Abstract
Mitochondria are dynamic organelles that change their architecture in normal physiological conditions. Mutations in genes that control mitochondrial fission or fusion, such as dynamin-related protein (Drp1), Mitofusins 1 (Mfn1) and 2 (Mfn2), and Optic atrophy 1 (Opa1), result in neuropathies or neurodegenerative diseases. It is increasingly clear that altered mitochondrial dynamics also underlie the pathology of other degenerative diseases, including Parkinson’s disease (PD). Thus, understanding mitochondrial distribution, shape, and dynamics in all cell types is a prerequisite for developing and defining treatment regimens that may differentially affect tissues. The majority of Drosophila genes implicated in mitochondrial dynamics have been studied in the adult indirect flight muscle (IFM). Here, we discuss the utility of Drosophila third instar larvae (L3) as an alternative model to analyze and quantify mitochondrial behaviors. Advantages include large muscle cell size, a stereotyped arrangement of mitochondria that is conserved in mammalian muscles, and the ability to analyze muscle-specific gene function in mutants that are lethal prior to adult stages. In particular, we highlight methods for sample preparation and analysis of mitochondrial morphological features.
Keywords: Drosophila, mitochondria, larval musculature
1. Introduction
Mitochondria have a long and storied history. The first observations that describe these intracellular organelles date back to the beginning of the1840s (Aubert, 1852; Butschli, 1871; Flemming, 1882; Henle, 1841; Kolliker, 1856, 1888). For over 150 years, the field of ‘mitochondriology’ has focused on multiple facets of mitochondrial composition and function. Only since the 1970s has the focus to mitochondrial fission/fusion and biogenesis shed new light on the role of these powerhouse organelles in biology (Ernster and Schatz, 1981). Most of the adenosine triphosphate (ATP) within cells is generated by mitochondria, and muscle cells in particular, require abundant amounts of ATP to carry out mechanically and energetically demanding functions, including muscle contraction, ion transport, protein synthesis, and other general metabolic roles (Nunnari and Suomalainen, 2012). The diversity in mitochondrial morphology, which varies widely in size from small, individual mitochondrion to highly interconnected, tubular networks, is influenced by regulated fission and fusion events that are collectively referred to as ‘mitochondrial dynamics.’ Multiple factors influence the ability of cells to rapidly adapt to intracellular or extracellular cues to regulate mitochondrial morphology, including cell type, organism, and environmental stressors (Rafelski, 2013).
1.2 The delicate balance between fission and fusion determines mitochondrial fate
The opposing processes of fission and fusion determine the architecture of the mitochondrial network within cells and influence the balance between organelle life and death. Fusion events are thought to bolster fitness, permitting normal and slightly damaged organelles to intermingle mitochondrial DNA (mtDNA), membrane components, and metabolic enzymes (see Twig, et al., 2011). While many of the signals that promote organelle fusion are not yet defined, it is clear that polarized mitochondria with normal or slightly reduced inner membrane potentials (ΔΨm) are competent to fuse with other mitochondria (Legros et al., 2002; Mattenberger et al., 2003). Recent findings also suggest that fusion mechanisms are maintained in mitochondria undergoing macroautophagy, thus enabling a constant supply of ATP during nutrient starvation (Gomes et al., 2011; Rambold et al., 2011).
Healthy mitochondria also undergo fission, for example, to segregate mitochondria into daughter cells during cell division (Mishra and Chan, 2014). In contrast, potentially dysfunctional mitochondria that have decreased or complete loss of ΔΨm, cannot fuse with other mitochondria and are fated to undergo fission, resulting in fragmented, depolarized mitochondria (Ishihara et al., 2003; Legros et al., 2002; Malka et al., 2005). This feature ensures that healthy mitochondria do not fuse with damaged organelles. Typically, these smaller, depolarized mitochondria are selectively targeted for mitophagy, a selective autophagic process that eliminates damaged mitochondria (Gomes and Scorrano, 2013; Shirihai et al., 2015; Tanaka et al., 2010; Twig and Shirihai, 2011).
Numerous proteins located in the outer (OMM) or inner (IMM) mitochondrial membranes are required for mammalian mitochondrial fission or fusion. The predominant regulator of mitochondrial fission is dynamin-related protein 1 (Drp1). Drp1 is an evolutionarily conserved GTPase that shares structural similarity to dynamin and is located predominantly in the cytoplasm (Bleazard et al., 1999; Smirnova et al., 2001). Upon signals that induce fission, Drp1 translocates to mitochondria, oligomerizes to form rings around the OMM that eventually results in the division into two separate organelles (Ingerman et al., 2005; Smirnova et al., 2001). Core proteins essential for mitochondrial fusion include the dynamin-like GTPase OMM components Mitofusin 1 (Mfn1) and 2 (Mfn 2), and the IMM protein Optic atrophy 1 (Opa1). The biological importance of these proteins is highlighted by human diseases resulting from mutations in these fission/fusion genes. Mutation in Drp1 lead to markedly elongated, tubular mitochondria and microcephaly that results in infant death (Waterham et al., 2007). Mutations in Mfn2 are associated with Charcot-Marie-Tooth Type 2A (CMT2A) neuropathy and dominant optic atrophy (DOA) is caused by lesions in Opa1 (Alexander et al., 2000; Delettre et al., 2000; Klein et al., 2011; Züchner et al., 2004).
Aside from mutations in the fission/fusion genes that result in the neuropathies mentioned above, alterations in other genes that influence mitochondrial dynamics are associated with neurodegenerative disorders, cardiometabolic diseases, and cancer (Archer, 2013; Chen and Chan, 2009; Liesa et al., 2009; Shirihai et al., 2015). Prevalent diseases linked to defects in mitochondrial behavior are Alzhemier’s disease and PD. Patients with PD, the second most common neurodegenerative disease, undergo a progressive loss of the dopaminergic neurons in the substantia nigra and symptoms include tremors, bradykinesia, postural instability, and rigidity (Postuma et al., 2015; Riekkinen et al., 1975). The majority of PD cases are sporadic, while approximately 10% of familial PD cases are caused by mutations in genes that encode for parkin, PTEN-induced kinase (PINK1), leucine-rich repeat kinase 2 (LRRK2), α-synuclein, and DJ-1 (Bentivoglio et al., 2001; Bonifati et al., 2003; Kitada et al., 1998; Polymeropoulos et al., 1997; Valente et al., 2001; van Duijn et al., 2001; Zimprich et al., 2004). While a number of recent excellent reviews discuss abnormal mitochondrial homeostasis as a key aspect in PD progression (Archer, 2013; Chen and Chan, 2009; Liesa et al., 2009; Shirihai et al., 2015), here we will discuss how the mitochondrial fission/fusion genes Drp1 and Mfn are linked to PD.
Mouse models of parkin and PINK1 deficiencies have subtle phenotypes that fail to adequately reproduce the common features observed in PD patients, whereas the corresponding mutants in flies show severe mitochondrial defects, predominantly in the adult flight muscle. PINK1 or parkin mutant flies display the same reduced lifespan, locomotor defects, and degenerative flight muscle with enlarged mitochondria and internal disrupted cristae (Clark et al., 2006; Greene et al., 2003; Park et al., 2006; Pesah et al., 2004; Yang et al., 2006). This swollen mitochondrial phenotype prompted studies to examine if PINK1 and parkin influence mitochondrial morphology using the fly model. Indeed, excess Drp1 or genetic loss of Opa1 or Marf (a fly homolog of Mfn 1 and Mfn 2), ameliorated the mitochondrial morphology and associated pathological phenotypes present in PINK1 or parkin mutants (Deng et al., 2008; Poole et al., 2008; Yang et al., 2008). Parkin is an E3-ubiquitin ligase that targets multiple substrates for degradation, one of which is Mfn. Consistent with this, loss of fly Parkin results in increased fusion events, presumably due to the accumulation of Mfn protein levels (Poole et al., 2010; Shiba-Fukushima et al., 2014; Ziviani et al., 2010). In mammalian cell culture experiments, loss of Parkin or PINK1 also results in increased Mfn protein levels (Gegg et al., 2010; Rakovic et al., 2011; Tanaka et al., 2010). Drp1 has also been shown to be a substrate for Parkin ubiquitination and protein degradation (Wang et al., 2011), although further studies are necessary to confirm these findings in vivo.
