SUMMARY
Type III-A CRISPR-Cas systems defend prokaryotes against viral infection using CRISPR RNA (crRNA)-guided nucleases that perform co-transcriptional cleavage of the viral target DNA and its transcripts. Whereas DNA cleavage is essential for immunity, the function of RNA targeting is unknown. Here we show that transcription-dependent targeting results in a sharp increase of viral genomes in the host cell when the target is located in a late-expressed phage gene. In this targeting condition, mutations in the active sites of the type III-A RNases Csm3 and Csm6 lead to the accumulation of the target phage mRNA and abrogate immunity. Csm6 is also required to provide defense in the presence of mutated phage targets, when DNA cleavage efficiency is reduced. Our results show that the degradation of phage transcripts by CRISPR-associated RNases ensures robust immunity in situations that lead to a slow clearance of the target DNA.
INTRODUCTION
Clustered, regularly interspaced, short, palindromic repeats (CRISPR) loci and their associated (cas) genes encode an adaptive immune system that protects bacteria and archaea from viral (phage) and plasmid infection (Barrangou et al., 2007; Marraffini and Sontheimer, 2008). The targets of immunity are specified by short spacer sequences that intercalate in between the CRISPR repeats (Bolotin et al., 2005; Mojica et al., 2005; Pourcel et al., 2005). New spacers are acquired during infection (Barrangou et al., 2007) from all regions of the invader’s genome (Datsenko et al., 2012; Heler et al., 2015; Paez-Espino et al., 2013) and are inserted into the CRISPR array by the Cas1-Cas2 complex (Arslan et al., 2014; Nunez et al., 2014; Nunez et al., 2015; Yosef et al., 2012). Each spacer sequence is transcribed and processed into a small antisense RNA (Brouns et al., 2008; Carte et al., 2008; Deltcheva et al., 2011), the CRISPR RNA (crRNA), which associates with RNA-guided Cas nucleases to specify a matching target in the genome of the invading phage or plasmid (Gasiunas et al., 2012; Jinek et al., 2012; Samai et al., 2015; Westra et al., 2012). It is believed that cleavage of the foreign DNA destroys the genetic material of the invader and stops the infection (Garneau et al., 2010).
Based on their cas gene content, CRISPR-Cas systems can be classified into five types: I–V (Makarova et al., 2015). Type III CRISPR-Cas systems display an elaborate targeting mechanism mediated by the Cas10-Csm (type III-A) or the Cas10-Cmr (type III-B) complexes. Transcription of the target sequence is required for immunity (Deng et al., 2013; Goldberg et al., 2014) and for the cleavage of the non-template strand of the target DNA (Samai et al., 2015). Type III Cas10 complexes are also capable of crRNA-guided RNA cleavage (Hale et al., 2009; Samai et al., 2015; Staals et al., 2014; Tamulaitis et al., 2014; Zhang et al., 2012). In the Cas10-Csm complex, conserved aspartate residues within the palm domain of Cas10 are required for DNA cleavage (Samai et al., 2015) and Csm3 contains the crRNA-guided RNase activity (Samai et al., 2015; Staals et al., 2014; Tamulaitis et al., 2014). These activities result in the co-transcriptional cleavage of the target DNA and its transcripts (Samai et al., 2015). While DNA targeting is essential for the clearance of foreign plasmids and DNA phages (Goldberg et al., 2014; Marraffini and Sontheimer, 2008; Samai et al., 2015), a role for transcript cleavage during immunity against these DNA elements has not been determined.
In addition to the Cas10 complexes, type III CRISPR-Cas loci code for Csm6 or Csx1 (Makarova et al., 2011b), proteins that contain a Higher Eukaryotes and Prokaryotes Nucleotide-binding (HEPN) domain (Anantharaman et al., 2013). Bioinformatic analysis of Csm6 and Csx1 suggested that they function as metal-independent RNases (Anantharaman et al., 2013). In Staphylococcus epidermidis, deletion of csm6 results in the disruption of type III-A CRISPR-Cas immunity against conjugative plasmids (Hatoum-Aslan et al., 2014). Since absence of Csm6 does not affect the biogenesis of crRNAs it has been proposed that this protein participates in the targeting of nucleic acids. However, Csm6 is not part of the Cas10-Csm complex that cleaves DNA molecules in vitro (Hatoum-Aslan et al., 2013; Samai et al., 2015; Staals et al., 2014; Tamulaitis et al., 2014) and its precise role in type III CRISPR-Cas immunity has not been explored.
We studied the role of RNA targeting for the type III-A CRISPR-Cas system of S. epidermidis. We first determined that Csm6 is an RNase not required for the destruction of the target DNA. We then explored the contribution of both RNases of this system, Csm3 and Csm6, to CRISPR immunity during phage infection. We show that transcription-dependent targeting results in a sharp increase of viral genomes in the host cell when the target is located in a late-expressed phage gene. In this targeting condition, mutations in the active sites of the type III-A RNases Csm3 and Csm6 lead to the accumulation of the target phage mRNA and abrogate immunity. Csm6 is also required to provide defense in the presence of mutated phage targets, when DNA cleavage efficiency is reduced. Our results show that the degradation of phage transcripts by CRISPR-associated RNases ensures robust immunity in situations that lead to a slow clearance of the target DNA.
RESULTS
Csm6 is an RNase not involved in type III-A DNA degradation
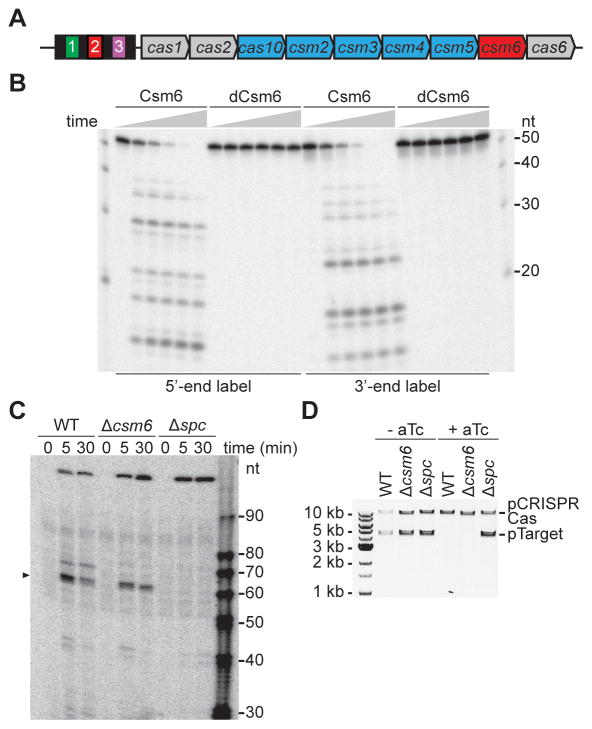
The type III-A CRISPR-Cas system of S. epidermidis harbors nine cas genes (Fig. 1A). cas1 and cas2 are present in most CRISPR-Cas systems and form a complex responsible for the integration of new spacers into the CRISPR array (Arslan et al., 2014; Nunez et al., 2015; Yosef et al., 2012). cas6 encodes for an endoribonuclease that processes the crRNA precursor into small crRNA guides in both type I (Brouns et al., 2008) and type III (Carte et al., 2008) systems. cas10, along with csm2, csm3, csm4 and csm5, encode a ribonucleoprotein complex characteristic of type III-A systems known as the Cas10-Csm complex (Hatoum-Aslan et al., 2013; Staals et al., 2014; Tamulaitis et al., 2014). csm6 is the only gene that has not been characterized in detail. A recent bioinformatics study of the HEPN family indicated that Csm6 is a member of this group, and may function as metal-independent RNase (Anantharaman et al., 2013). To investigate this, we expressed Csm6 in Escherichia coli and purified it to homogeneity, along with the putative active site double mutant R364A,H369A (Csm6R364A,H369A or “dead” Csm6, dCsm6). Incubation of Csm6 with ssRNAs radiolabeled at either end resulted in the degradation of the substrate by the wild-type Csm6, but not the active site double mutant (Fig. 1B). The reaction did not require any metal ion (Mg, Mn and EDTA were tested and obtained the same cleavage; data not shown). We obtained similar results with the individual mutants (Fig. S1A), using substrates of different sequences and lengths (Fig. S1B). Therefore these results confirm that Csm6 is a metal-independent, sequence-independent RNase. The Cas10-Csm complex also contains crRNA-guided RNase activity within the Csm3 subunit (Samai et al., 2015; Staals et al., 2014; Tamulaitis et al., 2014). To test if Csm6 RNase activity influences the RNA cleavage activity of the Cas10-Csm complex, we looked at the cleavage of target transcripts in vivo, using a previously developed primer extension assay (Samai et al., 2015) (Fig. S1C). We looked at the transcript cleavage products generated by the Cas10-Csm complex in strains carrying a wild-type, Δcsm6 or Δspc1 CRISPR-Cas system (Fig. 1C). No cleavage was detected in the absence of the spc1 crRNA guide and the same Csm3-dependent transcript cleavage product was detected in wild-type and Δcsm6 strains. These results indicate that the Csm6 sequence-independent RNase activity does not influence the crRNA-guided transcript cleavage performed by the Csm3 subunit of the Cas10-Csm complex.
