Abstract
Purpose
To compare different reference images selected for registration among chemical exchange saturation transfer (CEST) series.
Materials and Methods
Five normal volunteers and eight brain tumor patients were studied on a 3 Tesla scanner. Image registration was performed by choosing each of the acquired CEST saturation or unsaturation dynamic images as the reference. CEST images at 3.5 ppm (amide proton transfer, APT) were computed for each motion-corrected data set after main magnetic field inhomogeneity correction. A uniformity index was defined to quantify the efficacy of image registration using different reference images. Joint histograms and the structural similarity index (SSIM) were used to analyze the intrinsic image similarity between various dynamic images.
Results
Image registration increased the average uniformity index by 18% if the 3.5 ppm saturated image was selected as the reference image. However, registering to the unsaturated dynamic image reduced the uniformity index by 13% on average. The joint histogram analysis showed that the saturated dynamic images were highly similar (SSIM = 0.89 ± 0.01), and were considerably different from the unsaturated dynamic image (SSIM = 0.58 ± 0.03).
Conclusion
The selection of the 3.5 ppm dynamic image as the reference image generated the highest uniformity index for APT imaging though other saturated images were equally suited as reference images.
Keywords: CEST imaging, APT imaging, image registration, brain tumor
Introduction
Chemical Exchange Saturation Transfer (CEST) imaging (1) is an emerging MRI contrast mechanism that can provide valuable molecular level information through the perturbation of the exchange between biomolecules and water. This exchange process, typically occurring at various rates from tens of Hertz to thousands of Hertz, serves as a signal amplifier for the low-concentration biomolecules when detecting effects on the high-concentration water signal. CEST imaging has been shown to be useful in many human brain applications, such as tumors (2–4) and strokes (5,6).
CEST imaging typically requires unsaturated and saturated images to be repetitively acquired at various dynamic saturation frequencies, resulting in a relatively lengthy scan duration. Hence motion is not infrequent. To cope with motion artifacts, image registration has been widely used as an important preprocessing procedure to ensure high-quality CEST images (4–13). An essential step of image registration is the selection of a reference image against which other images are aligned. Interestingly, the unsaturated image was used as a reference image to register the other CEST images of a series in most previous studies (5,7–11), although other work did not report the choice (4,6,12) or used the anatomical “FLAIR” image (13). This work explores how to select a proper reference image for the registration of CEST images and aims at establishing a new registration guideline for this contrast imaging technique.
Materials and Methods
Data Acquisition
All human studies were approved by the local Institutional Review Board and were performed on a 3T Philips Achieva system (Best, The Netherlands) with a 32-channel head coil. Five normal volunteers and eight glioblastoma patients were recruited, and gave written, informed consent for CEST imaging. Cushion pads were inserted around the participant’s head to minimize any possible motion during experiments.
For normal volunteer studies, the CEST imaging contrast was reached using 32 Sinc-Gauss-shaped pulse elements with a total duration of 1.6 s and an effective B1 power of 2 μT. For image acquisition, the three-dimensional (3D) readout was used to avoid slice-order-dependent signal loss in the multislice scheme (14). Specifically, three different imaging sequences (turbo field echo, TFE; turbo spin echo, TSE; and gradient and spin echo, GRASE) were implemented with an identical field of view (FOV = 212×186×66 mm), acquisition resolution (2.2×2.2×4.4 mm), and “SENSE” factor (R = 2). Other key sequence-dependent parameters were, for TFE (turbo factor–number of echoes: 110; flip angle: 200), TSE (turbo factor: 55), and GRASE (spin echo factor: 22; echo planar factor: 7). The CEST acquisition was executed with nine dynamic images, i.e., at saturation frequency offsets of 3.0, −3.0, 3.5, 3.5, −3.5, −3.5, 4.0, and −4.0 ppm, as well as at unsaturation (−160 ppm), S0, an acquisition that is specific for amide proton transfer (APT) imaging (4).
As for main magnetic field (B0) inhomogeneity correction, a separate “WASSR” (15) sequence was deployed with an average saturation power of 0.5 μT, a saturation duration of 0.2 s, 26 dynamic frequencies from −1.9 ppm to 1.9 ppm in 0.125 ppm steps in addition to S0, and a TSE readout (same FOV and resolution as above; turbo factor: 220; duration: 4.4 min).