1.3 Examination of mitochondrial morphology in muscle tissue
Characterization of new mutants that may alter mitochondrial structure or function in muscle tissue requires a reliable method for quantitating organelle geometry. Computerized methods have been developed to rapidly assess mitochondrial dynamics in mammalian cell culture models (Chevrollier et al., 2012; Koopman et al., 2006; Leonard et al., 2015; Lihavainen et al., 2012; Peng et al., 2011; Reis et al., 2012). These methods have clear advantages, including automated fluorescence microscopy followed by high throughput computational binning of mitochondria into distinct morphological subclasses. However, this technology is not available to all research labs and is not ideal for in vivo studies. To address the significance of mitochondrial morphology in tissue, 3-dimensional (3-D) reconstructions using either confocal microscopy or electron microscopy provide evidence that the subsarcolemmal (SS) and intermyofibrillar (IMF) mitochondrial membranes physically interact in mouse and human skeletal muscle (Dahl et al., 2015; Picard et al., 2013). A recent paper extends these studies using quantitative 2-D transmission electron microscopy (TEM) to show that fusion indexes (Mfn2:Drp1 ratio) increase in aged muscle, possibly related to changes in mitochondrial morphology that contribute to mitochondrial dysfunction and sarcopenia (Leduc-Gaudet et al., 2015). The balance between fission and fusion seems to shift in favor of fission as myofibers differentiate. Elongated and interconnected mitochondria in myoblasts become more fragmented during myotube differentiation in the cardiomyocyte cell line H9c2 (Yi et al., 2012). Similarly, subsarcolemmal and longitudinal clusters of mitochondria are present in fast twitch muscles from young mice and become progressively smaller, rounder, and more abundant as myofibers age (Boncompagni et al., 2009). The mitochondria also undergo a positional shift and become restricted to I-bands as muscle maturation proceeds. Accordingly, there is a decrease in mitochondrial fission/fusion events (Eisner et al., 2014).
Mouse knockouts in the fission/fusion genes all result in embryonic lethality and mitochondrial abnormalities (Alavi et al., 2007; Chen et al., 2003; Davies et al., 2007; Ishihara et al., 2009; Wakabayashi et al., 2009). To examine the functional significance of these genes in striated tissues, conditional inactivation of Drp1, Mfn1, and Mfn2 have been performed. Mice deficient for Drp1 in both cardiac and skeletal tissue [muscle-specific (MS)-Drp1-KO] exhibit neonatal lethality due to dilated cardiomyopathy (Ishihara et al., 2015). Ultrastructural studies show enlarged mitochondria in heart tissue, but no observable abnormalities in skeletal muscle. It is possible that the early lethality of the MS-Drp1-KO pups precluded analysis of morphological defects that could accumulate with increase lifespan. Mice depleted for both Mfn1 and Mfn2 in fast-twitch skeletal muscle are smaller than their single KO counterparts, die at 6–8 weeks of age, and display fragmented mitochondria indicative of fusion defects (Chen et al., 2010). The high concentration of spherical mitochondria in the interfibrillar space also alters myofibril alignment. A haploinsufficient mouse model for the Opa1 gene (Opa1+/−) leads to increased mitochondrial size with abnormal cristae in both cardiac and skeletal tissue (Caffin et al., 2013; Piquereau et al., 2012). Thus, it is well-established that altering mitochondrial dynamics in contractile tissue has severe consequences for the organism.
2. Mitochondria in Drosophila muscle tissue
Drosophila has emerged as an excellent model to study mitochondrial dynamics. First, there are a variety of methods to create loss-of-function alleles or abrogate gene function using RNAi. In addition, the ‘simpler’ genome of the fly allows for the analysis of gene function without functional redundancy; two Mfn genes in mammals are replaced by Marf in Drosophila (note that another homolog of Mfn, called fuzzy onions, is testes-specific). Finally, numerous genes that affect either mitochondrial structure and/or function have already been characterized in Drosophila muscle tissue, providing a framework for the characterization of new genes that affect mitochondrial morphology.
A number of studies have successfully utilized Drosophila tissues to analyze the dynamic nature of mitochondria fission and fusion. Schneider 2 (S2) cells, a commonly used Drosophila cell line, are relatively small cells (diameter of ~15–20 μm) that can be grown as a semi-adherent monolayer or in suspension (Rogers and Rogers, 2008). Plating these cells on coverslips coated with concanavalin A (ConA) allows the cells to spread out for the visualization of cytosolic organelles or proteins. The tubular network of mitochondria is easily seen using this approach (Fig. 1A). A closely related cell line, called S2 receptor plus (S2R+) do not require ConA treatment, but can directly adhere to glass coverslips (Yanagawa et al., 1998). Both of these cell lines are amenable to gene inhibition using RNAi and have proven useful in identifying key regulators that alter the mitochondrial fission/fusion balance (Lutz et al., 2009; Pogson et al., 2014; Ziviani et al., 2010). S2 cells were originally derived from late embryonic stage Drosophila embryos (Schneider, 1972) and exhibit behaviors characteristic of phagocytic hemocytes (Rämet et al., 2002). Thus, examination of additional cell types is imperative to define specific mitochondrial behaviors and to understand how mitochondria are influenced in vivo by other tissues. Here we focus on Drosophila muscle tissue, where increased energy consumption requires mitochondria as a powerhouse for ATP production.
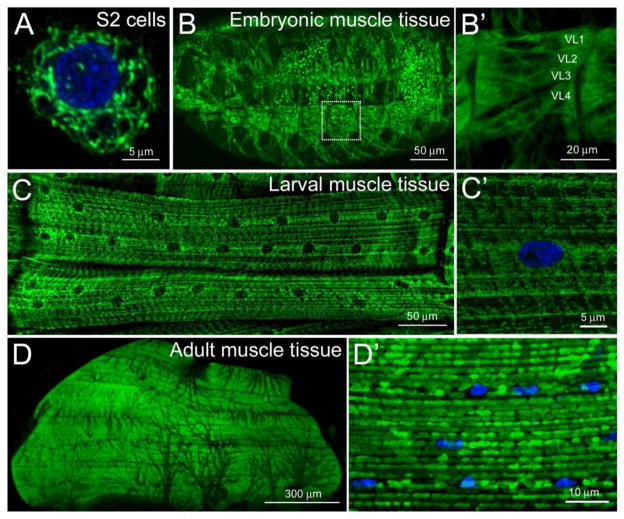
Figure 1. Mitochondrial distribution in Drosophila muscle tissues.
(A–D′) Confocal micrographs of mitochondria immunostained with an antibody against the IMM ATP synthase complex (green) in the indicated cell types. Nuclei are stained with DAPI (blue). (A) An assortment of mitochondrial shapes are observed in S2 cells plated on ConA. (B, B′) Mitochondria are abundant and uniformly distributed in embryonic muscle tissue (B). High magnification of the ventral longitudinal muscles 1–4 (also designated 6, 7, 12, and 13) shows an overall ubiquitous mitochondrial distribution with a slight accumulation at the myofiber ends (B′). (C, C′) In the contractile muscles of third instar larvae, ventral longitudinal muscles 1 and 2 (12 and 13) accumulate around nuclei and adopt an overall striated appearance (C). Tubular mitochondria can be viewed at the surface of the muscle (C′). Mitochondria appear homogeneous in the adult thoracic flight muscle (D) and adopt a round appearance at higher magnification (D′). All images were acquired using a Zeiss 700 confocal microscope and processed using ImageJ and Adobe Photoshop software. Scale bars are indicated.
