Summary
Mouse embryonic stem cells (ESCs) are maintained in a naive ground state of pluripotency in the presence of MEK and GSK3 inhibitors. Here, we show that ground-state ESCs express low Myc levels. Deletion of both c-myc and N-myc (dKO) or pharmacological inhibition of Myc activity strongly decreases transcription, splicing, and protein synthesis, leading to proliferation arrest. This process is reversible and occurs without affecting pluripotency, suggesting that Myc-depleted stem cells enter a state of dormancy similar to embryonic diapause. Indeed, c-Myc is depleted in diapaused blastocysts, and the differential expression signatures of dKO ESCs and diapaused epiblasts are remarkably similar. Following Myc inhibition, pre-implantation blastocysts enter biosynthetic dormancy but can progress through their normal developmental program after transfer into pseudo-pregnant recipients. Our study shows that Myc controls the biosynthetic machinery of stem cells without affecting their potency, thus regulating their entry and exit from the dormant state.
Graphical Abstract
Highlights
-
•
Myc-depleted ESCs exhibit reversible biosynthetic dormancy and proliferation arrest
-
•
Myc expression is strongly reduced in hormonally induced diapause embryos
-
•
Myc mutant ESCs and diapause embryos have similar expression signatures
-
•
Myc depletion induces a reversible, pluripotent diapause-like state in blastocysts
Myc inhibition in mouse blastocysts induces reversible biosynthetic dormancy mimicking hormonally controlled diapause without affecting the pluripotency capacity, suggesting the importance of Myc regulation in controlling entry and exit from stem cell dormancy during development.
Introduction
The Myc family of transcription factors comprises c-Myc, N-Myc, and L-Myc and has been implicated in the generation of a variety of human tumors. Enhanced Myc expression has been shown to contribute to several aspects of tumorigenesis (Adhikary and Eilers, 2005) including unrestricted proliferation (Eilers et al., 1991), inhibition of differentiation (Freytag and Geddes, 1992), cell growth and metabolism (Dang, 2013, Iritani and Eisenman, 1999, Johnston et al., 1999), reduction of cell adhesion (Arnold and Watt, 2001), and metastasis (Pelengaris et al., 2002). The role of Myc proteins in development has been widely investigated making use of gene targeting in mice: whereas L-myc knockout mice develop normally (Hatton et al., 1996), embryos lacking c-myc die before E10.5 due to hematopoietic and placental defects (Dubois et al., 2008, Trumpp et al., 2001), and N-myc-deficient embryos die before E11.5 displaying neuroectodermal and heart defects (Charron et al., 1992). Myc activity is essential for efficient cellular reprogramming (Wernig et al., 2008) and has complex roles in various stem and progenitor cell types (Laurenti et al., 2009). In the adult hematopoietic system, c-Myc controls the balance between hematopoietic stem cell (HSC) self-renewal and differentiation by regulating the interaction between HSCs and their niche (Wilson et al., 2004). Interestingly, only the highly quiescent, dormant HSCs survive the simultaneous deletion of both c-myc and N-myc in the hematopoietic system, while all the other hematopoietic cells are rapidly lost due to impaired proliferation, differentiation, and overt apoptosis (Laurenti et al., 2008, Laurenti et al., 2009). In ESCs grown in serum plus LIF (hereafter referred to as serum), Myc proteins have been suggested to sustain pluripotency by repressing the primitive endoderm master regulator Gata6 and to contribute to cell-cycle control by regulating the mir-17-92 miRNA cluster (Smith et al., 2010, Varlakhanova et al., 2010). However, ESCs cultured in serum exhibit heterogeneous expression of pluripotency markers, and only a fraction of these cells correlate with the pre-implantation epiblast, as shown by transcriptional profiling (Boroviak et al., 2014, Marks et al., 2012, Ying et al., 2008). In contrast, ESCs cultured in 2i plus LIF (hereafter referred to as 2i) are captured in a naive ground state of pluripotency and retain the essential features of the pluripotent epiblast cells (Boroviak et al., 2014). The precise function of Myc in naive ground-state ESCs and the role of Myc in the mouse epiblast remain elusive. Here, we use a combination of genetic, transcriptomic, and cellular analyses to show that Myc activity reversibly controls the biosynthetic and proliferative machineries of ground-state naive ESCs without affecting pluripotency and link these data on ESCs to the physiological status of dormant, diapaused embryos.
Results
Myc Is Essential for Proliferation but Not for Maintenance of the Core Pluripotency Network in Ground-State ESCs
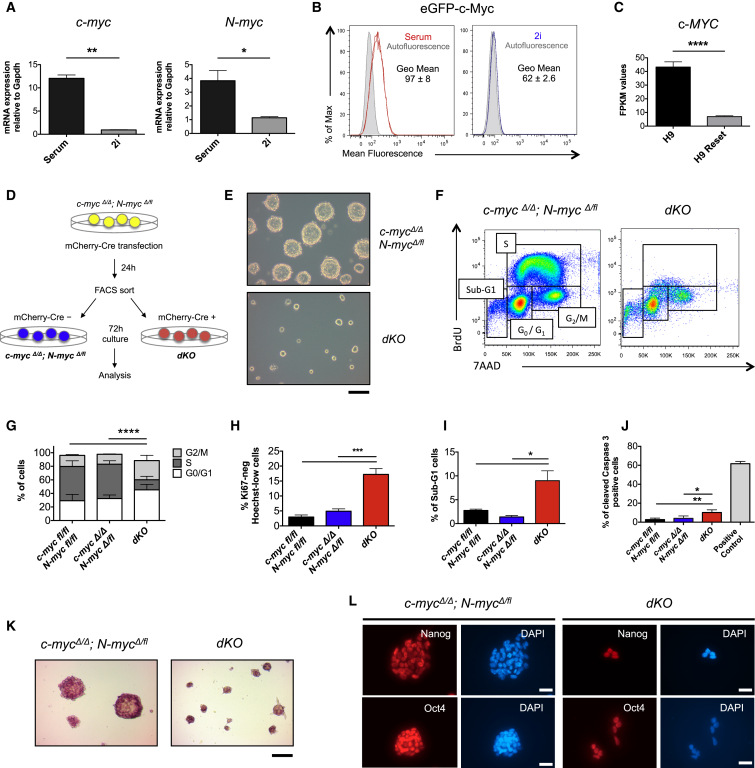
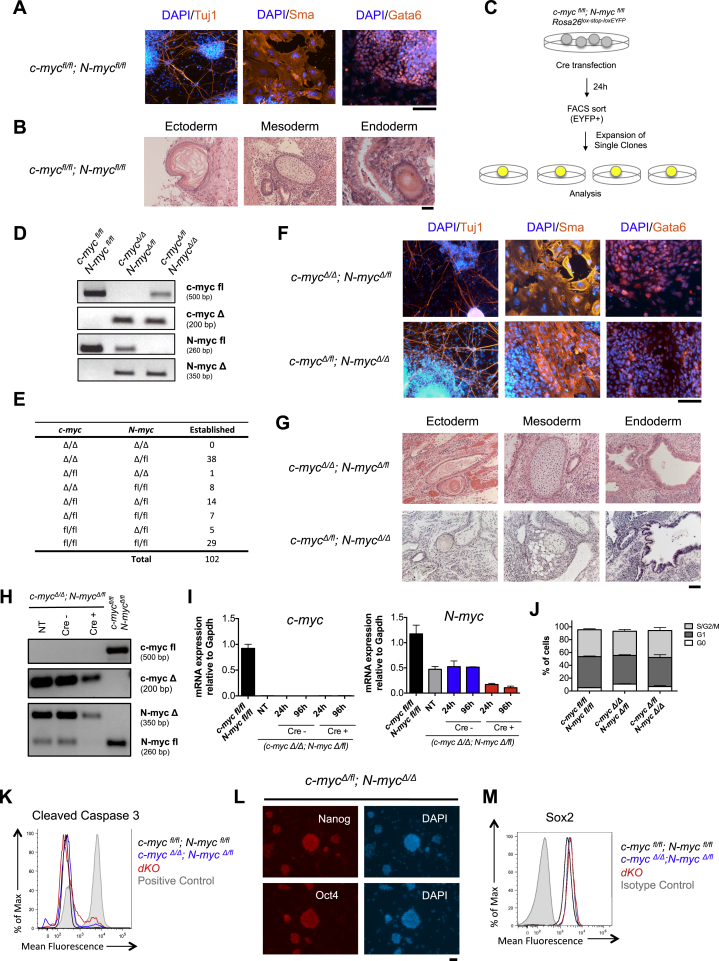
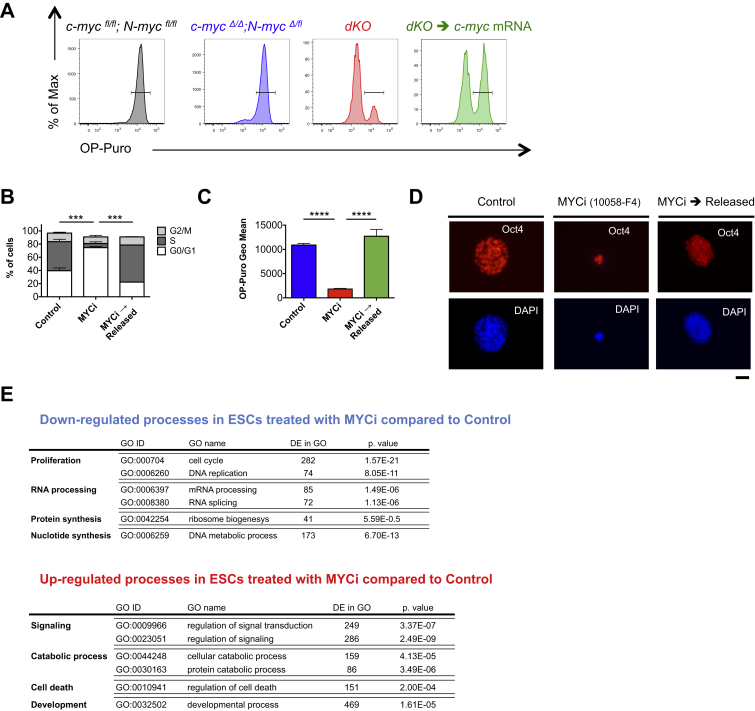
Mouse ESCs cultured in serum express significantly higher levels of c-myc and N-myc transcripts compared to ESCs grown in 2i (Figure 1A) (Marks et al., 2012). In agreement with this observation, a lower expression of c-Myc protein in 2i compared to serum was observed by flow cytometry using a c-MyceGFP knockin allele (Figure 1B) (Huang et al., 2008). Lower expression of c-MYC transcripts was also observed in human H9 ESCs that were reset to a naive state of pluripotency compared to their primed counterparts (Figure 1C) (Takashima et al., 2014). To genetically explore the function of Myc in naive ground-state ESCs, we derived ESC lines from mice homozygous for the c-myc and N-myc floxed alleles (c-mycfl/fl; N-mycfl/fl) (Knoepfler et al., 2002, Trumpp et al., 2001) as well as the Cre reporter gene Rosa26lox-stop-loxEYFP (Srinivas et al., 2001). All generated ESC lines had the capacity to differentiate into tissues derived from all three germ layers both in vitro and in vivo (Figures S1A and S1B). To induce the generation of c-myc and N-myc-null alleles (c-mycΔ and N-mycΔ, respectively), we transiently transfected c-mycfl/fl; N-mycfl/fl; Rosa26lox-stop-loxEYFP ESCs with a plasmid encoding a Cre recombinase and fluorescence-activated cell-sorted (FACS) EYFP+ cells followed by plating in 2i medium and expansion of single clones (Figure S1C). Although we could establish 102 clonal cell lines, no c-myc/N-myc double knockout (dKO) ESC clones could be derived, as all lines retained at least one allele of either c-mycfl or N-mycfl (Figures S1D and S1E). Single KO ESCs did not show any detectable phenotype as they formed dome-shaped colonies and were capable of multilineage differentiation both in vitro and in vivo (Figures S1F and S1G). These data show that neither c-myc nor N-myc alone are required for ESC maintenance in 2i and suggest functional redundancy between the two genes. To additionally eliminate the fourth “fl” allele, c-mycΔ/Δ; N-mycΔ/fl ESCs were transfected with a plasmid coding for an EF1α-driven mCherry-Cre fusion protein and mCherry positive (Cre+) and negative (Cre–) cells were FACS sorted and cultured in 2i (Figure 1D). We confirmed dKO ESCs in the Cre+ population 24 and 96 hr after transfection using both PCR and qRT-PCR analysis (Figures S1H and S1I). FACS analysis of the cell cycle showed that single deletion of either c-myc or N-myc did not affect the proliferative status of naive ground-state ESCs (Figure S1J). In contrast, 96 hr after Cre induction dKO ESCs ceased to proliferate and formed very small, dome-shaped colonies (Figure 1E). This was associated with the complete absence of DNA synthesis (Figures 1F and 1G), an increase in G0 arrested cells (Figure 1H), and a fraction of dKO ESCs undergoing apoptosis (Figures 1I, 1J, and S1K). These data demonstrate that, even in 2i conditions, low levels of c-Myc or N-Myc are required to prevent cell-cycle arrest of ground-state ESCs. Next, we addressed whether Myc-mediated cell-cycle arrest influences pluripotency and lineage differentiation. In 2i, both single KO and dKO ESCs expressed alkaline phosphatase (Figure 1K) and unexpectedly also displayed similar levels of Nanog, Oct4, and Sox2 (Figures 1L and S1L–S1M). These data provide evidence that c/N-Myc activity is specifically required for proliferation but dispensable for the expression of the core pluripotency factors in naive ground-state ESCs.
