Abstract
Purpose
To devise an improved Blood-Oxygen-Level-Dependent (BOLD) imaging protocol for cerebrovascular reactivity (CVR) measurement that can remove a known artifact of negative values.
Materials and Method
Theoretical and simulation studies were first performed to understand the biophysical mechanism of the negative CVR signals, through which improved BOLD sequence parameters were proposed. This was achieved by equating signal intensities between cerebrospinal fluid (CSF) and blood, via shortening the echo time (TE) of the BOLD sequence. Then, ten healthy volunteers were recruited to participate in an experimental study, in which we compared the CVR results of two versions of the optimized (“Opt1” and “Opt2”) protocols to that of the standard protocol at 3T. Two sessions were performed for each subject to test the reproducibility of all three protocols.
Results
Experimental results demonstrated that the optimized protocols resulted in elimination of negative-CVR voxels. Quantitative CVR results were compared across protocols, which show that the optimized protocols yielded smaller CVR values (Opt1: 0.16±0.01 %BOLD/mmHg CO2; Opt2: 0.15±0.01 %BOLD/mmHg CO2) than (p<0.001) the standard protocol (0.21±0.01 %BOLD/mmHg CO2), but the CNR was comparable (p=0.1) to the standard protocol. The coefficient-of-variation between repetitions was found to be 5.6±1.4%, 6.3±1.6% and 6.9±0.9% for the three protocols, but there were no significant differences (p=0.65).
Conclusion
Based on the theoretical and experimental results obtained from this study, we suggest that the use of a TE shorter than those used in fMRI is necessary in order to minimize negative artifact in CVR results.
Keywords: Cerebrovascular reactivity, CO2, Cerebrospinal fluid, blood vessels, Gas inhalation, Brain
Introduction
Cerebrovascular reactivity (CVR) measures the dilatory capacity of cerebral vasculature to a vasoactive stimulus (1-8). It is an important marker for vascular reserve and has crucial implications for cerebral vascular integrity and functionality (9). Compared to baseline perfusion and cerebral blood volume (CBV), CVR is thought to be a more specific marker for cerebrovascular diseases (10). Thus, availability of such a biomarker could be a useful tool in assessing and understanding many cerebral and cerebrovascular pathologies such as arteriovenous malformation (11), small vessel diseases (12), cerebral proliferative angiopathy (13), moyamoya disease (6,14), arterial stenosis(15), drug addictive condition (16), multiple sclerosis(15), cerebral gliomas(17) and Alzheimer’s disease (18,19). Additionally, it has been used to study the impact of pharmacological agents on cerebral vasculature (20) and has also been suggested to be important in presurgical planning of brain tumor patients (21). However, before this tool can be fully translated to clinical applications, optimization of imaging acquisition schemes is necessary.
The most commonly used approach to measure CVR is by applying a physiological maneuver to alter the arterial carbon dioxide (CO2) concentration (e.g. inhaling a small amount of CO2 which is a potent vasodilator), while continuously acquiring Blood-Oxygenation-Level-Dependent (BOLD) MR images (5,22-26). A logical expectation is that increase in arterial CO2 concentration results in an increase in BOLD signal (i.e. positive CVR) for healthy vasculature. A blunted response or, moreover, a decreased BOLD signal (i.e. negative CVR) would then indicate diseased vasculature. However, the interpretation of negative CVR data is not entirely straightforward (27-29). Mikulis et al. reported the observations of negative CVR in Moyamoya patients (6). However, even though the stenosis affects the right hemisphere only, negative CVR regions were also detected in the left hemisphere, particularly in the deep brain regions (6). Mandell et al. observed that negative CVR is present in the deep white matter of young, healthy individuals, close to posterior horns of the ventricle (28). The question is then whether this represents a true “stealing effect” occurred even in healthy subjects, or it represents an imaging artifact. Blockley et al. showed that negative CVR was mainly observed in the cerebrospinal fluid (CSF) rich ventricular region (29). The mechanistic reason for these observations was examined recently by Thomas et al.(27). It was shown that the presence of negative CVR in healthy brain is due to displacement of CSF by the dilation of blood vessels in the voxel. Because CSF signal is bright in a typical BOLD image, this reduction of CSF partial volume during vasodilation results in a decrease in BOLD signal, independent of any oxygenation changes. Since this negative signal change is due to physiological mechanisms other than perfusion or oxygenation increases, it is considered an artifact as far as CVR interpretation is concerned.
The goal of the present study is to use simulation and experimental approaches to optimize BOLD imaging parameters and remove the artifactually negative CVR effects.
