Abstract
The Notch protein is one of the most mechanistically direct transmembrane receptors – the intracellular domain contains a transcriptional regulator that is released from the membrane when engagement of the cognate extracellular ligand induces intramembrane proteolysis. We find that chimeric forms of Notch, in which both the extracellular sensor module and the intracellular transcriptional module are replaced with heterologous protein domains, can serve as a general platform for generating novel cell-cell contact signaling pathways. Synthetic Notch (synNotch) pathways can drive user-defined functional responses in diverse mammalian cell types. Because individual synNotch pathways do not share common signaling intermediates, the pathways are functionally orthogonal. Thus multiple synNotch receptors can be used in the same cell to achieve combinatorial integration of environmental cues, including Boolean response programs, multi-cellular signaling cascades, and self-organized cellular patterns. SynNotch receptors provide extraordinary flexibility in engineering cells with customized sensing/response behaviors to user-specified extracellular cues.
INTRODUCTION
In the emerging areas of synthetic biology and cell engineering, a fundamental goal is to be able to rationally change what extracellular cues a cell recognizes, as well as the resulting cellular response. Customized cell sensing/response pathways would be extremely useful for engineering therapeutic cells, allowing them to autonomously sense user-specified disease or injury signals, and to precisely deploy therapeutic or repair functions (Fischbach et al., 2013; Lienert et al., 2014; Slomovic et al., 2015). Customized cell sensing/response behaviors would also be useful tools for reporting on cell connectivity and environmental conditions. Novel cell-cell communication channels could also enable design of multicellular assemblies whose self-organization could be driven by specific cell-cell signaling networks. For these purposes we would like to have synthetic pathways for which input and output can be flexibly altered in a modular fashion. In addition, it would be ideal for such synthetic pathways to function orthogonally from endogenous pathways and one another, allowing for combinatorial input integration with little crosstalk.
Eukaryotic cells have evolved diverse transmembrane receptors that allow them to recognize extracellular molecules and induce intracellular responses. In most cases, the extracellular engagement of these receptors allosterically regulates an associated intracellular enzymatic activity (e.g. kinase or guanine nucleotide exchange factor) (Lim et al., 2014). The resulting enzyme and its substrates then transduce signals to various downstream modules, including transcriptional regulators that mediate global cellular response programs. It is challenging to rationally alter these complex enzyme-linked receptors and their downstream cascades in a way that leads to completely novel and orthogonal input/output linkages.
Thus, to generate synthetic pathways that would allow customizable sensing and response engineering, we turned to the Notch pathway, which is unique because of its very direct and simple mechanism of signal transduction (Kopan, 2002). Engagement of the Notch receptor with its ligand – Delta family proteins that are presented on the surface of partner cells – leads to intramembrane proteolysis (sequential proteolysis by ADAM metalloprotease and the gamma-secretase complex; Kopan and Ilagan, 2009). The induced cleavage of the receptor releases the intracellular fragment of Notch (Fig. 1A). This Notch intracellular domain is a transcriptional regulator that can only function when it is released from the membrane and can enter the nucleus to activate target genes that play key roles in cell-cell signaling during development (Artavanis-Tsakonas et al., 1999).
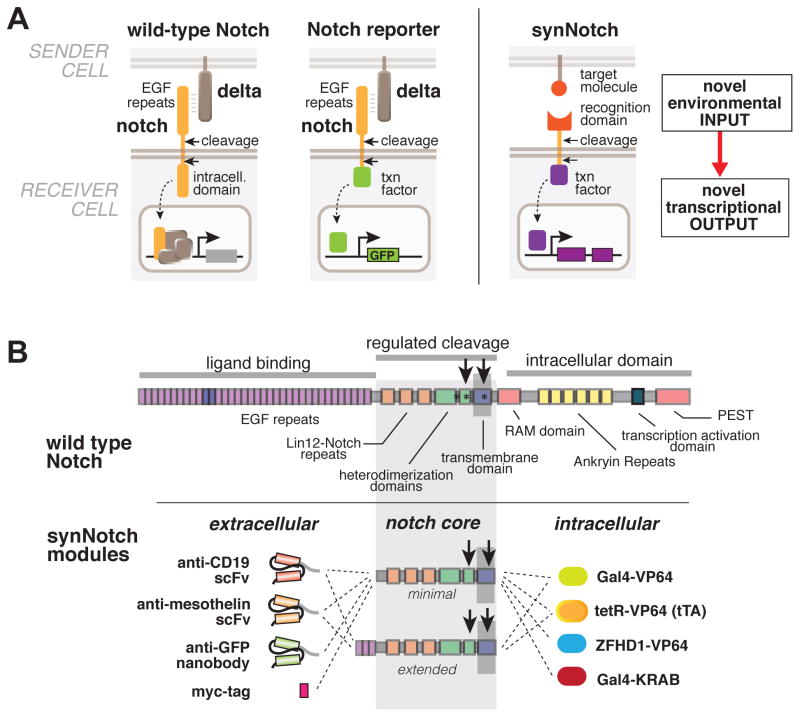
Figure 1. Modular Configuration of Synthetic Notch (SynNotch) Receptors.
(A) Conceptual design of synNotch receptor systems. Left: wild-type Notch has a large extracellular domain that binds to its ligand, Delta, expressed on opposing partner cells, and an intracellular transcriptional regulatory domain that is released by ligand induced cleavage. Arrows indicate the multiple proteolytic cleavage sites. Middle: Notch reporters have been built in which the intracellular domain is replaced by an orthogonal transcription factor. Right: in synNotch receptors both the extracellular and intracellular domains have been completely replaced, leaving only the small central regulatory region of Notch. Both novel inputs and outputs can be defined by using the synNotch architecture.
(B) Modularity of the synNotch platform: the input and output domains from Notch can be swapped with diverse domains. On the extracellular side, diverse recognition domains can be used (antibody based, or peptide tags are shown) and on the intracellular side, diverse effector can be used (transcriptional activators with different DNA-binding domains are shown, as well as a transcriptional repressor). See also Figure S1.
While the Notch proteolytic mechanism of sensing probably does not lead to significant signal amplification, it does appear to be quite flexible. Prior studies have demonstrated that the intracellular domain of Notch can be replaced with an artificial transcription factor (e.g. Gal4-VP64) to create a reporter of Notch activity (Lecourtois and Schweisguth, 1998; Struhl and Adachi, 1998) (Fig. 1A). Studies of the physical mechanism of Notch activation have also shown that the extracellular domain of Notch can be replaced by alternative domains (Gordon et al., 2015). The very direct mechanism of Notch signaling has also inspired the engineering of novel proteolytically induced receptors and reporter systems (Barnea et al., 2008; Daringer et al., 2014).
Given the apparent modularity of Notch receptors, we explored whether the Notch receptor could be used as a platform to generate synthetic signaling pathways in which both sensing and response were customized (Fig. 1A). Here we show that we can customize input sensing by swapping the extracellular recognition domain of these receptors, including the use of antibody-based domains (e.g. single-chain antibodies or nanobodies) to detect a wide range of user-specified cell surface proteins, such as disease antigens. Simultaneously, we can link these novel inputs to customized responses, by swapping the intracellular transcription domain and providing specific downstream effector target genes. The resulting synthetic Notch receptors only retain the minimal transmembrane core domain of the native Notch, which mediates control of proteolysis. These synthetic Notch receptors function in a range of cells, including immune cells and neurons.
Using synthetic Notch pathways, we can spatially direct induction of complex responses such as differentiation and pattern formation. We also show that synthetic Notch receptors, as long as they have distinct intracellular and extracellular domains, function orthogonally to one another, since they then share no common signaling intermediates. Thus, multiple synthetic Notch pathways can be deployed in the same cell, and can be used to engineer complex combinatorial sensing circuits. The flexibility of synthetic Notch receptors in engineering novel cell behaviors makes them powerful tools for constructing therapeutic cells (see companion paper, Roybal et al.), driving formation of complex multicellular patterns, or for modulating or reporting on cellular behavior in a complex in vivo milieu.