Figure 1. Csm6 is an RNase not involved in type III-A DNA degradation.
(A) S. epidermidis RP62a carries a type III-A CRISPR-Cas locus that harbors four repeats (black boxes), three spacers (colored boxes) and nine cas/csm genes. cas10 and csm2–5 (in blue) encode for the Cas10-Csm ribonucleoprotein complex which has crRNA-guided DNA and RNA cleavage activities. The function of csm6 (in red) is unknown. (B) Purified Csm6 and dCsm6 were incubated with a radiolabeled ssRNA substrate. The reaction proceeded for 1 hour and aliquots were taken at 0, 15, 30, 45 and 60 minutes for PAGE and phosphorimager visualization. (C) A 5′-radiolabeled primer is used to initiate reverse transcription of the target transcript, generating a 171 nt extension product in the absence of RNA cleavage. The target is located in the pTarget plasmid under the control of a tetracycline-inducible promoter; this plasmid was introduced in different strains carrying the wild-type, Δcsm6 or Δspc (non-targeting control) CRISPR-Cas systems. Total RNA for primer extension was extracted at different times after addition of the aTc transcription inducer. Primer extension products were separated by PAGE and detected by phosphorimaging; the products indicating transcript cleavage are marked with an arrowhead. (D) pTarget plasmid DNA was extracted from cells before and after 10 hours of treatment with aTc, testing the different CRISPR-Cas backgrounds described in panel (C). See also Fig. S1.
Genetic characterization of the S. epidermidis CRISPR-Cas system indicated that csm6 is required for immunity against plasmid conjugation (Hatoum-Aslan et al., 2014). Since DNA targeting is required for this immunity (Marraffini and Sontheimer, 2008), it was proposed that Csm6 could participate in the degradation of plasmid DNA. To test this we looked at the stability of the pTarget plasmid upon induction of target transcription with anhydrotetracycline (aTc) in different genetic backgrounds (Fig. 1D). The plasmid was stable in the absence of the spc1 crRNA and was equally degraded by the wild-type and Δcsm6 CRISPR-Cas systems. Corroborating this finding, we determined that csm6 is not required for targeting of chromosomal DNA (Fig. S1D–E). Finally, we tested the activity of Csm6 against ssDNA and dsDNA substrates in vitro, but failed to obtain any cleavage products (Fig. S1F). Altogether these results demonstrate that the RNase activity of Csm6 is not involved in DNA targeting.
Csm3 and Csm6 are required for the degradation of phage transcripts
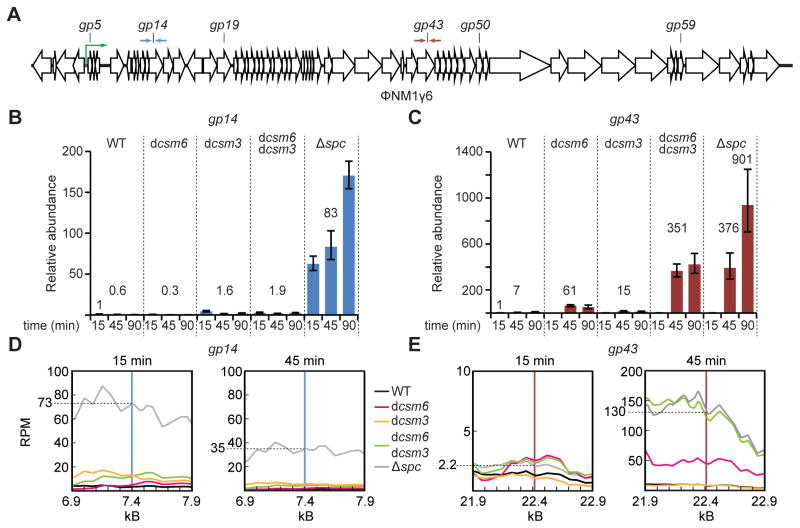
Together with previous reports, the above results show that the S. epidermidis type III-A CRISPR-Cas system encodes for two RNases: a sequence-specific, crRNA-guided endoribonuclease, Csm3, and another, sequence-independent ribonuclease, Csm6. To determine the role of these RNases in anti-viral immunity we explored their effect on phage transcripts during infection. To do this we transformed Staphylococcus aureus cells with different pCRISPR-Cas constructs (Hatoum-Aslan et al., 2013) and infected them with the staphylococcal lytic phage ΦNM1γ6 (Goldberg et al., 2014). Transcriptome analysis of S. aureus cells infected with ΦNM1γ6 in the absence of CRISPR-Cas immunity showed a gradual accumulation of transcripts after infection, with transcription of the first 12 kb of the genome during the first 15 minutes post-infection and requiring 30 minutes for the expression of the full set of lytic genes (Goldberg et al., 2014). At different times post-infection we extracted total RNA and measured the abundance of phage transcripts during type III-A CRISPR-Cas immunity in the presence or absence of Csm3 and/or Csm6 RNase activity using RT-qPCR. First we quantified phage RNA accumulation during the targeting of an early-transcribed gene, gp14 (encoding a protein involved in phage DNA replication), with primers that amplify the targeted region (~100 bp flanking the target sequence defined by the crRNA) at 15, 45 and 90 minutes post-infection (Fig. 2A). We compared a wild-type CRISPR-Cas system, a non-targeting control lacking the matching spacer sequence (Δspc), and systems lacking Csm3 RNase activity (csm3D32A or “dead” csm3, dcsm3) (Samai et al., 2015), Csm6 RNase activity (csm6R364A,H369A or dcsm6) or both (Fig. 2B). Transcript accumulation was minimal during gp14 targeting in the wild-type strain and increased dramatically in the absence of immunity, approximately a 130-fold increase at the 45-minute time-point (0.6 for wild-type vs 83 for Δspc). We did not detect any substantial accumulation of phage transcripts in the dcsm6, dcsm3 or dcsm3/dcsm6 strains. When we performed the same experiment but targeting the late-transcribed gp43 gene (encoding a phage capsid subunit) we observed minor differences between target transcript accumulation in wild-type, dcsm3 and dcsm6 strains (Fig. 2C). However, we detected a significant increase in viral transcripts in the dcsm3/dcsm6 double mutant, which accumulated similar levels of phage mRNA as infected cells that lack CRISPR-Cas immunity (Δspc) 45 minutes after infection. At 90 minutes post-infection we detected a two fold increase in the gp43 transcript level in Δspc cells compared to the dcsm3/dcsm6 double mutant, which is likely due to the presence of DNA cleavage and less phage propagation in the latter. We also performed RNA deep sequencing (RNA-seq) in infected cells with different genetic backgrounds. The results confirm the RT-qPCR data, showing that gp14 phage transcripts are not accumulated during the immune response of the different CRISPR-Cas systems (Fig. 2D), but that there is a substantial increase in gp43 transcript levels (similar to the Δspc control) in the dcsm3/dcsm6 double mutant at 45 minutes post-infection (Fig. 2E). Together, these results indicate that the RNase activity of either Csm3 or Csm6 is required to prevent the accumulation of phage transcripts when the type III-A CRISPR-Cas system targets late-, but not early-, expressed genes.