For patient studies, the same FOV, acquisition resolution and SENSE factor as those for normal volunteers were used except that only a 3D GRASE readout (14) was implemented with a CEST saturation duration of 0.8 s and the offset frequencies were 3.0, −3.0, 3.5, 3.5, 3.5, 3.5, −3.5, −3.5, −3.5, −3.5, 4.0, and −4.0 ppm, as well as S0 (saturation power turned off).
Image Processing and Analysis
First, acquired CEST images were co-registered deploying the “3dAllineate” function of AFNI (NIMH, Bethesda, MD) (16) with a six-degrees-of-freedom rigid body transformation and a weighted sinc interpolation method using different reference images. The reference image was successively evaluated using each of the dynamic images (the ±3.5 ppm dynamics were each averaged) and the mean of all dynamics, resulted in nine cases, including the original non-registered image with transformation parameters saved for each registered case. Qualitative joint histograms and the quantitative structural similarity index (SSIM) (17) between different dynamic images were used to analyze the underlying difference in contrast. In addition, three different cost functions were tested, which were normalized mutual information, correlation ratio, and mutual information. Second, the WASSR z-spectrum was fitted voxel-wise, with a 12th-order polynomial function and upsampled to a resolution of 1/128 ppm (1 Hz) (4). The frequency difference between the lowest point of the fitted z-spectrum and the nominal 0 ppm frequency was recorded as the B0 correction map. Third, the APT z-spectrum was linearly interpolated to a resolution of 1/128 ppm and the corrected values at ±3.5 ppm were sorted out based on the B0 offset frequency (4). Then, APT-weighted (APTw) images were generated by subtraction of the −3.5 ppm and 3.5 ppm images, and the APTw values were thresholded within −5% and 5% to remove outliers.
To evaluate the 27 different sets of APTw images for each of the three different readout sequences of each of the five normal volunteers quantitatively, a uniformity index (UI) was computed, based on the standards of the National Electrical Manufacturers Association used in a previous study (18), as:
| [1] |
where Yi is the intensity of the ith voxel, Ȳ is the mean intensity of all selected voxels, and N is the total number of selected voxels. The rationale was that a higher UI would be expected for better motion-corrected APTw images, which are presumably relatively uniform across the whole brain for normal volunteers at 3T. To select voxels only from the brain region, a brain mask was created based on the original non-registered S0 image, using the “BET” algorithm (19). When different reference images were used in co-registration, the brain mask was transformed accordingly based on the saved registration transformation parameters. However, the definition in Eq. [1] might cause negative UI values when Ȳ is small. To constrain the UI within 0 and 100 for APTw images, a scaling factor, R, was introduced:
| [2] |
For typical APTw images, Ȳ is a small positive value, and the minimal R value for ensuring positive UI values was ~4.9 in this study. Thus, a rounded value of 5 was chosen here. Note that the minimal value of R may be different for other data sets. Different choices of R values won’t affect the relative comparison of UI values, but a smaller R value is preferred for a bigger dynamic range in UI. All image-processing, except registration, was performed in Matlab (2013b, Mathworks, Natick, MA).
Statistical Analysis
SPSS (Version 16.0, Chicago, IL) was used for statistical analysis. A linear mixed model with a compound symmetry covariance type, followed by post hoc Bonferroni pairwise comparison, was used to analyze the impact of using different cost functions (random factor: data set index for each subject, each readout sequence, and each reference image; fixed factor: cost function index). For one cost function, the linear mixed model with the aforementioned settings was used to analyze the difference between different reference images (random factor: data set index for each subject and each readout sequence; fixed factor: reference image index). A p-value less than 0.05 was considered significant.
Results
There was no significant difference (three pairwise p-values = 1.000, 0.177 and 0.911) between different cost functions. However, the mutual information cost function generated an average of ~1% higher UI than others, and thus, was used thereafter.