Synthetic Notch receptors, with their customizable input/output function, add a powerful and flexible sensing functionality to the mammalian synthetic biology toolbox. The great variety of synthetic intracellular transcriptional circuits that have previously been engineered can now be linked to the outside of the cell and controlled by user-defined extracellular inputs.
RESULTS
Minimal Notch regulatory domain can be combined with novel extra- and intra-cellular domains to construct diverse synthetic receptors
Although the Notch receptor is a large multidomain protein, the regulatory core of the receptor is centered around the transmembrane region, where the proteolytic cleavages take place to release the intracellular transcriptional domain (Gordon et al., 2007). Prior studies indicate that the intracellular and extracellular domains of Notch can be replaced (Gordon et al., 2015; Lecourtois and Schweisguth, 1998; Struhl and Adachi, 1998). Thus, we reasoned that this regulatory core could potentially be used as a platform for engineering diverse synthetic receptors in which both the extracellular input domain and then intracellular output domain were simultaneously changed (Figure 1B).
We generated libraries of receptor molecules with different extracellular domains each coupled with a different intracellular domain, and expressed them in fibroblasts, along with their downstream response elements (Figure 1B). We found that these synthetic receptors are strongly activated by cell-cell contact with sender cells expressing the cognate ligand on their membrane and by surface-bound ligand (Figure S1A and Figure 2A). For example, a synthetic Notch receptor containing an extracellular single chain antibody (scFv) for the B cell surface antigen CD19, shows no basal transcriptional reporter activity, but can be potently activated by stimulating with CD19 expressing cells. Similarly strong activation is also observed by a synthetic Notch receptor containing an extracellular anti-GFP nanobody: cells expressing this receptor show strongly induced reporter transcription when stimulated with sender cells with surface expressed GFP (Fig. S1A). To determine if the activation of synNotch receptors occurred by a cleavage mechanism similar to endogenous Notch, we tested the effect of blocking the gamma-secretase protease using the drug N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT). Treatment with DAPT completely blocks synNotch activation (Figure 2A).
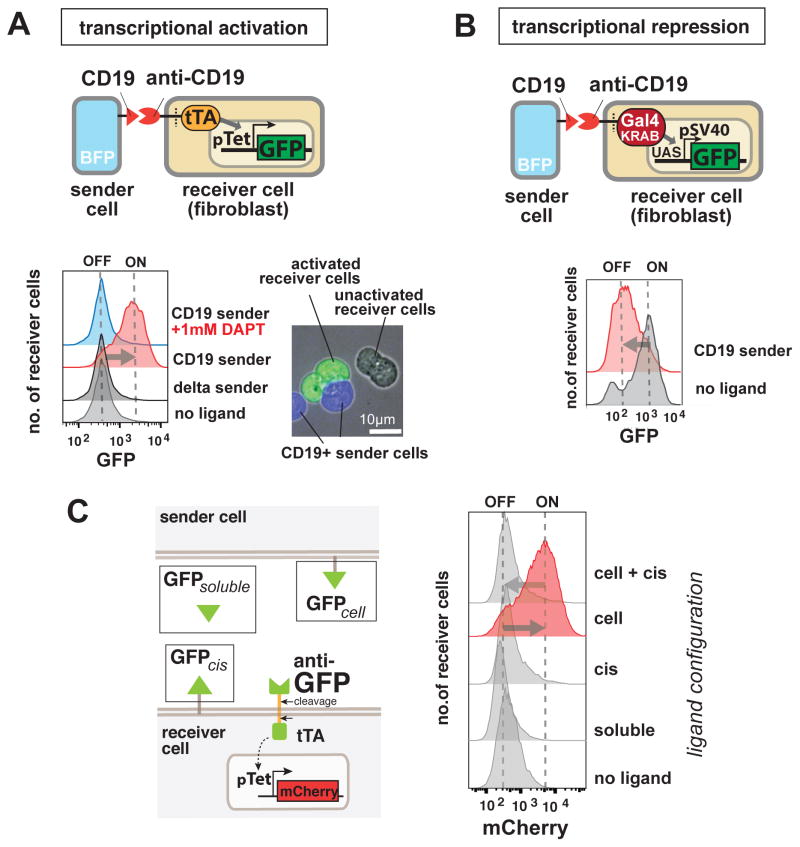
Figure 2. SynNotch Receptors can be used to Program Contact-Dependent Transcriptional Regulation.
(A) Synthetic Notch receptors can be used to detect endogenous disease antigens and induce the expression of a reporter gene. Mouse fibroblasts (L929 line) expressing anti-CD19/tTA synNotch are cultivated with K562 sender cells expressing Delta, CD19, or CD19 in the presence of the gamma-secretase inhibitor DAPT. FACS plots of the resulting GFP reporter intensity in receiver cells are shown. Inset shows an image of MDCK cells expressing the anti-CD19→ GFP synNotch, when co-cultivated for 24h with MDCK sender cells expressing CD19 (constitutively labeled with tagBFP). Only receiver cells in contact with (blue) sender cells activate the reporter and turn green.
(B) Mouse fibroblasts (L929 line) with anti-CD19 synNotch with a transcriptional repressor intracellular domain (Gal4-KRAB) are co-cultivated with K562 sender cells. The receiver cells constitutively express GFP downstream of a SV40/UAS combined promoter. FACS plot of receiver cells is shown, in presence of K562 sender cells with or without CD19 expression, as indicated in figure.
(C) Stimulation of mouse fibroblasts expressing anti-GFP synNotch with ligands in different formats. Anti-GFP synNotch receiver cells are stimulated for 1h with GFP either in soluble form, presented on a K562 sender-cell, or cis-presented on the receiver cell itself. The receiver cells show activation only when the ligand is present on an opposing surface and if they do not express the ligand in cis. The FACS data are recorded at 24h after the beginning of stimulation. See also Figure S2D for similar data on the anti-CD19 synNotch. FACS histograms include at least 10,000 cells for each condition.
A large number of the extracellular domains that we used yielded functional receptors (Fig. S1A), but for certain extracellular domains, we observed poor inducibility (e.g. α-integrin, E-cadherin, Ephrin), or high background activity, as was the case with a receptor with an scFv domain that recognized the tumor antigen mesothelin. We discovered that, by slightly extending the core Notch regulatory region to include one or more extracellular EGF-repeats, we could improve synthetic receptor function that would otherwise display high basal activation. The anti-mesothelin receptor displayed low basal activation when this extension was incorporated, but still was strongly activated upon stimulation (Figure S1B). In general, adding the EGF repeat decreased the basal activation of receptors, without reducing the induced state. Another way to control the output of these synthetic receptors is by changing the amount of receptor expressed, since the magnitude of induction correlates with expression (Fig. S1C).
The dose/response relationship between the ligand concentration and the receptor activation is graded, replicating a feature of the endogenous Notch signaling (Sprinzak et al., 2010) (Fig. S2A). We also characterized the dynamics of synNotch activation and found that significant transcriptional activity resulted from activation pulses as short as 1 hr (Fig. S2C). The synthetic receptor response is also reversible upon removal of the ligand expressing cells over a timescale of several hours (Fig. S2B), although the rate of decay will likely be a function of the stability of the specific effector proteins that are induced.
We tested the GFP-detecting synNotch receptor against ligands presented in different formats – soluble GFP, cell-surface expressed GFP (in trans – i.e. on opposing cell), and cis cell surface expressed (i.e. on same cell as the receptor) (Fig. 2C). We found that the synNotch receptor only transduces the signal when its ligand is presented on an opposing surface; no activation was detected when the cognate ligand was in solution or presented on the same surface as the receptor (cis). This is consistent with apparent need for the ligand to be presented in a manner that will exert force on the receptor. Interestingly, when the ligand is present in cis on the same cell surface that expresses the receptor, the synNotch receptors can no longer be activated by ligands presented in trans (Fig. 2C, Fig. S2D), mimicking a feature of native Notch known as cis-inhibition (Micchelli et al., 1997).