Figure 2. Csm3 and Csm6 are required for the degradation of phage transcripts.
(A) Schematic diagram of the ΦNM1γ6 genome in its linear (prophage) form, and the position of type III-A CRISPR-Cas targets used in this study. Supplemental Table S1 contains the full sequence of each target. The green arrow represents the rightwards promoter driving transcription of the lytic genes and defines the early expressed genes as those immediately downstream of this promoter. During the lytic cycle the genome is circular. The opposed arrows indicate the primers used for RT-qPCR experiments in panels B and C. (B) RT-qPCR performed on the ΦNM1γ6 gp14 transcript using total RNA collected at different times post-infection from cells carrying different type III-A CRISPR-Cas systems targeting the gp14 gene. Values for the rho gene were used for normalization. The normalized value for the measurement at 15 minutes in wild-type cells was set to 1 to obtain the relative abundance of the gp14 transcript for the rest of the data points (mean ± S.D. of four replicas). (C) Same as panel (B), but using CRISPR-Cas systems targeting the ΦNM1γ6 gp43 gene and measuring relative abundance of the gp43 transcript. (D) RNA-seq reads (Reads Per 500 bases of transcript per Million mapped reads, RPM) for transcripts in the vicinity of the gp14 target at 15 and 45 minutes post infection of cells harboring different mutations in the type III-A CRISPR-Cas system. Vertical blue line indicates target position. (E) Same as (D) but showing transcription levels in the gp43 target region. See also Fig. S2.
We also wanted to investigate the specificity of the RNase activity of Csm3 and Csm6 described above. We found no csm3- or csm6-specific degradation of the plasmid-born cat gene transcript (Fig. S2A) nor of the chromosomally expressed fabD and glyA mRNAs (Fig. S2B). In addition, overexpression of Csm6 did not result in a cell growth defect (Fig. S2C), which would be expected for a non-specific RNase. On the other hand, the specific degradation of phage transcripts extended for at least 1 kb at each side of the target site defined by the crRNA guide (Figs. S2E). RNA-seq of phage transcripts in wild-type, dcsm3, dcsm6 or dcsm3/dcsm6 cells corroborated these results (Fig. S2F). In addition it showed that there is a very low level of phage transcripts from sequences flanking the target region in dcsm3 cells, indistinguishable to the transcript levels of wild-type cells. On the other hand, an increased accumulation (similar to the Δspc control in the gp42 and gp52 regions), was observed for the dcsm6 mutant. These data suggest that Csm6, and not Csm3, is responsible for much of the transcript degradation outside of the target region. The mechanism by which the RNase activity of Csm6 is first localized to the Cas10-Csm transcript target remains to be elucidated.
Co-transcriptional type III CRISPR-Cas targeting leads to the accumulation of phage DNA
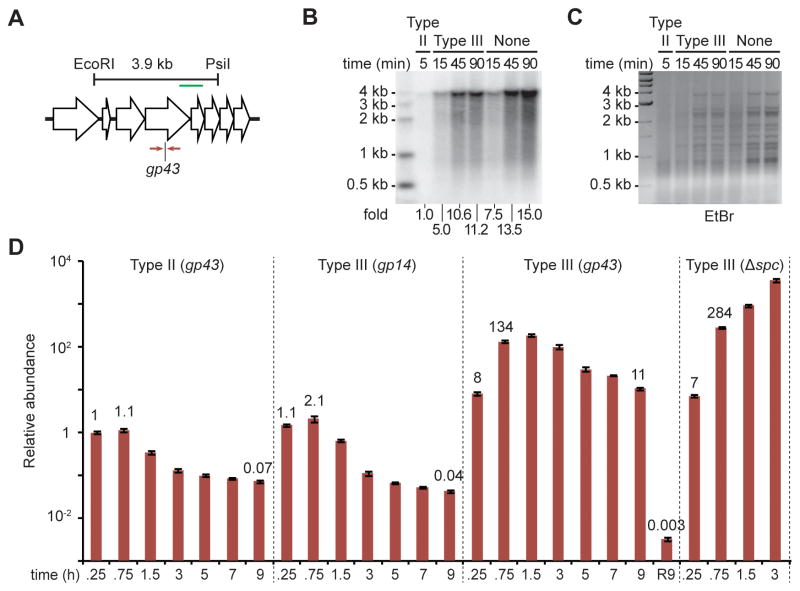
The transcription requirement for DNA cleavage of type III CRISPR-Cas systems implies that when a late-expressed gene is targeted, the phage infection cycle will proceed unchecked until the target is transcribed. This can take 15–30 minutes (Goldberg et al., 2014) and would allow the accumulation of both phage DNA and RNA that results from virus replication and transcription, respectively. Figure 2 shows that the type III-A RNases Csm3 or Csm6 prevent the accumulation of phage transcripts in this scenario, i.e. when the late-expressed gene gp43 is targeted; in Figure 3 we investigated the fate of phage DNA during type III-A CRISPR-Cas immunity. First we performed Southern blot analysis on cells infected with ΦNM1γ6 carrying different CRISPR-Cas systems targeting gp43 (Fig. 3A). As controls we looked at phage DNA in cells carrying a type II CRISPR-Cas system that targets gp43 (Goldberg et al., 2014) and in cells without CRISPR-Cas immunity. The expected 3.9 kb band that results from restriction digestion of the target site (Fig. 3A) was minimally detected in the presence of type II immunity (Fig. 3B). This is in agreement with previous findings that showed that phage DNA is immediately degraded upon injection by the type II Cas9 crRNA-guided nuclease (Garneau et al., 2010), which does not require target transcription for DNA cleavage (Gasiunas et al., 2012; Jinek et al., 2012). In cells with type III-A immunity on the other hand, the phage DNA accumulated over time to similar levels to those observed in the absence of CRISPR targeting (Fig. 3B). Ethidium bromide staining of the digested DNA also revealed a general accumulation of phage DNA (Fig. 3C). We corroborated this result using qPCR amplification of the gp43 target in different infection conditions (Fig. 3D). For quantification, the relative abundance of the gp43 qPCR product detected at 15 minutes post-infection in cells harboring the Streptococcus pyogenes type II-A CRISPR-Cas system was set as the reference point (a value of 1). In these cells the abundance of the gp43 qPCR product decreased rapidly with time, showing an efficient destruction of the viral DNA. In cells without CRISPR immunity (carrying a non-targeting, Δspc, type III-A CRISPR-Cas system) the abundance of the gp43 qPCR product increased dramatically with time, reflecting the progression of viral replication during the infectious cycle. Phage DNA clearance by type III CRISPR-Cas immunity strongly depended on the region targeted. When the early-expressed gene gp14 was targeted, the abundance of the gp43 qPCR product decreased with time similarly to the observations for type II targeting. In contrast, when the late-expressed gene gp43 was targeted the abundance of the qPCR product reached very high levels before it started a slow decrease. We obtained similar results when we measured phage DNA by qPCR using primers that amplify the gp14 gene (Fig. S3A), showing that DNA abundance at the target as well as at a distant locus is equally affected by CRISPR targeting. Since type III CRISPR-Cas immunity requires transcription across the target for efficient DNA cleavage, we considered the possibility that less transcription across the gp43 target, compared to transcription of the gp14 target, could be responsible for the accumulation of phage DNA observed. However, our RNA-seq data indicated that the level of transcription across the gp14 target at 15 minutes post-infection is similar or even lower to those of the gp43 target at 45 minutes post-infection, when this gene is expressed (compare 73 to 130 RPM for gp14 and gp43, respectively; Figs. 2D and 2E). Together these results revealed that the requirement of target transcription for type III CRISPR-Cas DNA cleavage results in the accumulation of phage DNA when a region that is expressed late in the infectious cycle is targeted. In vitro, an excess of target DNA prevents efficient cleavage by the Cas10-Csm complex (Fig. S3B). We performed a co-transcriptional DNA cleavage assay (Samai et al., 2015) using different complex:target molar ratios and we found target DNA cleavage at a 10:1, but not 1:1, ratio, suggesting that the accumulation of phage DNA observed during the targeting of a late-expressed gene is the result of inefficient DNA cleavage due to the excess of target DNA at this point of the viral infectious cycle. In spite of this significant difference in phage DNA accumulation and cleavage of the target DNA, we have previously shown that type III-A CRISPR-Cas immunity can protect the host from lysis regardless of the viral genomic region targeted (Goldberg et al., 2014).