Muscle formation occurs at two distinct stages in the Drosophila lifecycle (Weitkunat and Schnorrer, 2014). Muscles required for larval locomotion form through multiple rounds of myoblast fusion events in embryogenesis during stages 12–16 [7.5–16.5 h after egg laying (AEL)] (Abmayr et al., 2008; Haralalka and Abmayr, 2010; Schejter and Baylies, 2010). As shown in Fig. 1B, the mitochondria, stained with an antibody generated against mitochondrial Complex V (α-ATPase 5 α; Mitosciences), are evenly dispersed throughout the myoplasm. However, the relatively small size of these multinucleated, embryonic muscles are not ideal for a detailed subcellular examination of mitochondrial structure (Fig. 1B′). The assembly of the contractile apparatus and onset of muscle contraction near the end of embryogenesis allow for hatching into the first of three larval instar stages. All three larval instar stages undergo a rapid increase in organismal mass, mainly due to an increase in the size of polyploid cells, such as muscles, which retain the same general muscle patterning acquired during embryonic development. Thus, at the wandering third instar stage, each individual muscle cell is at its largest size and volume. The mitochondria in larval muscles are abundantly distributed along the length of the myofibers (Fig. 1C) and appear tubular in appearance when viewed at high magnification (Fig. 1C′).
The second phase of muscle development occurs in the pupal stage, where remodeling of existing larval scaffolds combined with de novo muscle biogenesis result in adult muscles required for flight, jumping, and mating (Weitkunat and Schnorrer, 2014). The IFMs of the adult thorax exhibit high metabolic activity and accordingly, are loaded with mitochondria (Fig. 1D). The observation that loss of parkin or PINK1 function results in abnormal mitochondrial morphology followed by degenerating IFMs (Clark et al., 2006; Greene et al., 2003; Park et al., 2006; Yang et al., 2006) has intensified the search for additional genes that influence mitochondrial structure and/or function in muscle tissue. While the IFMs are widely used as a model to study genes implicated in PD, mitochondria in WT muscles are typically round and analysis of mitochondrial shape is limited by the close apposition of the myofibrils and mitochondria (Fig. 1D′).
In our analysis of genes that affect mitochondrial structure and/or function, we and others have turned to the larger, L3 contractile muscles (Fig. 1C) as a paradigm to analyze genes that control mitochondrial dynamics (Debattisti et al., 2014) and to examine genes that are implicated in PD (Vos et al., 2012; Wang et al., 2015). Here we discuss approaches to assess the geometry of the mitochondrial network and to monitor the subcellular localization and dynamics of mitochondria in either fixed tissue followed by microscopy or live imaging of fluorescently-tagged proteins in Drosophila larval tissue.
2.1 Mitochondria distribution, or positioning
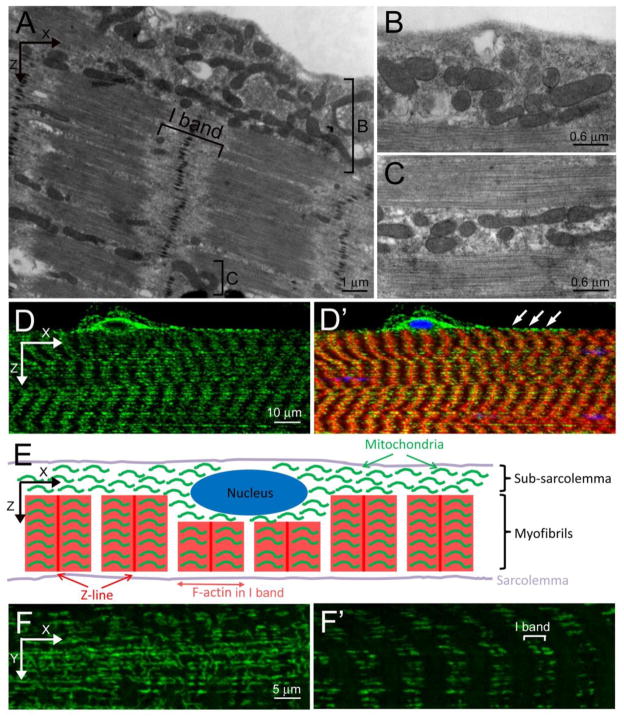
Mammalian skeletal muscle possesses an unequal distribution of mitochondria throughout muscle fibers. The SS mitochondria are clustered between the sarcolemma and myofibrils and the IMF mitochondria are scattered between myofibrils, but in tight register with the I-band, the region that surrounds the Z-line (Miledi and Slater, 1968; Müller, 1976; Picard et al., 2013). This asymmetric spatial organization is also mirrored in Drosophila L3 muscles when viewed by transmission electron microscopy (TEM) (Figs. 2A–C). A population of mitochondria accumulate in the SS space (Fig. 2B), while other mitochondria cluster between adjacent myofibrils (Fig. 2C). While TEM is excellent to examine the overall structure of an individual mitochondrion, this technique is neither cost nor time efficient to adequately address the spatial organization or abundance of mitochondria in multiple genetic backgrounds. Standard immunostaining techniques followed by confocal microscopy show a similar mitochondrial distribution with substantial clustering of mitochondria around myonuclei at the surface of the muscle and a tight association of mitochondria along the Z-line (arrowheads) in each sarcomeric unit (Fig. 2D–E). High magnification confocal images reveal an interconnected, tubular network for the SS mitochondria (Fig. 2F), while pairs of mitochondria align along the Z-line internally within muscle cells (Fig. 2F′).
Figure 2. Mitochondrial positioning in larval muscles.
(A–C) TEM images of L3 muscles. (A) Mitochondria are loosely organized in the subsarcolemmal space (bracket labeled B) and appear closer together between the myofibrils (bracket labeled C). Panels B and C are higher magnification images corresponding to the regions bracketed in A. (D, D′) Confocal micrographs of a lateral view of L3 muscle stained with anti-ATPase 5α to label mitochondria (green), sarcomeric actin (red) and nuclei (blue). (E) Schematic illustration of mitochondrial distribution in a Drosophila L3 muscle cell. (F, F′) Anti-ATPase 5α immunostaining of mitochondria (green) in the subsarcolemmal plane (F) or within the myofibers (F′). Scale bars are indicated.
2.2 Visualization and analysis of individual mitochondrion shape
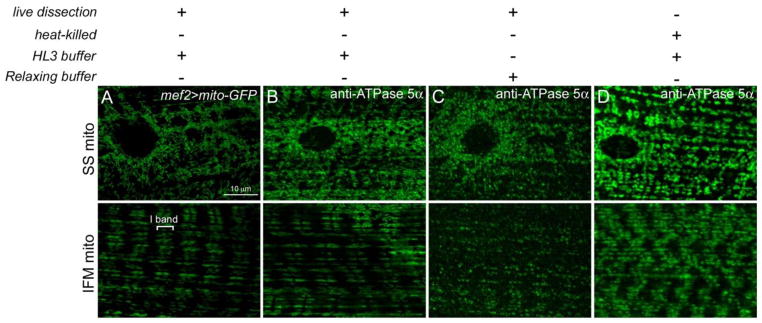
Live dissection of larvae that express green fluorescent protein (GFP) fused to a mitochondrial import signal (mito-GFP) in the muscle using the GAL4/UAS system (Brand and Perrimon, 1993) show long, tubular mitochondria near the muscle surface and an internal accumulation of elongated mitochondria consistent with I-band localization (Fig. 3A). An alternative technique that circumvents recombination of the UAS-mito-GFP organelle marker into mutant backgrounds is immunostaining with a mitochondrial-specific antibody. Live dissections, followed by fixation and tissue staining with an antibody against the ATPase/Complex V subunit (Fig. 3B) preserves the mitochondrial morphology observed in live preparations with mito-GFP (Fig. 3A). Dissection buffers and fixation conditions are an important consideration when preparing tissue for mitochondrial immunolocalization studies. Dissection of live larvae in HL3 buffer retains normal mitochondrial morphology (Fig. 3B), while performing these same larval fillets in relaxing buffer (Molnár et al., 2014; Vogler and Ocorr, 2009) alters both the shape and distribution of mitochondria (Fig. 3C). Moreover, heat-killing L3 individuals for immobilization prior to dissection is a common method in standard immunostaining protocols (Galko and Krasnow, 2004; Weaver and White, 1995; Yin et al., 2007). However, this can only be used for certain antigens and is not suitable for studying mitochondrial morphology. The mitochondria in heat-killed larvae lose their elongated shape and instead form ring-like structures (Fig. 3D).
Figure 3. Comparison of dissection methods to ensure proper mitochondrial morphology.
