Abstract
Four different well-defined IgG1 Fc glycoforms are proposed as a model system to examine important biological and physicochemical features for protein drug biosimilar analyses. The IgG1 Fc glycoforms were produced by yeast expression combined with in vitro enzymatic synthesis as a series of sequentially truncated, high mannose IgG1 Fc glycoforms with an anticipated range of biological activity and structural stability. Initial characterization with mass spectrometry, SDS-PAGE, SEC, and cIEF confirmed the glycoproteins are overall highly similar with the only major difference being glycosylation state. Binding to the activating Fc receptor FcγRIIIa was used to evaluate the potential biological activity of the IgG1 Fc glycoproteins. Two complementary methods utilizing biolayer interferometry (BLI), one with protein G immobilized IgG1 Fc and the other with streptavidin immobilized FcγRIIIa, were developed to assess FcγRIIIa affinity in kinetic binding studies. The HM-Fc and Man5-Fc were highly similar to one another with high affinity for FcγRIIIa, while GlcNAc-Fc had weak affinity, and the non-glycosylated N297Q-Fc had no measurable affinity for FcγRIIIa. These four IgG1 Fc glycoforms were also evaluated in terms of physical and chemical stability profiles, and then used as model system to mathematically assess overall biosimilarity, as described in a series of companion papers.
Keywords: IgG antibody, glycoprotein, glycosylation, receptors, biopharmaceuticals characterization, follow-on biologics, bio similar
Introduction
Protein therapeutics are inherently structurally complex biological drugs whose active components are not a single, well-defined molecule, but a mixture of similar molecules which can differ by type and extent of post-translational modification,1–5 chemical modifications,6–11 and three dimensional conformations.12,13 In addition, batch to batch variation of active components and production impurities can further complicate defining of the analytical characteristics of a protein drug.14 Because of this, the assessment of protein therapeutics in comparability studies or in the regulatory approval of biosimilars is not just a simple exercise in confirming the chemical structure of a single active chemical entity in the presence of excipients. Instead, the primary and higher order structures, physicochemical properties, and biological activities of the protein therapeutics must be analyzed. Often, even after extensive study, the relationship between safety and efficacy of a protein therapeutic and these analytical tests is not entirely clear because of the complexity of the mixtures in protein therapeutics and biological systems involved. In an effort to better understand how in vitro analytical tests can be utilized to determine similarity in biosimilar studies and analysis assessments, we have developed a series of well-defined IgG1 Fc glycoforms as a model system. The use of a series of well-defined glycoproteins in these studies should enable identification of important structural and biological features for comparability and biosimilarity analyses.
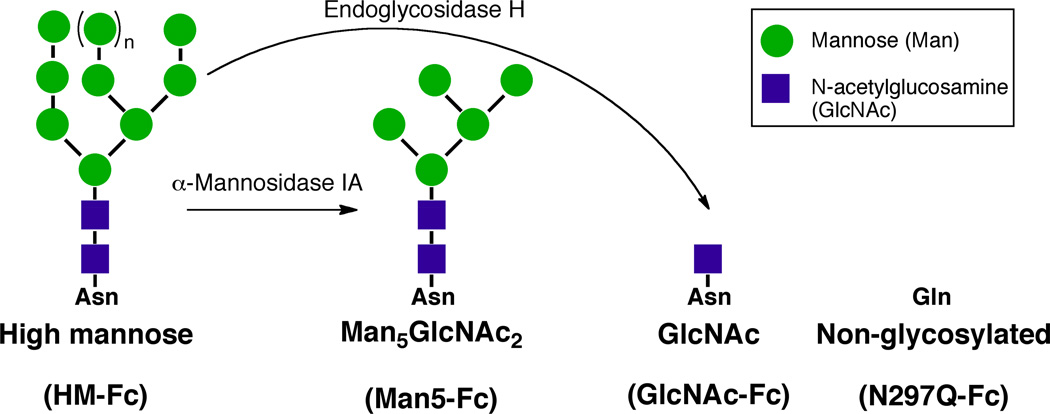
A series of IgG1 Fc glycoforms were chosen as the protein model system for biosimilarity analysis because as a fragment of full-length IgG1 it is a simpler system to study but contains the CH2 and CH3 constant domains which are present in all human IgG1 based mAb therapeutics. The Fc region is critical to antibody function in that it mediates effector functions such as antibody dependent cellular cytotoxicity (ADCC)15–24 and complement dependent cytotoxicity (CDC)25–27. The Fc region of an IgG1 is also important in antibody clearance because binding of the Fc region to the neonatal Fc receptor (FcRn) increases in vivo half-life28–31. In addition, N-linked glycosylation at asparagine 297 (N297) in the Fc region is known to modulate the biological activity19,32–39 and physical properties of IgG1 Fc,1,32,40–48 and this can be used to establish similarities and differences between the members of this model system. A series of sequentially truncated glycoforms of IgG1 Fc (Figure 1), which differ only in the size of the N-linked glycan at N297 or a single conservative amino acid mutation (N297Q), were chosen as members for the model system. Previous studies have indicated that these glycoforms would display a range of biological activities and physical properties1,32,45 that would be advantageous for our biosimilarity studies.
Figure 1.
Production of homogenous IgG1 Fc glycoforms. HM-Fc was expressed in glycoengineered P. pastoris and utilized as a precursor for generation of GlcNAc-Fc and Man5-Fc. N297Q-Fc was recombinantly expressed in a different yeast strain.
It was first necessary to develop laboratory production methods to produce sufficient quantities (≈100 mg each) of the well-defined IgG1 Fc glycoforms to conduct the wide range of analytical tests necessary for biosimilarity analysis. Presented here are the methods established to produce the glycoforms shown in Figure 1 through yeast expression, purification and in vitro enzymatic synthesis. Also presented in this work is the initial biochemical characterization of these glycoforms, the development of binding assays for IgG1 Fc binding to an Fc receptor using biolayer interferometry (BLI), and determination of the affinity of the different IgG1 Fc glycoforms for that Fc receptor as an initial evaluation of biological activity. Subsequent companion papers examine the physical and chemical stability profiles of these IgG1 Fc glycoforms, and the use of the resulting physicochemical data sets, to develop a mathematical algorithm for biosimilarity assessments.49–51
Materials and Methods
Materials
Yeast nitrogen base (YNB) was obtained from Sunrise Biosciences, and Bacto™ Tryptone and Yeast Extract was purchased from Becton Dickinson and Company (Franklin Lakes, NJ). Antifoam 204 was obtained from Sigma-Aldrich (St. Louis, MO). Certified ACS grade crystalline sucrose was purchased from Fisher Scientific (Pittsburg, PA). Boc-triglycine was supplied by Bachem Americas, Inc. (Torrance, CA). The enzymes PNGase F, Sortase, and B. thetaiotaomicron α-1,2-mannosidase (BT3990, B.t. α-1,2-mannosidase) were produced inhouse.52–55 Endoglycosidase H and restriction enzymes were obtained from New England Biolabs (Ipswich, MA). Protein G resin was produced by coupling protein G (recombinantly expressed in E. coli)56 with Sepharose® CL-4B (Sigma-Aldrich, St Louis, MO) using divinyl sulfone as a coupling reagent.57 General chemicals were purchased from Sigma-Aldrich and Fisher Scientific unless otherwise noted.
Production and initial characterization of IgG1 Fc Glycoforms
Expression of High Mannose IgG1 Fc (HM-Fc)
HM-Fc was expressed in a glycosylationdeficient strain of Pichia pastoris (an OCH1 and PNO1 deleted, IgG1 Fc expressing, SMD1168 strain of P. pastoris produced by Xiao et.al.58 was utilized) using glycerol (growth phase) and methanol (induction phase) as carbon sources in a NBS BioFlo 415 fermenter (Eppendorf). The starter culture (2 mL) was allowed to grow at 25°C for about 72 hrs in YPD media (1% yeast extract, 2% peptone, 5% glucose and zeocin 100 µg/ml). This starter culture was then inoculated into 250 mL of YPD media in a baffled shake flask and incubated for about 72 hrs. Before inoculation with the starter culture, the fermenter was filled with 7 liters of fermentation basal salts medium (BSM).59 The components of BSM per liter are 26.7 mL 85% phosphoric acid, 18.2 g potassium sulfate, 0.93 g calcium sulfate, 14.9 g magnesium sulphate heptahydrate, 4.13 g potassium hydroxide, and 40.0 g glycerol. Finally, water was added to the appropriate volume and the fermentation media was sterilized. After sterilization and cooling, the temperature was adjusted to 30°C and the pH of the BSM medium was adjusted to 6 with 28% Ammonium hydroxide. Next, PTM159 trace salts solution was prepared by mixing the following components (per L): 6.0 g copper sulfate pentahydrate, 0.08 g sodium Iodide, 3.0 g manganese sulfate monohydrate, 0.2 g Sodium Molybdate dihydrate, 0.02 g boric acid, 0.5 g cobalt(II) chloride, 20.0 g zinc chloride, 65.0 g Iron(II) sulfate heptahydrate, 0.2 g biotin and 5.0 mL sulfuric acid. This step was followed by adding 4.31 mL of filter-sterilized PTM1 trace salts/liter of BSM. Next, the dense 250 mL starter culture media was added into the fermentor. The culture media was allowed to grow until the batch glycerol was consumed, and an additional 300 mL of glycerol were then added to increase the total cell biomass. Dissolved oxygen was kept ≥ 30% throughout the fermentation processes. Once the added glycerol was consumed and the desired cell biomass was attained, a 100% methanol feeding was initiated to induce the AOX1 promoter for IgG1 Fc expression. The temperature was reduced to 25 °C for optimal IgG1 Fc expression prior to induction. Methanol induction was continued for approximately 72 hours. The resulting yeast suspension was pelleted by centrifugation at 12,390 × g for 20 min, and the supernatant was collected. Using this procedure, the amount of HM-Fc obtained after protein G affinity purification was approximately 50 mg/L. Four 7 liter fermentations were conducted to produce sufficient HM-Fc to develop the in vitro enzymatic synthesis used to make the Man5-Fc and GlcNAc-Fc glycoforms, and to generate material for the subsequent experiments described in this and the accompanying papers.
Expression of non-glycosylated mutant N297Q-IgG1 Fc (N297Q-Fc)
Spinner flask expression was utilized for the production of N297Q-Fc in a glycosylation-deficient strain of SMD1168 P. pastoris (Clone produced in previous study1) as follows. A starter culture of 2 mL was inoculated in YPD that contained 100 µg/mL Zeocin. This culture media was incubated at 25 °C for 72 hrs. The 2 mL culture was then used to inoculate a 50 mL of YPD/Zeocin culture, which was incubated with shaking at 25 °C for 72 hrs. The 50 mL culture was then used to inoculate 1 L of Buffered Glycerol-complex Media (BMGY)60 containing 0.00004% biotin and 0.004% histidine. After 48 hours, when the initial glycerol carbon source was metabolized, N297Q-Fc expression was induced by methanol feeding to a final concentration of 1 % (by addition of 50 mL of 20% methanol every 24 hrs.) for about 72 hrs. The same protocol used for harvesting HM-Fc was applied for harvesting of the N297Q-Fc supernatant. Using this procedure, the amount of N297Q-Fc obtained after protein G affinity purification was about 20 mg/L. A total of seven 1 L spinner flasks were grown to produce the 130 mg of N297Q-Fc used in the research described in this and the accompanying papers.
Purification of IgG1 Fc
Both HM- and N297Q-IgG1 Fc were purified using the same general procedure given below. The supernatant collected from yeast expression was filtered with 0.5 µm filter pads (Buon Vino Manufacturing) to remove particulates prior to protein G affinity chromatography. The protein G column (20 mL bed volume) was equilibrated with 20 mM potassium phosphate pH 6.0 in 10 column bed volumes (CV) and loaded with the filtered supernatant at pH 6.0. The column was subsequently washed with 20 mM potassium phosphate buffer, pH 6.0, containing 0.5 M NaCl (5 CV) and then 20 mM potassium phosphate buffer, pH 6.0, (5 CV). IgG1 Fc was eluted by using 100 mM glycine at pH 2.7. Eluted protein was collected in 20 mL fractions into tubes containing 4 mL of 1 M Tris pH 9.0 (200 µL of 1 M Tris pH 9.0 per mL of elution volume) to neutralize the acidic elution buffer. Fractions of eluted protein detected by UV absorbance at 280 nm were immediately dialyzed in 20 mM sodium phosphate buffer pH 7.0. HIC purification using phenyl sepharose™ high-performance resin (GE Healthcare) with a 125 mL column bed volume (packed in-house) and an ÄKTAmicro chromatographic system (GE Healthcare) were utilized to further purify the IgG1 Fc forms. The phenyl sepharose column column was pre-equilibrated with Buffer A (20 mM sodium phosphate, pH 7.0, containing 1M ammonium sulfate) for 5 CV. The protein G purified IgG1 Fc was dialyzed in buffer A and then loaded onto the phenyl sepharose column with a loading volume of 50 mL (concentration, 1 mg/mL). The chromatographic separation was then conducted with three gradient segments from 0 to 50% buffer B (20 mM sodium phosphate, pH 7.0): gradient segment 1 (0–25% B, 3.5 CV), gradient segment 2 (25–30% B, 2.6 CV), and gradient segment 3 (30–50% B, 6.7 CV). Collected fractions (10 mL) were analyzed by SDS-PAGE and mass spectrometry to check for purity and identity. Pure fractions were finally pooled and dialyzed in storage buffer (10% sucrose, 20 mM histidine pH 6.0) and frozen at −80°C in aliquots. The final collected sample pool was analyzed by SEC, SDS-PAGE, mass spectrometry, and cIEF characterization to confirm overall purity and quality. Samples containing pure fractions were concentrated to 0.2 mg/mL using Vivaflow 50, (10,000 MWCO, Sartorius Stedim Biotech). Using this procedure, approximately 445 mg of the HM-Fc glycoform and 118 mg of the N297Q-Fc non-glycosylated mutant were produced for use in synthesis of other glycoforms and biosimilar analysis studies.
In vitro enzymatic synthesis of the Man5-IgG1 Fc glycoform (Man5-Fc)
HM-IgG1 Fc was converted to Man5-IgG1 Fc in an in vitro enzymatic reaction using B.t. α-1,2-mannosidase (BT3990).54,55 It was found that the B.t. α-1,2-mannosidase used in this study had very low activity in the IgG1 Fc sample storage buffer which contained 10% (w/v) sucrose and 20 mM histidine buffer at pH 6.0. Because of this, prior to the reaction HM-Fc (125 mg) was dialyzed extensively in 10 mM MES buffer pH 6.6 to remove the sucrose and histidine. Next, HM-Fc was dialyzed in a reaction buffer containing 5 mM CaCl2, 150 mM NaCl, and 10 mM MES buffer pH 6.6 for 12 hrs. After dialysis the enzymatic reaction was started by adding 6.7 mg of the bacterial α-1,2-Mannosidase (BT3990). The reaction was incubated at room temperature for 48 hrs. The progress of the reaction was monitored by mass spectrometry, and the percentage of conversion to the Man5-Fc glycoform was estimated from the peak intensity (see supplemental table S1). Finally, the reaction mixture was purified using protein G affinity chromatography to remove unwanted impurities and excess enzyme using the same protocol as described in the purification of IgG1 Fc section above. The amount of Man5-Fc produced was 75 mg (60% yield), and the percentage of Man5-Fc in the final product was estimated to be 78% by mass spectrometry.