On the intracellular side, we show that we can use the synNotch platform to induce transcriptional repression as well as activation. We generated versions of the receptor with an intracellular transcriptional repressor domain (KRAB) fused to a Gal4 DNA-binding domain. The cells that express this synNotch receptor respond by down-regulating the reporter gene when co-cultivated with sender cells (Figure 2B).
We also inserted other intracellular domains into the synNotch platform, including Cre recombinases, and master transcription factors such as MyoD and Snail. While some of these showed regulated activity, in general these activities were quite low (data not shown), most likely because the synNotch receptor output domain functions stoichiometrically and does not, by itself, undergo amplification. Thus this receptor system seems to work most robustly with outputs that are intrinsically highly amplified.
Synthetic Notch receptors work in diverse primary cells: neurons and immune cells
To test if the synNotch receptor could function in diverse cells, we engineered a CD19-detecting synNotch receptor into a series of cell lines: epithelial MDCK cells, L929 and C3H mouse fibroblasts cell lines, HEK293 human epithelial cell lines, and Jurkat T cells. All of these cells showed clear induction of reporter gene activation upon synNotch receptor engagement (Fig. 3B, 4A–B, S1A). Importantly, we also showed that synNotch receptors could function in primary cells: primary hippocampal neurons that express the anti-CD19 synNotch can be induced to express a GFP reporter by contact with CD19-expressing sender cells (Figure 3A, Fig. S3). We also show in the accompanying paper that synNotch receptors function in primary human immune cells, and can act within the setting of live animals (Roybal et al.). These results indicate that a wide variety of cells have the necessary proteolytic machinery for synNotch activation.
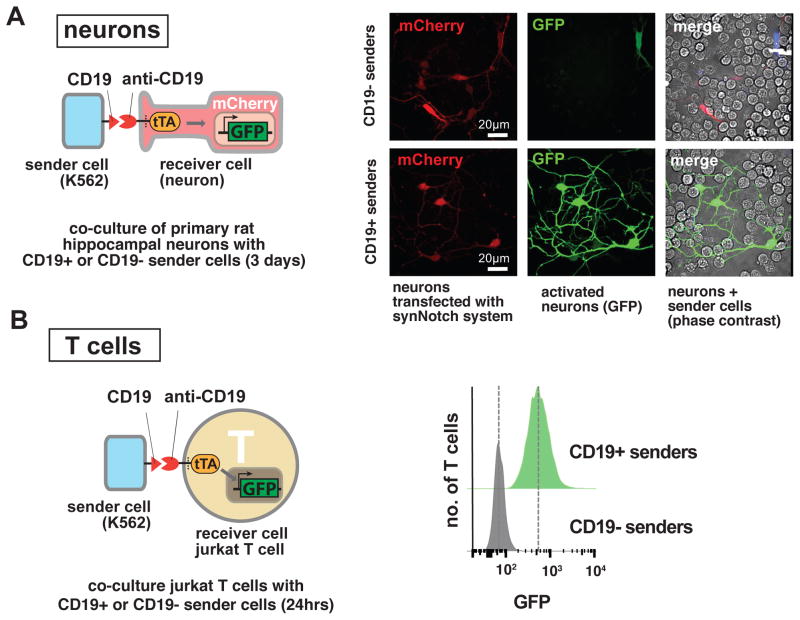
Figure 3. SynNotch Receptors Function in Diverse Cell Types, Including Neurons and Lymphocytes.
(A) Neurons. Primary hippocampal neurons were dissociated from E18 rat embryos and are nucleofected to express an anti-CD19 synNotch → GFP receptor and reporter. Neurons were plated on glass-bottom 35mm culture dish coated with Poly-D-Lysine and Laminin. 2 hours after neuron plating, sender cells (K562s) are added to the culture. Images are taken from live cells at day 4 after plating. On the right, representative images for neurons that are co-cultured with plain K562 cells (upper panel) or with CD19+ K562 sender cells (bottom panel) are shown. Neurons co-cultured with ligand presenting sender cells strongly induce GFP expression. See Figure S3 for quantification.
(B) T cell line. Jurkat T clonal cell line engineered to stably express an anti-CD19→ GFP synNotch receptor system. Data on the right show fluorescence of clonal Jurkat cell population upon stimulation with CD19+ or CD19- sender cells (K562s) at t=24h. T cells are activated only when they encounter cell with the cognate ligand. FACS histograms include at least 10,000 cells for each condition.
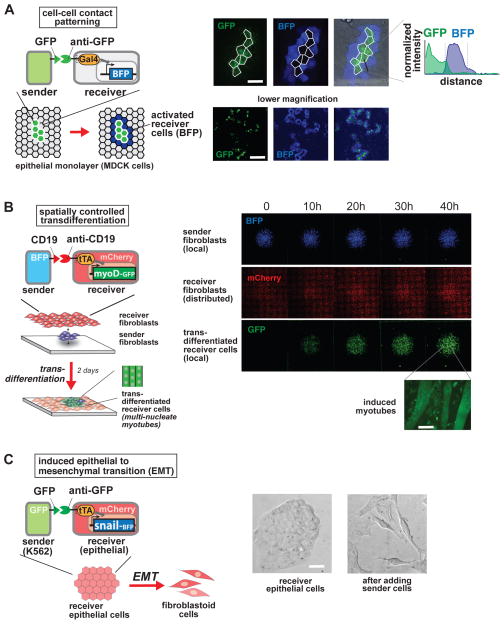
Figure 4. SynNotch Receptors Yield Spatial Control of Diverse Cellular Behaviors.
(A) Boundary detection in epithelial monolayer. Epithelial cells (MDCKs) are engineered as follows: sender cells express an extracellular GFP linked to a transmembrane domain; receiver cells are a clonal population that express the anti-GFP synNotch with LaG17 anti-GFP nanobody as extracellular domain, and Gal4-VP64 as intracellular domain, along with a UAS→BFP reporter construct. Sender cells are seeded into the receiver cell monolayer at a 1:50 ratio. Confocal images are taken 48h after plating of confluent monolayer. Representative pictures of high magnification and low magnification are shown, alongside with a representative line of intensity of BFP fluorescence over distance. Only receiver cells that are in contact with the green sender cells turn on the blue reporter, forming a ring around the sender cells. Receiver cells away from sender cells remain uninduced. Scale bars: higher magnification 20um; lower magnification 200um.
(B) SynNotch activation of a myogenesis master regulator (myoD) in fibroblasts induces transdifferentiation in a spatially controlled manner. C3H mouse fibroblasts are engineered as follows: sender cells express extracellular CD19 linked to a transmembrane domain, plus a tagBFP marker; receiver cells express the anti-CD19 synNotch with tTA intracellular domain, along with a TRE→myoD cassette and a constitutive mCherry marker. Sender fibroblasts (blue) are plated first in a limited region of the plate and allowed to adhere to the plate; after 1h the receiver cells (red) are plated to uniformly cover the entire glass plate. Images show a large area of the co-culture and are still-frames from a movie that span the first 48h after co-plating (See also Movie 1). GFP channel shows the induction of myoD-GFP in received cells in a region that overlaps with the blue channel (sender cells). Receiver cells away from sender cells remain uninduced, and provide an internal control for the experiment (See also Figure S4A). A higher magnification of the field for the green channel is shown on the right, showing the induction of multinucleate myotubes (scale bar = 50um).
(C) SynNotch can induce epithelial to mesenchymal transition in cultured epithelial cells. Epithelial cells (MDCKs) are engineered as follows: receiver cells express the anti-GFP synNotch with LaG17 anti-GFP nanobody as extracellular domain, and tTA as intracellular domain, along with a TRE→Snail-ires-BFP effector construct. Sender cells are GFP-expressing K562 cells. Representative bright field microscope images of epithelial cells before and after 48h from the addition of sender cells are shown (sender cells were removed before imaging). Scale bar = 20um. See Figure S4B for quantification and controls.