Methods
Theoretical study
Theoretical simulations in the present study were conducted in two steps, for two hypothetical voxels, respectively. In the first step, our goal was to identify all possible combinations of TR, TE and flip angle (FA) that could yield an artifact-free CVR result. This step was performed based on a hypothetical voxel containing CSF and blood, as previous work has shown that such negative CVR artifacts are most pronounced in ventricular voxels. The second step asks the question, among the possible parameters identified above, which ones have the least penalty, if any, on sensitivity in a typical parenchyma voxel. Thus, the second simulation was performed on a hypothetical voxel containing parenchyma.
To address our first aim, we used a two-compartment model to study the BOLD signal in the ventricular regions. In this model, the imaging voxel was assumed to contain a combination of CSF and blood, with volume fraction of fCSF and (1-fCSF), respectively. The total BOLD signal in the voxel is given by:
| [1] |
where SCSF and Sblood are MR signal per unit volume from CSF and blood compartment respectively. The unit-volume MR signal is in turn given by:
| [2] |
In the equation above, i could be CSF or blood. C is water density. TR, TE and FA represent repetition time, echo time and flip angle, respectively.
In an ideal BOLD-CVR experiment, the MR signal in Eq. [1] is assumed to be exclusively sensitive to . However, when SCSF and Sblood are not equal as in the case when conventional fMRI acquisition parameters are used, the total signal, S, is also affected by changes in fCSF. This was the reason for the negative CVR observed in previous reports. To make the signal insensitive to the “volume effect”, the following relationship needs to be satisfied:
| [3] |
One can therefore solve Eqs. [2] and [3] to identify imaging parameter sets that can yield CVR maps absent of negative artifacts. In Eqs. [2] and [3], the MR property parameters are considered known variables and their assumed values are listed in Table 1. The unknown variables are TR, TE, and FA. Since there are multiple unknown variables in the equations, the solution is not unique. Therefore, the outcome after completing the first aim of our simulations is a two-dimensional parameter (TR and TE) set within which the blood and CSF signals are equated. The associated FA is also obtained.
Table 1.
Parameters assumed in the simulation. C is the water density.
The second aim of the theoretical studies is to determine the imaging parameters that produce highest sensitivity and also meet the criteria of absence of negative artifact. We therefore calculated an index, contrast to noise ratio per unit time (CNR), for a hypothetical gray matter voxel, the signal of which takes the form of Eq. [2]. Using gray matter parameters assumed in Table 1, one can simulate the MR signal, Sgray, for any TR, TE, and FA. The CNR per unit time is given by:
| [4] |
In Eq. [4], SNR is replaced by Sgray assuming that the noise is independent of TR or TE. CVR is replaced by TE because they are linearly related to each other, for a given Δ. The division by square-root of TR allows the conversion to unit time value, i.e. number of images for a given scan duration is accounted for. CNR is computed for each imaging parameter set and the set that corresponds to maximum CNR value is determined.
The outcome after completing the second aim of our simulations is that, at each TR, an optimal TE that yields the highest CNR is calculated. Across different TRs, the value that gives the highest CNR could also be calculated. But note that TR is also constrained by other factors such as spatial coverage and number of slices.
Experimental study
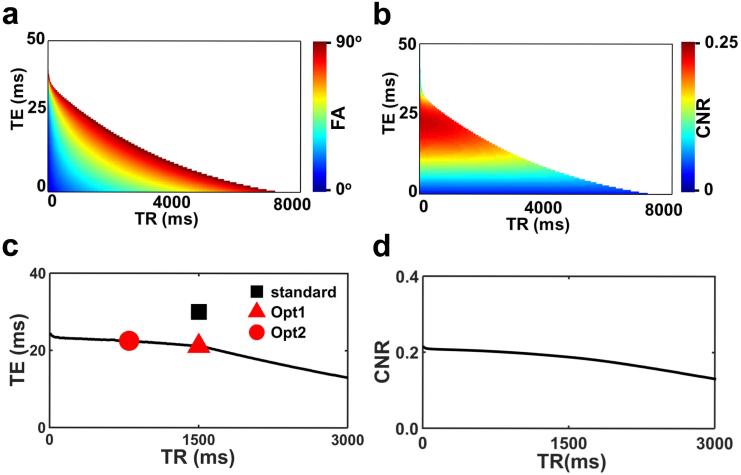
Experimental study was performed based on the results of the theoretical investigations. Specifically, within the two-dimensional parameter space of TE and TR, we chose three representative combinations. One protocol is the standard BOLD protocol, labeled as “standard”, which is a typical protocol used in fMRI and previous CVR studies (27) (black square in Figure 1c, TR/TE=1500ms/30ms). A second protocol, “Opt1”, is used to optimize TE and is a protocol using the same TR as the standard one but its TE is chosen to meet two criteria: 1) signals of blood and CSF are equated (thus no artifact of negative CVR); 2) CNR is the highest among all possible TEs at that TR. This protocol corresponds to the red triangle in Figure 1c. A third protocol, “Opt2”, is used to examine the TR dependence and is a protocol using a shorter TR of 800 ms with the TE chosen to again meet the two criteria described above. This corresponds to the red circle in Figure 1c. Note that each protocol takes 9 minutes to perform and, with test-retest assessment, this becomes 18 minutes. We have therefore limited the number of protocols experimentally examined to 3.