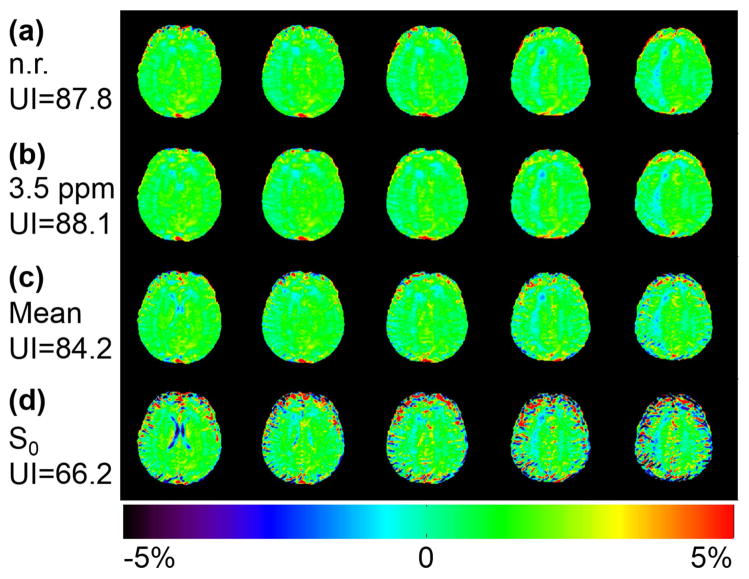
Figure 1 shows that the UI relatively increased by 0.3% after registering all dynamic images to the 3.5 ppm one (Fig. 1b), compared to that from the original non-registered images (Fig. 1a), for a relatively motion-free volunteer. Interestingly, the UI decreased slightly by 4% and largely by 25% after registering to the mean of all dynamic images (Fig. 1c) and the unsaturated S0 dynamic (Fig. 1d), respectively.
Figure 1.
Comparison of APT-weighted images on a mostly motion-free data set from a normal volunteer without and with registration using different reference images. APTw images from original non-registered data (a) and registered data with the 3.5 ppm dynamic image (b), the mean of all dynamics (c), and the S0 dynamic (d) as the reference images. The quality of the motion-free data set can only be preserved under a proper registration reference. The corresponding uniformity indices are shown on the left side and the color bar is shown at the bottom.
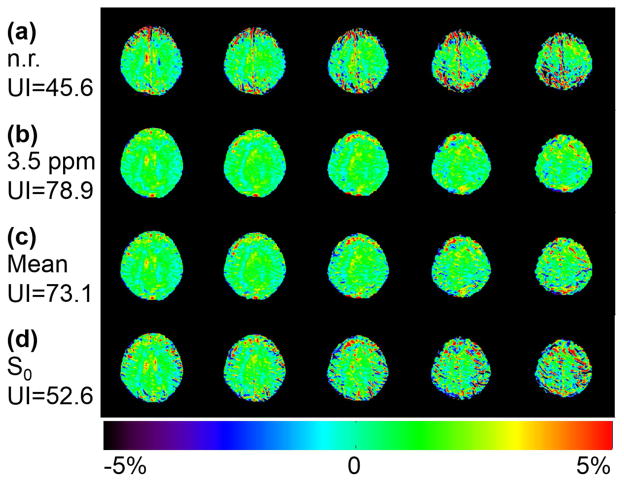
Figure 2 demonstrates that the quality of APTw images on a motion-corrupted data set can be substantially improved with post-processing image registration when referenced to a proper dynamic. Compared to the original motion-corrupted results (Fig. 2a), the UI was relatively increased by 73% and 60% when the 3.5 ppm dynamic (Fig. 2b) or the mean of all dynamics (Fig. 2c) were chosen as the reference, respectively. However, improvement was barely achieved when registering to the S0 dynamic (Fig. 2d).
Figure 2.
Comparison of APT-weighted images on a motion-corrupted data set from a normal volunteer without and with registration using different reference images. APTw images from original non-registered data (a) and registered data with the 3.5 ppm dynamic image (b), the mean of all dynamics (c), and the S0 dynamic (d) as the reference images. A proper registration reference is required to improve the quality of motion-corrupted data. The corresponding uniformity indices are shown on the left side and the color bar is shown at the bottom.