Figure 1.
Myc Is Required for Proliferation but Not for Maintenance of the Core Pluripotency Network in 2i-Cultured ESCs
(A and B) Ground-state ESCs express low levels of Myc.
(A) Mouse ESCs were cultured for more than eight passages in 2i or serum and c-myc and N-myc expression were quantified by qRT-PCR analysis and normalized to Gapdh. Values are shown as mean ± SEM.; n = 3.
(B) Flow cytometry analysis of eGFP-c-Myc expression in c-MyceGFP/eGFP reporter ESCs cultured for 72 hr in serum (red) or in 2i (blue). The gray histogram represents the autofluorescence signal from ESCs not expressing the eGFP-c-Myc reporter protein. Experiments were performed in triplicates, and values represent geometrical mean (Geo Mean) ± SD.
(C) c-MYC expression levels in H9 human ESCs and H9-reset cells. Bar graphs show FPKM (fragments per kilobase of exon per million reads mapped) values of three replicates and are represented as mean ± SD. Data are from Takashima et al. (2014).
(D–L) dKO ESCs undergo cell-cycle arrest but maintain the expression of the core pluripotency factors.
(D) Experimental workflow. c-mycΔ/Δ; N-mycΔ/fl ESCs were transfected with an EF1α-mCherry-Cre plasmid and sorted 24 hr later as mCherry negative (c-mycΔ/Δ; N-mycΔ/fl) and positive (dKO) cells. Sorted cells were plated in 2i medium and analyzed 72 hr after sort.
(E) Phase-contrast microscopy images showing the typical morphology of dKO ESCs. Scale bar, 100 μm.
(F) Representative flow cytometry plots of the cell-cycle profiles of c-mycΔ/Δ; N-mycΔ/fl and dKO ESCs. Incorporated BrdU and total DNA content (7AAD) are used to distinguish apoptotic (sub-G1) cells and cells in the G0/G1, S or G2/M phases.
(G) Quantitative analysis of the different cell-cycle phases as gated in (F). The bar charts indicate the mean ± SD; n = 4.
(H) Percentage of cells in the G0 phase (Hoechst low and intracellular Ki67neg) of the cell cycle as assessed by flow cytometry. Bar graphs indicate the mean ± SEM; n = 6.
(I) Quantitative analysis of apoptotic cells (sub-G1) as gated in (F). Values are shown as mean ± SEM; n = 4.
(J) Percentage of ESCs expressing cleaved Caspase 3, as assessed by flow cytometry. ESCs incubated for 10 min at 42°C were used as positive control. Values represent mean ± SD; n = 3.
(K) Alkaline phosphatase activity of c-mycΔ/Δ; N-mycΔ/fl and dKO cells cultured in 2i. Scale bar, 20 μm.
(L) Representative fluorescence staining showing Oct4 and Nanog expression in ESCs cultured in 2i. Scale bar, 50 μm.
Figure S1.
Myc Is Required for Proliferation but Not for Maintenance of the Core Pluripotency Network in 2i ESCs, Related to Figure 1
(A and B) c-mycfl/fl; N-mycfl/fl ESC lines are capable of multilineage differentiation.
(A) Fluorescence staining of in vitro differentiated ESCs for Tuj1, Sma and Gata6. Scale bar, 100 μm.
(B) Haematoxylin and eosin staining of teratomas derived from c-mycfl/fl; N-mycfl/fl ESCs. Representative images reveal structures derived from all the three germ layers. Scale bars, 50 μm (ectoderm and mesoderm) and 20 μm (endoderm).
(C) Experimental workflow. c-mycfl/fl; N-mycfl/flRosa26lox-stop-loxEYFP ESC were transduced with an EF1α-Cre plasmid and Cre positive (EYFP+) cells were FACS sorted and cultured in 2i, propagated as single clones and genotyped.
(D) PCR analysis of representative ESC clones for the c-mycflox, c-mycΔ, N-mycflox and N-mycΔ alleles.
(E) Number of clones obtained for each genotype.
(F and G) Lack of either c-myc or N-myc does not affect ESCs multilineage differentiation potential.
(F) Fluorescence staining for Tuj1, Sma and Gata6 of c-mycΔ/Δ; N-mycΔ//fl and c-mycΔ/fl; N-mycΔlΔ ESCs differentiated in vitro. Scale bar, 100 μm.
(G) Representative images of hematoxylin and eosin staining of teratomas from mycΔ/Δ; N-mycΔ//fl and c-mycΔ/fl; N-mycΔlΔ ESCs. Scale bar, 50 μm.
(H and I) c-mycΔ/Δ; N-mycΔ//fl ESCs were transfected with an EF1α-mCherry-Cre plasmid and mCherry positive (Cre+) and negative (Cre-) cells were FACS sorted and cultured in 2i. Not transfected (NT) cells were used as controls.
(H) PCR of the c-mycflox, c-mycΔ, N-mycflox and N-mycΔ alleles was performed 24h after transfection.
(I) qRT-PCR analysis for c-myc and N-myc was performed 24h and 96h after transfection. Transcript levels of c-myc and N-myc were normalized to Gapdh. Data are presented as the mean ± SEM of duplicate wells from a representative experiment.
(J) Lack of either c-myc or N-myc does not affect the proliferation of ESCs cultured in 2i. Cell cycle analysis of ESCs in the G0, G1 and S/G2/M phases as determined by Hoechst versus intracellular Ki67 staining. Bar graphs indicate the mean ± SD of two replicates, representative of two independent experiments.
(K) Representative flow cytometry plots of ESCs stained for cleaved Caspase 3. ESCs incubated for 10 min at 42°C were used as positive control.
(L) Representative images of c-mycΔ/fl; N-mycΔlΔ ESCs stained for Nanog or Oct4. Scale bar, 50 μm.
(M) FACS analysis of Sox2 expression in c-mycfl/fl; N-mycfllfl, c-mycΔ/Δ; N-mycΔ//fl and dKO ESCs.