Synthetic Notch receptors can induce cell fate changes in a spatially controlled manner
SynNotch receiver cells can detect whether a contacting cell is expressing the ligand or not. We tested if the contact-dependent feature of the signaling can be used to induce highly local, spatially controlled responses. Control of cell response with a short-range signaling channel, such as synNotch, would, in principle, allow the user to specify cell behavior in a highly localized manner within a multicellular tissue. We first tested whether synNotch receiver cells placed in an epithelial cell layer could be spatially induced to express a reporter gene. To do so, we plated a confluent layer of epithelial cells expressing the anti-GFP synNotch receptor (receivers), seeding the layer with a small number of GFP expressing cells (senders). As shown in Figure 4A, the BFP reporter gene is activated only in the receiver cells that are in direct contact with the GFP expressing sender cells, forming a blue ring pattern around each island of GFP cells. In this way, the monolayer of receiver cells is patterned in two cell populations, an activated population that highlights the boundary of the islands of sender cells, and an inactive population comprising all the cells in between the islands of sender cells.
We then wanted to test whether this synthetic cell-cell signaling could be used to regulate cell fate in a spatially controlled manner. We asked whether synNotch receptors could be used to induce transdifferentiation of fibroblasts to myoblasts, by inducing expression of MyoD, the prototypical master transcription factor for myogenesis (Davis et al., 1987). We engineered fibroblasts to express an anti-CD19 synNotch receptor with a tTa intracellular domain, along with the myoD gene expressed from the cognate tetracycline response element (TRE) promoter. We also engineered sender fibroblasts expressing the CD19 surface ligand. To test for spatial cell fate induction, we plated a small spot of sender cells, and then overlaid these with a confluent layer of the synNotch receiver cells. We found that only the synNotch receiver fibroblasts in contact with the cognate cells (blue cells in Fig. 4B) induced the expression of MyoD (detected via a fused GFP label). The induced expression of MyoD led to localized transdifferentiation and the formation of myotubes over the course of ~2 days (Figure 4B and S4A; see Supplemental Movie 1 for time-course). Thus we observed spatially directed differentiation into myotubes.
In another example, we demonstrated that synNotch could be used to control epithelial-to-mesenchymal transitions (EMT). Here we used the anti-GFP synNotch receptor to control expression of the EMT master regulator gene, Snail (Cano et al., 2000). In these experiments, epithelial receiver cells (MDCKs) have the anti-GFP synNotch that induces expression of Snail from a TRE promoter. We can induce EMT in these receiver cells by exposing them to cells expressing the cognate ligand – surface displayed GFP (Figure 4C and S4B). The observed dissociation of the receiver cells (Fig. 4C) is mirrored by a downregulation of cadherin expression (Fig. S4B). The induction of EMT in receiver cells is not observed when they are exposed to control sender cells that do not express surface GFP (Fig. S4B).
Synthetic Notch pathways are orthogonal to each other and native Notch
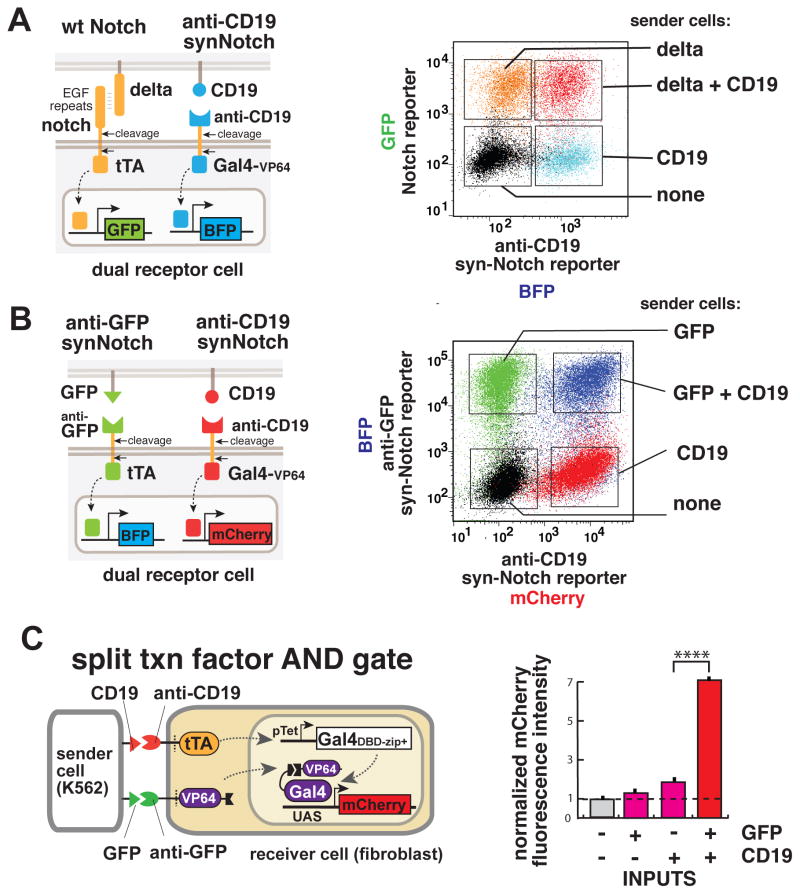
We wanted to explore whether multiple synNotch receptors could function within the same cell without crosstalk. Orthogonal function would allow a cell to elaborate different outputs according to the presence or absence of multiple inputs. We hypothesized that synNotch receptors might function orthogonally to one another because of their very simple mechanism of signaling – there are no common intermediates (e.g. an activated kinase or kinase substrates) that could yield cross-talk, as long as the intracellular transcriptional regulators in the different receptors are distinct. We first tested if there was any cross talk between the synNotch and the endogenous Notch signaling. We used one version of Notch containing the full endogenous extracellular domain (which recognizes the natural ligand Delta) and a tTa intracellular transcriptional domain. This receptor drives GFP expression. We also used a second synNotch receptor that recognizes CD19, and drives tagBFP expression (via intracellular Gal4-VP64 transcriptional domain). We engineered fibroblasts to express both receptor systems simultaneously. When we stimulated the receiver cells with only Delta, only the full-length-Notch reporter (GFP) was activated. When the same receiver cells were stimulated with sender cells expressing CD19 (the synNotch ligand), the receiver cells only induced the synNotch reporter tagBFP. Both reporters are induced with dual ligand stimulation. Thus the synNotch and the endogenous Notch pathways display independent but compatible activation (Figure 5A and S5A).
Figure 5. SynNotch Pathways are Orthogonal to One Another and Can be Used for Combinatorial Regulation.
(A) SynNotch and wild-type Notch activate orthogonal signaling pathways. L929 mouse fibroblasts receivers are engineered to express (i) the wild-type Notch receptor with a tTA intracellular domain and a TRE→GFP reporter, and (ii) a synNotch receptor with anti-CD19 extracellular domain and Gal4-VP64 intracellular domain, and a UAS→tagBFP reporter. The graph on the right shows the clonal population of receiver cells fluorescence signal for the BFP and the GFP reporters in different conditions: black - untreated; blue - stimulated with CD19 expressing senders; orange - stimulated with delta senders; and red - stimulated with sender cells expressing both CD19 and delta. Sender cells are mouse L929 fibroblasts. See Figure S5A for quantification.
(B) Multiple synNotch receptors are orthogonal to one another. L929 mouse fibroblast receiver cells are engineered to express (i) the anti-CD19 synNotch receptor with a tTA intracellular domain and a TRE→BFP reporter; and also (ii) the synNotch receptor with anti-CD19 extracellular domain and Gal4-VP64 intracellular domain, and a UAS→mCherry reporter. The graph on the right shows the receiver cell (clonal population) fluorescence signal for the BFP and the mCherry reporters in different conditions: black - untreated cells; red - stimulated with CD19 expressing sender cells; green - stimulated with GFP sender cells; and blue - stimulated with sender cells expressing both GFP and CD19. Sender cells are K562 cells. See Figure S5B for quantification.