In vitro enzymatic synthesis of the GlcNAc-IgG1 Fc glycoform (GlcNAc-Fc)
HM-IgG1 Fc was converted to the GlcNAc-IgG1 Fc glycoform using endoglycosidase H (Endo H). Endo H displayed full activity in the IgG1 Fc storage buffer (10 % (w/v) sucrose, 20 mM histidine pH 6.0), therefore it was possible to digest HM-IgG1 Fc directly without a dialysis step. HM-IgG1 Fc (100 mg) at a concentration of 0.2 mg/mL was incubated with Endo H (for every 1 mg of HMIgG1 Fc, 1000 U of Endo H enzyme were added, which corresponds to approximately 0.1 mg of Endo H per 100 mg HM-Fc) at room temperature for 24 hrs. The progress of the reaction was monitored by SDS-PAGE and mass spectrometry. The sample was analyzed by massspectrometry which showed a nearly quantitative reaction with the percentage of GlcNAc-Fc in the final product ≥99%.
LC-MS analysis IgG1 Fc glycoforms
Samples of IgG1 Fc glycoforms at a concentration of 0.2 mg/mL were first reduced with 10 mM dithiothreitol (DTT, Invitrogen) and then 30 µL were injected into the mobile phase of the LC. ESI spectra of the reduced samples were acquired on a Agilent 6520 Quadrupole Time-of-Flight (Q-TOF) system. The instrument was operated in positive ion mode, and a spectrum was acquired covering the mass range from 300–3000 m/z with an acquisition rate of 1 spectra/second. The samples were desalted on a reverse phase C4 column, 50 mm, 4.6 mm I.D. (Vydac 214 MS, 300 A pore size, 5µm particle size) using a Agilent 1200 series Liquid Chromatography system. The solvents used were A (99.9% H2O, 0.08% formic acid, 0.02% trifluoroacetic acid (TFA) and B (99.9% acetonitrile, 0.08% formic acid, 0.02% TFA). A gradient was developed from 5% B to 90% B in 7 min with a flow rate of 0.5 mL/min. Data was collected using Agilent MassHunter Acquisition software (Version B.02.00). Protein MW was calculated using the Maximum Entropy Deconvolution function and associated peak intensities of the IgG1 Fc glycoforms were obtained using Agilent MassHunter Qualitative Analysis software (Version B.03.01).
Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
For the reduced samples, each of the IgG1 Fc glycoforms (20 µg) were mixed with 2× TrisHCl SDS loading dye containing 100 mM DTT and incubated at 80 °C for 2 min. The reduced IgG1 Fc samples were then separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis using NuPAGE 4–12% Bis-Tris gradient gel (Life Technologies, Grand Island, NY) gels and a MES running buffer (Life Technologies). A similar method was followed for non-reduced samples of IgG1 Fc proteins except DTT was omitted during the incubation step. The running time for all the gels was 60 min at 150 V. Protein bands were visualized by staining with Coomassie blue R250 (Teknova, Hollister, CA) and destained with a mixture of 30% methanol, 10% acetic acid, and 60% ultrapure water. Gel images were recorded using an Alphaimager (Protein Simple, Santa Clara, CA) gel imaging system.
Size Exclusion High-Performance Liquid Chromatography (SEC)
Experiments were performed using a Shimadzu high-performance liquid chromatography system equipped with a temperature controlled auto sampler and a photodiode array detector capable of recording UV absorbance spectra from 200−400 nm. A Tosoh TSK-Gel Bioassist G3SWXL column (7.8 mm ID×30.0 cm L) and a corresponding guard column (TOSOH Biosciences, King of Prussia, Pennsylvania) were used for IgG1 Fc glycoform characterization. First, the SEC column was equilibrated for at least 10 CV with a mobile phase containing 200 mM sodium phosphate, pH 6.8 and a flow rate of 0.7 mL/min at 30 °C column temperature. Next, the column was calibrated using gel filtration molecular weight standards (Bio-Rad, Hercules, CA) before and after the runs of IgG1 Fc glycoform to ensure column and HPLC system integrity. All Fc samples were centrifuged at 14,000 g for 5 min before injection to remove insoluble protein aggregates. Protein samples at a concentration of 1 mg/mL were injected in a volume of 25 µL, and a 30 min run time was used for elution. Peaks quantification was carried out using LC solutions software (Shimadzu, Kyoto, Japan). The error bars for monomer content for all the four IgG1 Fc samples and soluble dimer aggregates (observed in N297Q IgG1 Fc) represent standard deviation (SD) of triplicate measurements.61,62
Capillary isoelectric focusing (cIEF)
The determination of isoelectric points (pIs) of all the IgG1 Fc glycoforms using capillary isoelectric focusing (cIEF) were performed with an iCE280 analyzer from Convergent Biosciences (now Protein Simple, Toronto, Canada) equipped with a microinjector. A FC Cartridge (Protein Simple, Toronto, Canada) with 50 mm, 100 µm I.D. fluorocarbon-coated capillary and built-in electrolyte tanks was used for focusing. The cartridge was calibrated by a hemoglobin standard (Protein Simple, Toronto, Canada) before and after focusing of IgG1 Fc samples to ensure its integrity. For focusing, a sample mixture was prepared where each of the IgG1 Fc glycoforms were mixed with urea (Fischer Scientific), methyl cellulose (Protein Simple, Toronto, Canada), sucrose (Pfanstiehl Inc., Waukegan, IL), N,N,N',N'-Tetramethylethane-1,2-diamine (Sigma-Aldrich, St Louis, MO) and Pharmalyte 3–10 (GE Healthcare Biosciences, Pittsburgh, PA). The final protein concentration in the sample mixture was 0.2 mg/mL. All the IgG1 Fc glycoforms were resolved using a pre-focusing time of 1 minute at 1500 V and a focusing time of 12,12,7.5 and 7 minutes was used for HM-Fc, N297Q-Fc, GlcNAc-IgG1Fc and Man5-Fc respectively at 3000 V. Observed peaks were calibrated using two pI markers with values of 5.84 and 8.18. The separation was monitored at 280 nm by a CCD detector. Quantitation of the peaks was done using Chromperfect® software. The error bars for pI values of all the four IgG1 Fc samples represent standard deviation (SD) of triplicate measurements.
Fc γ receptor IIIa binding assays
Production of a P. pastoris strain for expression of Fc γ receptor IIIa with a C-terminal sortase/histidine tag (FcγRIIIa-ST-H6)
The soluble region of the V158 polymorph of human FcγRIIIa was PCR-amplified from pPICzαA-FcγRIIIa58 using primers (forward 5’-ggcgccgaattcaaaagaatgcggactgaagatctc and reverse 5'gccgcgcgcgcggccgcttaatgatgatggtggtggtgtccacctccagtttctggcaatccaccaccttgagtgatggtgatgttcac) that added a sortase recognition site (ST) and hexahistidine tag (H6) to the 3’ end of the amplified FcγRIIIa DNA. The FcγRIIIa-ST-H6 PCR product was inserted into the methanol-inducible Pichia expression vector pPICzαA (Invitrogen, Carlsbad, CA) using the restriction sites EcoR I and Not I. The pPICzαA-FcγRIIIa-ST-H6 construct was confirmed by DNA sequencing, linearized using Sac I, and transformed into Pichia pastoris OCH1 deleted cells.58 Ten colonies were screened for levels of secreted FcγRIIIa-ST-H6 expression by growing the colonies in 2 mL culture tubes containing BMGY60 media +100 µg/mL Zeocin 100 U at 25°C and 250 rpm. Once they reached density, 0.5% (v/v) methanol was added once per day for three days. Relative levels of FcγRIIIa-ST-H6 in the media was determined by a dot blot using a mouse Anti-H6 primary antibody (Thermo Scientific, Rockford, IL) followed by a goat Anti-mouse IgG secondary antibody conjugated with alkaline phosphatase (Thermo Scientific, Rockford, IL). The colony that expressed the highest level of FcγRIIIa-ST-H6 was selected for 1 L spinner flask expression.
Expression and purification of FcγRIIIa-H6 and FcγRIIIa-ST-H6
FcγRIIIa-H6 and FcγRIIIa-ST-H6 were expressed in glycosylation deficient P. pastoris and Ni+2-NTA was used as described previously by Xiao et al.58 After Ni+2-NTA purification, approximately 30 mg/L of each receptor was obtained. The receptors were then further purified using hydrophobic interaction chromatography. A Hiprep™ phenyl FF (high sub) 16/10 column with a 20 mL bed volume was utilized with an ÄKTAmicro (GE Healthcare) to accomplish this. The column was preequilibrated with buffer A (1.5 M Ammonium sulfate,100 mM sodium phosphate for 10 CV) prior to loading. The Ni+2-NTA purified receptors were first dialyzed in 100 mM sodium phosphate pH 7.0 buffer for 12 hrs and then adjusted with ammonium sulfate to a final concentration of 1.5 M. The receptors, in 10 mg portions, were loaded onto the column with a loading volume of 5 mL. Due to the capacity of the column, 3 columns were necessary to purify all 30 mg of receptor obtained from Ni+2-NTA chromatography. The chromatographic separation involves three segments from 0 to 100% B (100 mM sodium phosphate pH 7.0): gradient segment 1 (0–25% B, 15 CV), gradient segment 2 (25–33% B, 15 CV), and gradient segment 3 (33–100% B, 12 CV). Collected fractions (5 mL) were characterized using SDS-PAGE to check for purity. Samples containing pure receptor were concentrated to 1 mg/mL using Vivaflow 50, (10,000 MWCO, Sartorius Stedim Biotech) and the amount obtained after purification for each receptor was approximately 18 mg/L.
Biotinylation of FcγRIIIa-ST-H6
Purified FcγRIIIa-ST-H6 was extensively dialyzed in 50 mM Tris-hydrochloride pH 7.5. Next, this receptor was dialyzed in a reaction buffer containing 50 mM Tris-hydrochloride pH 7.5, 150 mM sodium chloride. The sortase-mediated ligation reaction was carried out using a mixture containing 10 µM FcγRIIIa-ST-H6, 6 mM CaCl2, 1 mM GGG-linker-Biotin (compound 3, Fig. 6) and 5 µM sortase at room temperature. The reaction was terminated after 24 hours by adding excess EDTA to capture the Ca+2 required for activity of the sortase. Finally, the receptor was extensively dialyzed in PBS buffer to remove the unreacted GGG-linker-Biotin (3).
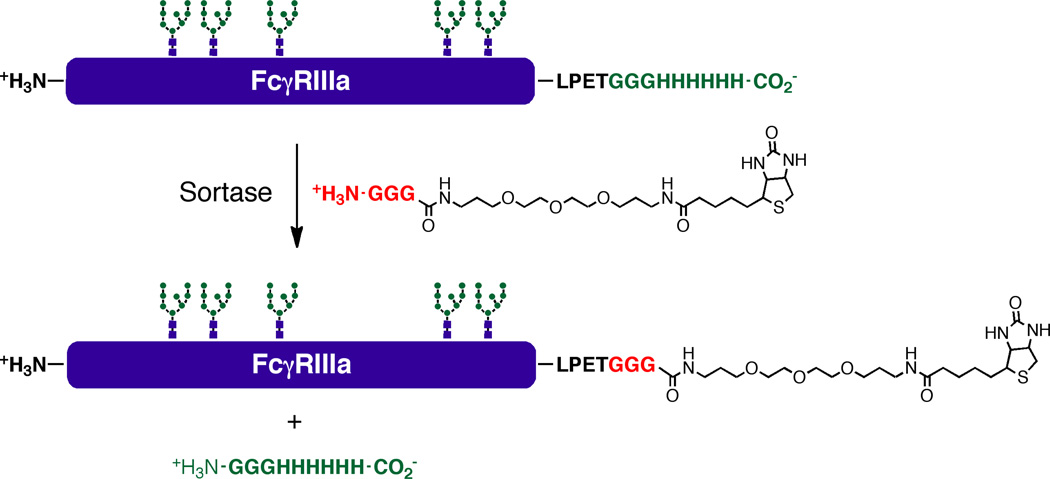
Figure 6.
Production of biotinylated FcγRIIIa using sortase-mediated ligation between FcγRIIIa-ST-H6 and GGG-linker-Biotin (compound 3 in the supporting information).
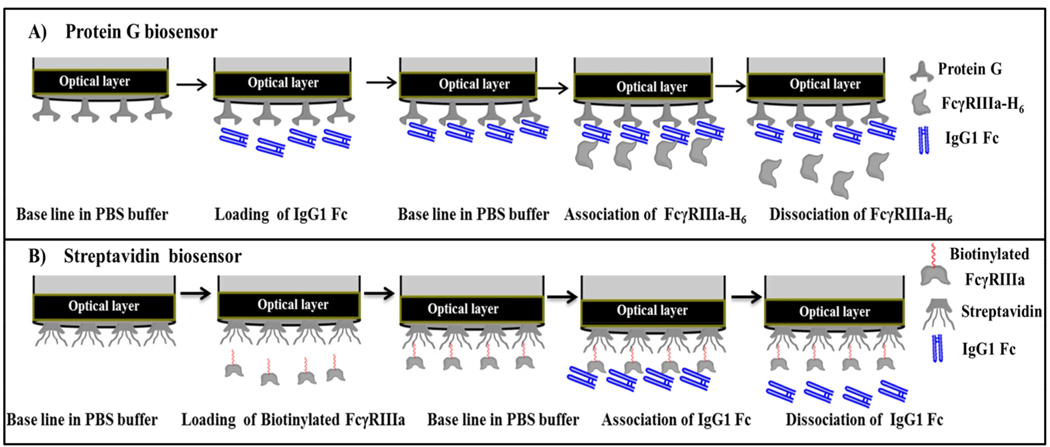
Analysis of the interaction of immobilized IgG1 Fc glycoforms with FcγRIIIa using biolayer interferometry (BLI)
The interactions of the different IgG1 Fc glycoforms with the FcγRIIIa-H6 were studied with biolayer interferometry using a BLITZ instrument (Fortebio, Menlo Park, CA) with protein G biosensor tips. Binding studies using this receptor were conducted as follows. The protein G biosensor tip was hydrated for 10 min with PBS buffer (150 mM NaCl, 50 mM sodium phosphate pH 7.4) and then incubated for 30 min with PBS kinetic buffer (PBS buffer containing 1 mg/mL casein as a blocking agent). Next, an initial baseline (30 s) was established with PBS kinetics buffer and then the protein G biosensor tips were loaded with the IgG1 Fc glycoforms at a concentration of 0.88 µM (120 s) to a response level of 2 nm. A new baseline (30 s) was then established and then the association (180 s) and dissociation (360 s) of FcγRIIIa-H6 was measured by dipping the biosensor into solutions of FcγRIIIa-H6 and PBS respectively. To determine the dissociation constant (KD) for the IgG1 Fc glycoforms, a range of FcγRIIIa-H6 concentrations from 50 nM-800 nM were tested for HM-Fc and Man5-Fc. For GlcNAc-Fc, the concentration range of FcγRIIIa-H6 tested was 200 nM to 1600 nM in twofold serial dilutions. For N297Q-Fc, no binding was observed at 20 µM FcγRIIIa-H6, the highest concentration of receptor tested. After each assay cycle, the biosensor tip was regenerated using two cycles of 10 mM HCl63 for 30 s and each time equilibrated using PBS kinetics buffer for 60 s. Data generated from the binding of the receptor to IgG1 Fc glycoforms were collected six times and globally fitted to a 1:1 binding model and analyzed using BLITZ Pro software.