(A–D) SS (top panels) and IMF (bottom panels) mitochondria in L3 muscle tissue in mef2-GAL4>mito-GFP (A) or WT larvae (B–D). (A, B) Live dissection of larvae in hemolymph-like (HL3) buffer (70 mM NaCl; 5 mM KCl; 1.5 mM CaCl2; 20 mM MgCl2; 10 mM NaHCO3; 5 mM trehalose; 115 mM sucrose; 5 mM HEPES; pH = 7.2) followed by fixation in 4% formaldehyde with GFP-tagged mitochondria under control of the muscle-specific mef2-GAL4 driver (A) or immunostaining with an anti-ATPase 5α antibody in WT larvae (B) both show longer, tubular mitochondria. (C) Performing larval fillets in relaxing solution (20 mM phosphate buffer, pH = 7.0; 5 mM MgCl2, 5 mM EGTA, 5 mM ATP), a common buffer used in muscle biology, altered the mitochondrial morphology and positioning. (D) Larvae were heat-killed in 65°C water, put on ice for 5 min, filleted and fixed in 4% formaldehyde prior to immunostaining with anti-ATPase 5α. The mitochondria form ring-like structures that differ in morphology from live dissected larvae. Scale bars are indicated.
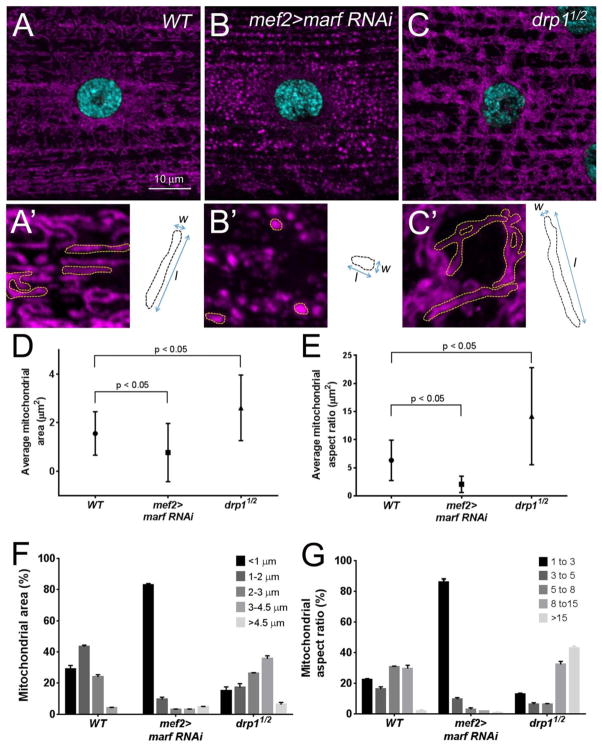
The large size of larval muscle cells makes them amenable to visualize the shape of single mitochondrion. The SS mitochondria, in relatively the same plane as the nuclei, are easier to quantify compared to the IFM mitochondria that appear more compact within sarcomeres. The majority of WT mitochondria are long and tubular (Fig. 4A,A′) and assume a fragmented appearance upon RNAi knockdown of the fusion gene marf (Fig. 4B,B′). In contrast, loss of Drp1 levels leads to longer, interconnected tubules (Fig. 4C,C′). These qualitative observations can be reproducibly measured for the comparison of mitochondrial networks amongst different genetic backgrounds. Two measurements are especially informative when classifying mitochondrial attributes: mitochondria size distribution and aspect ratio. Mitochondrial size measures the area encompassed by an individual mitochondrion and the aspect ratio is a determinant of the length/width proportion. The average mitochondrial area (Fig. 4D) and average mitochondrial aspect ratios (Fig. 4F) are decreased upon loss of marf and increased in a drp1 mutant background. As mitochondria are dynamic, a snapshot at any one time reveals variability in the ranges of both mitochondrial area (Fig. 4E) and the mitochondrial aspect ratio (Fig. 4G); inhibition of fusion by knockdown of marf skews nearly all mitochondria towards circular organelles, while decreasing fission shows a range of areas and sizes, all larger than controls. These approaches provide a reliable quantitative method for characterizing new mutants that alter mitochondrial fission or fusion.
Figure 4. Quantitation of mitochondrial shape.
(A–C′) Immunofluorescent images of the SS mitochondria (magenta) surrounding a single nuclei (blue) in L3 muscles. Individual organelles are outlined in yellow (A′-C′). (A, A′) Mitochondria in WT muscles exhibit a variety of shapes, being predominantly tubular. Knockdown of the fusion gene marf results in small, circular mitochondria (B, B′), while longer organelles are present upon loss of the fission gene drp1 (C,C′). (D–G) Quantitation of mitochondrial area illustrated by dotted black lines (shown in A′-C′) and the aspect ratio (mitochondrial length/width as shown in A′-C′). Representative images of mitochondrial immunostaining were taken at 63×/1.6× zoom (0.34 μm/step) around one nucleus from muscle 6 or 7. ImageJ was used to stack all images from the sarcolemmal surface and for subsequent mitochondrial morphology analysis. For quantification of mitochondrial area, individual mitochondrion were outlined using the freehand selection tool and the area (pixels2) was calculated using the measure function. The area was converted to μm2 based upon the magnification used for image acquisition (1 pixel2 = 0.0036 μm2). To calculate the aspect ratios, the freehand selection tool and measure function were used to determine the distance of the major [length (l)] and minor [width (w)] axes. The raw data was transferred into Microsoft Excel to calculate the aspect ratio (l/w). Computed values were imported into Prism5 and one-way ANOVA was used for statistical analysis (mean = +/− SEM). n = 180 individual mitochondrion for each genotype. (D, E) The average area of a mitochondria is decreased in marf RNAi and increased in drp1 mutant muscles. (F, G) This same trend is apparent for measurements of the mitochondrial aspect ratio. Scale bars are indicated.
2.3 Measurement of mitochondrial ΔΨm
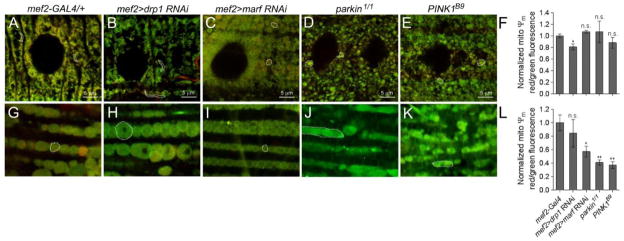
Often increased fission preceeds autophagy in muscle tissue. However, it cannot be assumed that all smaller mitochondria are destined for destruction. Use of a probe that monitors inner mitochondrial ΔΨm can be used as a readout for mitochondrial health. JC-1 is a cationic fluorescent dye whose accumulation is potential-dependent (Chazotte, 2011). The monomeric form of JC-1 emits at ~529 nm (green) and accumulates proportionally with the strength of the membrane potential. Further accumulation of the dye at higher concentrations or potentials forms red fluorescent aggregates with an emission maximum of ~590 nm (red). Thus, measurement of the relative red:green fluorescence signals is a reliable indicator of ΔΨm and overall viability. Quantitative analysis of JC-1 stained larval muscle reveals a steady state ratio of red (high ΔΨm):green (low ΔΨm) in mef2-GAL4 control muscle tissue (Fig. 5A) and a decrease upon RNAi knockdown of drp1 (Fig. 5B,F). Consistent with previous reports, loss of marf (Fig. 4B, 5C), parkin (Fig. 5D), or PINK1 (Fig. 5E), also result in altered mitochondrial morphology (Vos et al., 2012; Wang et al., 2015), but does not affect mitochondrial ΔΨm in larval tissue (Fig. 5F). We confirmed the functionality of JC-1 as a probe by analyzing the same genotypes in adult muscle tissue. Aside from loss of Drp1 (Fig. 5H), marf, parkin, and PINK1 mutants all exhibited decreased red:green fluorescence ratios indicative of decreased ΔΨm (Figs. 5I–L).
Figure 5. Measuring mitochondrial ΔΨm.