Loss of Myc in Ground-State ESCs Leads to Biosynthetic Quiescence
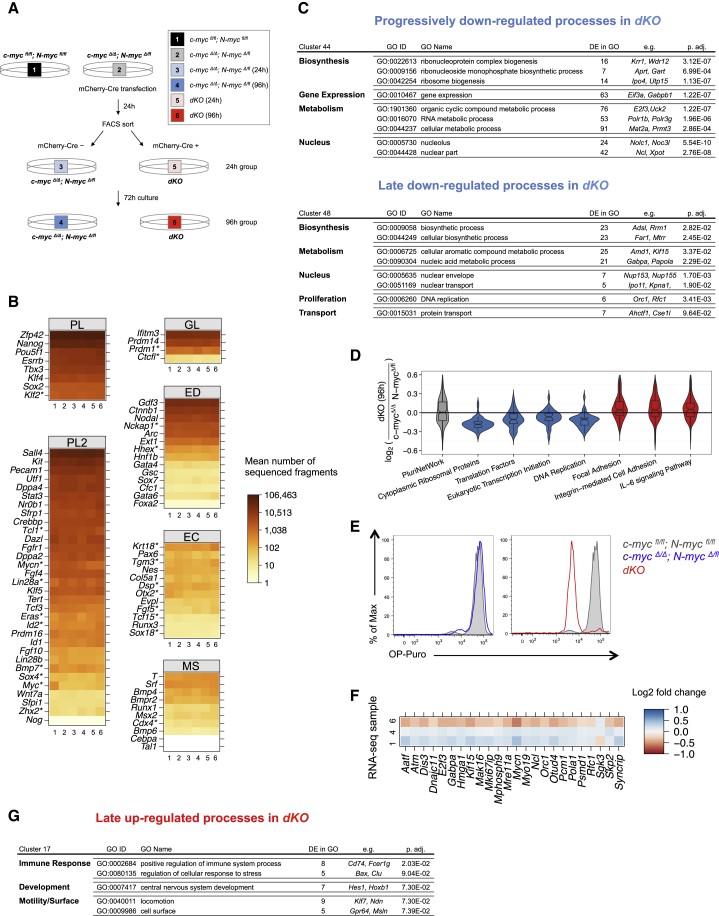
To further explore the consequences of c/N-myc deletion, we sequenced the transcriptome of ESCs 24 and 96 hr after Cre induction (Figures 2A and S2A; Table S1; Data S1). As expected, c-myc and N-myc downregulation was confirmed at both time points in the transcriptome data (Figure S2B). In line with the immunohistochemistry observations (Figure 1L), the transcript levels of the core and extended pluripotency networks (e.g., Zfp42, Nanog, Pou5f1 [Oct4], Esrrb, Tbx3) remained unaltered in dKO ground-state ESCs (Figures 2B and S2C) (Marks et al., 2012). Lineage-specific differentiation genes (germline, ectoderm, endoderm, and mesoderm) were mostly not upregulated (Figure 2B), demonstrating that arrested dKO ground-state ESCs maintain expression of the pluripotency network without upregulation of differentiation signatures. To identify genes and processes regulated upon c/N-myc deletion, we classified each gene based on its relative expression pattern in the different conditions with respect to its mean expression level across conditions (Figure S2D). Consistent with a function of Myc as both a transcriptional activator and repressor (Adhikary and Eilers, 2005, Walz et al., 2014), we found 215 genes significantly downregulated (clusters 44 + 48 + 60) and 165 genes upregulated (clusters 5 + 17 + 21) in dKO ESCs (false discovery rate [FDR] = 0.1; Table S2). Genes downregulated upon loss of Myc were enriched in Gene Ontology (GO) categories related to several metabolic and biosynthetic aspects of cellular physiology (clusters 44 and 48) including biosynthesis (ribosome biogenesis; e.g., Ipo4, Utp15), gene expression (e.g., Eif3a, Gabp1), metabolism (RNA metabolic process; e.g., Polr1b, Polr3g), nucleus (nuclear part; e.g., Ncl, Xpot), and proliferation (DNA replication; e.g., Orc1, Rfc1) (Figure 2C). Translation factors, transcription initiation, and DNA replication were also downregulated in dKO compared to c-mycΔ/Δ; N-mycΔ/fl ESCs (Figure 2D). To functionally assess the reduction in protein synthesis caused by loss of Myc activity, we made use of an alkyne analog of puromycin, O-propargyl-puromycin (OP-Puro) (Liu et al., 2012, Signer et al., 2014), that can diffuse through the plasma membrane of living cells, enter the ribosome acceptor sites and be incorporated into the C terminus of nascent polypeptide chains. OP-Puro can be fluorescently labeled with an azide-alkyne reaction, thereby allowing quantification of de novo protein synthesis. By using this assay, we show that OP-Puro incorporation is homogeneously decreased in dKO ESCs 96 hr after Cre induction, confirming the dramatic reduction of protein translation observed in the molecular signature (Figures 2E and S2E). Consistent with our cell-cycle analysis (Figures 1F and 1H), we found 24 genes related to proliferation to be differentially expressed in dKO ESCs (FDR = 0.1; Figure 2F). These include Pola1, the regulatory subunit of DNA polymerase, E2f3, known to be critical for normal cellular proliferation (Humbert et al., 2000) and Skp2, among others. Finally, processes related to immune response (positive regulation of immune system process; e.g., Cd74, Fcer1g), development (central nervous system development; e.g., Hes1, Hoxb1) and motility (locomotion; e.g., Klf7, Ndn) were over-represented among the genes upregulated in the dKO ESCs (cluster 17; Figure 2G). In agreement with previous studies in both HSCs and skin epidermis (Gebhardt et al., 2006, Wilson et al., 2004), integrin-mediated signaling processes were upregulated in dKO ESCs (Figure 2D). Pathways associated with survival and maintenance of ESCs such as IL-6 signaling (Nichols et al., 1994) were enriched in Myc-deficient ESCs (Figure 2D). Together, these data indicate that dKO ESCs display an increase in cell adhesion and processes associated with maintenance and survival and enter a state of biosynthetic quiescence, characterized by a strong reduction of protein and nucleic acid synthesis.
Figure 2.
Loss of Myc Activity Induces Cellular Dormancy in ESCs Cultured in 2i
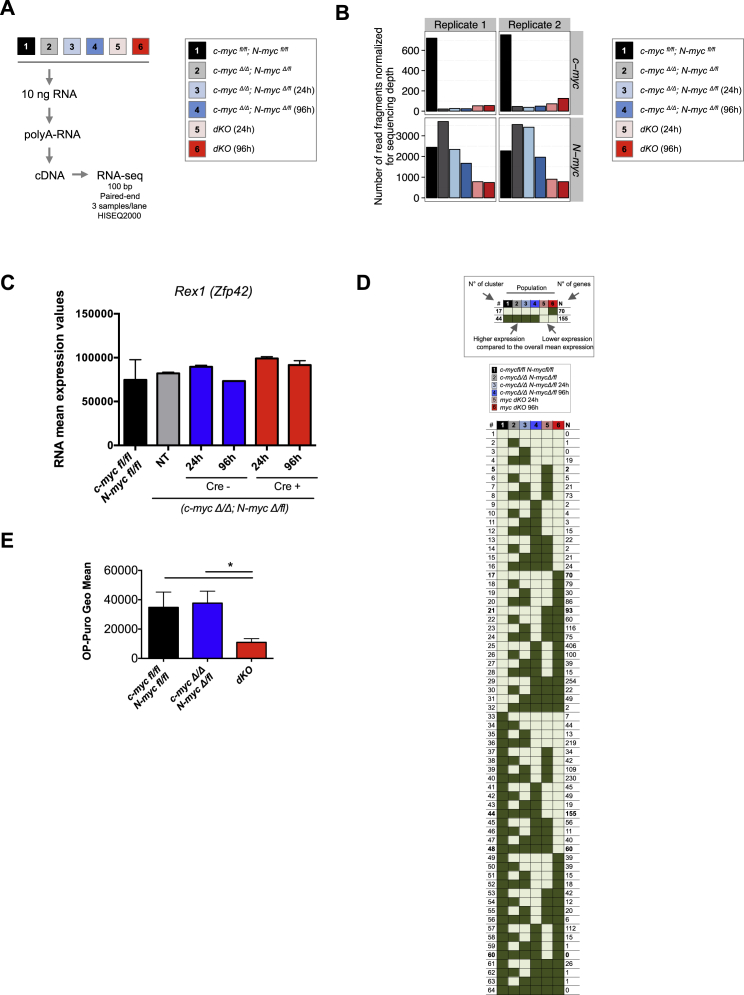
(A) Workflow of the RNA-seq analysis. Untransfected c-mycfl/fl; N-mycfl/fl (sample 1) and c-mycΔ/Δ; N-mycΔ/fl (sample 2) were used as controls. c-mycΔ/Δ; N-mycΔ/fl cells were transfected with an EF1α-mCherry-Cre plasmid and sorted 24 hr later as Cre negative (c-mycΔ/Δ; N-mycΔ/fl) and positive (dKO) cells. The samples were analyzed immediately after sort (24 hr after Cre transfection, samples 3 and 5) or plated in 2i and analyzed 72 hr after sort (96 hr after Cre transfection, samples 4 and 6). For each condition, two biological replicates in independent experiments were sequenced and analyzed.
(B) Heatmap representation of the mean number of sequenced fragments for the genes belonging to the different pluripotency and commitment gene signatures as defined by Marks et al. (2012). PL, pluripotency; PL2, extended pluripotency; GL, germline; ED, endoderm; EC, ectoderm; MS, mesoderm. A gene differentially expressed in at least one sample compared to another condition is indicated by an asterisk (FDR = 0.1).
(C) Overrepresented Gene Ontology (GO) categories in the genes progressively (at 24 and 96 hr; cluster 44) and lately (at 96 hr; cluster 48) downregulated in dKO ESCs.
(D) Violin representation of the distribution of log fold changes (base 2) of c-mycΔ/Δ; N-mycΔ/fl ESCs with respect to the dKO ESCs stratified by the biological pathways of the genes (as annotated by WikiPathways). Significantly downregulated pathways in dKO ESCs are depicted in blue, whereas upregulated processes are represented in red (FDR = 0.1).
(E) Translation activity of ESCs cultured in 2i. Representative FACS plots showing reduced OP-Puro incorporation in dKO ESCs 96 hr after Cre induction (red) compared to c-mycfl/fl; N-mycfl/fl (gray) and c-mycΔ/Δ; N-mycΔ/fl (blue) ESCs.
(F) Heatmap representation of the relative fold changes (base 2), with respect to the mean expression across conditions of the differentially expressed genes associated to the cell cycle.
(G) Over-represented GO categories in the genes upregulated in dKO ESCs (at 96 hr; cluster 17).
Figure S2.
Loss of Myc Activity Induces Cellular Dormancy in ESCs Cultured in 2i, Related to Figure 2
(A) RNA-seq workflow. Total RNA starting material was 10 ng. 100 bp paired–end libraries of two biological replicates for each condition were sequenced. We obtained 1x108 sequenced fragments per sample and detected the expression of more than 28,000 genes.
(B) Number of sequenced fragments of the genes c-myc and N-myc on each sample.
(C) RNA-seq expression levels of Rex1 (Zpf42) in dKO ESCs compared to the controls. Bar graphs are represented as mean ± SD.
(D) Gene expression clusters. For each of the genes detected to be differentially expressed, we estimated the relative fold change between each condition with respect to the mean expression across all the six conditions. The genes were grouped based on the signs of the relative fold change on each of the 64 (26) possible combinations. For instance, all the genes that showed a positive sign for the c-mycfl/fl; N-mycfl/fl and a negative sign for the rest of the conditions were grouped together.
(E) Quantification of OP-Puro incorporation in c-mycfl/fl; N-mycfl/fl, c-mycΔ/Δ; N-mycΔlfl and dKO ESCs. Values represent geometrical mean (Geo Mean) ± SEM.