Analysis of the interaction of IgG1 Fc glycoforms with immobilized FcγRIIIa using biolayer interferometry (BLI)
The interactions of the different IgG1 Fc glycoforms with the FcγRIIIa were studied with biolayer interferometry using a BLITZ instrument (Fortebio, Menlo Park, CA) with streptavidin (SA) biosensor tips. Prior to the binding experiment, IgG1 Fc samples were dialyzed in PBS buffer to remove the storage buffer (10% sucrose, 20 mM histidine, pH 6.0) and to adjust the pH to 7.4. The concentration of the samples after dialysis was 2.3 µM. Next, a stock solution of each glycoform in PBS kinetic buffer (PBS buffer containing 1 mg/mL casein) were prepared by adding casein (the stock solution of casein used was 10 mg/mL in PBS buffer), and PBS buffer. For HM-Fc and Man5-Fc, stock samples with a concentration of 1.6 µM were prepared and then serially diluted to prepare samples of 800 nM, 400 nM, 200 nM, 100 nM, and 50 nM concentrations by adding PBS kinetics buffer. For subsequent serial dilutions, PBS kinetic buffer containing 1 mg/mL of casein was used. Similarly, for GlcNAc-Fc a stock sample with a concentration of 2.0 µM was prepared and then used to prepare samples of 1600 nM, 800 nM, 400 nM, and 200 nM,by adding PBS kinetic buffer. For N297Q-Fc, after dialysis in PBS buffer, samples were first concentrated to 25 µM using an Amicon® Ultra-15 Centrifulgal Filter Device with a molecular weight cutoff of 10 kDa (EMD Millipore, Billerica, MA). Next, a solution containing 20 µM of N297Q-Fc in PBS kinetic buffer was prepared by dilution with PBS buffer and casein (added from a stock solution of 10 mg/mL casein in PBS buffer). This final sample concentration of 20 µM N297Q-Fc was then used for binding experiments without further dilution.
After sample preparation, binding studies were conducted as follows. First the streptavidin biosensor tip was hydrated for 10 min with PBS buffer and incubated with PBS kinetic buffer for 30 min. Next, the biotinylated FcγRIIIa-ST-H6 (0.1 µM) was immobilized onto the streptavidin biosensors to a response level of 0.4 nm and this step was followed by establishing an initial baseline (30 s) with PBS kinetic buffer. Then the association (180 s) and dissociation( 360 s) of the IgG1 Fc glycoforms were measured by dipping the biosensor into solutions of IgG1 Fc glycoforms and PBS respectively. After each assay cycle, the biosensor tip was regenerated using two cycles of 1 mM NaOH64 for 30 s and each time equilibrated with PBS kinetic buffer for 60 sec. To determine the dissociation constant (KD), the concentration range tested for HM-Fc and Man5-Fc in solution were 50 nM to 800 nM in twofold serial dilutions. The concentrations of GlcNAc-Fc tested in solution ranged from 200 nM to 1600 nM in twofold serial dilutions. For N297Q-Fc, no binding was observed with 20 µM N297Q-Fc in solution. Data generated from the binding of the receptor to IgG1 Fc glycoforms were collected six times and globally fitted to a 1:1 binding model and analyzed using BLITZ Pro software.
Results
Production and initial characterization of the four well-defined IgG1 Fc glycoforms
Expression and purification of high mannose IgG1 Fc (HM-Fc) and non-glycosylated mutant N297Q-IgG1 Fc (N297Q-Fc)
Both HM-Fc and N297Q-Fc were recombinantly expressed in glycosylation deficient strains derived from SMD1168 Pichia pastoris (Invitrogen, Carlsbad, CA). The expression strains were produced as described previously and have the genes OCH1 and PNO1 deleted to reduce the formation of higher order mannan structures and the addition of mannose-phosphorylation.58,1 The resulting yeast strains produce glycoproteins containing human-like high mannose N-linked glycans with some additional heterogeneous α-1,2-linked mannose residues added onto an initial Man8GlcNAc2 structure. Due to the large amount of HM-Fc required for production of the three glycosylated forms, HM-Fc, Man5-Fc, and GlcNAc-Fc, the HM-Fc was produced in a 10 liter fermentor using basal salts media supplemented with PTM1 trace salts solution. Protein expression was induced by methanol addition and yeast were harvested after approximately 3 days of induction. Compared to HM-Fc, relatively smaller amounts of N297Q-Fc were required (since it is not used to produce the other Fc glycoforms). Accordingly, N297Q-Fc was expressed in spinner flasks using BMGY media to generate cell mass, and 3 days of methanol induction to produce protein prior to harvest similar to the HMFc form. Typical yields from this expression system for the two IgG Fc proteins are summarized in the next section.
The same general purification procedure was utilized for both HM-Fc and N297Q-Fc. Yeast cells were removed by centrifugation and the resulting supernatant was filtered through 0.5 µm filters to remove remaining particulates prior to chromatography. Secreted IgG1 Fc was then isolated by protein G affinity chromatography. The average yield of HM-Fc from fermenter growth after protein G affinity chromatography was approximately 50 mg/L. The average yield of N297Q-Fc after spinner flask expression was approximately 20 mg/L. After protein G affinity chromatography there are still some residual yeast proteins remaining in both HM-Fc and N297Q-Fc. In the case of HM-Fc, incomplete glycosylation of the N297 site in yeast also results in macroheterogeneity of the glycosylation site.1,2,58,65 Because of this, the disulfide bonded HM-Fc dimer consists of three forms, a completely non-glycosylated form, a form that has glycosylation on only one chain of the dimer (mono-glycosylated), and a form that is glycosylated on both chains of the dimer (di-glycosylated). Hydrophobic interaction chromatography (HIC) using phenyl sepharose resin was utilized to remove residual yeast impurities for both proteins and to separate the di-glycosylated form of HM-Fc from its monoglycosylated and non-glycosylated forms. Because protein G purified HM-Fc is distributed between three different forms, the yield of di-glycosylated HM-Fc from HIC purification is lower than that of N297Q-Fc. Nonetheless, after HIC purification 445 mg of HM-Fc and 118 mg of N297Q-Fc were produced for further synthesis and studies. Hereafter HM-Fc will refer to the diglycosylated form of HM-IgG1 Fc obtained from yeast. Purified protein samples were pooled, dialyzed in storage buffer (10% sucrose, 20 mM histidine pH 6.0), concentrated or diluted to a final concentration of 0.2 mg/mL, and frozen at -80°C in aliquots for future use.
In vitro enzymatic synthesis of Man5-IgG1 Fc (Man5-Fc) and GlcNAc-IgG1 Fc (GlcNAc-Fc)
HM-Fc was converted into the Man5-Fc glycoform using B.t. α-1,2-mannosidase (BT3990).54,55 The outer mannose residues of high mannose N-linked glycans produced in glycosylation deficient yeast are α-1,2-linked to a core of five mannose residues that are α-1,3-and α-1,6-linked to one another and β-1,4-linked to the two N-acetylglucosamines attached to asparagine. Because of this, digestion the HM-Fc glycoform with a selective α-1,2-mannosidase will result in the formation of the Man5-Fc glycoform. Unfortunately, initial trial mannosidase reactions revealed that the B.t. α-1,2-mannosidase had very little activity in the sucrose containing storage buffer. Hence, before the reaction the starting material (HM-Fc) was extensively dialyzed to remove the storage buffer and exchange it into a buffer the α-1,2-mannosidase had higher activity in. Addition of mannosidase (6.7 mg) and incubation at room temperature for 2 days resulted in conversion of the HM-Fc into the Man5-Fc glycoform. Protein G affinity chromatography was utilized to remove the mannosidase enzyme from Man5-Fc, and the protein was dialyzed into storage buffer and adjusted to 0.2 mg/mL concentration. At total of 75 mg of Man5-Fc was produced from 125 mg of HM-Fc starting material. The low yield of Man5-Fc is likely due to the many extra dialysis steps and the protein G purification in this procedure.
The enzyme endoglycosidase H (Endo H) was utilized to generate further truncation of the high mannose glycan on HM-Fc. Endo H cleaves at the β-1,4 linkage between the two GlcNAc residues attached to asparagine in high mannose N-linked glycans. This leaves a single GlcNAc monosaccharide attached at the glycosylation site (Figure 1). Endo H has high activity in the sample storage buffer used to store HM-Fc, and this greatly simplified the production of the GlcNAc-Fc glycoform. A small amount (≈ 0.1 mg) of Endo H added to 100 mg HM-Fc in storage buffer resulted in a quantitative conversion into the GlcNAc-Fc glycoform after incubation at room temperature for 24 hours. Since Endo H was active in storage buffer and only a minute amount was added, GlcNAc-Fc was utilized without any further purification. Approximately 100 mg of GlcNAc-Fc was produced from 100 mg of HM-Fc starting material.
Analytical characterization of IgG1 Fc glycoforms
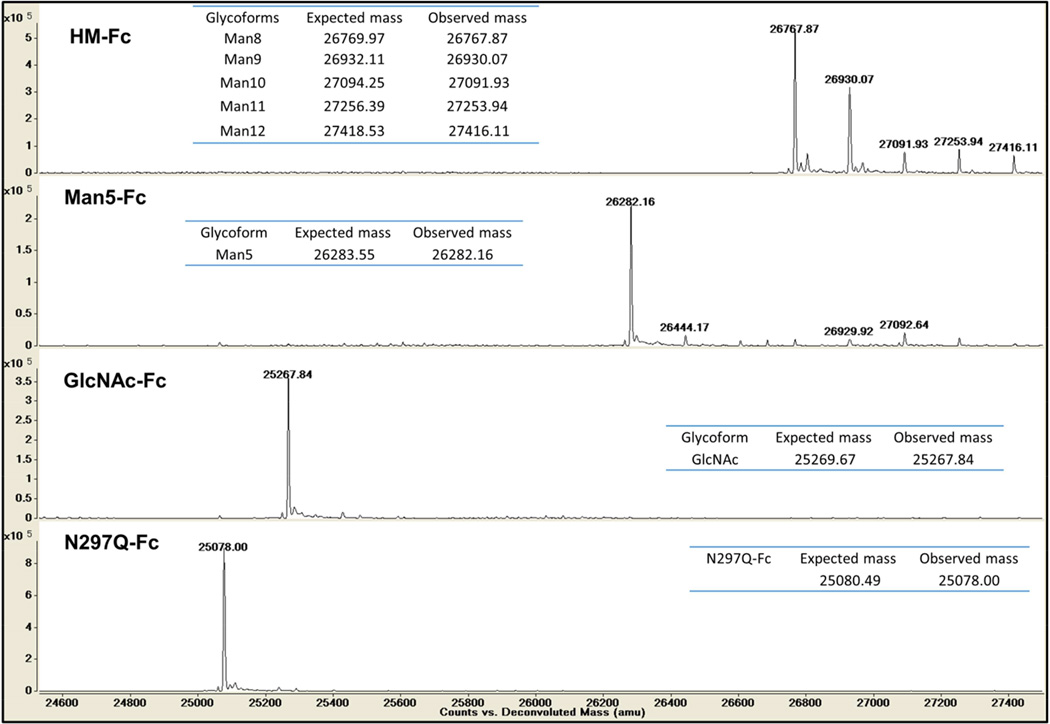
Figure 2 shows intact mass spectrometry data of the four glycoforms, HM-Fc, Man5-Fc, GlcNAc-Fc, and N297Q-Fc, with their respective expected and observed masses. Since the amino acid sequence of HM-Fc, Man5-Fc, and GlcNAc-Fc are identical, the differences in observed masses are mainly due to differences in the attached N-linked glycans. For the HM-Fc glycoform, glycosylation is heterogeneous with high mannose forms containing between 8 to 12 mannose residues with the major glycoform being the Man8GlcNAc2 form. The predominant forms of the truncated Man5-Fc and GlcNAc-Fc glycoforms on the other hand are largely one glycosylation state, with estimated abundance of 78% and 99% for the Man5 and GlcNAc forms respectively based on peak heights (see supplemental table S1). The N297Q-Fc form displays a single major peak corresponding to the non-glycosylated glutamine mutant as would be expected.
Figure 2.
Intact protein MS analysis of IgG1 Fc glycoforms under reducing conditions.