Figure 3. Co-transcriptional type III CRISPR-Cas targeting leads to the accumulation of phage DNA.
(A) Location of the EcoRI and PsiI restriction sites used to detect phage DNA via southern blot in panel (B). The green line indicates the location of the dsDNA probe used in this assay. The gp43 target and the primers (opposed arrows) used for qPCR in panel (D) are also shown. (B) Southern blot on total DNA extracted from cells treated with ΦNM1γ6 at different times after infection and digested with EcoRI and PsiI. Cells harboring type II-A or type III-A CRISPR-Cas systems programmed to target the gp43 gene, or without CRISPR-Cas immunity were infected. The intensity, relative to type II-A targeting, of the 3.9 kb phage fragment detected is reported. (C) Ethidium bromide gel used for southern blot shown in (B). (D) qPCR performed on the ΦNM1γ6 gp43 gene using total DNA collected at different times post-infection from cells carrying different CRISPR-Cas systems. Values for the rho gene were used for normalization. The normalized value for the measurement at 15 minutes in cells harboring a type II system was set to 1 to obtain the relative abundance of the gp43 transcript for the rest of the data points (mean ± S.D. of four replicas). The R9 time point indicates that cells were refreshed with new culture broth at 9 hours post-infection and were grown for an additional 9 hours before collection of DNA for qPCR. See also Fig. S3.
Degradation of phage transcripts by Csm3 and Csm6 enables type III CRISPR-Cas immunity targeting late-expressed genes
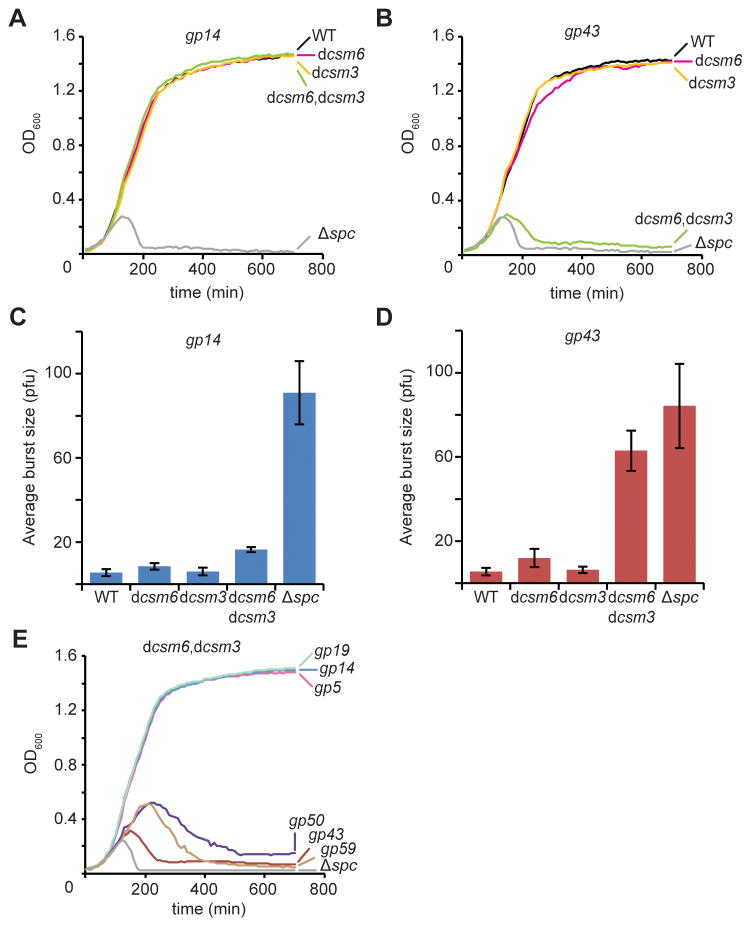
Previously we demonstrated that type III-A CRISPR-Cas immunity can protect the host from lysis regardless of the viral genomic region targeted (Goldberg et al., 2014). In light of our results above, we investigated whether the degradation of late-expressed transcripts mediated by Csm3 and Csm6 is also required for CRISPR-Cas immunity during the targeting of late genes. To this end, we compared lysis (measured by absorbance at OD600 of the bacterial culture) of infected cells carrying type III-A CRISPR-Cas systems targeting an early- or late-transcribed gene, gp14 or gp43, respectively. The mutants dcsm3, dcsm6 and dcsm3/dcsm6 were as effective as the wild-type CRISPR system to confer immunity via gp14 targeting (Fig. 4A), a result that demonstrates the sufficiency of DNA cleavage for viral clearance in this situation. In contrast, when gp43 was targeted the dcsm3/dcsm6 mutant failed to provide immunity (Fig. 4B), similarly to a no-targeting control (Δspc). A similar result was obtained when we measured the effect of type III-A CRISPR-Cas immunity on the propagation of the phage by determining the average burst size, i.e. the number of viral particles (counted as plaque forming units, pfu) released per infected cells (Figs. 4C–D). Both experiments indicate that the RNase activity of either Csm3 or Csm6 is required for immunity when targeting a late-, but not early-, expressed gene. To confirm this pattern we tested immunity mediated by type III-A dcsm3/dcsm6 mutant systems targeting two other early-transcribed (gp5 and gp19) and two other late-transcribed (gp50 and gp59) genes (Fig. 2A). We infected each strain with ΦNM1γ6 and looked for culture lysis over time (Fig. 4E). Whereas targeting of gp5, gp14 and gp19 produced efficient immunity, targeting of gp43, gp50 and gp59 resulted in the death of bacteria expressing inactive Csm3 and Csm6 RNases. These data demonstrate that the RNase activities of Csm3 and Csm6 are required for type III-A CRISPR-Cas immunity when the targets specified by the crRNA guide reside within late-expressed genes.
Figure 4. Degradation of phage transcripts by Csm3 and Csm6 enables type III CRISPR-Cas immunity targeting late-expressed genes.
(A) Staphylococci harboring different type III-A CRISPR-Cas systems targeting the gp14 gene were grown in liquid media and infected with ϕNM1γ6 phage (at 0 hours) with a multiplicity of infection of 5 viruses per bacteria. Optical density at 600 nm (OD600) was measured for the following 12 hours to monitor cell survival due to CRISPR immunity against the phage. Representative growth curves of at least three independent assays are shown. (B) Same as panel (A) but with the CRISPR-Cas systems programmed to target gp43. (C) The different infections performed in panel (A) were plated to enumerate plaque forming units (pfu) and calculate the average burst size. Mean ± S.D. of three replicas are reported. (D) Same as panel (C) but with the CRISPR-Cas systems programmed to target gp43. (E) Survival of cells (determined by measuring growth at OD600) carrying dcsm6/dcsm3 type III-A CRISPR-Cas systems targeting the different ϕNM1γ6 genes shown in Fig. 2A. Representative growth curves of at least three independent assays are shown.