(A–E, G–K) JC-1 staining of the mitochondria in larval muscles (A–E) or the adult musculature (G–K) in the indicated genotypes. Individual mitochondrion are outlined (dotted lines). (F,L) Quantitative measurements of the red (high ΔΨm):green (low ΔΨm) ratio in larvae or adult muscles. Live dissections of larvae or bisections of the adult thorax were performed in HL3 buffer and JC-1 (in HL3 buffer) was added at 1:800 dilution for 10 min at 25° C. Samples were washed twice and mounted for imaging. Fluorescence micrographs were taken at excitation wavelengths of 488 and 555 nm. The raw data was transferred into Microsoft Excel to calculate the red:green fluorescence ratios. Computed values were imported into Prism5 and a student-test was used for statistical analysis (mean = +/− SEM). n = 6 for each genotype. (n.s., not significant, * P<0.05, **P<0.01) Scale bars are indicated.
2.4 Analysis of mitochondrial dynamics
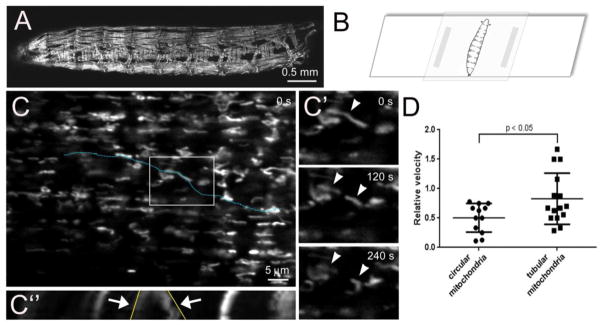
Immunostaining of mitochondria in fixed tissue provides limited information concerning the mitochondrial balance between fission and fusion. Imaging of mitochondria in live muscle tissue provides a method to analyze the dynamics, velocity, and directionality of mitochondrial movement. The dark exoskeleton of the adult cuticle does not easily allow for imaging of mitochondrial dynamics in the IFMs. Thus, we modified a method for imaging through the translucent cuticle of L3 animals to directly monitor mitochondrial movement in the musculature (Brechbiel and Gavis, 2008; Miller et al., 2005; Nienhaus et al., 2012). Crawling L3 larvae expressing UAS-mito-GFP under control of the mef2-GAL4 promoter (Fig. 6A) were placed on a microscope slide and covered with a 15–25% choroform:water solution for approximately 5 minutes to inhibit muscle contractions. The excess chloroform:water mixture was removed and a coverslip was placed on top of the larvae before image acquisition (Fig. 6B). Slight pressure against the edge of the coverslip allowed the larvae to roll for selection of the muscles to be imaged.
Figure 6. Live imaging of mitochondria in Drosophila larval muscles.
(A) Low magnification image of mef2-GAL4>mito-GFP larvae to visualize all muscles that express fluorescently-tagged mitochondria. (B) Larvae were immobilized in a 15–25% chloroform:water solution for 5 minutes and mounted on a glass slide for immediate imaging through the cuticle using a 63× oil lens. A Z-stack series (0.38 μm/interval) was acquired every 30 s for 4.5 minutes. Images were processed in Image J using the StackReg and PoorMan3Dreg plugins. (C) Projection of Z-stack images of dorsal oblique 4 (also called muscle 19) collected at t = 0 seconds (s). (C′) The boxed region in C at the indicated time points reveal both dynamic movement and fission events (arrowheads) of GFP-labeled mitochondria. (C″) Kymograph analysis of mitochondrial dynamics and motility along the linear ROI in C (blue dotted line). The x-axis in the kymograph illustrates the distance along the blue dotted line. Fission events are observed over time (y-axis, arrows) and shown in the XY plane in C′. The angle of the lines drawn along the contrast edge of the indicated mitochondria (yellow line) are proportional to the velocity. (F) In general, the circular mitochondria move less than tubular mitochondria in Drosophila skeletal muscle. Kymographs and relative velocities were generated using the Multiple Kymograph plugin and Velocity tsp macro in Image J. The raw data was imported into Graphpad Prism for the generation of graphs and a student-test was used for statistical analysis (mean = +/− SEM). Scale bars are indicated.
Analysis of individual mitochondria in series of x-y maximum projections at the indicated time points revealed a long, tubular mitochondrion splitting into two organelles (Fig. 6C, C′). Kymographs were generated and plotted as an x-time (t) scan to follow division of this mitochondrion over time. The x-axis represents the selected line of interest (blue dotted line in Fig. 6C) and the y-axis represents time. The resulting x-t image (Fig. 6C″) depicts the fission event in Fig. 6C′ and illustrates the relative displacement per time interval by tracing the angle of moving organelle after Kymograph analysis (yellow line). This type of analysis in multiple mitochondria revealed that tubular, elongated mitochondria appeared more dynamic than circular mitochondria (Fig. 6D).
3. Conclusion and future perspectives
The majority of Drosophila studies that focus on mitochondrial morphology have used S2 cells or the adult IFMs. In this article, we highlight the advantages of using the L3 musculature as a model for mitochondrial dynamics. First, L3 muscles are relatively large with multiple small nuclei, allowing for the easy visualization of mitochondria throughout the myoplasm. As muscle requires high mitochondrial ATP production, these tissues also tend to be sensitive to defects in mitochondrial structure and/or function. This feature, combined with the ability to easily manipulate gene function in the fly model, provide a platform for the identification and characterization of new genes essential in mitochondrial dynamics. Specifically, alterations in the elongated, tubular morphology of mitochondria in L3 muscles can be reproducibly quantified for changes in fission and/or fusion events, as illustrated by our analysis of marf and drp1 loss-of-function muscles. Moreover, live imaging through the larval cuticle allows for the analysis of mitochondrial dynamics over time. Finally, the conservation of muscle-specific mitochondrial distribution between Drosophila and mammals makes the fly an attractive genetic model to easily manipulate gene function, providing a platform for the identification and characterization of new genes essential in mitochondrial dynamics and human disease.
Drosophila larvae increase over ~200-fold in body mass from the L1 to the L3 stage. This dramatic growth is thought to be dependent on aerobic glycolysis rather than oxidative phosphorylation (Sen et al., 2013; Tennessen et al., 2011). Thus, while the L3 musculature appears to be a unique model for studying mitochondrial dynamics, it may not be suitable for all aspects of mitochondrial function as the mode of metabolism differs from adult muscle. This idea is supported by our observation that loss of marf, while altering mitochondrial morphology, does not alter mitochondrial ΔΨm in larval muscle tissue, but is reduced upon loss of Mfn/Marf levels in adult muscle (Fig. 5) and in other tissues (Bhandari et al., 2015; Chen et al., 2003; Song et al., 2015). We also noted that the overall ΔΨm appears lower in larval muscle than adult muscle and may not be as sensitive to changes upon loss of Marf, Parkin, and PINK1.
Several recent papers have examined the L3 musculature in marf, parkin, and PINK1 mutants to further understand the role of these proteins in maintaining mitochondrial morphology. In marf RNAi larval muscle, mitochondria are fragmented and cluster around the nuclei (Debattisti et al., 2014). Vos, et al., found that Vitamin K2 rescues PINK1 enlarged mitochondrial phenotypes (Vos et al., 2012). We also found abnormal, clustered mitochondrial phenotypes upon RNAi knockdown of marf and parkin in larval muscles (Wang et al., 2015) and we are currently using the larval musculature to identify new genes that genetically interact with parkin (Wang, et al., submitted). Of note, a recent paper highlights an additional feature of the Drosophila L3 musculature as a model in mapping the mitochondrial matrix proteome using an engineered ascorbate peroxidase (APEX) assay followed by mass spectrometry (Chen et al., 2015).
So why is the architecture of the mitochondrial network important in contractile muscle tissue? Aside from PD, dysfunctional mitochondria result in a heterogeneous group of disorders categorized as mitochondrial diseases that affect various numerous tissues in the body. Peripheral nerves, the cerebrum, and skeletal muscle are most commonly affected, possibly because these tissues tend to require more energy than other tissues (Finsterer, 2007). Loss of mitochondrial function is also thought to be a primary factor in the progressive decline in muscle mass and muscle strength (sarcopenia). Oxygen consumption and mitochondrial protein content decrease with age, in addition to an increase in oxidative stress and mitochondria-mediated apoptosis (Johnson et al., 2013; Peterson et al., 2012). Thus, a broad understanding of mechanisms that underpin mitochondrial structure and function will not only provide insight into mitochondrial-specific diseases, but also other diseases and ailments that suffer from altered mitochondrial states.