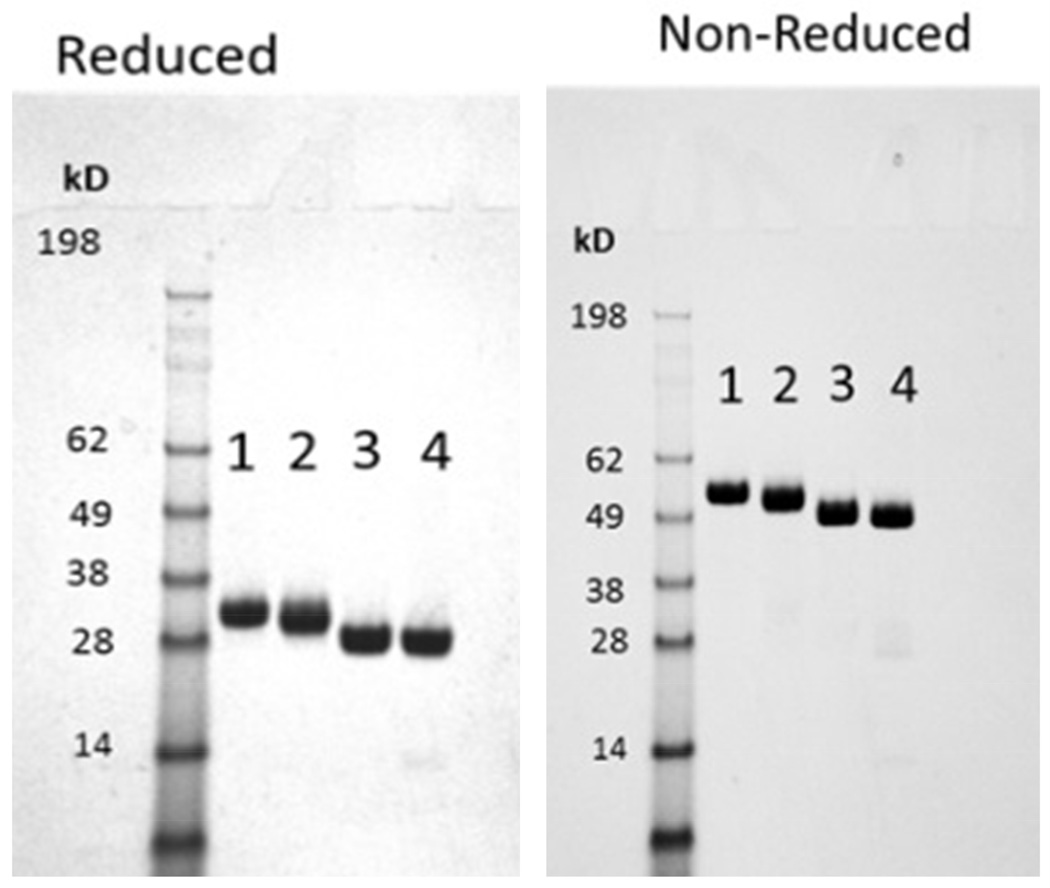
The four glycoforms were analyzed by SDS-PAGE under reducing and non-reducing conditions and the results are shown in Figure 3. Purity was estimated to be ~99% for each glycoform and no proteolysis or significant impurities were detected.66 The shift in migration between the reduced and non-reduced gels indicate intermolecular disulfide bonds are present forming dimers as would be expected in the hinge region. Also, no monomeric IgG1 Fc was detected in the non-reduced gel for any glycoform indicating that all of the IgG1 Fc is present in the dimerized state. Migration of the different glycoforms in both the reduced and non-reduced gels correlates with the size of the N-linked glycans attached, with slower migration occurring for forms with larger N-linked glycans.
Figure 3.
SDS-PAGE analysis of the four different IgG1 Fc glycoforms (1. HM; 2. Man5; 3. GlcNAc; 4. N297Q) under reduced and non-reduced conditions. The purified IgG1 Fc glycoforms showed ~99% purity under both conditions. The HM-Fc glycoform that has the highest molecular weight runs slowest among the four types followed by the Man5-Fc, GlcNAc-Fc, and N297Q-Fc.
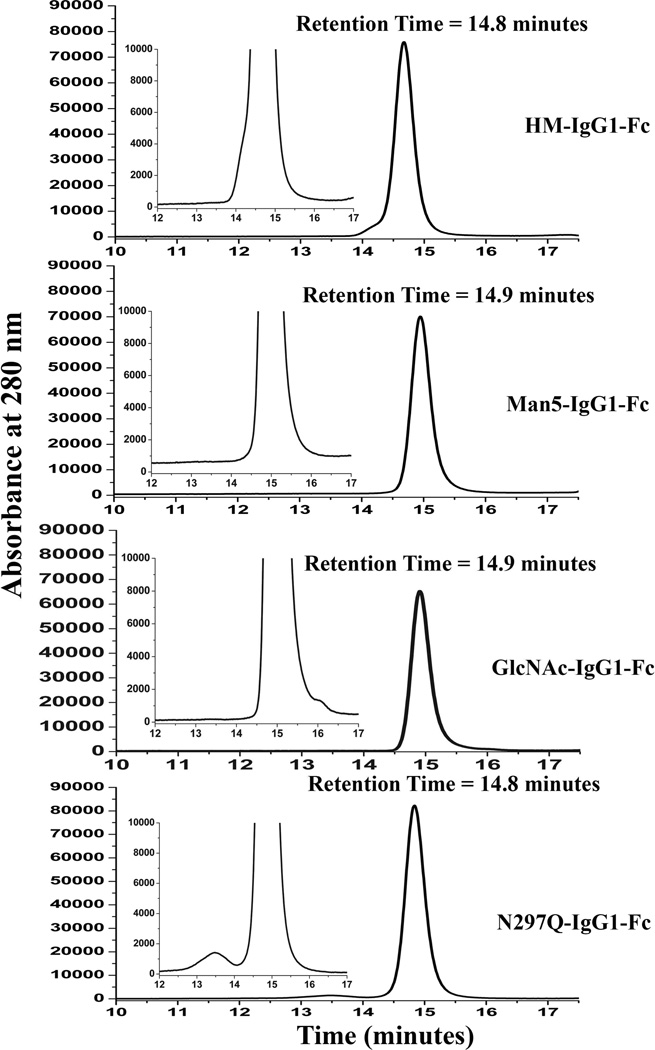
Size exclusion chromatography (SEC) was utilized to characterize the size and distribution of high-molecular weight species (HMWS) and low molecular weight species (LMWS) which potentially could be generated during production and purification of IgG1 Fc glycoforms. High molecular weight species could potentially be covalent aggregates, and low molecular weight species are related to fragments from the heavy chains.67,68 Representative SEC chromatograms of the IgG1 Fc glycoforms are shown in Figure 4 (also see Supplemental Table S2). All of the IgG1 Fc proteins eluted at ~15 min, which corresponded to a monomer based on the elution pattern of molecular weight standards. As illustrated in Figure 4, the results indicate all the IgG1 Fc glycoforms are monomeric (96–99%) with low levels of aggregates present across the IgG1 Fc samples (1–3 %). Low levels to essentially no higher molecular weight species (HMWS) were visible in the SEC chromatograms (less than estimated LOQ ~0.2%)69 of all the four glycoforms except for the N297Q-Fc, which showed some soluble dimers (~3%). Additionally, the SEC data also indicate low levels to essentially no low molecular weight species (LMWS) which is in agreement with SDS-PAGE characterization. All glycoforms were ≥98% monomeric except for the non-glycosylated form, which was greater than 96.4% monomeric. This difference is presumably due to the absence of glycosylation in N297Q-Fc. It has been reported that removal of glycosylation increases the aggregation propensity of IgGs.1,45,47
Figure 4.
Representative size exclusion chromatograms of the IgG1 Fc glycoforms. Results showed the following total monomer content (n=3; SD ~1.0%): HM-Fc 98.0% for HM-Fc, >99% purity for Man5-Fc and GlcNAc-Fc showed, and 96.7% for N297Q-Fc.
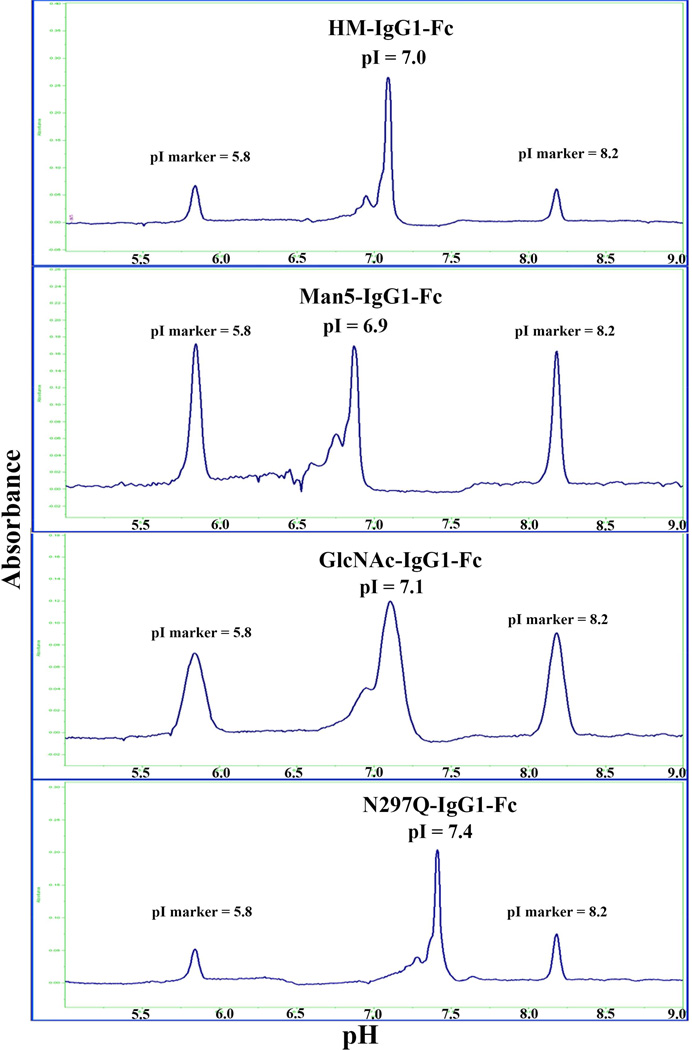
The charge distribution profiles of the four IgG1 Fc glycoforms were analyzed using cIEF. Representative electropherograms of HM-Fc, Man5-Fc, GlcNAc-Fc, and N297Q-Fc are shown in Figure 5 (also see Supplemental Table S3). All the electropherograms show one major peak, which after resolution gave pI values of 7.0, 6.9, 7.1 and 7.4 for HM-Fc, Man5-Fc, GlcNAc-Fc, and N297Q-Fc respectively. The theoretical isoelectric points (pI) of IgG1 Fc from the amino acid sequence is 6.9 which is in close agreement with the experimental pI values obtained given assay variability. Nonetheless, the slightly basic shift of the aglycosyated form, compared to other three glycoforms, could be due to the lack of oligosaccharides at the N297 site. Although there are no visible basic variants observed, there are minor acidic peaks/shoulders near the main peaks with the pI values ranging from 5.6–6.8. The observed heterogeneity could potentially be from chemical modifications in IgG1 Fc proteins (e.g., deamidation) during production and/or purification.
Figure 5.
Representative charge heterogeneity profiles of IgG1 Fc glycoforms as measured by capillary isoelectric focusing (cIEF). The isoelectric point (pI) of the main peak (n=3; SD ~0.1 pI units): pI of 7.0 for HM-Fc, pI of 6.9 for Man5-IgG1Fc,, pI of 7.1 for GlcNAc-Fc, and pI of 7.4 for N297Q-Fc.
In summary, these results demonstrate that each of the IgG1 Fc glycoforms is an overall well-defined glycoform species, with some minor charge heterogeneities and soluble aggregates present.
Evaluation of affinity for IgG1 Fc glycoforms with FcγRIIIa using biolayer interferometry (BLI)
Expression and purification of two forms of FcγRIIIa for use in binding assays
We have previously produced an expression strain for the production of the soluble domain of FcγRIIIa in yeast with a C-terminal histidine tag for affinity purification, reported in Xiao et al.58, and this strain was utilized to produce FcγRIIIa-H6 used in the binding assays with IgG1 Fc immobilized on protein G biosensors. To reverse the immobilization format and have the receptor immobilized, a new expression strain was also constructed that produces the soluble domain of FcγRIIIa in yeast with a combined C-terminal histidine and sortase tag. This new receptor form, FcγRIIIa-ST-H6, allows for affinity purification and the attachment of synthetic molecules, such as the synthetic biotin derivative used in this study, selectively to the C-terminus using sortase mediated ligation.53,70–72 Both receptors were expressed in a glycosylation-deficient strain of P. pastoris with glycerol and methanol as carbon sources. After centrifugation and filtration to remove yeast, Ni+2-NTA affinity chromatography was utilized to isolate the FcγRIIIa forms. The receptors were further purified by phenyl sepharose chromatography, and after isolation and concentration approximately 18 mg/L of purified protein were obtained for both forms of FcγRIIIa.
Selective C-terminal biotinylation of FcγRIIIa-ST-H6 using sortase mediated ligation
Sortase mediated ligation53,70–72 was utilized to attach biotin to the C-terminus of FcγRIIIa. Bacterial sortases, such as the SrtA of S. aureus, are enzymes that catalyze transpeptidase reactions to attach proteins containing sortase recognition sites to the peptidoglycan of bacterial cell walls. Sortases have been adapted for biotechnology applications by using the sortase reaction for immobilization and to attach synthetic molecules and labels selectively to the N- or C-termini of recombinant proteins. The requirements for sortase ligations are N-terminal poly-glycine containing peptides, such as those found in peptidoglycan, and C-terminal recognition peptide motifs such as LPETG, as in the SrtA mediated ligation used in this study.53,70–72 The C-terminal peptide is cleaved between threonine and glycine and a new peptide bond is formed between the N-terminal poly-glycine containing molecule and the C-terminal threonine. Accordingly, in order to selectively biotinylate FcγRIIIa-ST-H6 it was necessary to synthesize a form of biotin containing an N-terminal poly-glycine. In addition, to ensure strong binding to streptavidin and prevent steric hindrance with streptavidin interfering with the interaction of FcγRIIIa with IgG1 Fc, a long hydrophilic diamine linker was used to separate biotin from the C-terminus of FcγRIIIa. The synthesis of GGG-linker-Biotin (compound 3) is described in the supporting information for this paper (Figure S1), and the ligation of GGG-linker-Biotin to FcγRIIIa-ST-H6 is shown in Figure 6. The sortase reaction proceeded efficiently to attach biotin to the C-terminus of FcγRIIIa-ST-H6, and the resulting biotinylated FcγRIIIa was immobilized in binding assays with streptavidin biosensors.