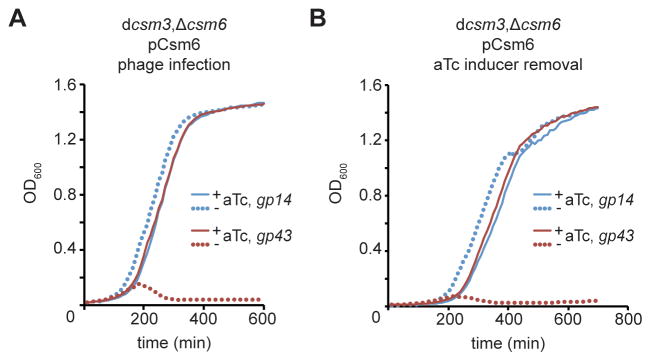
We showed that the accumulation of phage DNA that occurs prior to the targeting of late-expressed genes prevents the type III-A CRISPR-Cas system from rapidly clearing the phage DNA. In this scenario, we hypothesized that the degradation of phage transcripts by Csm3 and Csm6 limits further viral gene expression and the continuation of the lytic infectious cycle, which would otherwise compromise host cell viability. To test this, we designed an experiment to eliminate RNase activity 10 hours post-infection. We infected cells harboring dcsm3/Δcsm6 CRISPR-Cas systems targeting gp14 or gp43 and carrying the pCsm6 plasmid, which provides aTc-dependent expression of Csm6 (Fig. 5A). As expected from our previous results, in the absence of the inducer the cells targeting gp43, but not those targeting gp14, succumbed to phage infection. In the presence of aTc, both populations survived. The cells from these two populations were washed with fresh broth to eliminate aTc, and thus Csm6 expression, 10 hours after the addition of ΦNM1γ6 to the cultures. Cells were diluted in fresh broth with or without aTc and their growth was monitored by following absorbance at OD600 (Fig. 5B). While the growth of gp14-targeting cells was not affected by removal of Csm6, gp43-targeting cells were lysed by phage. This result demonstrates that without the degradation of phage transcripts by Csm6, the phage lytic cycle can continue in spite of DNA cleavage, leading to the death of the host cells.
Figure 5. Csm6 enables complete phage clearance during immunity against late-expressed genes.
(A) Cells harboring Δcsm6/dcsm3 type III-A CRISPR-Cas systems targeting gp14 or gp43 were complemented with the pCsm6 plasmid, which carries the csm6 gene under the control of a tetracycline-inducible promoter. Each strain was infected with ϕNM1γ6 in the presence or absence of the aTc (0.008 μg/ml); i.e. induction of Csm6 expression. Bacterial growth was monitored by measuring for OD600 for 10 hours. (B) The cells grown in the presence of aTc were collected, washed to remove the inducer and eliminate further expression of Csm6, and diluted (1:333) in fresh media without phage nor aTc. As a control an aliquot of the washed cells were re-inoculated in fresh media with aTc (0.008 μg/ml). Bacterial growth was monitored by measuring for OD600 for 12 hours.
Csm6 is required to provide immunity against viruses with target mutations
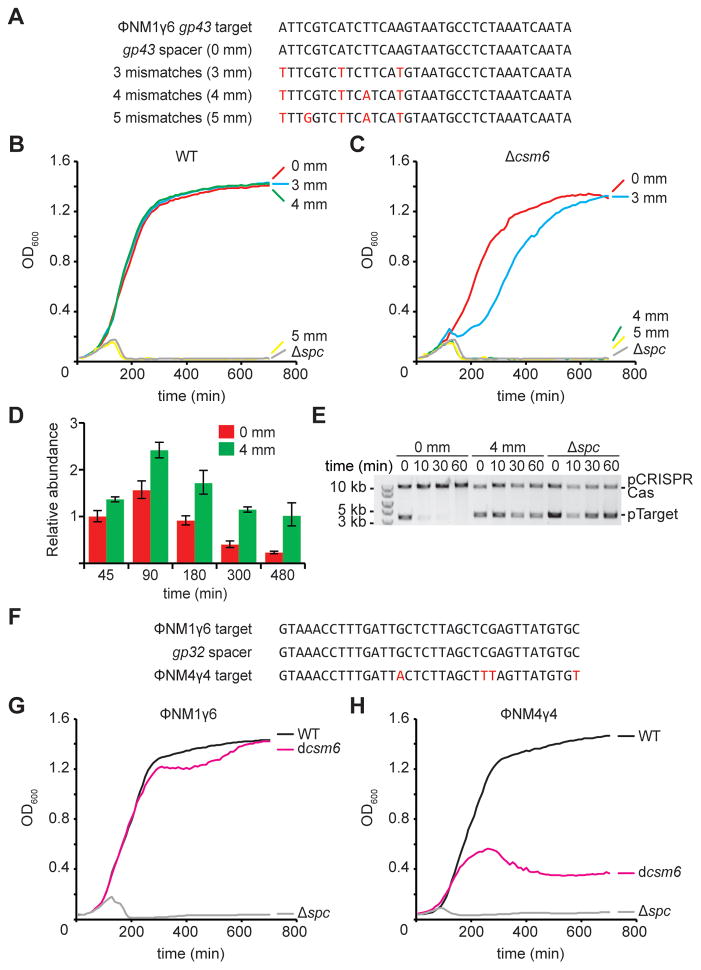
Our data shows that the targeting of a late-expressed gene leads to the accumulation of target DNA and that in this scenario the RNase activities of Csm3 or Csm6 are required to clear the phage transcripts and slow down the phage lytic cycle until all DNA targets are destroyed. A similar situation could present during infection with phages harboring target mismatches. In both type I and type II CRISPR-Cas immunity mutations in the target region lead to the escape of mutant phages due to a reduced crRNA-guided DNA cleavage. Type III CRISPR-Cas systems, however, seem much more tolerant of such mutations and are able to provide immunity even in the presence of several mismatches within the crRNA:target interaction (Goldberg et al., 2014; Manica et al., 2013). We speculated that, if target mutations result in inefficient DNA cleavage also during type III CRISPR-Cas immunity, the reported tolerance to mutations could be the result of phage transcript cleavage by the Csm3 and/or Csm6 RNase activity. To test this we introduced 3, 4 and 5 mismatches into the spacer sequence targeting the gp43 gene of ΦNM1γ6 (Fig. 6A). We infected hosts carrying these mutations and looked for the CRISPR immune response (Fig. 6B). Consistent with previous reports, type III CRISPR-Cas immunity protected staphylococci even in the presence of 3 and 4 mismatches (but not 5) between the crRNA guide and its target. We first tested if Csm6 was important for immunity in the presence of mismatches by performing infections in a Δcsm6 host (Fig. 6C). Consistent with Figure 4B, csm6 was not required to provide immunity when the phage carried a target with perfect homology. However, cells were not as protected in the presence of 3 mismatches and immunity was completely abrogated with 4 mismatches in the crRNA:target interaction. Protection in the presence of 4 mismatches required the RNase activity of Csm6 (Fig. S4A). In contrast, the RNase activity of Csm3 was not required for immunity in the presence of 4 mismatches (Fig. S4A). We used qPCR of the gp43 target to compare phage DNA accumulation during the course of infection of wild-type hosts carrying a perfectly matching or 4-mismatch spacer (Fig. 6D). We observed that indeed the presence of mismatches led to the accumulation of target phage DNA. This was corroborated by an anti-plasmid immunity assay similar to the one presented in Figure 1D using a 4-mismatch target (Figs. 6E and S4B). Target mismatches are not only present within a viral population but are very common between related phages. For example, we previously engineered a spacer matching the gp32 gene present in the staphylococcal phage ΦNM1γ6 (Goldberg et al., 2014) which has 4 mismatches to the same gene in the related phage ΦNM4γ4 (Bae et al., 2006; Heler et al., 2015) (Fig. 6F). Consistent with our results, type III-A CRISPR-Cas immunity against ΦNM1γ6 mediated by this spacer does not require the RNase activity of Csm6 (Fig. 6G). In contrast, whereas the wild-type CRISPR-Cas system tolerated the 4 mismatches and protected cells from ΦNM4γ4 infection, the dcsm6 mutant cells were susceptible to viral attack (Fig. 6H). Together these results show that Csm6 RNase activity is required to maintain immunity even in the presence of target mutations that decrease the efficiency of DNA targeting, a distinct property of type III systems.
Figure 6. Csm6 is required to provide immunity against viruses with target mutations.