(C) Cells engineered with two synNotch AND gate respond only when both the inputs are present. L929 mouse fibroblasts receivers are engineered to express: (i) the anti-CD19 synNotch receptor with a tTA intracellular domain and a TRE promoter that drives the expression of the DNA-binding domain (DBD) of Gal4 fused to a leucine zipper domain, and (ii) the synNotch receptor with anti-GFP extracellular domain and the VP64 transcriptional activation domain fused to a complementary leucine zipper as intracellular domain, and (iii) a Gal4-responsive promoter driving a red fluorescent protein (mCherry). The graph on the right shows the normalized mCherry fluorescence collected from a clonal population of receiver cells in co-culture with different sender cells (K562), that express either the two ligands alone (GFP or CD19), or both ligands together. Activation occurs only in the presence of both the inputs. Data shown are the median and coefficient of variance of at least 10,000 cells per condition.
We then tested whether two different synNotch pathways could function independently in the same cell. To do that, we engineered double synNotch receiver cells with both an anti-CD19 receptor and an anti-GFP receptor, each linked to a different intracellular transcription activation domain (Gal4-VP64 and tTA respectively) that in turn drives a distinct reporter fluorescent protein (mCherry and tagBFP respectively). In Figure 5B we show the results of stimulation of the double-synNotch receiver cells with cells expressing the two ligands CD19 and GFP separately or combined. When activated by CD19, only the anti-CD19-synNotch response is activated. Conversely, when activated by GFP-expressing sender cells, only the anti-GFP-synNotch response is activated. Importantly, when the cells are stimulated by both input ligands (CD19 and GFP) the two responses are activated together (Fig. 5B and S5B). Thus multiple synNotch pathways can work in the same cell as insulated and independent signal transduction pathways, enabling us to engineer cells that respond to multiple stimuli with distinct user-defined transcriptional programs.
Multiple synNotch receptors can be used to engineer cells that combinatorially integrate multiple inputs
We showed that multiple synNotch receptors can be used in the same cell to generate independent responses with no crosstalk. Thus we reasoned that we could use these receptors to engineer cells that integrate combinatorial environmental cues and respond only when certain dual criteria are met. In particular, we focused on the generation of cells that would respond only in the presence of two different contacting extracellular ligands (not responding to each ligand alone). We thus engineered cells that expressed both the anti-CD19 and anti-GFP synNotch receptors. But in this case, each receptor controls one half of a split transcription factor (Luan et al., 2006) – the anti-CD19 synNotch receptor drives expression of a Gal4 DNA binding domain fused to a leucine zipper dimerization domain, while the anti-GFP synNotch receptor contains as its intracellular domain the VP64 transcriptional activation domain fused to a complementary leucine zipper dimerization domain. To report on the presence of a fully functional Gal4 DBD-zipper:zipper-VP64 complex, these cells also contain a reporter gene (mCherry) under control a Gal4 UAS promoter (Fig. 5C). These double synNotch cells were stimulated with sender cells expressing either one or both of the input ligands. When a single input is presented to the receiver cells, no activation is visible (Figure 5C, columns 1–3). Only when the sender cells express both the antigens was the response in receiver cells induced (Figure 5C, last column). Thus we can use synNotch receptors in this type of split output circuit architecture to construct cellular AND-gate pathways that are dependent on the concomitant stimulation with two combinatorial inputs.
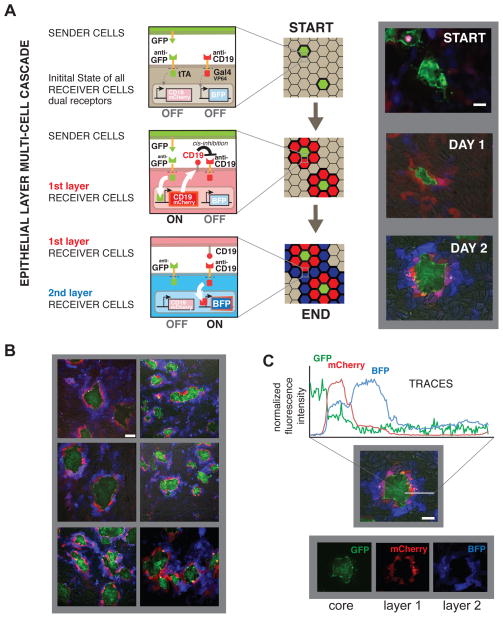
Engineering cascades of cell-cell signaling with multiple synNotch receptors
With multiple synNotch receptors, we should also be able to generate more complex pattern formation circuits involving cascades of cell-cell communication. Here we focused on inducing a self-organized multi-layer spatial pattern in epithelial cells. For this experiment, we constructed receiver cells expressing two different synNotch receptors that could potentially act in series – the first receptor induces the expression of the ligand for the second receptor. In particular, we used the anti-GFP-synNotch receptor to induce CD19 ligand expression (as well the reporter protein mCherry), and the anti-CD19-synNotch receptor to induce the reporter protein tagBFP.
To then start the induction of the cell-cell signaling cascade, GFP sender cells were seeded sparsely in a monolayer of the double synNotch receiver cells (Fig. 6). We observed that within the first day of plating the cells, the neighbors closest to the GFP sender cells become red (mCherry) reporting on the activation of the anti-GFP-synNotch (Fig. 6A, DAY 1). This first layer of receiver cells is also simultaneously inducing the expression of surface CD19. The second layer of receiver cells can then detect the CD19 that is now expressed on the first layer of receiver cells. In response, the second layer of receiver cells induces activation of the tagBFP reporter (via their anti CD19 synNotch receptor) (Figure 6A, DAY 2).
Figure 6. Multiple SynNotch Receptors can be used to Generate Multi-Layered Self-Organizing Epithelial Patterns.
Epithelial cells (MDCKs) are engineered as follows: sender cells express extracellular GFP linked to a transmembrane domain; receiver cells express two synNotch receptors: (i) the anti-GFP synNotch with tTA intracellular domain, along with a TRE→CD19-mCherry effector cassette; (ii) the anti-CD19 receptor with Gal4-VP64 intracellular domain, and a UAS→tagBFP reporter. Here, the first synNotch receptor, when stimulated, induced the expression of the ligand for the second synNotch receptor. (A) Representative images are shown for the epithelial layer of sender cells and a clonal population of receiver cells co-cultivated at a 1:50 ratio for 10h (START) 34h (DAY 1) and 58h (DAY2). Scale bar = 20um. (B) Multiple images of different fields of view of the co-culture at day 2. Scale bar = 50um (C) Representative quantification of the fluorescence signal as calculated from the fluorescence images for a pattern around sender cells at day 2. Scale bar = 20um.
This engineered cascade of cell-cell interactions ultimately generates a double-ring pattern of red first (mCherry), and then blue (tagBFP) receiver cells around the green sender cells (GFP) (Fig. 6B). In this way, two different cell types are dynamically produced in a spatially controlled manner within an otherwise identical population of receiver cells (Fig. 6C). Thus, multiple synNotch receptors can be used to engineer complex, multistep cell-cell signaling cascades that result in pattern formation, akin to what is observed in natural developmental programs.
DISCUSSION
Modularity of the Notch receptor and the mechanism of synNotch signaling
We have demonstrated that it is possible to engineer highly diverse forms of synthetic Notch receptors, for which one can change what contact ligands the cell detects, and what cellular responses occur upon engagement. Inputs are specified by what extracellular recognition domain is used, while outputs are determined by what intracellular transcriptional regulator is used and what effector genes this regulator drives. The wide variety of possible synNotch receptor architectures and domain combinations yields powerful and highly flexible tools for engineering cell-cell communication. We demonstrate that cells expressing the synNotch receptors are sensitive to new inputs and perform diverse user-defined actions when the cells are stimulated by these new inputs. SynNotch receptors can be used to drive diverse output functions, including transcriptional activation, repression, and differentiation. Moreover, because these interactions are, like the endogenous Delta-Notch interaction, dependent on direct cell-cell contact, the regulatory responses generated by synNotch receptors yield precise spatial control. SynNotch receptors work in a wide variety of cell types, consistent with the widespread expression of the required regulatory proteases that mediate Notch cleavage.