The Dormant State Induced by Myc Depletion Is Reversible
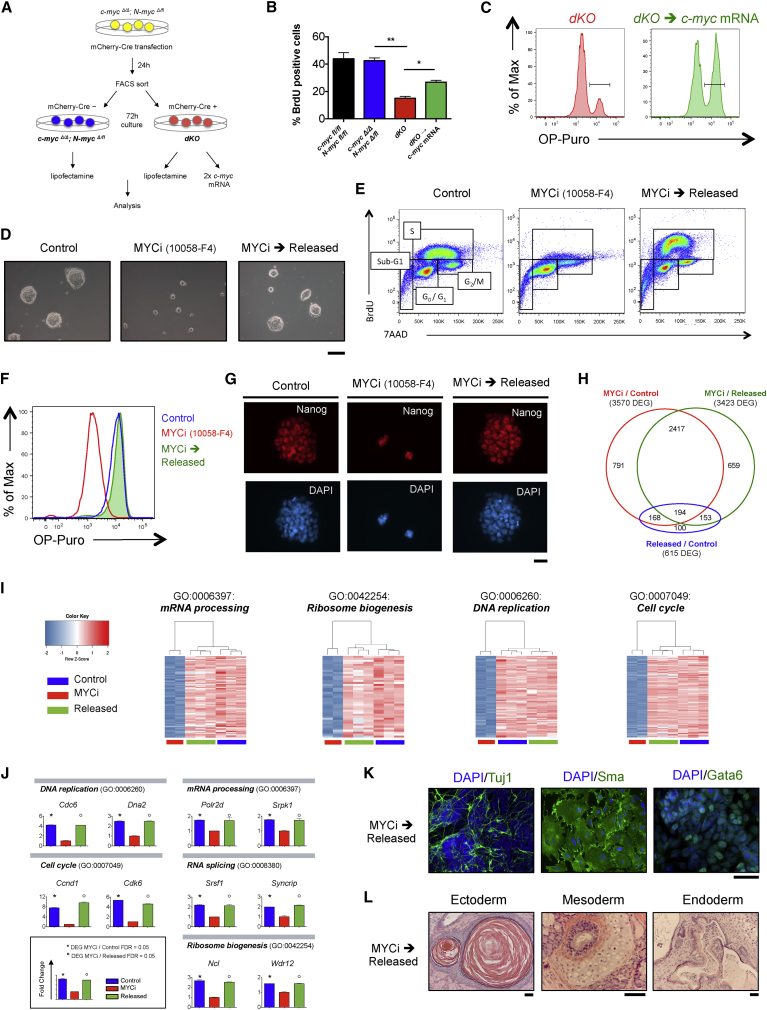
Maintenance of pluripotency in the absence of differentiation raised the question whether the proliferative and biosynthetic quiescence observed in the dKO ESCs represents a terminal or a reversible status. To approach this, we transfected dKO cells with synthetic c-myc mRNAs to transiently restore Myc activity (Figure 3A) (Warren et al., 2010). Upon expression of exogenous c-myc mRNAs, dKO ESCs re-entered the cell cycle, as shown by a significant increase in 5-bromo-2′- deoxyuridine (BrdU) incorporation (Figure 3B) and restoration of translational activity (Figures 3C and S3A). These results indicate that upon re-expression of c-myc by exogenous mRNAs, dKO ESCs transiently exit the status of biosynthetic dormancy and restore protein biosynthesis and DNA replication. The dimerization of Myc with Max, a helix-loop-helix leucine zipper protein, is necessary for the binding of Myc to the promoters of its target genes (Blackwood and Eisenman, 1991, Eilers and Eisenman, 2008). Low-molecular-weight compounds, such as 10058-F4, have been shown to inhibit both c-Myc and N-Myc activity by selective targeting of Myc-Max dimerization both in vitro and in vivo (Rahl et al., 2010, Zirath et al., 2013). In order to evaluate the long-term reversibility of the phenotype induced by Myc depletion, we treated naive ground-state ESCs with the Myc inhibitor 10058-F4 (MYCi) for 60 hr and analyzed the effect on cell proliferation, protein translation, and pluripotency. Entirely consistent with our previous results on dKO cells, mouse ESCs cultured in the presence of MYCi formed small, undifferentiated colonies (Figure 3D), which showed drastically reduced DNA and protein synthesis (Figures 3E, 3F, S3B, and S3C) but retained the expression of pluripotency factors such as Nanog and Oct4 (Figures 3G and S3D). Importantly, upon withdrawal of the inhibitor, these “released” cells (MYCi → released) rapidly exited the state of dormancy and reactivated both DNA and protein synthesis (Figures 3E, 3F, S3B, and S3C). We next compared the microarray expression profiles of ESCs cultured for 60 hr with the MYCi to the ones of control cells (control) or inhibitor-treated ESCs, 48 hr after MYCi withdrawal (released). This analysis revealed 3,570 differentially expressed genes (DEGs) between MYCi ESCs and the control group (FDR = 0.05, log2 fold change ± 0.5; Figure 3H; Table S3). Consistent with the GO terms enriched in the RNA sequencing (RNA-seq) analysis of the dKO cells (Figures 2C and 2G), processes related to cellular growth (e.g., mRNA processing; ribosome biogenesis), metabolism (e.g., DNA metabolic process) and proliferation (e.g., cell cycle; DNA replication) were strongly downregulated upon MYCi treatment. In contrast, processes related to signaling (e.g., regulation of signal transduction) and development (e.g., developmental process) were consistently upregulated (Figure S3E). Importantly, the comparison of the MYCi-treated with the released group showed rapid re-activation of all the biosynthetic and proliferative activities: genes and GO terms related to mRNA processing (e.g., Polr2d; Srpk1), RNA splicing (e.g., Syncript; Srsf1), ribosome biogenesis (e.g., Ncl; Wdr12), DNA replication (e.g., Cdc6; Dna2), and cell cycle (e.g., Ccnd1) were rapidly upregulated in the released group already 48 hr after withdrawal of the inhibitor (Figures 3I and 3J). Interestingly, Cdk6, recently shown to regulate exit from quiescence in human HSCs (Laurenti et al., 2015), was also rapidly induced upon exit from the Myc-induced dormant state in mouse ESCs (Figure 3J). Despite the transient depletion of Myc activity, ESCs retained their functional pluripotency and upon exit from the state of dormancy, MYCi-released ESCs were capable of both in vitro and in vivo differentiation toward the three germ layers (Figures 3K and 3L). In summary, our data indicate that treatment of ESCs with MYCi not only recapitulates the dKO phenotype at the molecular and functional level, but also demonstrates that the status of biosynthetic dormancy induced by transient loss of Myc activity in naive ground-state ESCs is reversible and occurs without compromising their stem cell identity and pluripotent potential.
Figure 3.
The Dormant State Induced by Myc Depletion Is Reversible
(A–C) Expression of exogenous c-myc mRNAs transiently rescues the dKO phenotype.
(A) Experimental workflow. c-mycΔ/Δ; N-mycΔ/fl ESCs were transfected with an EF1α-mCherry-Cre plasmid and sorted 24 hr later as mCherry negative (c-mycΔ/Δ; N-mycΔ/fl) and positive (dKO) cells. Sorted cells were plated in 2i medium and 72 hr after sort were transfected twice with c-myc mRNAs (dKO→c-myc mRNA). Control samples were treated with Lipofectamine only. Sample analysis was performed 8 hr after the second mRNA transfection.
(B) Quantitative analysis of BrdU incorporation measured by flow cytometry. The bar charts indicate the mean ± SEM; n = 3.
(C) Representative FACS plots of OP-Puro incorporation in dKO ESCs and dKO cells transfected with c-myc mRNAs (dKO→c-myc mRNA).
(D–L) Treatment of ESCs with the Myc inhibitor 10058-F4 (MYCi) recapitulates the dKO phenotype and shows molecular and functional reversibility of the state of biosynthetic dormancy induced by depletion of Myc activity. Mouse ESCs were analyzed after 60 hr of treatment with DMSO (Control) or MYCi. The MYCi → released group was treated with MYCi for 60 hr, followed by withdrawal of the inhibitor and culture in 2i medium for additional 48 hr.
(D) Phase-contrast microscopy images. Scale bar, 100 μm.
(E) Representative flow cytometry plots of the cell-cycle profiles based on the measurement of BrdU incorporation and total DNA content (7AAD).
(F) FACS analysis of OP-Puro incorporation to measure ESCs translational activity.
(G) Representative fluorescence staining for Nanog. Scale bar, 50 μm.
(H–J) Microarray expression analysis of mouse ESCs treated with MYCi.
(H) Venn diagram showing the number of differentially expressed genes (DEG) (log2 fc > ± 0.5; FDR = 0.05) between the different conditions and their overlaps.
(I) Clustered heatmaps of DEG in the indicated GO terms.
(J) Expression levels of representative DEG in the selected GO categories.
(K and L) MYCi-treated ESCs retain pluripotency.
(K) Fluorescence staining of in-vitro-differentiated ESCs for Tuj1, Sma, and Gata6. Scale bar, 100 μm.
(L) Hematoxylin and eosin staining of teratomas derived from ESCs treated with MYCi for 60 hr prior injection into immune-deficient mice. Scale bar, 50 μm.
Figure S3.
The Dormant State Induced by Myc Depletion Is Reversible, Related to Figure 3
(A) Representative FACS plots of OP-Puro incorporation in c-mycfl/fl; N-mycfl/fl (black), c-mycΔ/Δ; N-mycΔ/fl (blue), dKO (red) ESCs and dKO cells transfected with c-myc mRNAs (green). Experimental setting as described in Figure 3A.
(B–E) Mouse ESCs were analyzed after 60h of treatment with DMSO (Control) or MYCi. The MYCi→Released group was treated with MYCi for 60h, followed by withdrawal of the inhibitor and culture in 2i medium for additional 48h. (B) Quantitative analysis of the different cell cycle phases as gated in Figure 3E. The bar charts indicate the mean ± SD (C) Quantification of OP-Puro incorporation in the indicated experimental groups. Values represent geometrical mean (Geo Mean) ± SD. (D) Representative fluorescence staining for Oct4. Scale bar, 50 μm. (E) Microarray analysis of mouse ESCs treated with MYCi. List of selected processes down- or upregulated in the MYCi-treated group compared to the control (FDR 0.05).