Highlights.
Altered mitochondrial dynamics underlie the pathology of degenerative disease.
The Drosophila larval musculature is a good model to study mitochondrial behavior.
Mitochondrial muscle organization is conserved between fly and mammalian muscles.
Mitochondrial dynamics can be imaged and quantified in larval muscle tissue.
Acknowledgments
Thank you to members of the Geisbrecht laboratory, namely Nicole Green and David Brooks, for reading the manuscript and providing insightful comments.
Funding
This work was supported by NIH grant R01 AR060788 to ERG and a UMKC research grant KDF41 to ZW.
Abbreviations
- ATP
adenosine triphosphate
- AEL
after egg laying
- CMT2A
Charcot-Marie-Tooth Type 2A
- ConA
concanavalin A
- Drp1
dynamin-related protein
- DOA
dominant optic atrophy
- GFP
green fluorescent protein
- IFM
indirect flight muscle
- IMF
intermyofibrillar
- IMM
inner mitochondrial membrane
- LRRK2
leucine-rich repeat kinase 2
- L3
third instar larvae
- ΔΨm
mitochondrial membrane potential
- mtDNA
mitochondrial DNA
- Mfn1
Mitofusin 1
- Mfn2
Mitofusin 2
- Opa1
Optic atrophy 1
- OMM
outer mitochondrial membrane
- PD
Parkinson’s disease
- PINK1
PTEN-induced kinase
- S2
Schneider 2 cells
- S2R+
S2 receptor plus cells
- SS
subsarcolemmal
- TEM
transmission electron microscopy
- 3-D
3-dimensional
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abmayr SM, Zhuang S, Geisbrecht ER. Myoblast fusion in Drosophila. Methods Mol Biol. 2008;475:75–97. doi: 10.1007/978-1-59745-250-2_5. [DOI] [PubMed] [Google Scholar]
- Alavi MV, Bette S, Schimpf S, Schuettauf F, Schraermeyer U, Wehrl HF, Ruttiger L, Beck SC, Tonagel F, Pichler BJ, Knipper M, Peters T, Laufs J, Wissinger B. A splice site mutation in the murine Opa1 gene features pathology of autosomal dominant optic atrophy. Brain. 2007;130:1029–1042. doi: 10.1093/brain/awm005. [DOI] [PubMed] [Google Scholar]
- Alexander C, Votruba M, Pesch UE, Thiselton DL, Mayer S, Moore A, Rodriguez M, Kellner U, Leo-Kottler B, Auburger G, Bhattacharya SS, Wissinger B. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet. 2000;26:211–215. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- Archer SL. Mitochondrial dynamics--mitochondrial fission and fusion in human diseases. N Engl J Med. 2013;369:2236–2251. doi: 10.1056/NEJMra1215233. [DOI] [PubMed] [Google Scholar]
- Aubert H. Z Wiss Zool. 1852:388–399. [Google Scholar]
- Bentivoglio AR, Cortelli P, Valente EM, Ialongo T, Ferraris A, Elia A, Montagna P, Albanese A. Phenotypic characterisation of autosomal recessive PARK6-linked parkinsonism in three unrelated Italian families. Mov Disord. 2001;16:999–1006. doi: 10.1002/mds.10034. [DOI] [PubMed] [Google Scholar]
- Bhandari P, Song M, Dorn GW. Dissociation of mitochondrial from sarcoplasmic reticular stress in Drosophila cardiomyopathy induced by molecularly distinct mitochondrial fusion defects. J Mol Cell Cardiol. 2015;80:71–80. doi: 10.1016/j.yjmcc.2014.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boncompagni S, Rossi AE, Micaroni M, Beznoussenko GV, Polishchuk RS, Dirksen RT, Protasi F. Mitochondria are linked to calcium stores in striated muscle by developmentally regulated tethering structures. Mol Biol Cell. 2009;20:1058–1067. doi: 10.1091/mbc.E08-07-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M, van Dongen JW, Vanacore N, van Swieten JC, Brice A, Meco G, van Duijn CM, Oostra BA, Heutink P. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brechbiel JL, Gavis ER. Spatial regulation of nanos is required for its function in dendrite morphogenesis. Curr Biol. 2008;18:745–750. doi: 10.1016/j.cub.2008.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butschli O. Z Wiss Zool. 1871:402–415. 526–534. [Google Scholar]
- Caffin F, Prola A, Piquereau J, Novotova M, David DJ, Garnier A, Fortin D, Alavi MV, Veksler V, Ventura-Clapier R, Joubert F. Altered skeletal muscle mitochondrial biogenesis but improved endurance capacity in trained OPA1-deficient mice. J Physiol. 2013;591:6017–6037. doi: 10.1113/jphysiol.2013.263079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazotte B. Labeling mitochondria with JC-1. Cold Spring Harb Protoc. 2011;2011 doi: 10.1101/pdb.prot065490. [DOI] [PubMed] [Google Scholar]
- Chen CL, Hu Y, Udeshi ND, Lau TY, Wirtz-Peitz F, He L, Ting AY, Carr SA, Perrimon N. Proteomic mapping in live Drosophila tissues using an engineered ascorbate peroxidase. Proc Natl Acad Sci U S A. 2015;112:12093–12098. doi: 10.1073/pnas.1515623112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chan DC. Mitochondrial dynamics--fusion, fission, movement, and mitophagy--in neurodegenerative diseases. Hum Mol Genet. 2009;18:R169–176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, Chan DC. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrollier A, Cassereau J, Ferré M, Alban J, Desquiret-Dumas V, Gueguen N, Amati-Bonneau P, Procaccio V, Bonneau D, Reynier P. Standardized mitochondrial analysis gives new insights into mitochondrial dynamics and OPA1 function. Int J Biochem Cell Biol. 2012;44:980–988. doi: 10.1016/j.biocel.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- Dahl R, Larsen S, Dohlmann TL, Qvortrup K, Helge JW, Dela F, Prats C. Three-dimensional reconstruction of the human skeletal muscle mitochondrial network as a tool to assess mitochondrial content and structural organization. Acta Physiol (Oxf) 2015;213:145–155. doi: 10.1111/apha.12289. [DOI] [PubMed] [Google Scholar]
- Davies VJ, Hollins AJ, Piechota MJ, Yip W, Davies JR, White KE, Nicols PP, Boulton ME, Votruba M. Opa1 deficiency in a mouse model of autosomal dominant optic atrophy impairs mitochondrial morphology, optic nerve structure and visual function. Hum Mol Genet. 2007;16:1307–1318. doi: 10.1093/hmg/ddm079. [DOI] [PubMed] [Google Scholar]
- Debattisti V, Pendin D, Ziviani E, Daga A, Scorrano L. Reduction of endoplasmic reticulum stress attenuates the defects caused by Drosophila mitofusin depletion. J Cell Biol. 2014;204:303–312. doi: 10.1083/jcb.201306121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, Astarie-Dequeker C, Lasquellec L, Arnaud B, Ducommun B, Kaplan J, Hamel CP. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- Deng H, Dodson MW, Huang H, Guo M. The Parkinson’s disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci U S A. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisner V, Lenaers G, Hajnóczky G. Mitochondrial fusion is frequent in skeletal muscle and supports excitation-contraction coupling. J Cell Biol. 2014;205:179–195. doi: 10.1083/jcb.201312066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernster L, Schatz G. Mitochondria: a historical review. J Cell Biol. 1981;91:227s–255s. doi: 10.1083/jcb.91.3.227s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsterer J. Hematological manifestations of primary mitochondrial disorders. Acta Haematol. 2007;118:88–98. doi: 10.1159/000105676. [DOI] [PubMed] [Google Scholar]
- Flemming W. Leipzig, Zellsubstanz, Kern-und Zellteilung. 1882. [Google Scholar]
- Galko MJ, Krasnow MA. Cellular and genetic analysis of wound healing in Drosophila larvae. PLoS Biol. 2004;2:E239. doi: 10.1371/journal.pbio.0020239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes LC, Scorrano L. Mitochondrial morphology in mitophagy and macroautophagy. Biochim Biophys Acta. 2013;1833:205–212. doi: 10.1016/j.bbamcr.2012.02.012. [DOI] [PubMed] [Google Scholar]
- Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci U S A. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralalka S, Abmayr SM. Myoblast fusion in Drosophila. Exp Cell Res. 2010;316:3007–3013. doi: 10.1016/j.yexcr.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henle J. Leipzig, Allgemeine Anatomie. 1841. [Google Scholar]
- Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, Nunnari J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Jofuku A, Eura Y, Mihara K. Regulation of mitochondrial morphology by membrane potential, and DRP1-dependent division and FZO1-dependent fusion reaction in mammalian cells. Biochem Biophys Res Commun. 2003;301:891–898. doi: 10.1016/s0006-291x(03)00050-0. [DOI] [PubMed] [Google Scholar]
- Ishihara N, Nomura M, Jofuku A, Kato H, Suzuki SO, Masuda K, Otera H, Nakanishi Y, Nonaka I, Goto Y, Taguchi N, Morinaga H, Maeda M, Takayanagi R, Yokota S, Mihara K. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol. 2009;11:958–966. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- Ishihara T, Ban-Ishihara R, Maeda M, Matsunaga Y, Ichimura A, Kyogoku S, Aoki H, Katada S, Nakada K, Nomura M, Mizushima N, Mihara K, Ishihara N. Dynamics of mitochondrial DNA nucleoids regulated by mitochondrial fission is essential for maintenance of homogeneously active mitochondria during neonatal heart development. Mol Cell Biol. 2015;35:211–223. doi: 10.1128/MCB.01054-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ML, Robinson MM, Nair KS. Skeletal muscle aging and the mitochondrion. Trends Endocrinol Metab. 2013;24:247–256. doi: 10.1016/j.tem.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Klein CJ, Kimmel GW, Pittock SJ, Engelstad JE, Cunningham JM, Wu Y, Dyck PJ. Large kindred evaluation of mitofusin 2 novel mutation, extremes of neurologic presentations, and preserved nerve mitochondria. Arch Neurol. 2011;68:1295–1302. doi: 10.1001/archneurol.2011.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolliker A. Z Wiss Zool. 1856:311–325. [Google Scholar]
- Kolliker A. Z Wiss Zool. 1888:689–710. [Google Scholar]
- Koopman WJ, Visch HJ, Smeitink JA, Willems PH. Simultaneous quantitative measurement and automated analysis of mitochondrial morphology, mass, potential, and motility in living human skin fibroblasts. Cytometry A. 2006;69:1–12. doi: 10.1002/cyto.a.20198. [DOI] [PubMed] [Google Scholar]
- Leduc-Gaudet JP, Picard M, Pelletier FS, Sgarioto N, Auger MJ, Vallée J, Robitaille R, St-Pierre DH, Gouspillou G. Mitochondrial morphology is altered in atrophied skeletal muscle of aged mice. Oncotarget. 2015 doi: 10.18632/oncotarget.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros F, Lombès A, Frachon P, Rojo M. Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol Biol Cell. 2002;13:4343–4354. doi: 10.1091/mbc.E02-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard AP, Cameron RB, Speiser JL, Wolf BJ, Peterson YK, Schnellmann RG, Beeson CC, Rohrer B. Quantitative analysis of mitochondrial morphology and membrane potential in living cells using high-content imaging, machine learning, and morphological binning. Biochim Biophys Acta. 2015;1853:348–360. doi: 10.1016/j.bbamcr.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesa M, Palacín M, Zorzano A. Mitochondrial dynamics in mammalian health and disease. Physiol Rev. 2009;89:799–845. doi: 10.1152/physrev.00030.2008. [DOI] [PubMed] [Google Scholar]
- Lihavainen E, Mäkelä J, Spelbrink JN, Ribeiro AS. Mytoe: automatic analysis of mitochondrial dynamics. Bioinformatics. 2012;28:1050–1051. doi: 10.1093/bioinformatics/bts073. [DOI] [PubMed] [Google Scholar]
- Lutz AK, Exner N, Fett ME, Schlehe JS, Kloos K, Lämmermann K, Brunner B, Kurz-Drexler A, Vogel F, Reichert AS, Bouman L, Vogt-Weisenhorn D, Wurst W, Tatzelt J, Haass C, Winklhofer KF. Loss of parkin or PINK1 function increases Drp1-dependent mitochondrial fragmentation. J Biol Chem. 2009;284:22938–22951. doi: 10.1074/jbc.M109.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malka F, Guillery O, Cifuentes-Diaz C, Guillou E, Belenguer P, Lombès A, Rojo M. Separate fusion of outer and inner mitochondrial membranes. EMBO Rep. 2005;6:853–859. doi: 10.1038/sj.embor.7400488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattenberger Y, James DI, Martinou JC. Fusion of mitochondria in mammalian cells is dependent on the mitochondrial inner membrane potential and independent of microtubules or actin. FEBS Lett. 2003;538:53–59. doi: 10.1016/s0014-5793(03)00124-8. [DOI] [PubMed] [Google Scholar]
- Miledi R, Slater CR. Some mitochondrial changes in denervated muscle. J Cell Sci. 1968;3:49–54. doi: 10.1242/jcs.3.1.49. [DOI] [PubMed] [Google Scholar]
- Miller KE, DeProto J, Kaufmann N, Patel BN, Duckworth A, Van Vactor D. Direct observation demonstrates that Liprin-alpha is required for trafficking of synaptic vesicles. Curr Biol. 2005;15:684–689. doi: 10.1016/j.cub.2005.02.061. [DOI] [PubMed] [Google Scholar]
- Mishra P, Chan DC. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat Rev Mol Cell Biol. 2014;15:634–646. doi: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár I, Migh E, Szikora S, Kalmár T, Végh AG, Deák F, Barkó S, Bugyi B, Orfanos Z, Kovács J, Juhász G, Váró G, Nyitrai M, Sparrow J, Mihály J. DAAM is required for thin filament formation and Sarcomerogenesis during muscle development in Drosophila. PLoS Genet. 2014;10:e1004166. doi: 10.1371/journal.pgen.1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W. Subsarcolemmal mitochondria and capillarization of soleus muscle fibers in young rats subjected to an endurance training. A morphometric study of semithin sections. Cell Tissue Res. 1976;174:367–389. doi: 10.1007/BF00220682. [DOI] [PubMed] [Google Scholar]
- Nienhaus U, Aegerter-Wilmsen T, Aegerter CM. In-vivo imaging of the Drosophila wing imaginal disc over time: novel insights on growth and boundary formation. PLoS One. 2012;7:e47594. doi: 10.1371/journal.pone.0047594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, Chung J. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- Peng JY, Lin CC, Chen YJ, Kao LS, Liu YC, Chou CC, Huang YH, Chang FR, Wu YC, Tsai YS, Hsu CN. Automatic morphological subtyping reveals new roles of caspases in mitochondrial dynamics. PLoS Comput Biol. 2011;7:e1002212. doi: 10.1371/journal.pcbi.1002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesah Y, Pham T, Burgess H, Middlebrooks B, Verstreken P, Zhou Y, Harding M, Bellen H, Mardon G. Drosophila parkin mutants have decreased mass and cell size and increased sensitivity to oxygen radical stress. Development. 2004;131:2183–2194. doi: 10.1242/dev.01095. [DOI] [PubMed] [Google Scholar]
- Peterson CM, Johannsen DL, Ravussin E. Skeletal muscle mitochondria and aging: a review. J Aging Res. 2012;2012:194821. doi: 10.1155/2012/194821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M, White K, Turnbull DM. Mitochondrial morphology, topology, and membrane interactions in skeletal muscle: a quantitative three-dimensional electron microscopy study. J Appl Physiol (1985) 2013;114:161–171. doi: 10.1152/japplphysiol.01096.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piquereau J, Caffin F, Novotova M, Prola A, Garnier A, Mateo P, Fortin D, Huynh lH, Nicolas V, Alavi MV, Brenner C, Ventura-Clapier R, Veksler V, Joubert F. Down-regulation of OPA1 alters mouse mitochondrial morphology, PTP function, and cardiac adaptation to pressure overload. Cardiovasc Res. 2012;94:408–417. doi: 10.1093/cvr/cvs117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson JH, Ivatt RM, Sanchez-Martinez A, Tufi R, Wilson E, Mortiboys H, Whitworth AJ. The complex I subunit NDUFA10 selectively rescues Drosophila pink1 mutants through a mechanism independent of mitophagy. PLoS Genet. 2014;10:e1004815. doi: 10.1371/journal.pgen.1004815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci U S A. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Yu S, Vincow ES, Pallanck L. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS One. 2010;5:e10054. doi: 10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postuma RB, Berg D, Stern M, Poewe W, Olanow CW, Oertel W, Obeso J, Marek K, Litvan I, Lang AE, Halliday G, Goetz CG, Gasser T, Dubois B, Chan P, Bloem BR, Adler CH, Deuschl G. MDS clinical diagnostic criteria for Parkinson’s disease. Mov Disord. 2015;30:1591–1601. doi: 10.1002/mds.26424. [DOI] [PubMed] [Google Scholar]