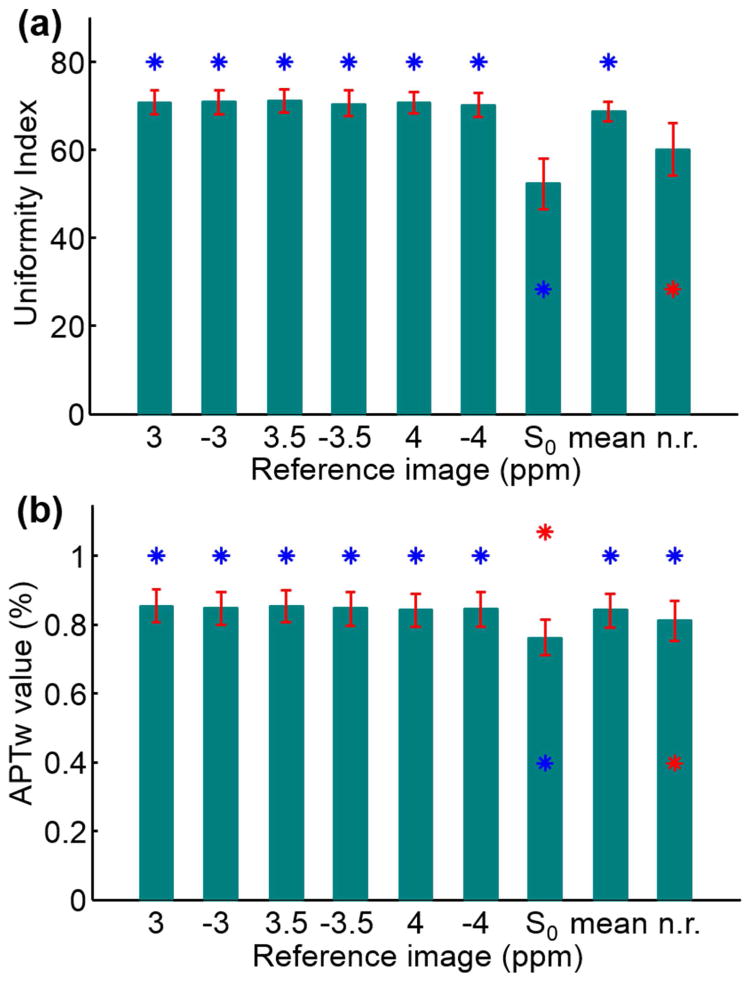
Figure 3 demonstrates that registering to the S0 dynamic generated a considerably lower mean UI than the other registered cases (by 24%, p-value ≤ 0.001) and the non-registered case (by 13%, p-value ≈ 1). Registering to the 3.5 ppm dynamic image generated the highest average UI and outperformed the non-registered case by 18% (p-value = 0.1) although this was barely different (≤ 1%) from the other dynamics at ±3 ppm, −3.5 ppm, and ±4 ppm (p-value ≈ 1). On the other hand, registering to the S0 dynamic reduced the mean APTw value by 10% compared to the other registered cases (p-value < 0.0001), and by 6% compared with the non-registered case (p-value = 0.018). Referencing to the 3.5 ppm dynamic resulted in a mean APTw value relatively 5% higher than the non-registered case (p-value = 0.12), and was ≤ 1% higher than the other registered cases (p-value ≈ 1).
Figure 3.
Uniformity indices (a) and APT-weighted values (b) (mean ± standard error) from all volunteers and all readout sequences by using 9 different reference images. The red star on top of each bar denotes a significant difference (p-value < 0.05) from non-registered data (indicated by a red star inside the bar). The blue star on top of each bar denotes a significant difference (p-value < 0.05) from data registered to the S0 dynamic (indicated by a blue star inside the bar).