Comparison of Expression Signatures between Myc-Depleted ESCs and the Diapause Epiblast
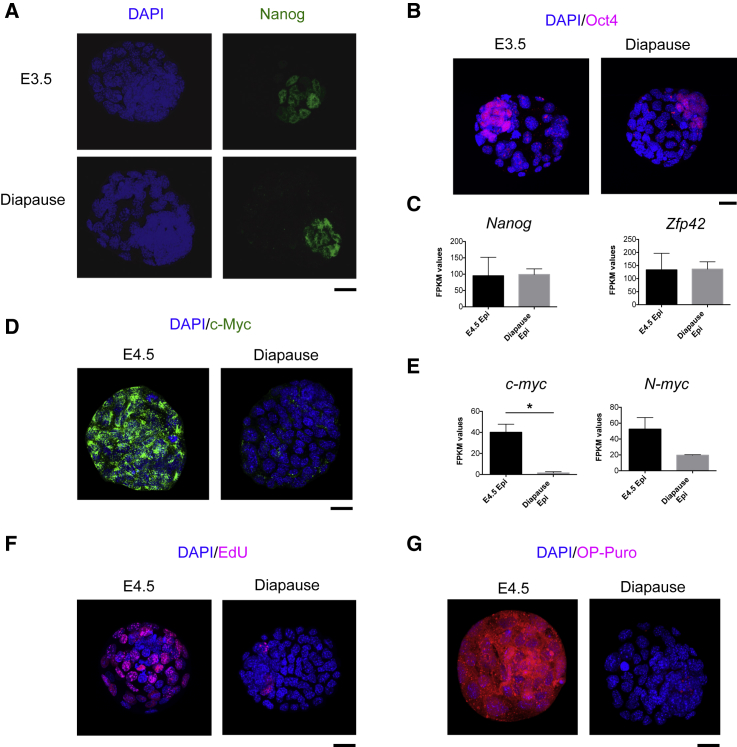
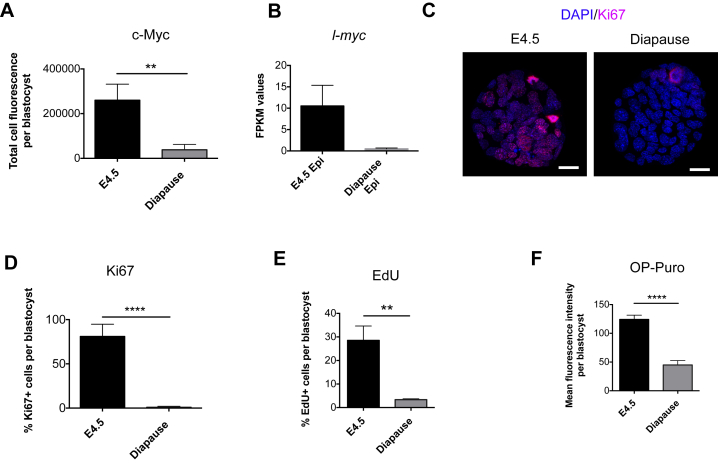
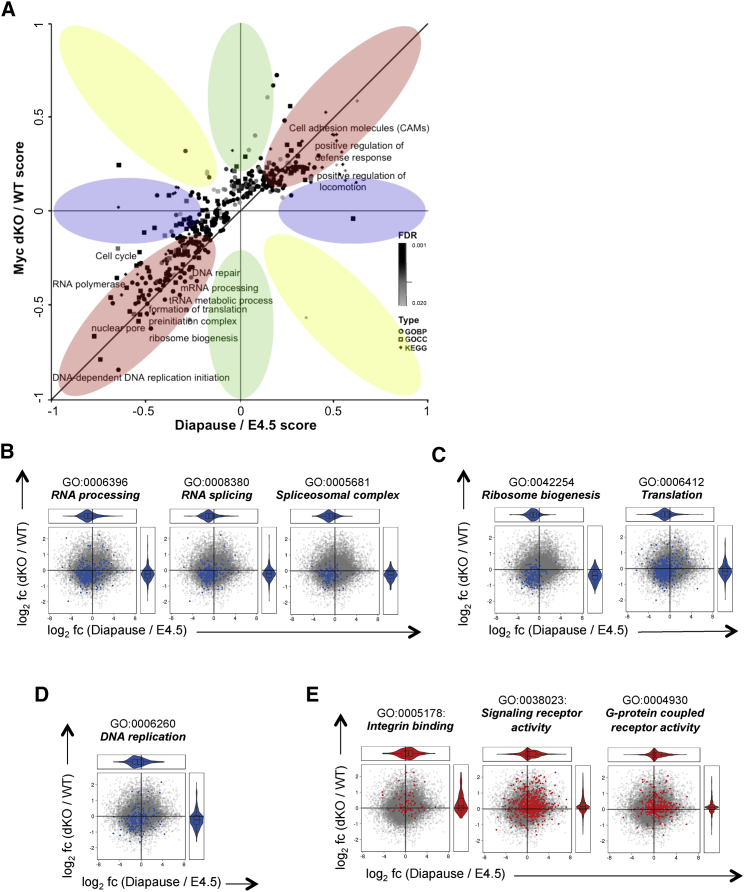
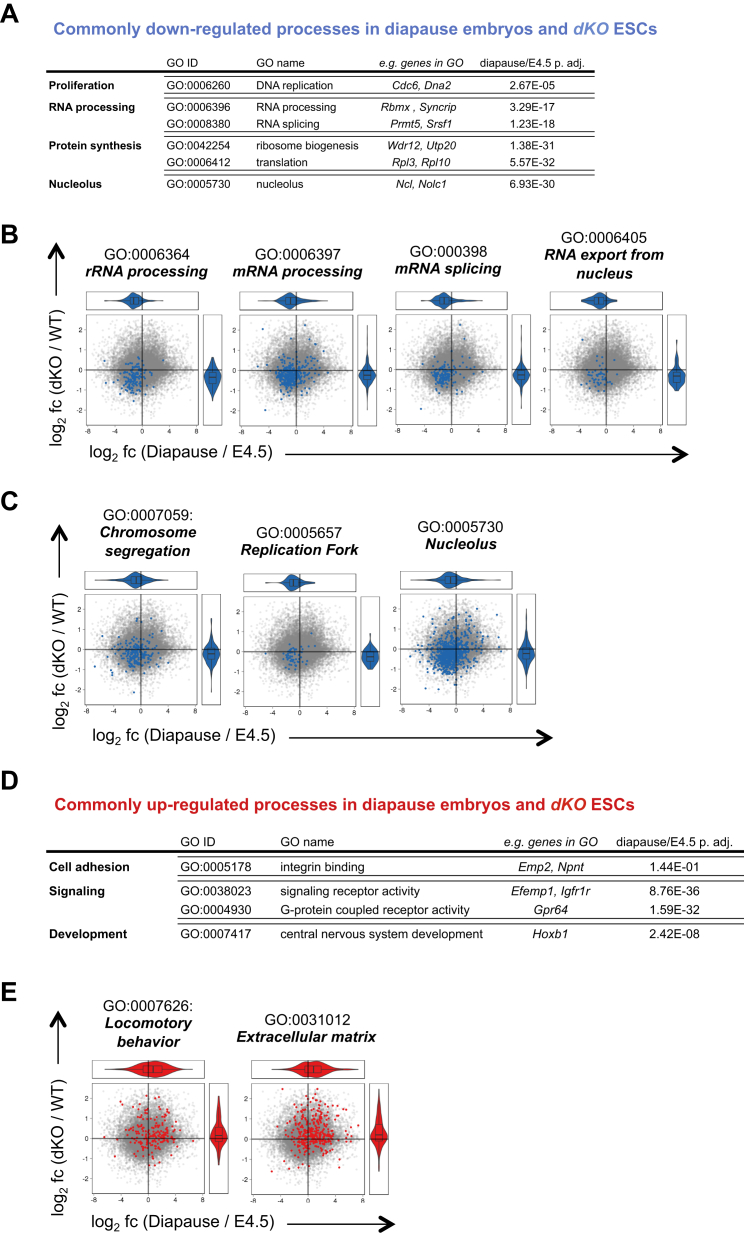
In numerous invertebrates and almost 100 mammalian species, embryonic development can be reversibly arrested as a strategy to enhance the reproductive fitness in response to changes in the living environment (Renfree and Shaw, 2000). In breast-feeding mice, delayed implantation, termed diapause, can occur in response to decreased levels of estrogen, which cause blastocysts to enter a transient dormant state of metabolic and proliferative quiescence (Hondo and Stewart, 2005, Mantalenakis and Ketchel, 1966). Our observation that the combined loss of c-myc and N-myc in ground-state ESCs induces biosynthetic dormancy raised the possibility that dKO ESCs may resemble a state equivalent to mammalian epiblast cells in diapause. To test this hypothesis, we experimentally induced delayed implantation by ovariectomizing E2.5 pregnant females and isolated diapaused embryos 4 days later. Additionally, we made use of recently generated transcriptome profiling data (RNA-seq) of diapaused and E4.5 pre-implantation epiblasts (Table S4) (Boroviak et al., 2015). As expected, diapaused embryos retained an active pluripotency network, as shown by the expression of Nanog, Oct4, and Zfp42 (Figures 4A–4C) (Batlle-Morera et al., 2008, Silva et al., 2009). Remarkably, the expression of c-Myc proteins and transcripts was low to undetectable during diapause (Figures 4D, 4E, S4A, and S4B), and Myc downregulation was associated with a significant decrease of Ki67 expression (Figures S4C and S4D) and reduced incorporation of both EdU (5-Ethynyl-2′-deoxyuridine) and OP-Puro (Figures 4F, 4G, S4E, and S4F). This reduction in DNA and protein synthesis functionally describes the state of biosynthetic and proliferative quiescence of diapaused embryos and remarkably resembles the functional phenotype of dKO ESCs. To evaluate the similarity of dKO ESCs to the diapause epiblast, we compared the molecular expression signatures of dKO cells to those of normal pre-implantation and diapaused epiblasts (Table S4) (Boroviak et al., 2015). Performing 2D GO enrichment analysis of RNA expression changes (comparing control ESCs/dKO ESCs and E4.5 normal/diapause epiblasts; Figure 5A), we found that genes from specific biological processes showed a strong positive correlation between both datasets. Specifically, processes related to gene expression (e.g., RNA processing, RNA splicing, RNA export from nucleus), protein synthesis (e.g., ribosome biogenesis, translation), and proliferation (e.g., DNA replication, replication fork, chromosome segregation) were robustly downregulated in both datasets (Figures 5B–5D and S5A–S5C). In contrast, processes related to cell adhesion (e.g., Integrin binding) and signaling (e.g., signaling receptor activity, G protein coupled receptor activity) were commonly upregulated (Figures 5E, S5D, and S5E). These expression profiling data strongly indicate that the transcriptomic changes occurring upon Myc depletion in naive ground-state ESCs closely correlate with the ones observed during induction of diapause.
Figure 4.
Translation, Proliferation, and Myc Expression Are Reduced in Diapause Embryos
(A–C) Expression of pluripotency markers in the diapaused embryos. Confocal microscope images of peri-implantation (E3.5) and diapause embryos immunostained for Nanog (A) or Oct4 (B). Scale bar, 20 μm. (C) RNA-seq data based on Boroviak et al. (2015) showing expression levels of Nanog and Zfp42 in E4.5 and diapause embryos. Bar graphs show FPKM (fragments per kilobase of exon per million reads mapped) values and are represented as mean ± SEM. E4.5 epiblast (n = 3); diapause epiblast (n = 2).
(D and E) Diapause embryos have reduced Myc expression.
(D) Fluorescence staining for c-Myc in E4.5 and diapause embryos. Scale bar, 20 μm. Images are representative of two independent experiments.
(E) Expression levels of c-myc and N-myc in the epiblast of pre-implantation (E4.5) and diapause embryos. RNA-seq data based on Boroviak et al. (2015). E4.5 epiblast (n = 3); diapause epiblast (n = 2). Values are represented as mean ± SEM.
(F and G) Diapause embryos are characterized by reduced DNA and protein synthesis. Representative confocal microscope images showing EdU (F) and OP-Puro (G) incorporation in E4.5 and diapause embryos. Scale bar, 20 μm.
Figure S4.
Translation, Proliferation, and Myc Expression Are Reduced in Diapause Embryos, Related to Figure 4
(A) c-Myc protein expression in E4.5 and diapause embryos. Quantification of confocal microscopy images as in Figure 4D.
(B) Expression levels of l-myc in the epiblast of E4.5 and diapause embryos. RNA-seq data based on (Boroviak et al., 2015). Bar graphs show FPKM values and are represented as mean ± SEM. E4.5 epiblast (n = 3); diapause epiblast (n = 2).
(C) Fluorescence staining for Ki67 (red) and DAPI (blue) in E4.5 and diapause embryos. Scale bar, 20 μm.
(D) Quantification of (C). Values are represented as mean ± SD.
(E) EdU incorporation in E4.5 and Diapause embryos. Quantification of confocal microscopy images as in Figure 4F.
(F) OP-Puro incorporation in E4.5 and Diapause embryos. Quantification of Figure 4G.
Figure 5.
Correlation of Expression Signatures between Myc-Depleted ESCs and Diapause Epiblasts
(A) 2D GO enrichment analysis of RNA expression changes of dKO/c-mycfl/fl; N-mycfl/fl (WT) ESCs and Diapause/E4.5 epiblasts. Dataset is from Boroviak et al. (2015). Red regions correspond to concordantly higher or lower expression. Blue and green regions correspond to lower or higher expression in one direction, but not in the other. Terms in yellow regions show anti-correlating behavior. Analysis was performed using the Perseus software according to Cox and Mann (2008). FDR = 0.02.