Analysis of binding of Immobilized IgG1 Fc glycoforms to FcγRIIIa using biolayer interferometry (BLI)
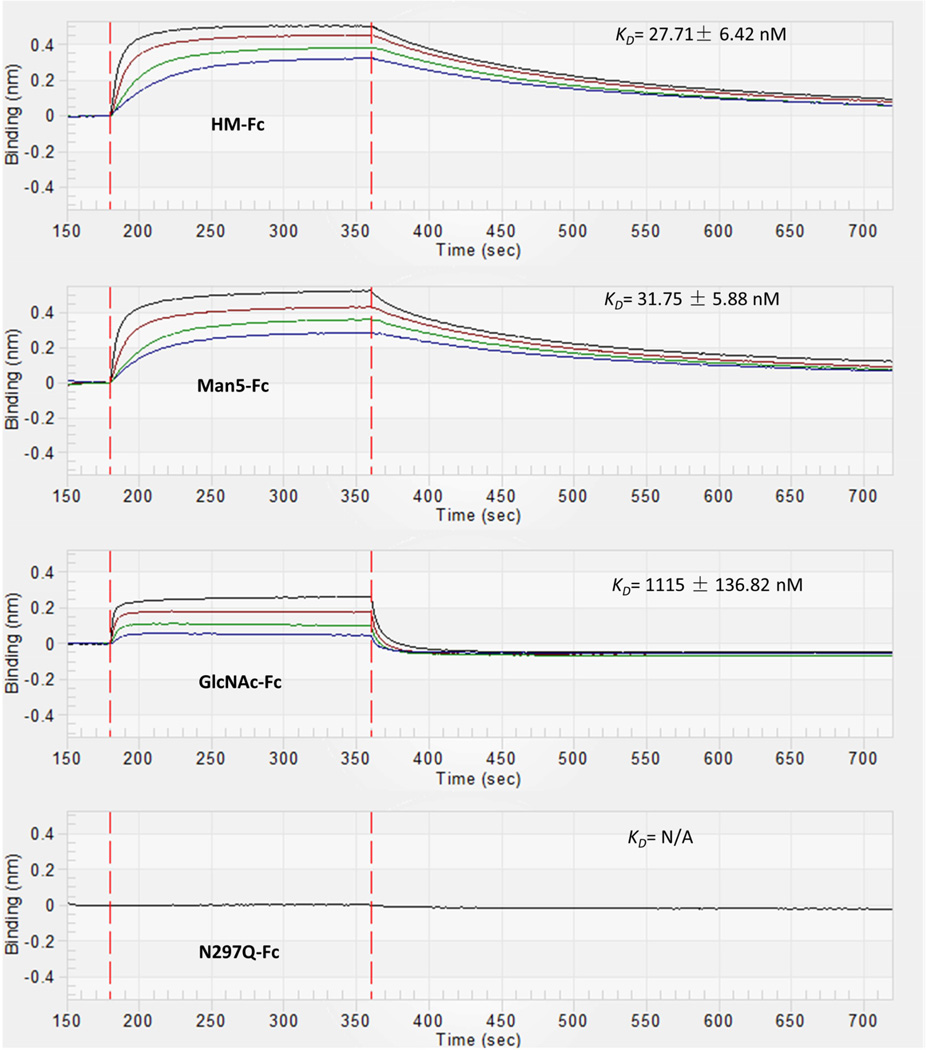
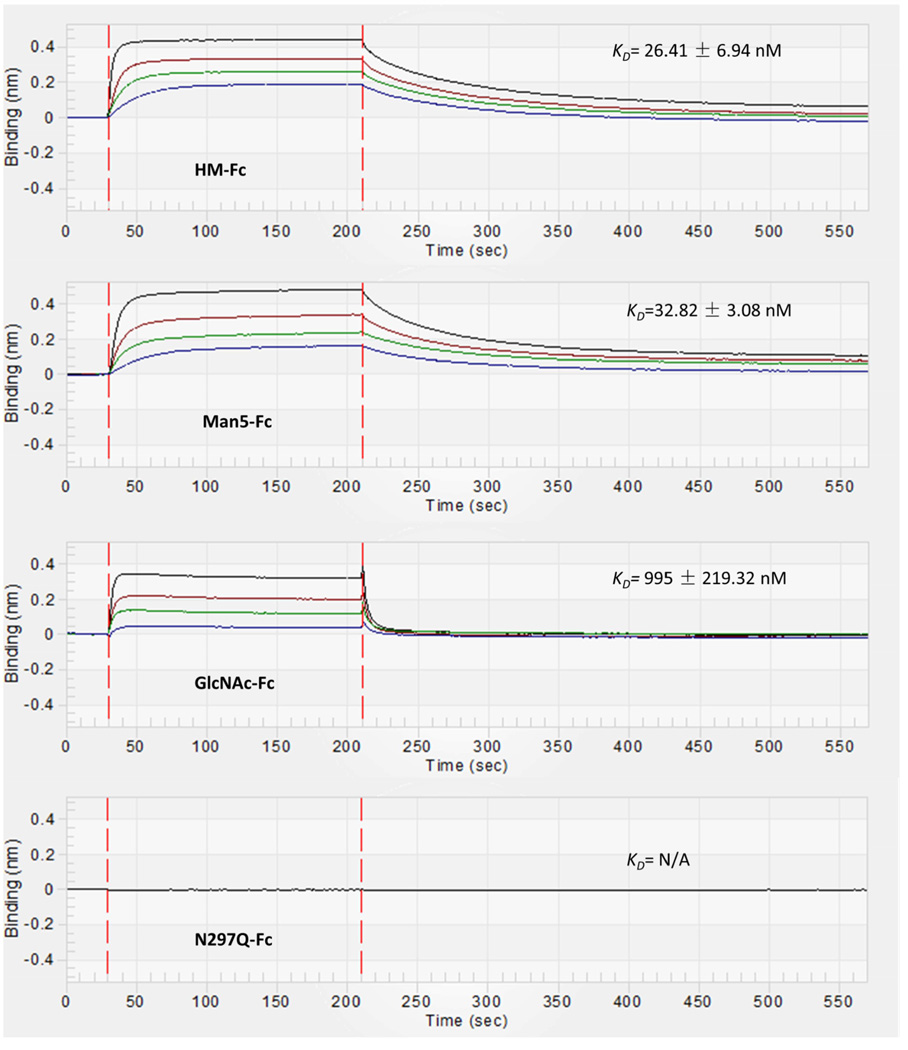
The affinity of FcγRIIIa in solution for immobilized IgG1 Fc glycoforms was studied using BLI with protein G biosensors. The format for this binding study is shown in Figure 7A. Protein G biosensors were first loaded with IgG1 Fc glycoforms to a response level of 2 nm using 0.8 µM solutions of the various IgG1 Fc glycoforms. Then a baseline was established and the association of FcγRIIIa was observed by dipping the biosensor into solutions of different concentrations of FcγRIIIa. After the Fc-FcγRIIIa complex had formed on the biosensor, dissociation of FcγRIIIa was observed by placing the biosensor in a solution of PBS kinetic buffer. The kinetic rate constants for association, ka, and dissociation, kd, were then obtained by fitting the resulting curves to a 1:1 binding model which is consistent with previous biochemical and structural studies of this interaction.73 The equilibrium dissociation constant was determined by dividing kd by ka for each glycoform. Representative sensorgrams for each glycoform (HM-Fc, Man5-Fc, GlcNAc-Fc, and N297Q-Fc) are displayed in Figure 8, and the results for these binding experiments are given in Table 1 (Supplemental Figure S4 also displays representative sensograms with curve fits added). HM-Fc and Man5-Fc have similar high affinity for FcγRIIIa as would be expected for high mannose IgG1 Fc glycoforms, with KD's of 27.7 nM and 31.8 nM respectively.1,2,74 In contrast, the GlcNAc-Fc glycoform has signficantly weaker binding to FcγRIIIa with a KD of 1115 nM, and this agrees with recent studies of this glycoform.75 Interestingly, the association rate constants (ka) are very similar for HM-Fc, Man5-Fc, and GlcNAc-Fc, such that the driving factor behind the GlcNAc-Fc's weaker affinity for FcγRIIIa is its significantly faster dissociation rate (kd). This similarity in ka values and significant differences in kd and KD values is illustrated in Figure S3. No interaction was observed between FcγRIIIa and the non-glycosylated N297Q-Fc form at the highest concentration of receptor tested (20 µM).
Figure 7.
Steps followed in kinetic binding experiments using biolayer interferometry. A) Top panel shows steps followed for measuring binding kinetics using protein G biosensors, where IgG1 Fc glycoforms were immobilized and FcγRIIIa-H6 was present in solution. B) Bottom panel shown steps involved in binding kinetics using streptavidin biosensor, where biotinylated FcγRIIIa was immobilized and IgG1 Fc glycoforms were present in solution.
Figure 8.
Representative BLI binding data for the interaction of FcγRIIIa with IgG1 Fc glycoforms immobilized on protein G biosensors. The binding curves correspond to a range of receptor concentrations in solution. For HM-Fc and Man5-Fc data is shown for FcγRIIIa at concentrations of 800 nM, 400 nM, 200 nM, 100 nM which corresponds with the curves from top to bottom. For GlcNAc-Fc data is shown for FcγRIIIa at concentrations of 1600 nM, 800 nM, 400 nM and 200 nM which corresponds with the binding curves from top to bottom. For N297Q-Fc a single concentration of 20 µM was tested where no binding was observed.
Table 1.
Kinetic parameters obtained for binding of FcγRIIIa with IgG1 Fc glycoforms. Binding kinetics were measured at 25°C. The data shows kinetic association rate ka, kinetic dissociation rate kd, and equilibrium dissociation constant KD. These data are averaged values of six independent experiments.
| Glycoforms | Immobilized protein |
Immobilization technique |
Average ka × 10−5 (1/Ms) |
Average kd × 103 (1/s) |
Average KD (nM) |
|---|---|---|---|---|---|
| HM-Fc | IgG1 Fc | Protein G | 1.9 ± 0.3 | 5.4 ± 1.0 | 27.7 ± 6.4 |
| FcγRIIIa | Streptavidin | 3.0 ± 0.6 | 7.9 ± 1.4 | 26.4 ± 6.9 | |
| Man5-Fc | IgG1 Fc | Protein G | 1.6 ± 0.1 | 5.1 ± 0.9 | 31.8 ± 5.9 |
| FcγRIIIa | Streptavidin | 2.2 ± 0.2 | 7.3 ± 0.3 | 32.8 ± 3.1 | |
| GlcNAc-Fc | IgG1 Fc | Protein G | 1.2 ± 0.2 | 140.0 ± 9.1 | 1115.0 ± 136.8 |
| FcγRIIIa | Streptavidin | 1.6 ± 0.3 | 161.3 ± 21.8 | 995.0 ± 219.3 | |
| N297Q-Fc | IgG1 Fc / FcγRIIIa | Protein G / Streptavidin | * | * | * |
For N297Q, there was no detectable binding at the highest concentration tested (20 µM) for both methods.
Analysis of binding of IgG1 Fc glycoforms to immobilized FcγRIIIa using biolayer interferometry (BLI)
The affinity of the IgG1 Fc glycoforms in solution for immobilized FcγRIIIa was studied using BLI with streptavidin biosensors. The format for this binding study is shown in Figure 7B. Streptavidin biosensors were first loaded with C-terminally biotinylated FcγRIIIa to a response level of 0.4 using 0.1 µM biotinylated FcγRIIIa. Next a baseline was established and the association of IgG1 Fc glycoforms was observed by dipping the biosensors into solutions of different concentrations of IgG1 Fc glycoforms. After the complex had formed on the biosensor, dissociation of the IgG1 Fc glycoforms was observed by dipping the biosensor into of PBS kinetic buffer. Kinetic rate constants and equilibrium dissociation constants were determined as described for the protein G method above. Representative sensorgrams for each glycoform (HM-Fc, Man5-Fc, GlcNAc-Fc, and N297Q-Fc) are displayed in Figure 9, and the results for these binding experiments are given in Table 1 (Supplemental Figure S5 also displays representative sensograms with curve fits added). Perhaps not surprisingly, the affinities determined for this receptor immobilized binding format were very similar to the results observed for the IgG1 Fc immobilized binding format. HM-Fc and Man5-Fc showed similar high affinity for FcγRIIIa with KD's of 26.4 nM and 32.8 nM respectively.1,2,74 Also, the GlcNAc-Fc glycoform displayed much weaker binding with a KD of 995 nM, and this weak affinity was again due to a larger dissociation rate constant (kd) for GlcNAc-Fc relative to the other glycoforms. Finally, the N297Q-Fc form displayed no affinity for immobilized FcγRIIIa even at 20 µM N297Q-Fc.
Figure 9.
Representative BLI binding data for the interaction of IgG1 Fc glycoforms with FcγRIIIa immobilized on streptavidin biosensors. The binding curves correspond to a range of IgG1 Fc glycoform concentrations in solution. For HM-Fc and Man5-Fc data is shown for IgG1 Fc glycoforms at concentrations of 800 nM, 400 nM, 200 nM, 100 nM which corresponds with the curves from top to bottom. For GlcNAc-Fc data is shown for GlcNAc-Fc at concentrations of 1600 nM, 800 nM, 400 nM and 200 nM which corresponds with the binding curves from top to bottom. For N297Q-Fc a single concentration of 20 µM was tested where no binding was observed.
Discussion
Production and initial characterization of well-defined IgG1 Fc glycoforms
As a first step to develop a model system for biosimilar analysis studies, it was desirable to not only have a group of related well-defined proteins that differ from one another in specific attributes, but also to be able to produce relatively large amounts of those proteins, on an academic laboratory scale, to provide sufficient material to conduct biosimilarity assessments through a combination of biochemical, physicochemical and biological tests. To achieve this, glycosylation deficient yeast (OCH1/PNO1 deleted P. pastoris derived from strain SMD1168) that express human-like, high mannose type N-linked glycosylation were utilized to produce the IgG1 Fc glycoproteins and the non-glycosylated mutant used in this and the accompanying studies. IgG1 Fc was secreted into culture media, and after centrifugation and filtration the IgG1 Fc was captured and purified using protein G affinity chromatography. Residual yeast impurities and, in the case of HM-Fc, removal of mono- and non-glycosylated IgG1 Fc forms was accomplished using HIC chromatography. The non-glycosylated N297Q-Fc mutant (Figure 1) was utilized directly as a negative control for the effects of glycosylation in these studies. The glycosylated HM-Fc glycoform (Figure 1) is a member of this model system and also serves as the starting material for the production of the remaining two glycoforms of the model system. The HM-Fc glycoform was treated with B.t. α-1,2-mannosidase and Endo H to produce the Man5-Fc and GlcNAc-Fc glycoforms respectively (Figure 1). The HM-Fc, Man5-Fc, and GlcNAc-Fc glycoforms form a well-defined series where HM-Fc contains the largest N-linked glycan and the remaining glycoforms are sequentially truncated to give the intermediate glycosylation state of Man5-Fc, and the minimally glycosylated state of GlcNAc-Fc. This series of glycoproteins with decreasing N-linked glycan size all derived from the same precursor should be useful in elucidating effects of glycosylation on the biological and physicochemical properties of IgG1 Fc.
In terms of ease of production, the HM-Fc and N297Q-Fc are both derived directly from fermentation, and therefore more easily accessed than the other two glycoforms. Because N297Q-Fc has no glycosylation site occupancy heterogeneity, its HIC purification is simplified compared to the HM-Fc form which requires separation of di-, mono-, and non-glycosylated forms. After HIC purification of both forms, 445 mg of HM-Fc and 118 mg of N297Q-Fc were produced for this study from approximately 28 and 7 liters of growth media respectively. Conversion of 100 mg of HM-Fc into GlcNAc-Fc using Endo H (approximately 0.1 mg) was straightforward and nearly quantitative, allowing relatively easy access to 100 mg of the GlcNAc-Fc glycoform. Production of the Man5-Fc glycoform was more problematic since it was discovered that the B.t. α-1,2-mannosidase had little activity in sucrose containing storage buffer after the HM-Fc had been transferred into the storage buffer. This required additional dialysis steps to transfer the HM-Fc into a different buffer to conduct the B.t. α-1,2-mannosidase reaction. In addition, a larger amount of B.t. α-1,2-mannosidase (6.7 mg) relative to Endo H (0.1 mg) was required for the reaction to produce Man5-Fc, and this made it necessary to purify the Man5-Fc glycoform by an additonal protein G affinity chromatography step after the mannosidase reaction. All of these factors combined to result in a 60% yield for conversion of HM-Fc into Man5-Fc and only 75 mg of the Man5-Fc glycoform being produced. The amount of Man5-Fc produced was sufficient to conduct this and the accompanying studies, but the procedure to produce this glycoform could be optimized in the future to produce more of this glycoform in a higher yield. Taken together, these methods allow easy access to the four well-defined glycoforms from Figure 1 in quantities sufficient to enable a wide range of biosimilar analysis and stability studies.