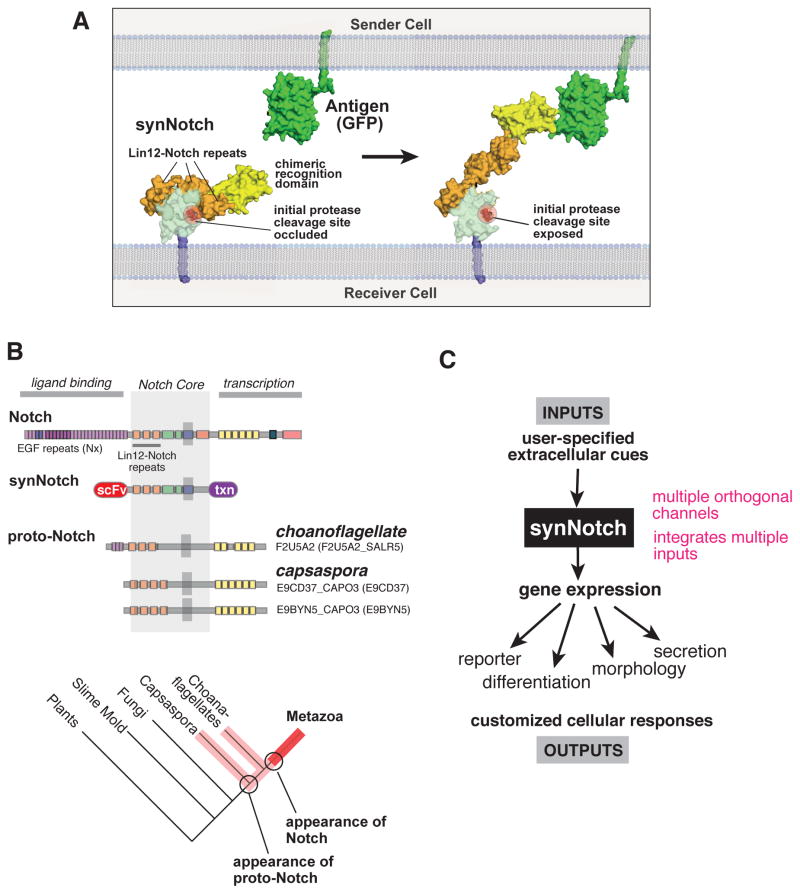
These new receptors highlight the extraordinary functional modularity of the Notch receptor. Endogenous Notch is composed of three parts: an extracellular ligand-binding domain, a regulatory domain (including the transmembrane region) where the core signal transduction occurs, and the intracellular effector domain. Work by Blacklow and colleagues suggests that transduction in the core Notch regulatory region occurs through a physical pulling mechanism (Gordon et al., 2007; 2015). Within the core regulatory region, directly adjacent to the transmembrane region, are three Lin-12 Notch Repeats (LNRs), which in the basal state bind and shield the ADAM/TACE protease cleavage site (the initial of two protease cleavage events required to ultimately release the intracellular domain of Notch). Pulling forces generated by binding of the extracellular domain to a ligand presented on an opposing cell surface are thought to release the LNR domains, exposing the ADAM/TACE protease cleavage site. The protease, which is constitutively active in the membrane, can then initiate the cleavage and activation of Notch.
One of the sources of the pulling force in Notch/Delta is thought to be the endocytosis of the Delta ligand in the sender cells (Pratt et al., 2011; Yamamoto et al., 2010). It is unclear whether ligand endocytosis is required for synNotch receptor activation, since surface bound ligands are sufficient for activation, and we do not observe the need to include endocytosis motifs in the ligand constructs. It is possible, however, that endocytosis does in some cases contribute to stronger activation of certain synNotch receptors.
We hypothesize that in synNotch receptors, the autoinhibition within the regulatory region is maintained, but that engagement of the chimeric extracellular domain to a surface displayed ligand is sufficient to pull and displace this autoinhibition, exposing the cleavage site (Fig. 7A). Given this model, it is not surprising that for some extracellular recognition domains, it may be necessary to optimize the receptor by extending or contracting the linker between the extracellular and regulatory domains – proper accessibility and force extension to the regulatory domain will likely have some geometric constraints. Nonetheless, we show that extracellular domains ranging from a single chain antibody to a small peptide epitope, can function as the synNotch recognition domain.
Figure 7. Modularity of SynNotch Receptors Expands Sensing/Response Engineering of Mammalian Cells.
(A) Putative mechanism of activation of the synNotch receptors. As shown by Blacklow and others (Gordon et al., 2007), in native Notch, the LNR domains mask the protease cleavage site in the unbound conformation (left). When the ligand engages the receptor, mechanical force is thought to expose this protease site, initiating the multi-step process leading to release of the intracellular transcriptional domain (right). We propose that when the extracellular domain is replaced by a novel recognition domain (here an anti-GFP nanobody is shown), then proper cell-cell engagement of the cognate ligand (GFP on the opposing cell) can exert a similar force to expose the Notch core to proteolysis.
(B) Alignment of LNR-containing molecules from different metazoan and pre-metazoan species shows that full length Notch (with delta binding domain) is only found in metazoans, but that proto-Notch genes that lack the delta binding domain are found in pre-metazoans such as choanoflagellates and capsaspora (Gazave et al., 2009; King et al., 2008).
(C) The modularity of the synNotch receptor platform allows the user to specify the extracellular cues the cells now respond to, as well as the cellular responses that are induced downstream of receptor activation.
Evolution of Notch signaling
It is striking that despite this remarkable modularity, there is not a wide range of naturally occurring Notch receptor homologs that mimic the diversity of synNotch receptors that we can construct. Unlike tyrosine kinase receptors (of which humans have ~50 diverse family members), there are only 1–4 closely related versions of Notch in most metazoan species (Blume-Jensen and Hunter, 2001; Kovall and Blacklow, 2010). Thus although this receptor has a high degree of potential functional modularity, this modularity appears to have been almost completely untapped by evolution, at least in species that have been sequenced (there are, however, many mechanisms to modulate both Delta and Notch). It is possible that since Notch-Delta signaling functions only through highly localized direct cell contact, there was actually less need for a highly diversified set of non-crossreacting ligand-receptor pairs, as is the case of soluble signaling systems. Notch-Delta can and is heavily reused by diverse cell pairs during development.
We have noted that pre-multicellular organisms, such as choanoflagellates and capsaspora, have highly simplified versions of Notch that are similar to the core of synNotch (Fig. 7B) – in particular they lack the large extracellular domain of metazoan Notch that is necessary to recognize Delta (Gazave et al., 2009). These pre-metazoan species also lack Delta, leading to the hypothesis that perhaps other quite different interactions might regulate this proto-Notch molecule. It has been speculated that the evolution of the modern Notch-Delta signaling pair was one of the drivers of modern metazoan multicellularity (Gazave et al., 2009; King et al., 2008). It is interesting that going back to a core framework that is similar to the extremely simplified ancestral Notch, we are able to then re-elaborate the system using alternative extracellular and intracellular domains to design highly diverse user-defined cell-cell interactions.
New capabilities in engineering custom extracellular sensing and response programs
A major goal in synthetic biology is the predictive forward design and control of cell behavior. Because of the highly modular nature of transcriptional regulatory components, many synthetic biology efforts in the last decade have focused on engineering very sophisticated transcriptional circuits (Purnick and Weiss, 2009). On the other hand, tissue engineers have focused on engineering extracellular scaffolds or stimulation protocols that can influence cell fate and behavior by tapping into through endogenous cell surface receptors (Huch and Koo, 2015; Sasai, 2013; Webber et al., 2014). What has been missing is an easily customizable way to link changes in intracellular signal processing to user-defined extracellular signals. Linking novel extracellular inputs to intracellular decisions is extremely challenging because there are limited ways to transmit extracellular information across the plasma membrane. This task is particularly daunting if one wants to build a system that is orthogonal to endogenous signaling receptors.