(B–E) Scatterplots of pathways significantly changed both in the comparison dKO/WT (FDR = 0.1) and in Diapause/E4.5 (FDR = 0.1). The gray dots represent the set of all genes. Genes belonging to the specific pathways are highlighted in blue (downregulated processes) or red (upregulated processes). The x axis shows the DESeq2-moderated log2 fold change (log2 fc) between the diapause compared to the E4.5 epiblast; the y axis shows the DESeq2-moderated log2 fc between the dKO ESCs compared to c-mycfl/fl; N-mycfl/fl (WT) cells. The violin plots at the margins show the marginal distributions of fold changes of the genes in the highlighted pathways.
Figure S5.
Correlation of Expression Signatures between Myc Depleted ESCs and Diapause Epiblasts, Related to Figure 5
(A) Selected processes commonly downregulated in diapause embryos and dKO ESCs compared to E4.5 epiblast and c-mycfl/fl; N-mycfl/fl (WT) ESCs respectively (FDR 0.1).
(B and C) Scatterplots of pathways significantly downregulated both in the comparison dKO / WT (FDR 0.1) and in Diapause / E4.5 (FDR 0.1).
(D) Selected processes commonly upregulated in diapause embryos and dKO ESCs compared to E4.5 epiblast and WT ESCs respectively (FDR 0.1).
(E) Scatterplots of pathways significantly upregulated both in the comparison dKO / WT (FDR 0.1) and in Diapause / E4.5 (FDR 0.1).
Inhibition of Myc Activity in Mouse Blastocysts Induces a Diapause-like State of Dormancy
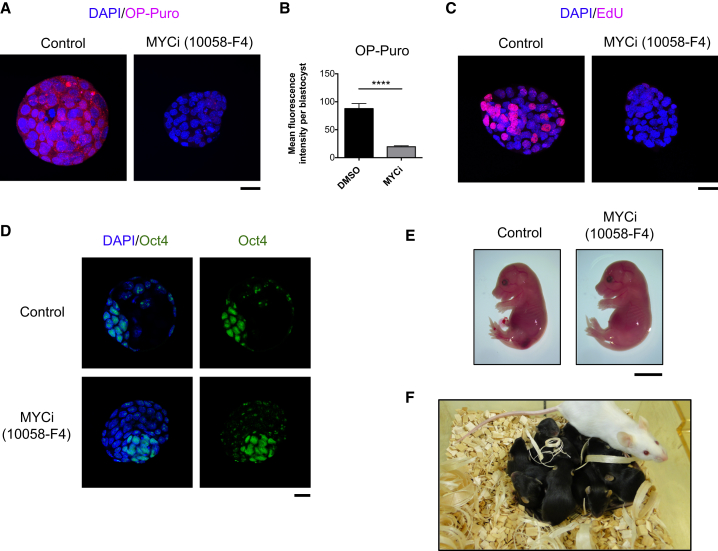
To investigate whether reduction of Myc activity can induce a diapause-like state in normal, pre-implantation embryos, we isolated E3.75–E4.0 blastocysts and cultured them in the presence of the MYCi for 18 hr. This resulted in a striking reduction of both de novo protein synthesis (Figures 6A and 6B) and cellular proliferation (Figure 6C). Importantly, pluripotency markers such as Oct4 were still expressed upon Myc inhibition (Figure 6D), highlighting the maintenance of the stem cell circuitry in the epiblast of Myc-inhibited embryos. Next, we assessed the ability of those Myc-inhibited, diapause-like blastocysts to resume development upon withdrawal of MYCi by transferring MYCi-treated embryos into the uterus of pseudo-pregnant foster mothers. MYCi-treated blastocysts yielded morphologically normal embryos at E15.5 and live born pups (Figures 6E and 6F). These findings not only show that Myc is downregulated in diapause embryos, but also demonstrate that transient reduction of Myc activity is sufficient to drive embryos into a reversible diapause-like state, functionally characterized by strong reduction of proliferation and protein synthesis while a fully functional pluripotent developmental program is maintained.
Figure 6.
Inhibition of Myc Activity Induces a Diapause-like State in Mouse Blastocysts
E3.75–E4.0 embryos were collected from pregnant females and treated for 18 hr with MYCi or DMSO (Control).
(A) Representative confocal microscope images of OP-Puro incorporation. Scale bar, 20 μm.
(B) Quantification of (A). Values are represented as mean ± SEM. MYCi (n = 6); Control (n = 7).
(C) Representative images of EdU incorporation. Scale bar, 20 μm.
(D) Confocal microscopy images of embryos immunostained for Oct4. Scale bar, 20 μm.
(E and F) MYCi-treated and control blastocysts were transplanted into the uteri of pseudo-pregnant foster mothers.
(E) Representative images of E15.5 embryos derived from MYCi-treated or control blastocysts. This result is representative of two independent experiments. MYCi (n = 24 embryos E15.5); control (n = 11 embryos E15.5). Scale bar, 0.5 cm.
(F) Representative picture of the litter generated from the transplant of MYCi-treated blastocysts into pseudo-pregnant foster mothers. The result is representative of two independent experiments. In the MYCi condition, totally nine litters were born from nine embryo-transferred foster mothers (totally 64 live-born mice).
Discussion
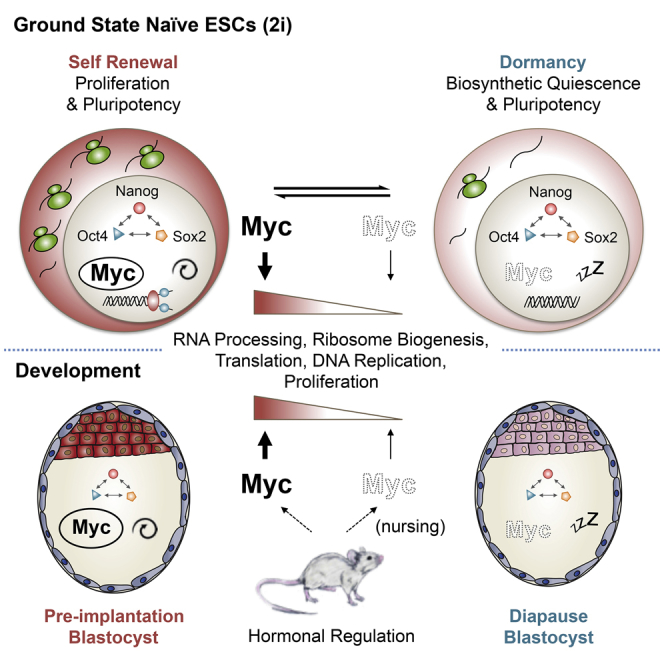
The stem cell self-renewal comprises two biological processes, namely, cellular growth-proliferation linked to inhibition of differentiation. The latter is synonymous with the capacity to maintain stem cell pluripotency or multipotency in at least one of the daughter cells after cell division. By using both a genetic model and an inhibitor of Myc activity, our data demonstrate that, while Myc controls biosynthetic activity and cellular proliferation of naive ground-state ESCs, the maintenance of pluripotency and stem cell identity is independent of Myc. In the absence of Myc activity, maintenance of pluripotency and the associated lack of differentiation are only observed in 2i-cultured ground-state ESCs and in the blastocyst. These findings suggest that the effects of Myc are highly linked to the microenvironmental signals naturally present in the uterus and recapitulated in vitro by the 2i culture. Thus, the two key components of ESC self-renewal can be separated into a Myc-controlled biosynthetic program and a Myc-independent pluripotency program. These results strongly support findings in other cell types, including cancer cells (Carroll et al., 2015, Dang, 2013, Dang, 2015), and are in agreement with a recently described Myc-centered transcriptional module (Kim et al., 2010). Diapause is a unique biological phenomenon, developed in many invertebrates (e.g., butterflies, C. elegans) and mammals (e.g., rodents, deer) as a reproductive strategy to survive predictable, unfavorable environmental conditions, such as temperature extremes, drought, or reduced food availability (Renfree and Shaw, 2000). In the mouse, hormonally mediated diapause of pre-implantation embryos occurs if the mother still nurses pups of the previous litter (Hondo and Stewart, 2005, Mantalenakis and Ketchel, 1966). During diapause, embryos or organisms undergo a reversible biosynthetic/metabolic dormancy, in which certain cells must retain their properties of pluri-/multipotency to ensure reactivation of development at later stages. The molecular mechanisms governing mouse embryonic diapause are largely unsolved and key factors regulating embryonic dormancy have yet to be identified (Hondo and Stewart, 2005). In this study, we identify Myc as a master regulator of RNA processing, protein synthesis, and proliferation both in 2i-cultured ESCs and in the pre-implantation embryo and propose the depletion of Myc activity as an important signal for the execution of the diapause program. Myc expression is indeed strongly reduced during the physiological and reversible state of embryonic dormancy, and cellular processes controlled by Myc in ESCs, such as RNA splicing, DNA and protein synthesis, and cellular proliferation, are dramatically affected during diapause. The observation that transient inhibition of Myc activity in pre-implantation blastocysts is on its own sufficient to trigger a diapause-like state of biosynthetic suspension without affecting pluripotent identity or compromising the developmental program is remarkable. Upon release of Myc inhibition, ground-state ESC re-enter into the cell cycle and diapause-like embryos progress through their normal developmental program. Yet, finely tunable genetic gain-of-function models will be needed to assess whether sustained Myc expression would prevent entry into diapause and provide additional evidence that Myc activity is sufficient to exit from the dormant state in vivo. Biosynthetic and energetic demands are intimately coupled. Diapaused embryos are characterized by a robust downregulation of biosynthetic processes, which is likely associated with a reduced bioenergetic activity. Metabolomics studies will need to explore whether the type or only the extent of energy production is different in normal and diapause embryos and how Myc interferes with this process (Hsieh et al., 2015, Takubo et al., 2013). In summary, our findings suggest pluripotent stem cell dormancy as a Myclow state and identify differential Myc activity as a key event in orchestrating the complex and rapid changes occurring during entry and exit from diapause.
Successful implantation results from the reciprocal interaction between an implantation-competent blastocyst and the receptive uterus (Wang and Dey, 2006). Among other factors, the estrogen-induced cytokine LIF, secreted first by the uterine glands and then by the surrounding stroma, is crucial for the preparation of the uterus for implantation. Females lacking a functional LIF gene are fertile, but their blastocysts fail to implant and can only continue embryogenesis if transferred to wild-type pseudo-pregnant recipients (Chen et al., 2000, Stewart et al., 1992). Similarly, embryos lacking gp130, part of the LIF receptor complex, cannot resume development after undergoing diapause (Nichols et al., 2001), confirming the essential role of LIF/Stat3 signaling within the diapause program. Although it has been suggested that c-Myc lies downstream of LIF/Stat3 in ESCs cultured in serum (Cartwright et al., 2005), the connection between the two pathways remains controversial in ESCs and may depend on culture conditions (Hall et al., 2009, Martello et al., 2013). To identify the mechanisms controlling Myc activity in normal and diapaused epiblasts within their natural uterine microenvironment, future studies will need to elucidate the complex signaling interactions between LIF/Stat3 and Myc downstream of estrogens.