(A) Introduction of mutations (in red) in the spacer targeting the gp43 phage gene that generate 3, 4 or 5 mismatches in the crRNA:target region. (B) Staphylococci harboring a wild-type III-A CRISPR-Cas system targeting the gp43 gene in the presence of different crRNA:target mismatches were grown in liquid media and infected with ϕNM1γ6 phage (at 0 hours) with a multiplicity of infection of 5 viruses per bacteria. Optical density at 600 nm (OD600) was measured for the following 12 hours to monitor cell survival. Representative growth curves of at least three independent assays are shown. (C) Same as panel (B) but with cells harboring a CRISPR-Cas locus without csm6. (D) qPCR performed on the ΦNM1γ6 gp43 gene using total DNA collected at different times post-infection from cells carrying CRISPR-Cas systems targeting in the presence (4 mm) or absence (0 mm) of crRNA:target mismatches. Values for the rho gene were used for normalization to obtain the relative abundance of the gp43 gene for each data point (mean ± S.D. of four replicas). (E) pTarget plasmid DNA, harboring the gp43 target under the control of a tetracycline-inducible promoter, was extracted from cells harboring a type III-A CRISPR-Cas system without a spacer (Δspc) or with a gp43-targeting spacer with or without mismatches (4 mm or 0 mm, respectively), at different times after treatment with aTc. Plasmid DNA was visualized by agarose gel electrophoresis followed by ethidium bromide staining. (F) The gp32 spacer has a complete match in the ϕNM1γ6 genome but presents four mismatches in the ϕNM4γ4 phage. (G) Staphylococci harboring a wild-type, Δcsm6 or Δspc type III-A CRISPR-Cas system with the gp32 spacer were grown in liquid media and infected with ϕNM1γ6 phage (at 0 hours) with a multiplicity of infection of 5 viruses per bacteria. Optical density at 600 nm (OD600) was measured for the following 12 hours to monitor cell survival due to CRISPR immunity against the phage. Representative growth curves of at least three independent assays are shown. (H) Same as (G) but following infection with phage ϕNM4γ4. See also Fig. S4.
DISCUSSION
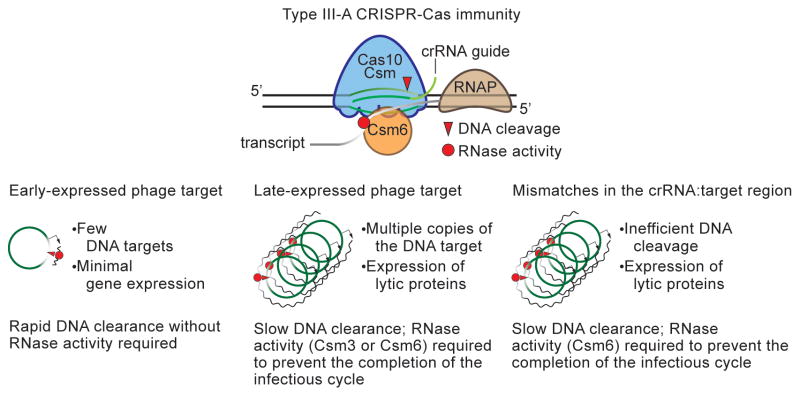
Type III CRISPR-Cas systems cleave both the genome and the transcripts of invaders (Peng et al., 2015; Samai et al., 2015). Whereas DNA cleavage is fundamental for CRISPR immunity against these invaders, a role for the RNase activity of these systems has been elusive. A recent report showed that the Cas10-Csm complex could prevent the propagation of ssRNA phages in E. coli (Tamulaitis et al., 2014). However, these types of viruses are rare in the prokaryotic world (Koonin et al., 2015) and CRISPR spacer sequences that match RNA viruses have not yet been found. In addition, it has been argued that due to the high mutation rate of RNA viruses (about three orders of magnitude above that of dsDNA viruses), maintaining long-term immunity against these invaders would require an extremely rapid acquisition of new spacer sequences (Weinberger and Gilmore, 2015; Weinberger et al., 2012). Therefore a role for type III CRISPR-Cas system providing immunity against RNA viruses in a natural setting remains to be determined. Here we showed a role for transcript degradation in the immunity mechanism against dsDNA phages, predicted to be the most common type of viruses that infect prokaryotes (Koonin et al., 2015), provided by the type III-A CRISPR-Cas system of S. epidermidis. We found that these systems encode two RNases, Csm3 and Csm6, that are required to provide immunity against dsDNA phages when the crRNA guide of the Cas10-Csm complex matches late-, but not early-, expressed genes. We propose that, as a consequence of the co-transcriptional DNA cleavage activity of type III CRISPR-Cas systems, which cannot cleave the viral DNA until the target is transcribed, the targeting of late phage genes results in the accumulation of viral genomes. In this scenario the RNase activities of the type III-A CRISPR-Cas system are required to degrade phage transcripts and prevent the completion of the viral infectious cycle until all the viral genomes are cleared (Fig. 7). In contrast, when an early-transcribed gene is targeted, DNA cleavage occurs shortly after genome injection. In this case the endonuclease activity of the Cas10-Csm complex is sufficient to clear the virus; the infectious cycle does not proceed further and the degradation of phage transcripts is not necessary to prevent phage propagation.
Figure 7. A model for the requirement for transcript degradation during type III CRISPR-Cas immunity.
The type III-A Cas10-Csm complex performs co-transcriptional cleavage of the target DNA and its transcripts. Within this complex, Cas10 contains the DNase activity and Csm3 is an RNase. Csm6 is another type III-A RNase that degrades target transcripts. This molecular mechanism of immunity allows for the rapid attack of the viral genome when early-expressed targets are specified by the crRNA guide, which leads to fast and efficient degradation of the invader’s genetic material and the clearance of the infection without the need of RNase activity. In contrast, the targeting of a late-expressed gene allows viral replication and transcription before DNA cleavage can occur. The accumulated genomes are not cleared efficiently by the endonuclease activity of the Cas10 complex and the degradation of phage transcripts by Csm3 or Csm6 is required to prevent the completion of the infectious cycle and the lysis of the host cell. Similarly, the presence of crRNA:target mismatches within the phage population prevents efficient DNA cleavage that also leads to the accumulation of phage genomes in the cell. In this scenario the Csm6 RNase is required for transcript degradation and survival.
All the staphylococcal phages characterized so far belong to the Caudovirales order and are mainly temperate Siphoviridae (Deghorain and Van Melderen, 2012), such as ΦNM1. A small number belong to the Podoviridae or Myoviridae family. Regardless of the different genomic organizations and infectious cycles, all of these phages present a tight regulation of gene expression (Kwan et al., 2005) and therefore different sets of genes are transcribed at different times post-infection. We believe then that the ability of transcript degradation by type III-A CRISPR-Cas systems would be important to provide immunity against most classes of dsDNA viruses when the crRNA guide targets genes that are expressed late in the infectious cycle. Target selection occurs during the “adaptation” phase of CRISPR-Cas immunity, when new spacer sequences from an invading phage are incorporated into the CRISPR array (Heler et al., 2014). Although little is known about the acquisition of spacers by type III CRISPR-Cas systems, type I and II systems incorporate spacers matching all regions of the viral genome (Datsenko et al., 2012; Heler et al., 2015; Paez-Espino et al., 2013), without any noticeable bias towards early or late genes. If such bias is also absent during spacer acquisition by type III CRISPR-Cas systems, the RNase activity would be necessary to confer immunity to all bacteria that incorporate a spacer specifying a late-expressed gene.
Our studies showed that either Csm3 or Csm6 RNase activity is required for immunity when the target of the Cas10-Csm complex is located in a late-expressed gene. Mutations in the active sites of either of these genes are not sufficient to disrupt immunity. However, in the presence of target mismatches that lead to the accumulation of phage DNA, the RNase activity of Csm6, but not that of Csm3, is required for immunity. This is an important function that distinguishes the type III from the type I and II CRISPR immune response. Whereas type I and II CRISPR immunity is very sensitive to mutations in the target sequence (especially in the “seed” region of the target) (Gasiunas et al., 2012; Jinek et al., 2012; Westra et al., 2012; Wiedenheft et al., 2011), type III immunity is unusually tolerant of such mutations (Goldberg et al., 2014; Manica et al., 2013), allowing the targeting of “escape” or related viruses. In all three systems, target mutations prevent efficient DNA cleavage, however in type III systems, transcript degradation by Csm6 results in robust immunity, presumably by stalling the progression of the phage lytic cycle and allowing for more time for phage DNA clearance (Fig. 7). Here we found that the most dramatic results were obtained in the presence of 4 mismatches, but we suspect that this number may vary for different target sequences. Mismatches may not be the only condition in which one or the other RNase activity of type III systems is required. We speculate that in other scenarios that lead to the accumulation of phage DNA in the cell, such as infection by phages with particular propagation cycles, by phages that introduce modifications to their nucleic acids, or in selected habitats (type III systems are predominant in thermophilic archaea (Makarova et al., 2011a)), one or the other RNase activity may be more important to allow for complete DNA clearance.
