C, Tseng E, Gleason M, Zarin P, Tian M, Alt FW. Antibody class switching mediated by yeast endonuclease-generated DNA breaks. Science. 2007;315:377–381. doi: 10.1126/science.1136386. [DOI] [PubMed] [Google Scholar]
- Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.