At the cellular level, our findings may not be restricted to the embryo but also apply to dormant, somatic stem cells where the quiescent state is known to protect them from mutations that might occur following replicative stress and result in aging, stem cell loss, or in the transition toward a cancerous state (Cheung and Rando, 2013, Latil et al., 2012, Sosa et al., 2014, Wagner et al., 1993, Wilson et al., 2008, Wilson et al., 2009). In the hematopoietic system, for instance, the exit of HSCs from dormancy triggered by interferon-α is associated to the upregulation of c-Myc protein (Ehninger et al., 2014). Additionally, upon conditional deletion of both c-myc and N-myc in the bone marrow, the only maintained blood cells are dormant HSCs, while non-stem cells undergo rapid apoptosis (Laurenti et al., 2008, Laurenti et al., 2009). Dormancy is therefore a transient stem cell state, molecularly defined by extremely low levels of Myc expression, where the most primitive cells within the stem cell compartment can be stored as a transiently quiescent reservoir to preserve genomic integrity and be protected from microenvironmental hazards (Baldridge et al., 2010, Essers et al., 2009, Flach et al., 2014, Walter et al., 2015, Wilson et al., 2008). This phenomenon might not be exclusive to normal stem cells but similarly be exploited in cancer, for example, by malignant metastatic stem cells following invasion into a new tissue environment (Oskarsson et al., 2014, Sosa et al., 2014). In tumor cells with stem-like character, environmentally mediated physiological downregulation or pharmacological inhibition of Myc may thus induce tumor cell dormancy instead of apoptosis.
Experimental Procedures
Other procedures and reagents details are provided in the Supplemental Experimental Procedures.
Mice
Six- to 12-week-old mice were used throughout the study. C57BL/6 mice were purchased from Harlan Laboratories. Teratoma assays were performed in NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice. c-mycfl/fl; N-mycfl/fl; RosaEYFPfl/fl (Knoepfler et al., 2002, Srinivas et al., 2001, Trumpp et al., 2001) and c-MyceGFP/eGFP (Huang et al., 2008) mice were bred in-house in the animal facility of the DKFZ, Heidelberg, Germany. All mice were maintained at the DKFZ under specific pathogen-free (SPF) conditions in individually ventilated cages (IVCs). Animal procedures were performed according to protocols approved by the German authorities, Regierungspräsidium Karlsruhe (Nr. Z110/02, DKFZ 299 and G135/15).
Embryonic Stem Cell Derivation and Culture
Mouse ESCs were derived from 8- to 10-week-old c-mycfl/fl; N-mycfl/fl; RosaEYFPfl/fl (Knoepfler et al., 2002, Srinivas et al., 2001, Trumpp et al., 2001) or c-MyceGFP/eGFP (Huang et al., 2008) mice. Females were injected intraperitoneally with 7U of pregnant mare serum gonadotropin (PMSG) followed by 7 U of human chorionic gonadotropin (hCG), injected 47 hr later. Females were then paired with males and checked for vaginal plug the following morning, considered as 0.5 days post-coitum (dpc). 2.5-dpc embryos were flushed from the oviducts into a plastic dish with M2 medium (Sigma, M7167). Embryos were washed ten times in M2 medium. To remove the zona pellucida, embryos were incubated shortly in Tyrode’s solution (Sigma, T1788-100ML) and transferred again to M2 medium. Finally, each embryo was cultured on a layer of mitotically inactivated mouse embryonic fibroblasts (MEFs). ESCs were routinely propagated on 0.1% gelatin-coated plates, without feeders or serum, in N2B27 medium (NDiff N2B27 base medium, Stem Cell Sciences, SCS-SF-NB-02) supplemented with the small-molecule inhibitors PD0325901 (Stemgent, 04-0006, 1 μM) and CHIR99021 (Stemgent, 04-0004, 3 μM), LIF (Millipore, ESG1106, 1,000 U/ml), and β-mercaptoethanol (Gibco, 31350-010, 0.05 mM). Cells were replated every 3 days following dissociation with StemPro Accutase (Gibco, A11105-01). In the experiments with serum, ESCs were cultured on MEFs in knockout DMEM medium (Gibco, 10829) supplemented with L-glutamine (Gibco, 25030-081, 2 mM), LIF (Millipore, ESG1106, 1,000 U/ml), β-mercaptoethanol (Gibco, 31350-010, 0.05 mM), and 15% ESC-tested FBS (Pansera ES, Pan Biotech, 2602-P271303). In the experiments with the small-molecule inhibitor 10058-F4 (MYCi) (Calbiochem, 475956), MYCi was dissolved in 2i medium at a final concentration of 64 μM. As a control, DMSO alone was added at the same final volume as MYCi.
Induction of Diapause
Six- to 10-week-old female mice were injected intraperitoneally with 7 U of pregnant mare serum gonadotropin (PMSG) followed by 7 U of human chorionic gonadotropin (hCG), injected 48 hr later. The females were paired with males and checked for vaginal plug the following morning, considered as 0.5 days postcoitum (dpc). At 2.5 dpc, the females were ovariectomized, and diapause embryos were isolated by uterine flush 4 days later at 6.5 dpc. The control group was not operated and females were sacrificed at 3.5 or 4.5 dpc for embryo isolation.
Treatment of Mouse Blastocysts with the Myc Inhibitor 10058-F4 and Embryo Transfer
E3.75–E4.0 embryos were collected from superovulated C57BL/6 pregnant females and incubated for 18 hr with the Myc inhibitor (MYCi) 10058-F4 (Calbiochem, 475956, final concentration 55 μM) in EmbryoMax KSOM medium + amino acids (Merck Millipore, MR121-D). The control group was treated with DMSO alone, added at the same final volume as the inhibitor. Treated embryos were transferred into the uterus of CD1 pseudo-pregnant foster mothers and analyzed at E15.5 or after birth.
Analysis of OP-Puro and EdU Incorporation Using the Click-iT Technology
To measure protein synthesis, in-vitro-cultured ESCs or mouse embryos were incubated for 1 hr in 2i medium or EmbryoMax KSOM medium + amino acids (Merck Millipore, MR121-D), respectively, supplemented with O-propargyl-puromycin (OP-Puro) (Jena Biosciences, NU-931-05, 50 μM final concentration). After harvesting, the samples were fixed for 15 min at room temperature in PBS supplemented with 4% paraformaldehyde (Electron Microscopy Sciences, 19208) and then permeabilized in PBS supplemented with 1% BSA and 0.1% saponin for 5 min at room temperature. The copper-catalyzed azide-alkyne cycloaddition (CuAAC) was performed using an Alexa 647-azide (Life Technologies, A10277, 5 μM final concentration) and the Click-iT Cell Reaction Buffer Kit (Life Technologies, C10269) according to the manufacturer’s instructions. For the analysis of EdU incorporation, isolated embryos were incubated in EmbryoMax KSOM medium + amino acids (Merck Millipore, MR121-D) supplemented with EdU (10 μM final concentration) for 80 min. EdU labeling and staining of the embryos were performed using the Click-iT Plus EdU Alexa Fluor 647 Flow Cytometry Assay Kit (Life Technologies, C10634) according to the manufacturer’s instructions.
Statistical Analysis
Data were processed using Prism 6 (GraphPad Software). All analyses were performed using two-tailed Student’s t tests (unless otherwise specified). Statistical significance is indicated by ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; and ∗∗∗∗p < 0.0001.
Acknowledgments
We thank Corinna Klein, Katja Müdder, Melanie Neubauer, Adriana Przybylla, and Markus Sohn for technical assistance. We would like to thank Gelo Victoriano De la Cruz and the DKFZ Flow Cytometry Core facility for their assistance and Dr. Kurt Reifenberg, Dr. Michaela Socher, Anja Rathgeb, and all members of the DKFZ Laboratory Animal Core Facility for excellent animal welfare and husbandry. Support by Damir Krunic, Manuela Brom, and the DKFZ Light Microscopy facility is gratefully acknowledged. We thank the microarray unit of the DKFZ Genomics and Proteomics Core Facility for providing the Illumina Whole-Genome Expression analysis and related services. We thank Erika Krückel for help with all administrative matters and all colleagues of the A.T.’s and HI-STEM laboratories for helpful discussions. The authors would like to thank Dr. Henk Stunnenberg (Nijmegen, NL) for helpful discussions during the initial phase of the project. R.S. was a member of the Hartmut Hoffmann-Berling International Graduate School (HBIGS). This work was supported by the FOR2033 and SFB873 funded by the Deutsche Forschungsgemeinschaft (DFG), the Dietmar Hopp Foundation (all to A.T.), and the Wellcome Trust (to A.S.).
Published: February 11, 2016
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, five figures, four tables, and one data file and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2015.12.033.
Accession Numbers
The accession number for the RNA-seq data reported in this paper is ArrayExpress: E-MTAB-3386. The accession number for the microarray data is GEO: GSE74337.
Supplemental Information
Each column of the matrix represents a sample and each row represents one gene. The matrix is filled with the number of reads that were tabulated to each gene based on the genomic coordinates of both the read alignments to the reference genome and the annotation of the genes.