Initial successes in receptor engineering have come from focused efforts often based on surgical mutational changes. Recently, more modular solutions for engineering new extracellular sensing have emerged. For example, chimeric antigen receptors (CARs) have been developed that link an extracellular antigen sensing antibody domain with internal signaling domains from the T cell receptor. With the CAR system, the researchers are now able to link novel user-specified disease antigen inputs to synthetic activation of the native T-cell activation program - these engineered T cells can kill tumor cells that express the cognate antigen, and have proven to be clinically effective for B cell cancers (Miller and Sadelain, 2015). Despite the power of CARs, these still represent changing only the INPUT that the cell senses - the OUTPUT is still the full T cell activation program.
The synNotch receptors therefore bring engineering of extracellular sensing by mammalian cells to a new level. Not only can one use them to flexibly detect new inputs, but because the output domain and response are also highly modular, the overall response that can be generated can also be highly customizable. We show in the accompanying manuscript (Roybal et al.) how this flexibility can be used to engineer novel, highly customized T cell response behaviors. Overall we believe that synNotch receptors may prove to be very useful in therapeutic cells, such as engineered immune cells and in regenerative medicine, both of which require specific sensing of environmental signals linked to precise cellular responses. These receptors may also prove to be useful tools for mapping cell interactions during development or in the brain. The fact that we can generate many orthogonal versions makes them highly extensible for mapping multiple interactions. These synthetic receptors may also prove to be useful tools for systematically perturbing particular cell-cell interactions to better understand complex developmental processes.
MATERIAL AND METHODS
SynNotch Receptor and Response Element Construct Design
SynNotch receptors were built by fusing the CD19 scFv (Grupp et al., 2013), mesothelin scFv (Chowdhury et al., 1998), LaG17 (lower affinity), or LaG16_2 (high affinity) nanobody (Fridy et al., 2014) to the mouse Notch1 (NM_008714) minimal regulatory region (Ile1427 to Arg1752) or extended regulatory region (Pro1390 to Arg1752) and Gal4-VP64, Gal4-KRAB (Zalatan et al., 2015), tTA (gift from Miki Ebisuya) or ZFHD1 (N205 nLV EF-1a-ZFHD1-link-FKBP-HA <T2A> HP1aCS-Frbx2-V5-PGK-Blast was a gift from Jerry Crabtree (Addgene plasmid # 44017), (Hathaway et al., 2012)). All synNotch receptors contain an N-terminal CD8α signal peptide (MALPVTALLLPLALLLHAARP) for membrane targeting and a myc-tag (EQKLISEEDL) for easy determination of surface expression with α-myc A647 (cell-signaling #2233). For a list of aminoacid sequences of the constructs used, see Table S1. Full length Notch with tTA intracellular domain and Delta are from plasmids from the Ebisuya Lab (Matsuda et al., 2012). All the receptors were cloned into a modified pHR’SIN:CSW vector containing a SFFV promoter. The pHR’SIN:CSW vector was also modified to make the response element plasmids. Five copies of the Gal4 DNA binding domain target sequence (GGAGCACTGTCCTCCGAACG), seven copies of the tetracycline responsive element (TCCCTATCAGTGATAGAGA) and 4 copies of the ZFHD1 responsive elements (TAATGATGGGCG) were cloned 5′ to a minimal CMV promoter. The codon optimized cDNA sequence coding for mouse Snail (NP_035557.1), mouse myoD (NP_034996.2), was cloned into a MCS downstream of the indicated inducible promoter and 5′ of the indicated fluorescent protein reporter. All constructs were cloned via In fusion cloning (Clontech #ST0345). The sequences for the split Gal4 transcription factor are from (Luan et al., 2006).
Lentivirus Production
Lentivirus was produced by cotransfecting the pHR plasmids and vectors encoding packaging proteins (pMD2.G and p8.91) using the Fugene 6 HD transfection reagent in HEK293-T cells plated in 6-well plates at approximately 70% confluence. Viral supernatants were collected 2 days after transfection and 0.45 μm filtered. Supernatant was used for transduction immediately, stored at 4 degrees for up to 2 weeks, or kept at −80 degrees for long-term storage.
Cell Lines
L929 mouse fibroblast cells (ATCC# CCL-1), HEK293 cells, C3H/10T1/2 Clone 8 (ATCC# CCL-226) were cultured in 10% fetal bovine serum (UCSF Cell Culture Facility) in DMEM (Invitrogen) with penicillin, streptomycin and glutamine. MDCK cells were a gift from the Mostov Lab at UCSF; they were cultured in MEM (UCSF Cell Culture Facility) with 5% fetal bovine serum (UCSF Cell Culture Facility). K562s were lentivirally transduced to stably express human CD19 or mesothelin. K562s were also transduced to stably express surface GFP (GFP fused to the PDGFR transmembrane domain). All cell lines were sorted for expression of the transgenes.
All cell culture were maintained in a humidified incubator at 37% with 5% CO2, and at 37°C with 5% CO2 in a humidified incubator.
For viral transduction, cells were plated in 6-well dishes to achieve approximately 10% confluence at the time of infection. For lentiviral transduction, 10–100 μl of each virus’ supernatant was added directly to cells, with polybrene to increase infection efficiency. Viral media was replaced with normal growth media 24 hr post infection. Cells were sorted for coexpression of each component of the pathways via fluorescence-activated cell sorting on a FacsAria2 (Beckton-Dickinson), by staining for the appropriate tag with fluorescently tagged antibody where needed. A bulk-sorted population consisting of fluorescence positive cells was used for experiment, unless otherwise noted.
For single cell clonal population establishment, single cell were sorted in 96 wells plates starting from populations of cells infected with lentiviral particles for the relevant expression constructs; the sorting was performed with a FACS ARIA II.
Neuron Culture
Rat hippocampus neurons are dissected and disassociated from E18 rat embryos, and are transected with Lonza Amaxa nucleofector before plating on Poly-D-lysine and Laminin coated glass bottomed cell culture dish. Ligand expressing or plain (control) K562 cells are plated together with neurons in neural culture medium (neural basal medium supplemented with Glutamax, B27 and 5%FBS). Microscope images are taken 4 days after plating.
GFP Purification
For soluble ligand stimulation experiments, green fluorescent protein (GFP) was purified as an N-terminal hexahistidine fusion protein. To express protein, BL21(T1R) E. coli cells were grown to an OD of 0.6 from a fresh transformation, chilled to 18°C, induced with 0.8 mM IPTG, and allowed to express overnight. The proteins were purified by Ni-NTA affinity chromatography following the manufacturer’s (Qiagen) instructions. The protein was further purified by gel-filtration chromatography on a Superdex S200 10/300 equilibrated in PBS. The pure fractions were concentrated, aliquoted, flash frozen, and stored at −80 C until use.
Western Blots
For Western Blots, 5×10^6 MDCK cells were plated in wells of a 6-well plate; at appropriate time points after stimulation, wells were washed once with ice-cold PBS and lysed in 150 μl ice-cold lysis buffer (RIPA buffer, Pierce 89900, and freshly added protease inhibitors from Roche Applied Science Cat. #04906837001) and incubated on ice with occasional shaking for 5–10 min. Cells and lysis buffer were scraped off each well, collected in 1.5 ml Eppendorf tubes and centrifuged at 13,000 g at 4°C for 15 min. 90 μl were transferred to screw-top Eppendorf tubes and stored at −80°C.
SDS sample buffer (Novex NuPAGE with 2.5% 2-mercaptoethanol) was added and samples were boiled for 5 min and loaded immediately onto gels for western blot processing. Lysates were loaded into 4–12% Bis-Tris, 15 well, gels in MOPS buffer (Novex, Life Technologies) and run at 90V/140V for a total of 1 hr. Gels were transferred for 1 hr onto nitrocellulose membranes using a Trans-Blot semi-dry transfer apparatus (Bio-Rad). Membranes were blocked in Odyssey blocking buffer (Li-Cor) for > 1 hr, exposed to diluted primary antibodies in Odyssey buffer for 2 hr at room temperature or overnight at 4°C, washed in TBST, and exposed to IRDye (Li-Cor) fluorescent anti-rabbit (800CW, 1:15,000) and anti-mouse (680CT, 1:20,000) secondary antibodies in Odyssey buffer for 1.5 hr at room temperature. Membranes were then imaged on a Li-Cor imaging station and band intensities quantified in ImageJ. Primary antibodies used were anti-E-cadherin (Cell Signaling #3195 diluted 1:1000) and anti-alpha-tubulin (Cell Signaling #3873, diluted 1:2000).