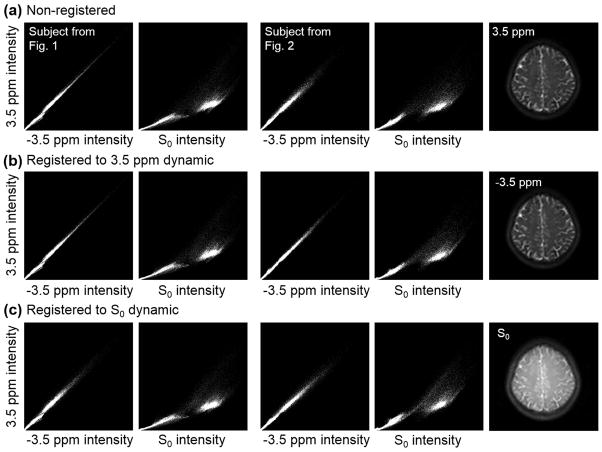
Figure 4 illustrates that the image contrast (or image intensity distribution feature) was quite similar between the 3.5 ppm and −3.5 ppm dynamics, as reflected by the close alignment with the diagonal in the joint histograms. A high similarity of image contrast was observed in each pair between the ±3 ppm, ±3.5 ppm and ±4 ppm dynamic images (not shown). However, the contrast of the 3.5 ppm dynamic was considerably different from that of the S0 dynamic, as shown by the large deviation and dispersion from the diagonal in the joint histograms. The contrast revealed by qualitative joint histograms was consistent with that assessed by the quantitative SSIM, which was 0.89 ± 0.01 between the 3.5 ppm and −3.5 ppm images, and 0.58 ± 0.03 between the 3.5 ppm and S0 images, respectively, for all normal volunteers. For the relatively motion-free case that provided the data shown in Fig. 1, the dispersion around the diagonal in joint histograms was preserved after registering to the 3.5 ppm dynamic (first column, Fig. 4b), compared to the original case (first column, Fig. 4a), but was enlarged after registering to the S0 dynamic (first column, Fig. 4c). However, for the motion-corrupted case that provided the data shown in Fig. 2, image registration could reduce dispersion in joint histograms when a proper reference image (the 3.5 ppm dynamic) was chosen (third column, Fig. 4b), but it could not reduce dispersion if the S0 dynamic was selected (third column, Fig. 4c).
Figure 4.
Joint histograms between different dynamic images from two volunteers shown in Figs. 1 and 2 (calculated from the middle image slice). The 3.5 ppm dynamic was compared with the −3.5 ppm dynamic (columns 1 and 3), and to the S0 dynamic (columns 2 and 4) for three registering cases: original non-registering (a), registering to the 3.5 ppm dynamic (b), and registering to the S0 dynamic (c). Raw non-registered images corresponding to Fig. 2 are shown in the fifth column. Larger deviation and dispersion from the diagonal line in a joint histogram means greater dissimilarity of the contrast of underlying raw images.
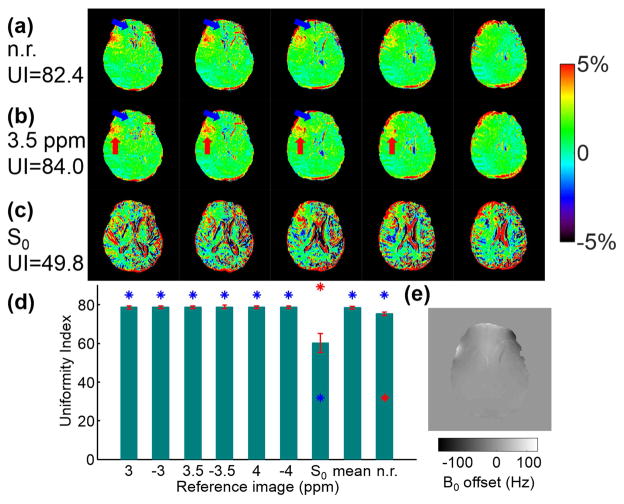
Figure 5 plots APTw images from a glioblastoma patient without (Fig. 5a) and with image registration (Fig. 5b and 5c). Although a uniform APTw map in the whole brain was not expected for brain tumor patients, a higher UI was still obtained, with a better motion-corrected APTw map. The quantitative UIs from all patient data (Fig. 5d) showed a highly similar trend with those from volunteer data (Fig. 3a). Registering to the 3.5 ppm dynamic image improved the visual quality as indicate by blue arrays compared to the non-registered case. On the contrary, registering to the S0 dynamic image yielded markedly degraded APTw maps as shown in Fig. 5c compared to those shown in Fig. 5a and 5b.
Figure 5.