References
- Adhikary S., Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- Arnold I., Watt F.M. c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr. Biol. 2001;11:558–568. doi: 10.1016/s0960-9822(01)00154-3. [DOI] [PubMed] [Google Scholar]
- Baldridge M.T., King K.Y., Boles N.C., Weksberg D.C., Goodell M.A. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature. 2010;465:793–797. doi: 10.1038/nature09135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle-Morera L., Smith A., Nichols J. Parameters influencing derivation of embryonic stem cells from murine embryos. Genesis. 2008;46:758–767. doi: 10.1002/dvg.20442. [DOI] [PubMed] [Google Scholar]
- Blackwood E.M., Eisenman R.N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Boroviak T., Loos R., Bertone P., Smith A., Nichols J. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat. Cell Biol. 2014;16:516–528. doi: 10.1038/ncb2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak T., Loos R., Lombard P., Okahara J., Behr R., Sasaki E., Nichols J., Smith A., Bertone P. Lineage-Specific Profiling Delineates the Emergence and Progression of Naive Pluripotency in Mammalian Embryogenesis. Dev. Cell. 2015;35:366–382. doi: 10.1016/j.devcel.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll P.A., Diolaiti D., McFerrin L., Gu H., Djukovic D., Du J., Cheng P.F., Anderson S., Ulrich M., Hurley J.B. Deregulated Myc requires MondoA/Mlx for metabolic reprogramming and tumorigenesis. Cancer Cell. 2015;27:271–285. doi: 10.1016/j.ccell.2014.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright P., McLean C., Sheppard A., Rivett D., Jones K., Dalton S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- Charron J., Malynn B.A., Fisher P., Stewart V., Jeannotte L., Goff S.P., Robertson E.J., Alt F.W. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes Dev. 1992;6(12A):2248–2257. doi: 10.1101/gad.6.12a.2248. [DOI] [PubMed] [Google Scholar]
- Chen J.R., Cheng J.G., Shatzer T., Sewell L., Hernandez L., Stewart C.L. Leukemia inhibitory factor can substitute for nidatory estrogen and is essential to inducing a receptive uterus for implantation but is not essential for subsequent embryogenesis. Endocrinology. 2000;141:4365–4372. doi: 10.1210/endo.141.12.7855. [DOI] [PubMed] [Google Scholar]
- Cheung T.H., Rando T.A. Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol. 2013;14:329–340. doi: 10.1038/nrm3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Dang C.V. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb. Perspect. Med. 2013;3:3. doi: 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C.V. Web of the extended Myc network captures metabolism for tumorigenesis. Cancer Cell. 2015;27:160–162. doi: 10.1016/j.ccell.2015.01.004. [DOI] [PubMed] [Google Scholar]
- Dubois N.C., Adolphe C., Ehninger A., Wang R.A., Robertson E.J., Trumpp A. Placental rescue reveals a sole requirement for c-Myc in embryonic erythroblast survival and hematopoietic stem cell function. Development. 2008;135:2455–2465. doi: 10.1242/dev.022707. [DOI] [PubMed] [Google Scholar]
- Ehninger A., Boch T., Uckelmann H., Essers M.A., Müdder K., Sleckman B.P., Trumpp A. Posttranscriptional regulation of c-Myc expression in adult murine HSCs during homeostasis and interferon-α-induced stress response. Blood. 2014;123:3909–3913. doi: 10.1182/blood-2013-10-531038. [DOI] [PubMed] [Google Scholar]
- Eilers M., Eisenman R.N. Myc’s broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M., Schirm S., Bishop J.M. The MYC protein activates transcription of the alpha-prothymosin gene. EMBO J. 1991;10:133–141. doi: 10.1002/j.1460-2075.1991.tb07929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers M.A., Offner S., Blanco-Bose W.E., Waibler Z., Kalinke U., Duchosal M.A., Trumpp A. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- Flach J., Bakker S.T., Mohrin M., Conroy P.C., Pietras E.M., Reynaud D., Alvarez S., Diolaiti M.E., Ugarte F., Forsberg E.C. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 2014;512:198–202. doi: 10.1038/nature13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freytag S.O., Geddes T.J. Reciprocal regulation of adipogenesis by Myc and C/EBP alpha. Science. 1992;256:379–382. doi: 10.1126/science.256.5055.379. [DOI] [PubMed] [Google Scholar]
- Gebhardt A., Frye M., Herold S., Benitah S.A., Braun K., Samans B., Watt F.M., Elsässer H.P., Eilers M. Myc regulates keratinocyte adhesion and differentiation via complex formation with Miz1. J. Cell Biol. 2006;172:139–149. doi: 10.1083/jcb.200506057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Guo G., Wray J., Eyres I., Nichols J., Grotewold L., Morfopoulou S., Humphreys P., Mansfield W., Walker R. Oct4 and LIF/Stat3 additively induce Krüppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell. 2009;5:597–609. doi: 10.1016/j.stem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Hatton K.S., Mahon K., Chin L., Chiu F.C., Lee H.W., Peng D., Morgenbesser S.D., Horner J., DePinho R.A. Expression and activity of L-Myc in normal mouse development. Mol. Cell. Biol. 1996;16:1794–1804. doi: 10.1128/mcb.16.4.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondo E., Stewart C.L. Profiling gene expression in growth-arrested mouse embryos in diapause. Genome Biol. 2005;6:202. doi: 10.1186/gb-2004-6-1-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh A.L., Walton Z.E., Altman B.J., Stine Z.E., Dang C.V. MYC and metabolism on the path to cancer. Semin. Cell Dev. Biol. 2015;43:11–21. doi: 10.1016/j.semcdb.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.Y., Bredemeyer A.L., Walker L.M., Bassing C.H., Sleckman B.P. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur. J. Immunol. 2008;38:342–349. doi: 10.1002/eji.200737972. [DOI] [PubMed] [Google Scholar]
- Humbert P.O., Verona R., Trimarchi J.M., Rogers C., Dandapani S., Lees J.A. E2f3 is critical for normal cellular proliferation. Genes Dev. 2000;14:690–703. [PMC free article] [PubMed] [Google Scholar]
- Iritani B.M., Eisenman R.N. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc. Natl. Acad. Sci. USA. 1999;96:13180–13185. doi: 10.1073/pnas.96.23.13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L.A., Prober D.A., Edgar B.A., Eisenman R.N., Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Woo A.J., Chu J., Snow J.W., Fujiwara Y., Kim C.G., Cantor A.B., Orkin S.H. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler P.S., Cheng P.F., Eisenman R.N. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latil M., Rocheteau P., Châtre L., Sanulli S., Mémet S., Ricchetti M., Tajbakhsh S., Chrétien F. Skeletal muscle stem cells adopt a dormant cell state post mortem and retain regenerative capacity. Nat. Commun. 2012;3:903. doi: 10.1038/ncomms1890. [DOI] [PubMed] [Google Scholar]
- Laurenti E., Varnum-Finney B., Wilson A., Ferrero I., Blanco-Bose W.E., Ehninger A., Knoepfler P.S., Cheng P.F., MacDonald H.R., Eisenman R.N. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 2008;3:611–624. doi: 10.1016/j.stem.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti E., Wilson A., Trumpp A. Myc’s other life: stem cells and beyond. Curr. Opin. Cell Biol. 2009;21:844–854. doi: 10.1016/j.ceb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Laurenti E., Frelin C., Xie S., Ferrari R., Dunant C.F., Zandi S., Neumann A., Plumb I., Doulatov S., Chen J. CDK6 levels regulate quiescence exit in human hematopoietic stem cells. Cell Stem Cell. 2015;16:302–313. doi: 10.1016/j.stem.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Xu Y., Stoleru D., Salic A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc. Natl. Acad. Sci. USA. 2012;109:413–418. doi: 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantalenakis S.J., Ketchel M.M. Frequency and extent of delayed implantation in lactating rats and mice. J. Reprod. Fertil. 1966;12:391–394. doi: 10.1530/jrf.0.0120391. [DOI] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A.F., Smith A., Stunnenberg H.G. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G., Bertone P., Smith A. Identification of the missing pluripotency mediator downstream of leukaemia inhibitory factor. EMBO J. 2013;32:2561–2574. doi: 10.1038/emboj.2013.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Chambers I., Smith A. Derivation of germline competent embryonic stem cells with a combination of interleukin-6 and soluble interleukin-6 receptor. Exp. Cell Res. 1994;215:237–239. doi: 10.1006/excr.1994.1338. [DOI] [PubMed] [Google Scholar]
- Nichols J., Chambers I., Taga T., Smith A. Physiological rationale for responsiveness of mouse embryonic stem cells to gp130 cytokines. Development. 2001;128:2333–2339. doi: 10.1242/dev.128.12.2333. [DOI] [PubMed] [Google Scholar]
- Oskarsson T., Batlle E., Massagué J. Metastatic stem cells: sources, niches, and vital pathways. Cell Stem Cell. 2014;14:306–321. doi: 10.1016/j.stem.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelengaris S., Khan M., Evan G.I. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109:321–334. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- Rahl P.B., Lin C.Y., Seila A.C., Flynn R.A., McCuine S., Burge C.B., Sharp P.A., Young R.A. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfree M.B., Shaw G. Diapause. Annu. Rev. Physiol. 2000;62:353–375. doi: 10.1146/annurev.physiol.62.1.353. [DOI] [PubMed] [Google Scholar]
- Signer R.A., Magee J.A., Salic A., Morrison S.J. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509:49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Nichols J., Theunissen T.W., Guo G., van Oosten A.L., Barrandon O., Wray J., Yamanaka S., Chambers I., Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.N., Singh A.M., Dalton S. Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell Stem Cell. 2010;7:343–354. doi: 10.1016/j.stem.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa M.S., Bragado P., Aguirre-Ghiso J.A. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat. Rev. Cancer. 2014;14:611–622. doi: 10.1038/nrc3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C.L., Kaspar P., Brunet L.J., Bhatt H., Gadi I., Köntgen F., Abbondanzo S.J. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature. 1992;359:76–79. doi: 10.1038/359076a0. [DOI] [PubMed] [Google Scholar]
- Takashima Y., Guo G., Loos R., Nichols J., Ficz G., Krueger F., Oxley D., Santos F., Clarke J., Mansfield W. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takubo K., Nagamatsu G., Kobayashi C.I., Nakamura-Ishizu A., Kobayashi H., Ikeda E., Goda N., Rahimi Y., Johnson R.S., Soga T. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell. 2013;12:49–61. doi: 10.1016/j.stem.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpp A., Refaeli Y., Oskarsson T., Gasser S., Murphy M., Martin G.R., Bishop J.M. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- Varlakhanova N.V., Cotterman R.F., deVries W.N., Morgan J., Donahue L.R., Murray S., Knowles B.B., Knoepfler P.S. myc maintains embryonic stem cell pluripotency and self-renewal. Differentiation. 2010;80:9–19. doi: 10.1016/j.diff.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A.J., Meyers C., Laimins L.A., Hay N. c-Myc induces the expression and activity of ornithine decarboxylase. Cell Growth Differ. 1993;4:879–883. [PubMed] [Google Scholar]
- Walter D., Lier A., Geiselhart A., Thalheimer F.B., Huntscha S., Sobotta M.C., Moehrle B., Brocks D., Bayindir I., Kaschutnig P. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 2015;520:549–552. doi: 10.1038/nature14131. [DOI] [PubMed] [Google Scholar]
- Walz S., Lorenzin F., Morton J., Wiese K.E., von Eyss B., Herold S., Rycak L., Dumay-Odelot H., Karim S., Bartkuhn M. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014;511:483–487. doi: 10.1038/nature13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Dey S.K. Roadmap to embryo implantation: clues from mouse models. Nat. Rev. Genet. 2006;7:185–199. doi: 10.1038/nrg1808. [DOI] [PubMed] [Google Scholar]
- Warren L., Manos P.D., Ahfeldt T., Loh Y.H., Li H., Lau F., Ebina W., Mandal P.K., Smith Z.D., Meissner A. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M., Meissner A., Cassady J.P., Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Wilson A., Murphy M.J., Oskarsson T., Kaloulis K., Bettess M.D., Oser G.M., Pasche A.C., Knabenhans C., Macdonald H.R., Trumpp A. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18:2747–2763. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A., Laurenti E., Oser G., van der Wath R.C., Blanco-Bose W., Jaworski M., Offner S., Dunant C.F., Eshkind L., Bockamp E. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- Wilson A., Laurenti E., Trumpp A. Balancing dormant and self-renewing hematopoietic stem cells. Curr. Opin. Genet. Dev. 2009;19:461–468. doi: 10.1016/j.gde.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirath H., Frenzel A., Oliynyk G., Segerström L., Westermark U.K., Larsson K., Munksgaard Persson M., Hultenby K., Lehtiö J., Einvik C. MYC inhibition induces metabolic changes leading to accumulation of lipid droplets in tumor cells. Proc. Natl. Acad. Sci. USA. 2013;110:10258–10263. doi: 10.1073/pnas.1222404110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Each column of the matrix represents a sample and each row represents one gene. The matrix is filled with the number of reads that were tabulated to each gene based on the genomic coordinates of both the read alignments to the reference genome and the annotation of the genes.