Whereas Csm3 is part of the Cas10-Csm complex and cleaves sequences that are specified by the crRNA guide (Samai et al., 2015; Staals et al., 2014; Tamulaitis et al., 2014), Csm6 is not part of the complex and has sequence-independent activity. Although these mechanisms of RNA cleavage are considerably different, both are sufficient to degrade phage transcripts and facilitate type III-A CRISPR-Cas immunity in the conditions tested in this study. How Csm6 achieves specificity for phage transcripts is not known. One possible mechanism to restrict Csm6 activity to the Cas10-Csm target would be the existence of a biophysical interaction between them during targeting. Additional work focused on Csm6 will address its specificity and its role during plasmid conjugation, two intriguing properties of this RNase.
While Csm6 is associated with type III-A CRISPR-Cas systems, some type III-B systems harbor a Csm6 ortholog, Csx1 (Makarova et al., 2011a; Makarova et al., 2011b; Makarova et al., 2015), which also belongs to the HEPN family (Anantharaman et al., 2013). Based on our results obtained for Csm6 and the sequence similarity between these genes, we propose that Csx1 also provides sequence-independent RNase activity to the type III-B CRISPR-Cas systems. Our work also predicts that Csx1 and the Cmr4 crRNA-guided RNA nuclease (the Csm3 ortholog) within the Cas10-Cmr complex (Benda et al., 2014; Hale et al., 2014; Ramia et al., 2014) may be required for the targeting of late-expressed genes in dsDNA phages by these systems. Future experiments will consider this and other intriguing aspects of these elaborate immune systems.
Experimental procedures
Bacterial strains and growth conditions
Cultivation of E. coli was done in LB medium (BD) or Terrific Broth medium (Fisher Scientific) at 37 °C. Whenever applicable, media were supplemented with 100 μg/ml ampicilin or 34 μg/ml chloramphenicol to ensure plasmid maintenance.
All in vivo experiments were performed in S. aureus RN4220 (Kreiswirth et al., 1983). Cultivation of S. aureus RN4220 was done in TSB medium (BD) at 37 °C. Whenever applicable, media were supplemented with chloramphenicol or erythromycin at 10 μg/ml to ensure plasmid maintenance. When appropriate, anhydrotetracycline (aTc) was used at a concentration of 0.25 μg/ml (unless otherwise indicated) to initiate transcription from the Ptet promoter.
Plasmid Cloning
Plasmid DNA preparation
Plasmid DNA was purified from 2 to 6 ml of E. coli DH5α or S. aureus RN4220 overnight cultures. For preparation from S. aureus cultures, cells were pelleted, re-suspended in 100 μl TSM buffer (50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 0.5 M sucrose) then treated with 5 μl lysostaphin (2 mg ml−1) at 37 °C for 1 h before treatment with plasmid miniprep reagents from Qiagen. Purification used Qiagen or EconoSpin columns.
Purification of Csm6
Purification was performed via Ni-NTA affinity chromatography. See Supplemental Experimental procedures for details.
Csm6 RNA cleavage assay
RNA cleavage reactions were performed at 37°C with 1 μM of 5′-radiolabeled (R55 and R24) and 3′-radiolabeled ssRNA (R55) substrates and 10 μM of wild type or mutant Csm6. The reaction was carried out in reaction buffer containing 50 mM Tris-HCl [pH7.5], 30 mM NaCl, 2 mM DTT and 1% glycerol. Reaction mixtures were withdrawn at specified time intervals and subsequently quenched with 90% formamide and 50 mM EDTA. Reaction products were separated by denaturing PAGE and the gel was visualized by phosphorimaging. The 5′-radiolabeled decade RNA ladder (Life Technologies) was used as a size marker.
Csm6 DNA cleavage assay
DNA cleavage reactions were performed at 37°C for up to 2 hours with 1 μM of 5′-radiolabeled ssDNA (PS362) and dsDNA (PS362/PS363) substrates and 10 μM of wild type Csm6. The reaction was carried out in reaction buffer containing 50 mM Tris-HCl [pH7.5], 10 mM MgCl2, 30 mM NaCl, 2 mM DTT and 1% glycerol. Reaction mixtures were withdrawn at specified time intervals and subsequently quenched with 90% formamide and 50 mM EDTA. Reaction products were separated by denaturing PAGE and the gel was visualized by phosphorimaging. The 5′-radiolabeled 10 bp DNA ladder (Promega) was used as a size marker.
Transcription Coupled DNA Cleavage
Performed as previously described (Samai et al., 2015). See Supplemental Experimental procedures for details.
Preparation of electrocompetent S. aureus cells
Preparation of S. aureus RN4220 competent cells and DNA transformation was performed as previously described (Goldberg et al., 2014).
Phage infections and plate reader growth curves
Infection of S. aureus RN4220 cells with bacteriophage ΦNM1γ6 or ΦNM4γ4 was performed as described previously (Goldberg et al., 2014). See Supplemental Experimental procedures for details.
Measurement of average burst size
RN4220 cells with appropriate CRISPR-Cas plasmids were grown in TSB supplemented with 5mM CaCl2 and appropriate antibiotics to an OD600 of 0.3–0.5. Cells were infected by ΦNM1γ6 at MOI = 0.1 for 5 minutes. Cells were immediately washed in TSB twice at 4°C and re-suspended in equal initial volume. An aliquot of cells were spotted on heart infusion (BD) soft agar plates with a sensitive lawn (i.e. RN4220). The rest of the cells were incubated at 37°C for another 75 minutes before an aliquot of cells were spotted on a sensitive lawn. Agar plates were incubated at 37°C for 16–20 hours before plaques were enumerated. Average burst size was calculated as the ratio of plaques formed at 80 minutes to plaques formed at 5 minutes for each strain of interest.
Total RNA Extraction
Performed as previously described (Samai et al., 2015). See Supplemental Experimental procedures for details.
Total DNA Extraction
Performed as previously described (Samai et al., 2015). See Supplemental Experimental procedures for details.
Plasmid-curing assay
Performed as previously described (Samai et al., 2015). See Supplemental Experimental procedures for details.
Primer extension
Primer extension assays were performed as previously described (Hatoum-Aslan et al., 2011) using primer A248.
qPCR
Cells were infected by ΦNM1γ6 (MOI=5) during early log phase (at OD600 of 0.3–0.4). qPCR was performed using Fast SYBR® Green Master Mix (Life technologies) and 7900HT Fast Real-Time PCR System (Applied Biosystems). For RNA samples, total RNA was treated with DNase I (Sigma-Aldrich). 1 μg of DNase I treated RNA samples were subjected to reverse transcription using M-MuLV Reverse Transcriptase (NEB) and 100 ng of random hexamer (Invitrogen) according to the NEB protocol. The resulting cDNA was diluted 5 times as stocks. 500 nM of primers were used and 0.2 μl of the cDNA stock was used as template for a 10 μl reaction according to the Fast SYBR® Green Master Mix protocol. For DNA samples, 25 ng of total DNA were used as template. The house-keeping rho gene was used as endogenous control for normalization (Theis et al., 2007). Primers used for amplification are shown in Table S2.
RNA sequencing
Total RNA was treated with DNase I (Sigma-Aldrich) and subjected to TruSeq Stranded mRNA Library Prep Kit (Illumina) without rRNA depletion and Illumina NextSeq. Reads were aligned to reference genomes using Bowtie and sorted using Samtools. Using a custom script, sorted reads were accessed via Pysam, normalized as reads per million values, and plotted as the average over consecutive windows of 500 base pairs using matplotlib tools for IPython.