Initial characterization of the glycosylation state of the model glycoforms was conducted using intact protein mass spectrometry. Mass spectrometry was used to confirm the type of glycosylation on each glycoform and determine if there were any significant proteolytic products present (Figure 2). None of the IgG1 Fc glycoforms showed any evidence of internal proteolysis, and all were found to be in forms containing their complete amino acid sequences including C-terminal lysine residues. The intact protein mass spectra also allowed evaluation of the glycosylation state of the IgG1 Fc glycoforms. The N297Q-Fc and GlcNAc-Fc glycoforms were highly homogeneous with single peaks corresponding to no glycosylation with a glutamine mutation and a single GlcNAc attached to asparagine respectively. The HM-Fc glycoform obtained directly from yeast was heterogenously glycosylated with high mannose type glycans ranging in size from Man8GlcNAc2 to Man12GlcNAc2, with the form containing Man8GlcNAc2 being the major form at approximately 49% of its glycosylation sites based on peak intensities (supporting information, Supplemental Table S1). The mannosidase treated Man5-Fc glycoform is much more homogeneous and estimated to contain Man5GlcNAc2 on 78% of its glycosylation sites based on peak intensities (supporting information, Supplemental Table S1). The remainder of the glycoform peaks observed in the Man5-Fc mass spectra correspond to small amounts of incompletely cleaved high mannose glycosylated forms, indicating the mannosidase reaction could be further optimized in the future. The mass spectrometry results demonstrate well-defined differences between the model IgG1 Fc glycoforms based on the size or absence of N-linked glycosylation.
Additional biochemical characterization was conducted using SDS-PAGE, size exclusion chromatography (SEC), and capillary isoelectric focusing (cIEF). This was done to further define the chemical state and purity of the IgG1 Fc glycoforms and to establish a baseline prior to the chemical and physical stability studies described in the companion papers in this series. Reducing and non-reducing SDS-PAGE (Figure 3) display an absence of proteolytic fragments and show high purity for each glycoform. The electrophoretic mobility of the IgG1 Fc glycoforms shown in Figure 3 was also consistent with slower migration for the larger branched N-linked glycoproteins with HM-Fc migrating slowest, Man5-Fc migrating intermediately, and the GlcNAc-Fc and N297Q-Fc forms migrating fastest.76 In addition, the non-reduced SDS-PAGE indicated complete intermolecular disulfide bond formation corresponding to 100% dimerized form for all glycoforms. As illustrated in Figure 4, the SEC results indicate the IgG1 Fc glycoforms are mostly monomeric (96–99%) with low levels of aggregates present across the IgG1 Fc samples (1–3 %). Low levels to essentially no higher molecular weight species (HMWS) were visible in the SEC chromatograms (less than estimated LOQ ~0.2%) of all the four glycoforms except for the N297Q-Fc, which showed some soluble dimers (~3%). Additionally, the SEC data also indicate low levels to essentially no low molecular weight species which is in agreement with SDS-PAGE characterization. In summary, the Fc glycoforms were ≥98% monomeric by SEC except for the non-glycosylated form, which was 96.4% monomeric. This difference is presumably due to the absence of glycosylation in N297Q-Fc. It has been reported that removal of N-linked glycans in the Fc region increases the aggregation propensity of IgG Fcs and IgGs.1,45
Analysis of the isoelectric points for the four glycoforms by cIEF identified a difference between the N297Q-Fc form and the other forms, with the N297Q-Fc form having a pI of 7.4 compared to 7.0, 6.9, and 7.1 for HM-Fc, Man5-Fc, and GlcNAc-Fc respectively (Figure 5). Based on MS analysis described in the companion paper in this series (see Mozziconacci et al, “Comparative evaluation of the chemical stability of four well-defined IgG1-Fc glycoforms”)50, the only Asn residue that displayed measurable deamidation in the four non-stressed Fc samples was N315 (0.5–2.0%), but the levels were similar for the Man5 glycoform and the N297Q aglycosylated form (~0.5%; data not shown) yet their pI values were different. Thus, Asn deamidation is not the likely cause of the observed differences. Moreover, the N-linked glycans present on the glycoforms are neutral glycans and are not charged. Interestingly, it has been noted by our laboratory and others that cation exchange chromatography can be used to separate glycosylated and non-glycosylated proteins from one another even when only neutral oligosaccharides are present on the glycosylation sites. The elution order observed from cation exchage chromatography in the cases of separation of glycosylated and non-glycosylated IL-1ra and di-glycosylated IgG from mono- and non-glycosylated IgG are consistent with the nonglycosylated forms having higher isoelectric points.2,3,77 It may be that the measured pI differences arise from different interactions of the IgG1 Fc glycoforms within the polymeric cartridge containing ampholytes used in cIEF, rather than actual pI differences. The root cause of the observed pI differences appears to be complex and will be a topic for future studies.
Biological evaluation of well-defined IgG1 Fc glycoforms
Potential biological activities of the IgG1 Fc glycoforms in our model system were assessed using binding to an activating Fc receptor, FcγRIIIa, as a measure of the potential to activate antibody dependent effector functions. In full-length antibodies, simultaneous binding of the antigen binding regions of an antibody to a polyvalent antigen and the antibody Fc region to FcγRIIIa can activate antibody dependent cell-mediated cytotoxicity (ADCC).78 FcγRIIIa is also important in the function of natural killer cells, since it is the only Fc receptor expressed on natural killer cells (NK cells).79 ADCC and NK cells are both believed to be important in the function of many therapeutic mAbs.79,80 Additionally, FcγRIIIa is sensitive to the type of glycosylation present on the Fc region of antibodies, having high affinity to antibodies that do not contain core-linked fucose, and like most Fc receptors, lower affinity for truncated N-linked glycans.80 This makes FcγRIIIa useful in distinguishing the different IgG1 Fc glycoforms in our model system.
The interactions of the IgG1 Fc glycoforms with FcγRIIIa were assessed with kinetic binding studies using biolayer interferometry (BLI). Observation of binding using surface techniques such as BLI or surface plasmon resonance (SPR) generally do not require large amounts of material to conduct binding studies, and this is an advantage for biosimilarity analysis studies where many samples may be required to be analyzed. Kinetic binding studies using BLI or SPR allow determination of kinetic rate constants for association (ka) and dissociation (kd), and by taking the ratio of the kinetic rate constants, equilibrium dissociation constants (KD) can be determined. One drawback of surface binding techniques is that one binding partner, receptor or ligand, needs to be immobilized to conduct the experiment (Figure 7). Choice of which component, receptor or ligand, to immobilize and method of immobilization can both affect the outcome of the experiment. In addition, from the perspective of conducting experiments with samples of formulated drug products, the format of binding assays can have practical effects on how the assays are conducted and what kind of information is obtained. To gain a better understanding of how binding format affects the results of binding studies, we developed two formats for studying the interaction of IgG1 Fc glycoforms with FcγRIIIa (Figure 7), one with the IgG1 Fc immobilized and one with the receptor immobilized.
For a BLI binding format where the IgG1 Fc glycoforms were immobilized (Figure 7A), we decided to use protein G biosensors for immobilization of IgG1 Fc glycoforms. Protein A and/or protein G have previously been used in SPR binding studies of IgG interactions with Fc receptors, and immobilization using these proteins have many advantages.81,82 Since protein G binds tightly to IgGs over a wide pH range (≈ pH 5–8), protein G biosensor tips can be loaded under a variety of formulation conditions without having to adjust the buffer conditions of samples to promote immobilization. In addition, once IgG1 Fc (or full-length IgG) has been immobilized onto a protein G biosensor, binding experiments can be conducted in buffers optimized for receptor binding by simply dipping the IgG1 Fc loaded protein G biosensor into receptor solutions with optimized buffer conditions. Also, protein G biosensors can be regenerated by acidic treatment much the way protein A and protein G affinity resins are eluted during affinity chromatography, allowing for relatively easy reuse of protein G biosensors. A disadvantage of using protein G biosensors to immobilize IgG1 Fc in binding studies may arise during stability studies when there is the potential of the formation of damaged forms of IgG1 Fc which would no longer be capable of binding to protein G, but which could still retain the ability to bind to Fc receptors. Such proteins would not be measured in this assay format. Also, this assay format is relatively insensitive to the actual concentration of IgG1 Fc glycoforms in samples, since to obtain reproducible binding curves the biosensors are loaded to the same approximate level, and because of this changes in concentrations in the samples may be difficult to detect. Nevertheless, this assay format provides valuable information about the kinetics and thermodynamics of the interaction of IgG1 Fc glycoforms with FcγRIIIa.
Figure 8 shows representative binding curves and Table 1 summarizes the resulting rate constants and dissociation constants obtained for the interaction of immobilized IgG1 Fc glycoforms with FcγRIIIa in solution using this binding format. As can be seen in Table 1, both HM-Fc and Man5-Fc have high affinity for FcγRIIIa (KD's of 28 and 32 nM respectively), GlcNAc-Fc showed much lower affinity (KD of 1115 nM), and N297Q-Fc had no observable affinity for FcγRIIIa (all measurements were conducted at 25°C). Examination of the rate constants for the different interactions reveals that the main factor in the GlcNAc-Fc glycoform's lower affinity is due to significantly faster dissociation of the complex once it forms relative to HM-Fc and Man5-Fc, and this can be seen in the magnitudes of the dissociation rate constants, kd (Table 1), and the ratios of kinetic and thermodynamic values when compared to HM-Fc values (supporting information Supplemental Figure S3).
For a BLI binding format where FcγRIIIa is immobilized and IgG1 Fc glycoforms were in solution (Figure 7B), we utilized selectively biotinylated FcγRIIIa and streptavidin biosensors. Streptavidin biosensors were chosen for immobilization of FcγRIIIa because the high affinity of non-fucosylated IgG1 Fcs, such as HM-Fc and Man5-Fc (KD ≈ 30 nM), required either covalent or significantly higher affinity immobilization to prevent artifacts related to dissociation of receptors during binding measurements. The high affinity of streptavidin for biotin (KD ≈ 10−14 M)83 was appropriate for this application, therefore we developed a novel method to selectively biotinylate the C-terminus of FcγRIIIa using sortase mediated ligation. This required constructing an expression strain of FcγRIIIa which contained a C-terminal sortase tag, expression and purification of the sortase tagged FcγRIIIa, synthesis of a triglycine containing biotin derivative (compound 3, supporting information), and sortase mediated ligation to produce the C-terminally biotinylated FcγRIIIa, and these methods are described in the supporting information. An advantage of using an immobilized FcγRIIIa binding format such as that shown in Figure 7B is that binding measurements can be made directly in solutions of IgG1 Fc glycoforms. This type of format is also potentially more sensitive to changes in IgG1 Fc concentration since measurements are done with the samples directly in solution rather than in an immobilized format.
A disadvantage of this binding format is that buffer conditions present in samples must be compatible with receptor binding measurements. The affinity of many receptors is pH dependent, so formulation pH could affect binding measurements. In addition, high concentrations of sugars, such as the 10% sucrose in our sample storage buffer, can have significant effects on the kinetics and thermodynamics of binding measurements. Because of these factors, in our experiments it was necessary to transfer the IgG1 Fc glycoforms from storage buffer (10% sucrose, 20 mM histidine pH 6.0) to kinetics buffer (PBS buffer containing 1 mg/mL casein as a blocking agent) by dialysis, prior to binding measurements. This added a step to the binding measurements and has the potential to skew results if there is loss of sample or changes in concentration during the transfer of samples to the new buffer. The results of the receptor immobilized binding format are shown in Figure 9 and Table 1. The binding results for the receptor immobilized format (Figure 7B) are highly similar to the IgG1 Fc immobilized format (Figure 7A), suggesting the extra dialysis step in the receptor immobilized format did not affect the binding measurement significantly. Both HM-Fc and Man5-Fc displayed high affinity for FcγRIIIa with KD's of 26 and 33 nM respectively, while the GlcNAc-Fc had lower affinity at 995 nM, and the N297Q-Fc form had no observable affinity (all measurements were conducted at 25°C). The driving factor in the difference between the high affinity HM-Fc and Man5-Fc and the lower affinity GlcNAc-Fc was again faster dissociation of the complex (see Table 1 and supporting information Supplemental Figure S3). Using these methods, the standard deviations for determining KD varies between the different forms, with larger overall standard deviations occurring for the weaker binding GlcNAc-Fc glycoform. If on the other hand, the relative standard deviation (RSD) is considered, it is relatively consistent between all forms varying from 9.4% to 26.3% RSD with no clear indication of higher or lower RSD for weak vs. strong binding glycoforms or one binding format vs. the other. These results suggest that these two different binding formats are complementary in these experiments using well-defined IgG1 Fc glycoforms.
In subsequent papers in this series, these model IgG1 Fc glycoforms are utilized to assess how glycosylation alters the chemical stability profiles (see Mozziconacci et al, “Comparative evaluation of the chemical stability of four well-defined IgG1-Fc glycoforms”), and physical stability properties (see More et al, “Correlating the impact of well-defined oligosaccharide structures on the physical stability profiles of IgG-Fc glycoforms”) of these glycoproteins. The physical stability data sets are then used as a model data set to develop a mathematical algorithm, based on data mining and machine learning tools, for potential use for biosimilarity assessments (see Kim JH et al, “Biosimilarity assessments of model IgG1-Fc glycoforms using a machine learning approach”).
In summary, we have presented here the production and biological evaluation of four well-defined IgG1 Fc glycoforms as the initial steps of developing a model system for biosimilarity analysis. The methods developed here allow relatively easy access to the four well-defined IgG1 Fc glycoforms (HM-Fc, Man5-Fc, GlcNAc-Fc and N297Q-Fc) on a laboratory scale in quantities sufficient to enable a wide variety of biosimilarity analyses. Initial characterization of the glycoforms demonstrate that they are well-defined glycoproteins which vary significantly only in their glycosylation state, or in the case of non-glycosylated N297Q-Fc, in the presence of a conservative N to Q amino acid mutation. The four IgG1 Fc glycoforms form a set of sequentially truncated glycoproteins which can be used to assess the effects of glycosylation on biological activity and protein physicochemical stability. Potential biological activity of the IgG1 Fc glycoforms was evaluated by using BLI to study the interaction of the glycoforms with the activating Fc receptor FcγRIIIa. Two BLI binding formats were developed, one utilizing protein G biosensors to immobilize IgG1 Fc, and one utilizing C-terminally biotinylated FcγRIIIa produced by a novel method to immobilize FcγRIIIa on streptavidin biosensors. The two assay formats resulted in complementary information about the affinity of the IgG1 Fc glycoforms for FcγRIIIa (Table 1) and can be applied to future studies of full-length antibodies, antibody-drug conjugates and antibodies with more complex glycosylation than the types studied here. From the perspective of developing a model system for biosimilarity analysis, the binding studies also identified members of the model system that exhibited highly similar biological activity and those with distinct differences. The HM-Fc and Man5-Fc glycoforms exhibited highly similar affinity for FcγRIIIa (approximate KD's of 27 and 32 nM respectively), while GlcNAc-Fc had much weaker affinity (KD of around 1000 nM), and N297Q-Fc displayed no affinity at the highest concentrations (20 µM) tested. These similarities and distinct differences in biological activity may be useful in identifying physical and chemical features that correlate with changes in biological activity and these and other aspects of this model system will be reported in the accompanying papers in this series.49–51 In addition, in future studies, we plan to implement additional in vitro enzymatic synthetic steps to produce more complex, well-defined IgG1 Fc glycoforms containing fucose and sialic acid into this model system for future biosimilarity analyses.