- Rafelski SM. Mitochondrial network morphology: building an integrative, geometrical view. BMC Biol. 2013;11:71. doi: 10.1186/1741-7007-11-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci U S A. 2011;108:10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis Y, Bernardo-Faura M, Richter D, Wolf T, Brors B, Hamacher-Brady A, Eils R, Brady NR. Multi-parametric analysis and modeling of relationships between mitochondrial morphology and apoptosis. PLoS One. 2012;7:e28694. doi: 10.1371/journal.pone.0028694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riekkinen P, Rinne UK, Pelliniemi TT, Sonninen V. Interaction between dopamine and phospholipids. Studies of the substantia nigra in Parkinson disease patients. Arch Neurol. 1975;32:25–27. doi: 10.1001/archneur.1975.00490430047006. [DOI] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC. Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nat Protoc. 2008;3:606–611. doi: 10.1038/nprot.2008.18. [DOI] [PubMed] [Google Scholar]
- Rämet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416:644–648. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- Schejter ED, Baylies MK. Born to run: creating the muscle fiber. Curr Opin Cell Biol. 2010;22:566–574. doi: 10.1016/j.ceb.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972;27:353–365. [PubMed] [Google Scholar]
- Sen A, Damm VT, Cox RT. Drosophila clueless is highly expressed in larval neuroblasts, affects mitochondrial localization and suppresses mitochondrial oxidative damage. PLoS One. 2013;8:e54283. doi: 10.1371/journal.pone.0054283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba-Fukushima K, Inoshita T, Hattori N, Imai Y. PINK1-mediated phosphorylation of Parkin boosts Parkin activity in Drosophila. PLoS Genet. 2014;10:e1004391. doi: 10.1371/journal.pgen.1004391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirihai OS, Song M, Dorn GW. How Mitochondrial Dynamism Orchestrates Mitophagy. Circ Res. 2015;116:1835–1849. doi: 10.1161/CIRCRESAHA.116.306374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Mihara K, Chen Y, Scorrano L, Dorn GW. Mitochondrial fission and fusion factors reciprocally orchestrate mitophagic culling in mouse hearts and cultured fibroblasts. Cell Metab. 2015;21:273–285. doi: 10.1016/j.cmet.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JM, Baker KD, Lam G, Evans J, Thummel CS. The Drosophila estrogen-related receptor directs a metabolic switch that supports developmental growth. Cell Metab. 2011;13:139–148. doi: 10.1016/j.cmet.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Shirihai OS. The interplay between mitochondrial dynamics and mitophagy. Antioxid Redox Signal. 2011;14:1939–1951. doi: 10.1089/ars.2010.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Bentivoglio AR, Dixon PH, Ferraris A, Ialongo T, Frontali M, Albanese A, Wood NW. Localization of a novel locus for autosomal recessive early-onset parkinsonism, PARK6, on human chromosome 1p35-p36. Am J Hum Genet. 2001;68:895–900. doi: 10.1086/319522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duijn CM, Dekker MC, Bonifati V, Galjaard RJ, Houwing-Duistermaat JJ, Snijders PJ, Testers L, Breedveld GJ, Horstink M, Sandkuijl LA, van Swieten JC, Oostra BA, Heutink P. Park7, a novel locus for autosomal recessive early-onset parkinsonism, on chromosome 1p36. Am J Hum Genet. 2001;69:629–634. doi: 10.1086/322996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler G, Ocorr K. Visualizing the beating heart in Drosophila. J Vis Exp. 2009 doi: 10.3791/1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos M, Esposito G, Edirisinghe JN, Vilain S, Haddad DM, Slabbaert JR, Van Meensel S, Schaap O, De Strooper B, Meganathan R, Morais VA, Verstreken P. Vitamin K2 is a mitochondrial electron carrier that rescues pink1 deficiency. Science. 2012;336:1306–1310. doi: 10.1126/science.1218632. [DOI] [PubMed] [Google Scholar]
- Wakabayashi J, Zhang Z, Wakabayashi N, Tamura Y, Fukaya M, Kensler TW, Iijima M, Sesaki H. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol. 2009;186:805–816. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Song P, Du L, Tian W, Yue W, Liu M, Li D, Wang B, Zhu Y, Cao C, Zhou J, Chen Q. Parkin ubiquitinates Drp1 for proteasome-dependent degradation: implication of dysregulated mitochondrial dynamics in Parkinson disease. J Biol Chem. 2011;286:11649–11658. doi: 10.1074/jbc.M110.144238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZH, Rabouille C, Geisbrecht ER. Loss of a Clueless-dGRASP complex results in ER stress and blocks Integrin exit from the perinuclear endoplasmic reticulum in Drosophila larval muscle. Biol Open. 2015;4:636–648. doi: 10.1242/bio.201511551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- Weaver TA, White RA. headcase, an imaginal specific gene required for adult morphogenesis in Drosophila melanogaster. Development. 1995;121:4149–4160. doi: 10.1242/dev.121.12.4149. [DOI] [PubMed] [Google Scholar]
- Weitkunat M, Schnorrer F. A guide to study Drosophila muscle biology. Methods. 2014;68:2–14. doi: 10.1016/j.ymeth.2014.02.037. [DOI] [PubMed] [Google Scholar]
- Yanagawa S, Lee JS, Ishimoto A. Identification and characterization of a novel line of Drosophila Schneider S2 cells that respond to wingless signaling. J Biol Chem. 1998;273:32353–32359. doi: 10.1074/jbc.273.48.32353. [DOI] [PubMed] [Google Scholar]
- Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang Y, Wang JW, Yang L, Beal MF, Vogel H, Lu B. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci U S A. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ouyang Y, Yang L, Beal MF, McQuibban A, Vogel H, Lu B. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci U S A. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, Weaver D, Eisner V, Várnai P, Hunyady L, Ma J, Csordás G, Hajnóczky G. Switch from ER-mitochondrial to SR-mitochondrial calcium coupling during muscle differentiation. Cell Calcium. 2012;52:355–365. doi: 10.1016/j.ceca.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin VP, Thummel CS, Bashirullah A. Down-regulation of inhibitor of apoptosis levels provides competence for steroid-triggered cell death. J Cell Biol. 2007;178:85–92. doi: 10.1083/jcb.200703206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A, Müller-Myhsok B, Farrer M, Leitner P, Sharma M, Hulihan M, Lockhart P, Strongosky A, Kachergus J, Calne DB, Stoessl J, Uitti RJ, Pfeiffer RF, Trenkwalder C, Homann N, Ott E, Wenzel K, Asmus F, Hardy J, Wszolek Z, Gasser T. The PARK8 locus in autosomal dominant parkinsonism: confirmation of linkage and further delineation of the disease-containing interval. Am J Hum Genet. 2004;74:11–19. doi: 10.1086/380647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci U S A. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, Parman Y, Evgrafov O, Jonghe PD, Takahashi Y, Tsuji S, Pericak-Vance MA, Quattrone A, Battaloglu E, Polyakov AV, Timmerman V, Schröder JM, Vance JM, Battologlu E. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]