Comparison of APT-weighted images from a brain tumor patient without and with registration using different reference images. APTw images were calculated from original non-registered data (a) and registered data with the 3.5 ppm dynamic (b) and the S0 dynamic (c) as the reference image. Quantitative uniformity indices (mean ± standard error) obtained from eight patients using nine different reference images for registration are shown in part (d). A B0 map used for calculating the APTw image (middle slice in part a) is shown in part (e). The corresponding uniformity indices for parts (a–c) are shown on the left side and the color bar is shown on the right side. A better uniformity was observed in non-tumor regions after registration, as indicated by the blue arrows. The red arrows indicate tumor regions.
Discussion
Image registration is an important preprocessing step to correct motion artifacts and to yield high-quality CEST maps for clinical use. CEST maps can be substantially different in terms of quantitative UIs when different reference images are chosen during registration. In contrast to the use of S0 as the reference image, as described in many reports (5,7–11), we showed here, in five normal volunteers and eight brain tumor patients, that S0 is actually the worst reference image while the 3.5 ppm dynamic generates a higher, though not statistically significantly different, average UI than other saturated dynamics for APT tumor imaging.
The underlying image contrast was highly similar between image pairs from the ±3 ppm, the ±3.5 ppm, or the ±4 ppm dynamics. However, the S0 dynamic has a significantly different intrinsic contrast from the others, which caused the image registration algorithm to generate inaccurate results when the S0 image was chosen as the reference image. A similar situation was reported in image registration for diffusion imaging where images with very high b-values (≫ 1100 s/mm2) would not be properly registered due to the pronounced dissimilarity of intrinsic contrast to low b-value images (20). By choosing the 3.5 ppm dynamic as the reference image, it could produce good alignment with other spectrally close dynamics. Registering the S0 image to the 3.5 ppm dynamic would not cause noticeable errors in the final results, in practice, since the S0 image was only used for normalization, whereas the subtraction of the 3.5 ppm and −3.5 ppm images was more critical. Thus, we suggest choosing the 3.5 ppm dynamic as the reference image.
There were a few limitations in the current study. First, only inter-dynamic motion correction was performed. There is potential concern that one dynamic image might be corrupted severely by intra-dynamic motion and lead to the failure of image registration. For such motion artifacts, the best approach is likely to derive correction from k-space raw data rather than from reconstructed images. Second, the WASSR images were not registered to CEST images. Likely due to the distinct image contrast, poor results were frequently obtained at the edge slices when registering the WASSR images to the CEST images. Thus, we chose not to register WASSR images to the CEST images. This did not cause problems, in practice, since the B0 map varies rather slowly in space. Third, only a limited number of saturation frequencies were implemented due to the long scan time of 3D imaging. For a wide range of frequencies, the image contrast would likely differ considerably. In such cases, one could choose different reference images sequentially rather than a single fixed one, as suggested for registration in diffusion imaging (20). Fourth, only AFNI was used with a rigid-body motion correction algorithm. However, a recent study (21) showed AFNI performed equally well with other popular tools, such as FSL and SPM. As for the motion correction algorithm, it is possible that more advanced motion correction algorithms may not be affected by different selection of reference images or may not require a reference image explicitly. Fifth, only a limited number of patients with one specific disease were recruited for this study, though the methodology should be applicable to other situations as well.
In conclusion, the image contrast of the unsaturated dynamic image differs considerably from that of other saturated dynamics; thus, it should not be chosen as the reference image for image registration in CEST imaging or at least APT imaging. Although any saturated image would be equally suited as the reference image, the 3.5 ppm dynamic image generated the highest uniformity index when chosen as the reference image for APT imaging.
Acknowledgments
We thank Ms. Mary McAllister for editorial assistance.