Supplementary Material
Acknowledgements
The authors wish to acknowledge primary support by grant NIPTE U01-KS-2014 and NIH grant NIGMS R01 GM090080. Funding for this work was also made possible, in part, by the Food and Drug Administration through grant 1U01FD005285-01, views expressed in this publication do not necessarily reflect the official policies of the Department of Health and Human Services nor does any mention of trade names, commercial practices, or organization imply endorsement by the United States Government. The authors also wish to acknowledge financial support of S.Z.O. via NIH biotechnology training grant 5-T32-GM008359. The authors thank the following laboratories for generously donating plasmids and enzyme expression strains: the DiMarchi laboratory at Indiana University for providing the sortase expression plasmid, the Gilbert laboratory at Newcastle University for providing the B.t. α-1,2-mannosidase expression plasmid, and the Lott laboratory at Massey University for providing a PNGase F overexpressing strain.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting Information
Supporting information including experimental procedures for synthesis of GGG-linker-Biotin (3), supplemental data tables, and supporting figures is available online from the Journal of Pharmaceutical Sciences website.
Uncategorized References
- 1.Alsenaidy MA, Okbazghi SZ, Kim JH, Joshi SB, Middaugh CR, Tolbert TJ, Volkin DB. Physical Stability Comparisons of IgG1-Fc Variants: Effects of N-Glycosylation Site Occupancy and Asp/Gln Residues at Site Asn 297. Journal of Pharmaceutical Sciences. 2014;103(6):1613–1627. doi: 10.1002/jps.23975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ha S, Ou Y, Vlasak J, Li Y, Wang S, Vo K, Du Y, Mach A, Fang Y, Zhang N. Isolation and characterization of IgG1 with asymmetrical Fc glycosylation. Glycobiology. 2011;21(8):1087–1096. doi: 10.1093/glycob/cwr047. [DOI] [PubMed] [Google Scholar]
- 3.Wang S, Ionescu R, Peekhaus N, Leung J-y, Ha S, Vlasak J. Separation of post-translational modifications in monoclonal antibodies by exploiting subtle conformational changes under mildly acidic conditions. Journal of Chromatography A. 2010;1217(42):6496–6502. doi: 10.1016/j.chroma.2010.08.044. [DOI] [PubMed] [Google Scholar]
- 4.Walsh CT, Garneau-Tsodikova S, Gatto GJ. Protein Posttranslational Modifications: The Chemistry of Proteome Diversifications. Angewandte Chemie International Edition. 2005;44(45):7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 5.Farley AR, Link AJ. Chapter 40 Identification and Quantification of Protein Posttranslational Modifications. In: Richard RB, Murray PD, editors. Methods in Enzymology. ed. Academic Press; 2009. pp. 725–763. [DOI] [PubMed] [Google Scholar]
- 6.Fischer S, Hoernschemeyer J, Mahler H-C. Glycation during storage and administration of monoclonal antibody formulations. European Journal of Pharmaceutics and Biopharmaceutics. 2008;70(1):42–50. doi: 10.1016/j.ejpb.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 7.Mozziconacci O, Kerwin BA, Schöneich C. Exposure of a Monoclonal Antibody, IgG1, to UVLight Leads to Protein Dithiohemiacetal and Thioether Cross-Links: A Role for Thiyl Radicals? Chemical Research in Toxicology. 2010;23(8):1310–1312. doi: 10.1021/tx100193b. [DOI] [PubMed] [Google Scholar]
- 8.Gaza-Bulseco G, Faldu S, Hurkmans K, Chumsae C, Liu H. Effect of methionine oxidation of a recombinant monoclonal antibody on the binding affinity to protein A and protein G. Journal of Chromatography B. 2008;870(1):55–62. doi: 10.1016/j.jchromb.2008.05.045. [DOI] [PubMed] [Google Scholar]
- 9.Cohen SL, Price C, Vlasak J. β-Elimination and Peptide Bond Hydrolysis: Two Distinct Mechanisms of Human IgG1 Hinge Fragmentation upon Storage. Journal of the American Chemical Society. 2007;129(22):6976–6977. doi: 10.1021/ja0705994. [DOI] [PubMed] [Google Scholar]
- 10.Xiang T, Lundell E, Sun Z, Liu H. Structural effect of a recombinant monoclonal antibody on hinge region peptide bond hydrolysis. Journal of Chromatography B. 2007;858(1–2):254–262. doi: 10.1016/j.jchromb.2007.08.043. [DOI] [PubMed] [Google Scholar]
- 11.Hambly DM, Banks DD, Scavezze JL, Siska CC, Gadgil HS. Detection and Quantitation of IgG 1 Hinge Aspartate Isomerization: A Rapid Degradation in Stressed Stability Studies. Analytical Chemistry. 2009;81(17):7454–7459. doi: 10.1021/ac901258g. [DOI] [PubMed] [Google Scholar]
- 12.Houde D, Peng Y, Berkowitz SA, Engen JR. Post-translational Modifications Differentially Affect IgG1 Conformation and Receptor Binding. Molecular & Cellular Proteomics. 2010;9(8):1716–1728. doi: 10.1074/mcp.M900540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berkowitz SA, Engen JR, Mazzeo JR, Jones GB. Analytical tools for characterizing biopharmaceuticals and the implications for biosimilars. Nat Rev Drug Discov. 2012;11(7):527–540. doi: 10.1038/nrd3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhlmann M, Covic A. The protein science of biosimilars. Nephrology Dialysis Transplantation. 2006;21(suppl 5):v4–v8. doi: 10.1093/ndt/gfl474. [DOI] [PubMed] [Google Scholar]
- 15.Putnam WS, Prabhu S, Zheng Y, Subramanyam M, Wang Y-MC. Pharmacokinetic, pharmacodynamic and immunogenicity comparability assessment strategies for monoclonal antibodies. Trends in Biotechnology. 2010;28(10):509–516. doi: 10.1016/j.tibtech.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Jones AJS, Papac DI, Chin EH, Keck R, Baughman SA, Lin YS, Kneer J, Battersby JE. Selective clearance of glycoforms of a complex glycoprotein pharmaceutical caused by terminal N-acetylglucosamine is similar in humans and cynomolgus monkeys. Glycobiology. 2007;17(5):529–540. doi: 10.1093/glycob/cwm017. [DOI] [PubMed] [Google Scholar]
- 17.Shinkawa T, Nakamura K, Yamane N, Shoji-Hosaka E, Kanda Y, Sakurada M, Uchida K, Anazawa H, Satoh M, Yamasaki M, Hanai N, Shitara K. The Absence of Fucose but Not the Presence of Galactose or Bisecting N-Acetylglucosamine of Human IgG1 Complex-type Oligosaccharides Shows the Critical Role of Enhancing Antibody-dependent Cellular Cytotoxicity. Journal of Biological Chemistry. 2003;278(5):3466–3473. doi: 10.1074/jbc.M210665200. [DOI] [PubMed] [Google Scholar]
- 18.Raju TS. Terminal sugars of Fc glycans influence antibody effector functions of IgGs. Current Opinion in Immunology. 2008;20(4):471–478. doi: 10.1016/j.coi.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Okazaki A, Shoji-Hosaka E, Nakamura K, Wakitani M, Uchida K, Kakita S, Tsumoto K, Kumagai I, Shitara K. Fucose Depletion from Human IgG1 Oligosaccharide Enhances Binding Enthalpy and Association Rate Between IgG1 and FcγRIIIa. Journal of Molecular Biology. 2004;336(5):1239–1249. doi: 10.1016/j.jmb.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Yamane-Ohnuki N, Kinoshita S, Inoue-Urakubo M, Kusunoki M, Iida S, Nakano R, Wakitani M, Niwa R, Sakurada M, Uchida K, Shitara K, Satoh M. Establishment of FUT8 knockout Chinese hamster ovary cells: An ideal host cell line for producing completely defucosylated antibodies with enhanced antibody-dependent cellular cytotoxicity. Biotechnology and Bioengineering. 2004;87(5):614–622. doi: 10.1002/bit.20151. [DOI] [PubMed] [Google Scholar]
- 21.Mori K, Kuni-Kamochi R, Yamane-Ohnuki N, Wakitani M, Yamano K, Imai H, Kanda Y, Niwa R, Iida S, Uchida K, Shitara K, Satoh M. Engineering Chinese hamster ovary cells to maximize effector function of produced antibodies using FUT8 siRNA. Biotechnology and Bioengineering. 2004;88(7):901–908. doi: 10.1002/bit.20326. [DOI] [PubMed] [Google Scholar]
- 22.Schuster M, Umana P, Ferrara C, Brunker P, Gerdes C, Waxenecker G, Wiederkum S, Schwager C, Loibner H, Himmler G, Mudde GC. Improved Effector Functions of a Therapeutic Monoclonal Lewis Y-Specific Antibody by Glycoform Engineering. Cancer Research. 2005;65(17):7934–7941. doi: 10.1158/0008-5472.CAN-04-4212. [DOI] [PubMed] [Google Scholar]
- 23.Niwa R, Natsume A, Uehara A, Wakitani M, Iida S, Uchida K, Satoh M, Shitara K. IgG subclass-independent improvement of antibody-dependent cellular cytotoxicity by fucose removal from Asn297-linked oligosaccharides. Journal of Immunological Methods. 2005;306(1–2):151–160. doi: 10.1016/j.jim.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Shields RL, Lai J, Keck R, O'Connell LY, Hong K, Meng YG, Weikert SHA, Presta LG. Lack of Fucose on Human IgG1 N-Linked Oligosaccharide Improves Binding to Human FcγRIII and Antibody-dependent Cellular Toxicity. Journal of Biological Chemistry. 2002;277(30):26733–26740. doi: 10.1074/jbc.M202069200. [DOI] [PubMed] [Google Scholar]
- 25.Raju TS. Impact of Fc Glycosylation on Monoclonal Antibody Effector Functions and Degradation by Proteases. In: Shire SJ, Gombotz W, Bechtold-Peters K, Andya J, editors. Current Trends in Monoclonal Antibody Development and Manufacturing, ed. ed. New York: Springer; 2010. pp. 249–269. [Google Scholar]
- 26.Hodoniczky J, Zheng YZ, James DC. Control of Recombinant Monoclonal Antibody Effector Functions by Fc N-Glycan Remodeling in Vitro. Biotechnology Progress. 2005;21(6):1644–1652. doi: 10.1021/bp050228w. [DOI] [PubMed] [Google Scholar]
- 27.Raju TS, Jordan RE. Galactosylation variations in marketed therapeutic antibodies. mAbs. 2012;4(3):385–391. doi: 10.4161/mabs.19868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki T, Ishii-Watabe A, Tada M, Kobayashi T, Kanayasu-Toyoda T, Kawanishi T, Yamaguchi T. Importance of Neonatal FcR in Regulating the Serum Half-Life of Therapeutic Proteins Containing the Fc Domain of Human IgG1: A Comparative Study of the Affinity of Monoclonal Antibodies and Fc-Fusion Proteins to Human Neonatal FcR. The Journal of Immunology. 2010;184(4):1968–1976. doi: 10.4049/jimmunol.0903296. [DOI] [PubMed] [Google Scholar]
- 29.Wang W, Vlasak J, Li Y, Pristatsky P, Fang Y, Pittman T, Roman J, Wang Y, Prueksaritanont T, Ionescu R. Impact of methionine oxidation in human IgG1 Fc on serum half-life of monoclonal antibodies. Molecular Immunology. 2011;48(6–7):860–866. doi: 10.1016/j.molimm.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 30.Wang W, Lu P, Fang Y, Hamuro L, Pittman T, Carr B, Hochman J, Prueksaritanont T. Monoclonal Antibodies with Identical Fc Sequences Can Bind to FcRn Differentially with Pharmacokinetic Consequences. Drug Metabolism and Disposition. 2011;39(9):1469–1477. doi: 10.1124/dmd.111.039453. [DOI] [PubMed] [Google Scholar]
- 31.Vaccaro C, Zhou J, Ober RJ, Ward ES. Engineering the Fc region of immunoglobulin G to modulate in vivo antibody levels. Nat Biotech. 2005;23(10):1283–1288. doi: 10.1038/nbt1143. [DOI] [PubMed] [Google Scholar]
- 32.Mimura Y, Church S, Ghirlando R, Ashton PR, Dong S, Goodall M, Lund J, Jefferis R. The influence of glycosylation on the thermal stability and effector function expression of human IgG1-Fc: properties of a series of truncated glycoforms. Molecular Immunology. 2000;37(12–13):697–706. doi: 10.1016/s0161-5890(00)00105-x. [DOI] [PubMed] [Google Scholar]
- 33.Kaneko Y, Nimmerjahn F, Ravetch JV. Anti-Inflammatory Activity of Immunoglobulin G Resulting from Fc Sialylation. Science. 2006;313(5787):670–673. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]
- 34.Anthony RM, Nimmerjahn F, Ashline DJ, Reinhold VN, Paulson JC, Ravetch JV. Recapitulation of IVIG Anti-Inflammatory Activity with a Recombinant IgG Fc. Science. 2008;320(5874):373–376. doi: 10.1126/science.1154315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okazaki A, Shoji E, Nakamura K, Wakitani M, Uchida K, Kakita S, Tsumoto K, Kumagai I, Shitara K. Thermodynamic and kinetic effects of human IgG1 defucosylation on IgG1-FcγRIIIa interaction. In: Iijima S, Nishijima K-I, editors. Animal Cell Technology: Basic & Applied Aspects. ed. Netherlands: Springer; 2006. pp. 47–53. [Google Scholar]
- 36.Mori K, Iida S, Yamane-Ohnuki N, Kanda Y, Kuni-Kamochi R, Nakano R, Imai-Nishiya H, Okazaki A, Shinkawa T, Natsume A, Niwa R, Shitara K, Satoh M. Non-fucosylated therapeutic antibodies: the next generation of therapeutic antibodies. Cytotechnology. 2007;55(2–3):109–114. doi: 10.1007/s10616-007-9103-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamane-Ohnuki N, Satoh M. Production of therapeutic antibodies with controlled fucosylation. mAbs. 2009;1(3):230–236. doi: 10.4161/mabs.1.3.8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Böhm S, Schwab I, Lux A, Nimmerjahn F. The role of sialic acid as a modulator of the anti-inflammatory activity of IgG. Semin Immunopathol. 2012;34(3):443–453. doi: 10.1007/s00281-012-0308-x. [DOI] [PubMed] [Google Scholar]