Image Analysis
To quantify the profiles of fluorescence intensity in microscope images, the function of NIS Elements has been used to average the intensity in a rectangular selection with the width of 3 pixels.
Statistical Analysis
Statistical significance was determined by Student’s t-test (two-tailed). All statistical analysis and curve fitting was performed with Prism 6 (Graphpad) and p values are reported (*** = p ≤ 0.001, **** = p ≤ 0.0001).
Supplementary Material
Acknowledgments
We would like to thank members of the Lim lab for helpful discussions and comments on the manuscript. We thank K. McNally and Joan Garbarino for technical assistance, and R. Nicoll for neurons. This work was supported by a Jane Coffin Childs Memorial Fund Postdoctoral Fellowship A121505 (K.T.R.), a Human Frontiers of Science Program (HFSP) and a European Molecular Biology Organization (EMBO) Postdoctoral Fellowship (L.M.), and NIH grants K99 1K99EB021030 (L.M.), PN2 EY016546, P50FM081879, R01 GM055040, R01 CA196277, and the Howard Hughes Medical Institute (W.A.L).
Footnotes
AUTHOR CONTRIBUTIONS
L.M., K.T.R., and W.A.L conceived and designed the experiments. L.M. and K.T.R. performed the experiments and analyzed the data. X.X. performed the experiments with primary neurons. R.M.G. helped with the protein engineering. S.M.C. performed protein purification and the evolutionary analysis. M.T. provided guidance with transdifferentiation experiments. L.M., K.T.R. and W.A.L. wrote and edited the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Barnea G, Strapps W, Herrada G, Berman Y, Ong J, Kloss B, Axel R, Lee KJ. The genetic design of signaling cascades to record receptor activation. Proc Natl Acad Sci USa. 2008;105:64–69. doi: 10.1073/pnas.0710487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor Snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nature Cell Biology. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Chowdhury PS, Viner JL, Beers R, Pastan I. Isolation of a high-affinity stable single-chain Fv specific for mesothelin from DNA-immunized mice by phage display and construction of a recombinant immunotoxin with anti-tumor activity. Pnas. 1998;95:669–674. doi: 10.1073/pnas.95.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daringer NM, Dudek RM, Schwarz KA, Leonard JN. Modular Extracellular Sensor Architecture for Engineering Mammalian Cell-based Devices. ACS Synth Biol. 2014;3:892–902. doi: 10.1021/sb400128g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Fischbach MA, Bluestone JA, Lim WA. Cell-based therapeutics: the next pillar of medicine. Sci Transl Med. 2013;5:179ps7–179ps7. doi: 10.1126/scitranslmed.3005568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridy PC, Li Y, Keegan S, Thompson MK, Nudelman I, Scheid JF, Oeffinger M, Nussenzweig MC, Fenyö D, Chait BT, et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nature Methods. 2014;11:1253–1260. doi: 10.1038/nmeth.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazave E, Lapébie P, Richards GS, Brunet F, Ereskovsky AV, Degnan BM, Borchiellini C, Vervoort M, Renard E. Origin and evolution of the Notch signalling pathway: an overview from eukaryotic genomes. BMC Evol Biol. 2009;9:249–27. doi: 10.1186/1471-2148-9-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- Gordon WR, Zimmerman B, He L, Miles LJ, Huang J, Tiyanont K, McArthur DG, Aster JC, Perrimon N, Loparo JJ, et al. Mechanical Allostery: Evidence for a Force Requirement in the Proteolytic Activation of Notch. Developmental Cell. 2015;33:729–736. doi: 10.1016/j.devcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grupp SA, Kalos M, Barrett D, Aplenc R, Porter DL, Rheingold SR, Teachey DT, Chew A, Hauck B, Wright JF, et al. Chimeric Antigen Receptor–Modified T Cells for Acute Lymphoid Leukemia. New England Journal of Medicine. 2013;368:1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hathaway NA, Bell O, Hodges C, Miller EL, Neel DS, Crabtree GR. Dynamics and Memory of Heterochromatin in Living Cells. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M, Koo BK. Modeling mouse and human development using organoid cultures. Development. 2015;142:3113–3125. doi: 10.1242/dev.118570. [DOI] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R. Notch: a membrane-bound transcription factor. J Cell Sci. 2002;115:1095–1097. doi: 10.1242/jcs.115.6.1095. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MXG. The Canonical Notch Signaling Pathway: Unfolding the Activation Mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall RA, Blacklow SC. Chapter Two - Mechanistic Insights into Notch Receptor Signaling from Structural and Biochemical Studies. Elsevier Inc; 2010. [DOI] [PubMed] [Google Scholar]
- Lecourtois M, Schweisguth F. Indirect evidence for Delta-dependent intracellular processing of notch in Drosophila embryos. Curr Biol. 1998;8:771–774. doi: 10.1016/s0960-9822(98)70300-8. [DOI] [PubMed] [Google Scholar]
- Lienert F, Lohmueller JJ, Garg A, Silver PA. Synthetic biology in mammalian cells: next generation research tools and therapeutics. Vol. 15. Nature Publishing Group; 2014. pp. 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim W, Mayer B, Pawson T. Cell Signaling. Taylor & Francis; 2014. [Google Scholar]
- Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006;52:425–436. doi: 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Koga M, Nishida E, Ebisuya M. Synthetic signal propagation through direct cell-cell interaction. Sci Signal. 2012;5:ra31–ra31. doi: 10.1126/scisignal.2002764. [DOI] [PubMed] [Google Scholar]
- Micchelli CA, Rulifson EJ, Blair SS. The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development. 1997;124:1485–1495. doi: 10.1242/dev.124.8.1485. [DOI] [PubMed] [Google Scholar]
- Miller JFAP, Sadelain M. The Journey from Discoveries in Fundamental Immunology to Cancer Immunotherapy. Cancer Cell. 2015;27:439–449. doi: 10.1016/j.ccell.2015.03.007. [DOI] [PubMed] [Google Scholar]
- Pratt EB, Wentzell JS, Maxson JE, Courter L, Hazelett D, Christian JL. The cell giveth and the cell taketh away An overview of Notch pathway activation by endocytic trafficking of ligands and receptors. Acta Histochemica. 2011;113:248–255. doi: 10.1016/j.acthis.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnick PEM, Weiss R. The second wave of synthetic biology: from modules to systems. 2009:1–13. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- Sasai Y. Cytosystems dynamics in self-organization of tissue architecture. Nature. 2013;493:318–326. doi: 10.1038/nature11859. [DOI] [PubMed] [Google Scholar]
- Slomovic S, Pardee K, Collins JJ. Synthetic biology devices for in vitro and in vivo diagnostics. Pnas. 2015:201508521–201508527. doi: 10.1073/pnas.1508521112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzak D, Lakhanpal A, LeBon L, Santat LA, Fontes ME, Anderson GA, Garcia-Ojalvo J, Elowitz MB. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature. 2010;465:86–90. doi: 10.1038/nature08959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- Webber MJ, Khan OF, Sydlik SA, Tang BC, Langer R. A Perspective on the Clinical Translation of Scaffolds for Tissue Engineering. Ann Biomed Eng. 2014;43:641–656. doi: 10.1007/s10439-014-1104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Charng W-L, Bellen HJ. Chapter Five - Endocytosis and Intracellular Trafficking of Notch and Its Ligands. Elsevier Inc; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, et al. Engineering Complex Synthetic Transcriptional Programs with CRISPR RNA Scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.