Grant Support:
NIH Grants: R01 CA166171, R01 EB009731, R01 NS083435 and R21 EB015555
References
- 1.Ward K, Aletras A, Balaban R. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST) J Magn Reson. 2000;143(1):79–87. doi: 10.1006/jmre.1999.1956. [DOI] [PubMed] [Google Scholar]
- 2.Jia G, Abaza R, Williams JD, et al. Amide proton transfer MR imaging of prostate cancer: a preliminary study. J Magn Reson Imaging. 2011;33(3):647–654. doi: 10.1002/jmri.22480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dula AN, Arlinghaus LR, Dortch RD, et al. Amide proton transfer imaging of the breast at 3 T: establishing reproducibility and possible feasibility assessing chemotherapy response. Magn Reson Med. 2013;70(1):216–224. doi: 10.1002/mrm.24450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou J, Zhu H, Lim M, et al. Three-dimensional amide proton transfer MR imaging of gliomas: Initial experience and comparison with gadolinium enhancement. J Magn Reson Imaging. 2013;38(5):1119–1128. doi: 10.1002/jmri.24067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tee YK, Harston GWJ, Blockley N, et al. Comparing different analysis methods for quantifying the MRI amide proton transfer (APT) effect in hyperacute stroke patients. NMR Biomed. 2014;27(9):1019–1029. doi: 10.1002/nbm.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tietze A, Blicher J, Mikkelsen IK, et al. Assessment of ischemic penumbra in patients with hyperacute stroke using amide proton transfer (APT) chemical exchange saturation transfer (CEST) MRI. NMR Biomed. 2014;27(2):163–174. doi: 10.1002/nbm.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hua J, Jones CK, Blakeley J, Smith SA, van Zijl P, Zhou J. Quantitative description of the asymmetry in magnetization transfer effects around the water resonance in the human brain. Magn Reson Med. 2007;58(4):786–793. doi: 10.1002/mrm.21387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones CK, Polders D, Hua J, et al. In vivo three-dimensional whole-brain pulsed steady-state chemical exchange saturation transfer at 7 T. Magn Reson Med. 2012;67(6):1579–1589. doi: 10.1002/mrm.23141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones CK, Huang A, Xu J, et al. Nuclear Overhauser enhancement (NOE) imaging in the human brain at 7T. Neuroimage. 2013;77:114–124. doi: 10.1016/j.neuroimage.2013.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheidegger R, Wong ET, Alsop DC. Contributors to contrast between glioma and brain tissue in chemical exchange saturation transfer sensitive imaging at 3Tesla. Neuroimage. 2014;99:256–268. doi: 10.1016/j.neuroimage.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tee YK, Donahue MJ, Harston GW, Payne SJ, Chappell MA. Quantification of amide proton transfer effect pre- and post- gadolinium contrast agent administration. J Magn Reson Imaging. 2014;40(4):832–838. doi: 10.1002/jmri.24441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Togao O, Yoshiura T, Keupp J, et al. Amide proton transfer imaging of adult diffuse gliomas: correlation with histopathological grades. Neuro Oncol. 2014;16(3):441–448. doi: 10.1093/neuonc/not158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakata A, Okada T, Yamamoto A, et al. Grading glial tumors with amide proton transfer MR imaging: different analytical approaches. J Neurooncol. 2015;122(2):339–348. doi: 10.1007/s11060-014-1715-8. [DOI] [PubMed] [Google Scholar]
- 14.Zhu H, Jones CK, van Zijl P, Barker PB, Zhou J. Fast 3D chemical exchange saturation transfer (CEST) imaging of the human brain. Magn Reson Med. 2010;64(3):638–644. doi: 10.1002/mrm.22546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim M, Gillen J, Landman BA, Zhou J, van Zijl P. Water saturation shift referencing (WASSR) for chemical exchange saturation transfer (CEST) experiments. Magn Reson Med. 2009;61(6):1441–1450. doi: 10.1002/mrm.21873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29(3):162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: from error visibility to structural similarity. Image Processing, IEEE Transactions on. 2004;13(4):600–612. doi: 10.1109/tip.2003.819861. [DOI] [PubMed] [Google Scholar]
- 18.Goerner FL, Duong T, Stafford RJ, Clarke GD. A comparison of five standard methods for evaluating image intensity uniformity in partially parallel imaging MRI. Med Phys. 2013;40(8):082302. doi: 10.1118/1.4816306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rohde G, Barnett A, Basser P, Marenco S, Pierpaoli C. Comprehensive approach for correction of motion and distortion in diffusion-weighted MRI. Magn Reson Med. 2004;51(1):103–114. doi: 10.1002/mrm.10677. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann M, Carpenter TA, Williams GB, Sawiak SJ. A survey of patient motion in disorders of consciousness and optimization of its retrospective correction. Magn Reson Imaging. 2015;33(3):346–350. doi: 10.1016/j.mri.2014.11.004. [DOI] [PubMed] [Google Scholar]