- 39.Mimura Y, Sondermann P, Ghirlando R, Lund J, Young SP, Goodall M, Jefferis R. Role of Oligosaccharide Residues of IgG1-Fc in FcγRIIb Binding. Journal of Biological Chemistry. 2001;276(49):45539–45547. doi: 10.1074/jbc.M107478200. [DOI] [PubMed] [Google Scholar]
- 40.Zheng K, Bantog C, Bayer R. The impact of glycosylation on monoclonal antibody conformation and stability. mAbs. 2011;3(6):568–576. doi: 10.4161/mabs.3.6.17922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghirlando R, Lund J, Goodall M, Jefferis R. Glycosylation of human IgG-Fc: influences on structure revealed by differential scanning micro-calorimetry. Immunology Letters. 1999;68(1):47–52. doi: 10.1016/s0165-2478(99)00029-2. [DOI] [PubMed] [Google Scholar]
- 42.Solá RJ, Griebenow K. Effects of glycosylation on the stability of protein pharmaceuticals. Journal of Pharmaceutical Sciences. 2009;98(4):1223–1245. doi: 10.1002/jps.21504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaguchi Y, Nishimura M, Nagano M, Yagi H, Sasakawa H, Uchida K, Shitara K, Kato K. Glycoform-dependent conformational alteration of the Fc region of human immunoglobulin G1 as revealed by NMR spectroscopy. Biochimica et Biophysica Acta (BBA) - General Subjects. 2006;1760(4):693–700. doi: 10.1016/j.bbagen.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 44.Liu D, Ren D, Huang H, Dankberg J, Rosenfeld R, Cocco MJ, Li L, Brems DN, Remmele RL. Structure and Stability Changes of Human IgG1 Fc as a Consequence of Methionine Oxidation. Biochemistry. 2008;47(18):5088–5100. doi: 10.1021/bi702238b. [DOI] [PubMed] [Google Scholar]
- 45.Alsenaidy MA, Kim JH, Majumdar R, Weis DD, Joshi SB, Tolbert TJ, Middaugh CR, Volkin DB. High-Throughput Biophysical Analysis and Data Visualization of Conformational Stability of an IgG1 Monoclonal Antibody After Deglycosylation. Journal of Pharmaceutical Sciences. 2013;102(11):3942–3956. doi: 10.1002/jps.23730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li CH, Narhi LO, Wen J, Dimitrova M, Wen Z-q, Li J, Pollastrini J, Nguyen X, Tsuruda T, Jiang Y. Effect of pH, Temperature, and Salt on the Stability of Escherichia coli- and Chinese Hamster Ovary Cell-Derived IgG1 Fc. Biochemistry. 2012;51(50):10056–10065. doi: 10.1021/bi300702e. [DOI] [PubMed] [Google Scholar]
- 47.Kayser V, Chennamsetty N, Voynov V, Forrer K, Helk B, Trout BL. Glycosylation influences on the aggregation propensity of therapeutic monoclonal antibodies. Biotechnology Journal. 2011;6(1):38–44. doi: 10.1002/biot.201000091. [DOI] [PubMed] [Google Scholar]
- 48.Krapp S, Mimura Y, Jefferis R, Huber R, Sondermann P. Structural Analysis of Human IgG-Fc Glycoforms Reveals a Correlation Between Glycosylation and Structural Integrity. Journal of Molecular Biology. 2003;325(5):979–989. doi: 10.1016/s0022-2836(02)01250-0. [DOI] [PubMed] [Google Scholar]
- 49.More AS, Toprani VM, Okbazghi SZ, Kim JH, Joshi SB, Middaugh CR, Tolbert TJ, Volkin DB. Correlating the impact of well-defined oligosaccharide structures on physical stability profiles of IgG1-Fc glycoforms. Journal of Pharmaceutical Sciences. doi: 10.1016/j.xphs.2015.10.014. accepted. [DOI] [PubMed] [Google Scholar]
- 50.Mozziconacci O, Okbazghi S, More AS, Volkin DB, Tolbert T, Schöneich C. Comparative evaluation of the chemical stability of four well-defined IgG1 Fc glycoforms. Journal of Pharmaceutical Sciences. doi: 10.1016/j.xphs.2015.10.024. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim JH, Joshi SB, Tolbert TJ, Middaugh CR, Volkin DB, Hall AS. Biosimilarity Assessments of Model IgG1-Fc Glycoforms Using a Machine Learning Approach. Journal of Pharmaceutical Sciences. doi: 10.1016/j.xphs.2015.10.013. accepted. [DOI] [PubMed] [Google Scholar]
- 52.Loo T, Patchett ML, Norris GE, Lott JS. Using Secretion to Solve a Solubility Problem: High-Yield Expression in Escherichia coli and Purification of the Bacterial Glycoamidase PNGase F. Protein Expression and Purification. 2002;24(1):90–98. doi: 10.1006/prep.2001.1555. [DOI] [PubMed] [Google Scholar]
- 53.Mao H, Hart SA, Schink A, Pollok BA. Sortase-Mediated Protein Ligation: A New Method for Protein Engineering. Journal of the American Chemical Society. 2004;126(9):2670–2671. doi: 10.1021/ja039915e. [DOI] [PubMed] [Google Scholar]
- 54.Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron EA, Pudlo NA, Porter NT, Urs K, Thompson AJ, Cartmell A, Rogowski A, Hamilton BS, Chen R, Tolbert TJ, Piens K, Bracke D, Vervecken W, Hakki Z, Speciale G, Munoz-Munoz JL, Day A, Pena MJ, McLean R, Suits MD, Boraston AB, Atherly T, Ziemer CJ, Williams SJ, Davies GJ, Abbott DW, Martens EC, Gilbert HJ. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature. 2015;517(7533):165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu Y, Suits MDL, Thompson AJ, Chavan S, Dinev Z, Dumon C, Smith N, Moremen KW, Xiang Y, Siriwardena A, Williams SJ, Gilbert HJ, Davies GJ. Mechanistic insights into a Ca2+-dependent family of α-mannosidases in a human gut symbiont. Nat Chem Biol. 2010;6(2):125–132. doi: 10.1038/nchembio.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao J, Tolbert TJ. Synthesis of Polymerizable Protein Monomers for Protein-Acrylamide Hydrogel Formation. Biomacromolecules. 2009;10(7):1939–1946. doi: 10.1021/bm900339q. [DOI] [PubMed] [Google Scholar]
- 57.Porath J. General methods and coupling procedures. Methods in Enzymology. 1974;34:13–30. [Google Scholar]
- 58.Xiao J, Chen R, Pawlicki MA, Tolbert TJ. Targeting a Homogeneously Glycosylated Antibody Fc To Bind Cancer Cells Using a Synthetic Receptor Ligand. Journal of the American Chemical Society. 2009;131(38):13616–13618. doi: 10.1021/ja9045179. [DOI] [PubMed] [Google Scholar]
- 59.Zhang W, Inan M, Meagher M. Fermentation strategies for recombinant protein expression in the methylotrophic yeastPichia pastoris. Biotechnol Bioprocess Eng. 2000;5(4):275–287. [Google Scholar]
- 60.Choi B-K, Bobrowicz P, Davidson RC, Hamilton SR, Kung DH, Li H, Miele RG, Nett JH, Wildt S, Gerngross TU. Use of combinatorial genetic libraries to humanize N-linked glycosylation in the yeast Pichia pastoris. Proceedings of the National Academy of Sciences. 2003;100(9):5022–5027. doi: 10.1073/pnas.0931263100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manikwar P, Majumdar R, Hickey JM, Thakkar SV, Samra HS, Sathish HA, Bishop SM, Middaugh CR, Weis DD, Volkin DB. Correlating excipient effects on conformational and storage stability of an IgG1 monoclonal antibody with local dynamics as measured by hydrogen/deuterium-exchange mass spectrometry. J Pharm Sci. 2013;102(7):2136–2151. doi: 10.1002/jps.23543. [DOI] [PubMed] [Google Scholar]
- 62.Alsenaidy MA, Okbazghi SZ, Kim JH, Joshi SB, Middaugh CR, Tolbert TJ, Volkin DB. Physical stability comparisons of IgG1-Fc variants: effects of N-glycosylation site occupancy and Asp/Gln residues at site Asn 297. J Pharm Sci. 2014;103(6):1613–1627. doi: 10.1002/jps.23975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cooper MA. Label-Free Biosensors: Techniques and Applications. first edition. Cambridge University Press; 2009. [Google Scholar]
- 64.Bruhns P, Iannascoli B, England P, Mancardi DA, Fernandez N, Jorieux S, Daeron M. Specificity and affinity of human Fcγ receptors and their polymorphic variants for human IgG subclasses. Blood. 2009;113(16):3716–3725. doi: 10.1182/blood-2008-09-179754. [DOI] [PubMed] [Google Scholar]
- 65.Choi B-K, Warburton S, Lin H, Patel R, Boldogh I, Meehl M, d’Anjou M, Pon L, Stadheim T, Sethuraman N. Improvement of N-glycan site occupancy of therapeutic glycoproteins produced in Pichia pastoris. Appl Microbiol Biotechnol. 2012;95(3):671–682. doi: 10.1007/s00253-012-4067-3. [DOI] [PubMed] [Google Scholar]
- 66.Liu H, Gaza-Bulseco G, Chumsae C, Newby-Kew A. Characterization of lower molecular weight artifact bands of recombinant monoclonal IgG1 antibodies on non-reducing SDS-PAGE. Biotechnology Letters. 2007;29(11):1611–1622. doi: 10.1007/s10529-007-9449-8. [DOI] [PubMed] [Google Scholar]
- 67.Franey H, Brych SR, Kolvenbach CG, Rajan RS. Increased aggregation propensity of IgG2 subclass over IgG1: Role of conformational changes and covalent character in isolated aggregates. Protein Science. 2010;19(9):1601–1615. doi: 10.1002/pro.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vlasak J, Ionescu R. Fragmentation of monoclonal antibodies. mAbs. 2011;3(3):253–263. doi: 10.4161/mabs.3.3.15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bond MD, Panek ME, Zhang Z, Wang D, Mehndiratta P, Zhao H, Gunton K, Ni A, Nedved ML, Burman S, Volkin DB. Evaluation of a dual-wavelength size exclusion HPLC method with improved sensitivity to detect protein aggregates and its use to better characterize degradation pathways of an IgG1 monoclonal antibody. Journal of Pharmaceutical Sciences. 2010;99(6):2582–2597. doi: 10.1002/jps.22034. [DOI] [PubMed] [Google Scholar]
- 70.Tsukiji S, Nagamune T. Sortase-Mediated Ligation: A Gift from Gram-Positive Bacteria to Protein Engineering. Chem Bio Chem. 2009;10(5):787–798. doi: 10.1002/cbic.200800724. [DOI] [PubMed] [Google Scholar]
- 71.Pritz S, Wolf Y, Kraetke O, Klose J, Bienert M, Beyermann M. Synthesis of Biologically Active Peptide Nucleic Acid–Pepdde Conjugates by Sortase-Mediated Ligation. The Journal of Organic Chemistry. 2007;72(10):3909–3912. doi: 10.1021/jo062331l. [DOI] [PubMed] [Google Scholar]
- 72.Proft T. Sortase-mediated protein ligation: an emerging biotechnology tool for protein modification and immobilisation. Biotechnology Letters. 2010;32(1):1–10. doi: 10.1007/s10529-009-0116-0. [DOI] [PubMed] [Google Scholar]
- 73.Zhang Y, Boesen CC, Radaev S, Brooks AG, Fridman W-H, Sautes-Fridman C, Sun PD. Crystal Structure of the Extracellular Domain of a Human FcγRIII. Immunity. 2000;13(3):387–395. doi: 10.1016/s1074-7613(00)00038-8. [DOI] [PubMed] [Google Scholar]
- 74.Kanda Y, Yamada T, Mori K, Okazaki A, Inoue M, Kitajima-Miyama K, Kuni-Kamochi R, Nakano R, Yano K, Kakita S, Shitara K, Satoh M. Comparison of biological activity among nonfucosylated therapeutic IgG1 antibodies with three different N-linked Fc oligosaccharides: the high-mannose, hybrid, and complex types. Glycobiology. 2007;17(1):104–118. doi: 10.1093/glycob/cwl057. [DOI] [PubMed] [Google Scholar]
- 75.Subedi Ganesh P, Hanson Quinlin M, Barb Adam W. Restricted Motion of the Conserved Immunoglobulin G1 N-Glycan Is Essential for Efficient FcγRIIIa Binding. Structure. 2014;22(10):1478–1488. doi: 10.1016/j.str.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Magnelli PE, Bielik AM, Guthrie EP. Identification and Characterization of Protein Glycosylation using Specific Endo- and Exoglycosidases. Journal of Visualized Experiments : JoVE. 2011;(58):3749. doi: 10.3791/3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hamilton BS, Brede Y, Tolbert TJ. Expression and characterization of human glycosylated interleukin-1 receptor antagonist in Pichia pastoris. Protein expression and purification. 2008;59(1):64–68. doi: 10.1016/j.pep.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 78.Presta LG, Shields RL, Namenuk AK, Hong K, Meng YG. Engineering therapeutic antibodies for improved function. Biochemical Society transactions. 2002;30(4):487–490. doi: 10.1042/bst0300487. [DOI] [PubMed] [Google Scholar]
- 79.Seidel UJE, Schlegel P, Lang P. Natural killer (NK) cell mediated antibody-dependent cellular cytotoxicity (ADCC) in tumour immunotherapy with therapeutic antibodies. Frontiers in Immunology. 2013;4 doi: 10.3389/fimmu.2013.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Abes R, Teillaud J-L. Impact of Glycosylation on Effector Functions of Therapeutic IgG. Pharmaceuticals. 2010;3(1):146–157. doi: 10.3390/ph3010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kuramochi T, Igawa T, Tsunoda H, Hattori K. Humanization and Simultaneous Optimization of Monoclonal Antibody. In: Steinitz M, editor. Human Monoclonal Antibodies. ed. Humana Press; 2014. pp. 123–137. [DOI] [PubMed] [Google Scholar]
- 82.Champion T, Beck A. Capture of the Human IgG1 Antibodies by Protein A for the Kinetic Study of h-IgG/Fc?R Interaction Using SPR-Based Biosensor Technology #. T Glycosylation Engineering of Biopharmaceuticals. (ed.) 2013:331–343. doi: 10.1007/978-1-62703-327-5_21. [DOI] [PubMed] [Google Scholar]
- 83.Holmberg A, Blomstergren A, Nord O, Lukacs M, Lundeberg J, Uhlén M. The biotin-streptavidin interaction can be reversibly broken using water at elevated temperatures. ELECTROPHORESIS. 2005;26(3):501–510. doi: 10.1002/elps.200410070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.