Southern blot
20 μg of total DNA prepared from infected cells were digested with restriction enzymes EcoRI and PsiI for 5 hours and resolved on a 1% argarose gel. DNA fragments were transferred from the gel via capillary action to a Hybond membrane (GE Healthcare) using alkaline transfer (Sambrook et al., 1989). Probes for the upstream and downstream fragments were produced via PCR of ΦNM1γ6 DNA using primers W863/W864 and W865/W866, respectively, and α-32P-dATP in addition to regular dNTPs. Hybridization was performed at 65°C overnight in Church buffer (Sambrook et al., 1989).
Supplementary Material
Acknowledgments
We would like to thank Andrew Varble, Gregory Goldberg and Nora Pyenson for critical reading of the manuscript. We are grateful to Gregory Goldberg for initial demonstrations of the csm6-independent chromosomal targeting. We thank the Rockefeller University Genomics Resource Center for performing the RNA-seq experiments. P.S. is supported by a Helmsley Postdoctoral Fellowship for Basic and Translational Research on Disorders of the Digestive System at The Rockefeller University. L.A.M is supported by the Rita Allen Scholars Program, an Irma T. Hirschl Award, a Sinsheimer Foundation Award and a NIH Director’s New Innovator Award (1DP2AI104556-01).
Footnotes
Author contributions. WJ performed all the experiments in this paper. PS purified wild-type and mutant versions of Csm6 and performed biochemical assays. LAM and WJ wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anantharaman V, Makarova KS, Burroughs AM, Koonin EV, Aravind L. Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol Direct. 2013;8:15. doi: 10.1186/1745-6150-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arslan Z, Hermanns V, Wurm R, Wagner R, Pul U. Detection and characterization of spacer integration intermediates in type I-E CRISPR-Cas system. Nucleic Acids Res. 2014;42:7884–7893. doi: 10.1093/nar/gku510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae T, Baba T, Hiramatsu K, Schneewind O. Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol Microbiol. 2006;62:1035–1047. doi: 10.1111/j.1365-2958.2006.05441.x. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Benda C, Ebert J, Scheltema RA, Schiller HB, Baumgartner M, Bonneau F, Mann M, Conti E. Structural model of a CRISPR RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4. Mol Cell. 2014;56:43–54. doi: 10.1016/j.molcel.2014.09.002. [DOI] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- Deghorain M, Van Melderen L. The Staphylococci phages family: an overview. Viruses. 2012;4:3316–3335. doi: 10.3390/v4123316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Garrett RA, Shah SA, Peng X, She Q. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol Microbiol. 2013;87:1088–1099. doi: 10.1111/mmi.12152. [DOI] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg GW, Jiang W, Bikard D, Marraffini LA. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature. 2014;514:633–637. doi: 10.1038/nature13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Cocozaki A, Li H, Terns RM, Terns MP. Target RNA capture and cleavage by the Cmr type III-B CRISPR-Cas effector complex. Genes Dev. 2014;28:2432–2443. doi: 10.1101/gad.250712.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum-Aslan A, Maniv I, Marraffini LA. Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. Proc Natl Acad Sci USA. 2011;108:21218–21222. doi: 10.1073/pnas.1112832108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum-Aslan A, Maniv I, Samai P, Marraffini LA. Genetic Characterization of Antiplasmid Immunity through a Type III-A CRISPR-Cas System. J Bacteriol. 2014;196:310–317. doi: 10.1128/JB.01130-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum-Aslan A, Samai P, Maniv I, Jiang W, Marraffini LA. A ruler protein in a complex for antiviral defense determines the length of small interfering CRISPR RNAs. J Biol Chem. 2013;288:27888–27897. doi: 10.1074/jbc.M113.499244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heler R, Marraffini LA, Bikard D. Adapting to new threats: the generation of memory by CRISPR-Cas immune systems. Mol Microbiol. 2014;93:1–9. doi: 10.1111/mmi.12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heler R, Samai P, Modell JW, Weiner C, Goldberg GW, Bikard D, Marraffini LA. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015;519:199–202. doi: 10.1038/nature14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Krupovic M, Yutin N. Evolution of double-stranded DNA viruses of eukaryotes: from bacteriophages to transposons to giant viruses. Ann N Y Acad Sci. 2015;1341:10–24. doi: 10.1111/nyas.12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiswirth BN, Lofdahl S, Betley MJ, O’Reilly M, Schlievert PM, Bergdoll MS, Novick RP. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- Kwan T, Liu J, DuBow M, Gros P, Pelletier J. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc Natl Acad Sci U S A. 2005;102:5174–5179. doi: 10.1073/pnas.0501140102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011a;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011b;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manica A, Zebec Z, Steinkellner J, Schleper C. Unexpectedly broad target recognition of the CRISPR-mediated virus defence system in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 2013;41:10509–10517. doi: 10.1093/nar/gkt767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Nunez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat Struct Mol Biol. 2014;21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez JK, Lee AS, Engelman A, Doudna JA. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature. 2015;519:193–198. doi: 10.1038/nature14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Espino D, Morovic W, Sun CL, Thomas BC, Ueda K, Stahl B, Barrangou R, Banfield JF. Strong bias in the bacterial CRISPR elements that confer immunity to phage. Nature communications. 2013;4:1430. doi: 10.1038/ncomms2440. [DOI] [PubMed] [Google Scholar]
- Peng W, Feng M, Feng X, Liang YX, She Q. An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res. 2015;43:406–417. doi: 10.1093/nar/gku1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- Ramia NF, Spilman M, Tang L, Shao Y, Elmore J, Hale C, Cocozaki A, Bhattacharya N, Terns RM, Terns MP, et al. Essential structural and functional roles of the Cmr4 subunit in RNA cleavage by the Cmr CRISPR-Cas complex. Cell Rep. 2014;9:1610–1617. doi: 10.1016/j.celrep.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samai P, Pyenson N, Jiang W, Goldberg GW, Hatoum-Aslan A, Marraffini LA. Co-transcriptional DNA and RNA Cleavage during Type III CRISPR-Cas Immunity. Cell. 2015;161:1164–1174. doi: 10.1016/j.cell.2015.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Staals RH, Zhu Y, Taylor DW, Kornfeld JE, Sharma K, Barendregt A, Koehorst JJ, Vlot M, Neupane N, Varossieau K, et al. RNA Targeting by the Type III-A CRISPR-Cas Csm Complex of Thermus thermophilus. Mol Cell. 2014;56:518–530. doi: 10.1016/j.molcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamulaitis G, Kazlauskiene M, Manakova E, Venclovas C, Nwokeoji AO, Dickman MJ, Horvath P, Siksnys V. Programmable RNA Shredding by the Type III-A CRISPR-Cas System of Streptococcus thermophilus. Mol Cell. 2014;56:506–517. doi: 10.1016/j.molcel.2014.09.027. [DOI] [PubMed] [Google Scholar]
- Theis T, Skurray RA, Brown MH. Identification of suitable internal controls to study expression of a Staphylococcus aureus multidrug resistance system by quantitative real-time PCR. J Microbiol Methods. 2007;70:355–362. doi: 10.1016/j.mimet.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Weinberger AD, Gilmore MS. A CRISPR View of Cleavage. Cell. 2015;161:964–966. doi: 10.1016/j.cell.2015.05.003. [DOI] [PubMed] [Google Scholar]
- Weinberger AD, Wolf YI, Lobkovsky AE, Gilmore MS, Koonin EV. Viral diversity threshold for adaptive immunity in prokaryotes. MBio. 2012;3:e00456–00412. doi: 10.1128/mBio.00456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, van Erp PB, Kunne T, Wong SP, Staals RH, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, van Duijn E, Bultema J, Waghmare S, Zhou K, Barendregt A, Westphal W, Heck A, Boekema E, Dickman M, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci USA. 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, Reimann J, Cannone G, Liu H, Albers SV, et al. Structure and Mechanism of the CMR Complex for CRISPR-Mediated Antiviral Immunity. Mol Cell. 2012;45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.