Figure 1.
Simulation results from the theoretical study. (a) All possible combinations of TR, TE, and FA values that can equate the CSF and blood signal intensities in the BOLD image. The TR and TE values are represented on the x and y axis, respectively, and the FA value is represented by color scale. Note that, the imaging parameter space is divided into 2 distinct regions. The white region indicates that such TR and TE combination will not meet the signal equivalence criterion regardless the FA value. These results are obtained from step 1 of the simulation. (b) Corresponding CNR per unit time in a hypothetical parenchyma voxel. These results are obtained from step 2 of the simulation. (c) Optimal TE value as a function of TR. The symbols represent TR/TE combinations tested in the experiment. A total of three protocols were used: two using the optimized protocols with different TRs (red symbols) and one using the standard protocol (black symbol). With this strategy, both the TE and TR dependence of the CNR are examined. (d) CNR as a function of TR. For each TR, the optimal TE (and associated FA) was used to compute the CNR.
The order of the three protocols was randomized for each subject. Two sessions were performed for each subject to test the reproducibility of the three protocols. The two sessions were carried out on the same day, but the participant was allowed to come out of the scanner and take a break before entering it again. The total gas-challenge scan duration for each participant was approximately 60 minutes, divided into two sessions of approximately 30 minutes each.
All experiments were performed at 3T (Philips Healthcare, Best, The Netherlands) using a body coil for transmission and a 32 channel head coil for receiving. The protocol has been approved by the Institutional Review Board (IRB) of UT Southwestern Medical Center and all volunteers gave written informed consent before participating. The volunteers were all fitted with foam pads to reduce head motion. Ten healthy volunteers (age 29±8 years, 6 Female & 4 Male) were recruited to participate in this study.
The CVR measurement was performed using a CO2-inhalation paradigm described previously (5). The subject was fitted with a nose clip and a mouthpiece to allow mouth breathing. The subject inspired room air and the hypercapnic gas in an interleaved fashion (50 sec CO2 followed by 70 sec room air, repeated four times with additional 45 sec room air inhalation at the end). The composition of the hypercapnic gas was 5% CO2, 74% N2 and 21% O2. A research assistant was always present inside the scanner room during the experiment to monitor the subject and switch the valve that controls delivery of hypercapnic gas and room air. End-tidal CO2 (Et-CO2), which measures the concentration of CO2 in the lungs and represents a surrogate for concentration of CO2 in the arterial blood, was recorded for the entire duration of the scan using a capnograph device (Capnogard, Model 1265, Novametrix Medical Systems, CT). The duration of each CVR scan was 9 minutes.
Other BOLD imaging parameters were: field-of-view (FOV)=220×220 mm2, matrix=64×64, thickness=5 mm, no gap between slices, the slice number was 28 for the “standard” and “Opt1” protocols, but was 18 for the “Opt2” protocol due to a shorter TR used. Although Opt2 only provides partial brain coverage with 18 slices, we included this protocol in our testing because future studies using fast imaging techniques such as multiband EPI could provide more coverage at this TR.
Data analysis
BOLD data were processed using the Statistical Parametric Mapping (SPM) software, (University College London, UK) and in-house MATLAB (MathWorks, Natick, MA) scripts. Pre-processing included realignment of BOLD images, normalization to Montreal-Neurological-Institute (MNI) template EPI images with a resolution of 2×2×2 mm3, and finally smoothing using a 6mm full width half maximum (FWHM) Gaussian kernel.
BOLD signal lags behind the EtCO2 time course, which is the input function to the cerebral vasculature. The Et-CO2 time-course was therefore shifted by a delay so that it is matched with the timing of the BOLD time-course. This delay represents the transit time for the blood to travel from the lung to the heart then to the brain, and it was computed on a subject-by-subject basis based on an algorithm described previously (5).
For each subject, a General Linear Model (GLM) was used to calculate voxel-by-voxel CVR in units of % BOLD signal change per mmHg CO2. In this model, the Et-CO2 time-course was the independent variable and the BOLD time-course was the dependent variable. Group-level one sample t test was then performed to identify voxels with significantly negative CVR using a family-wise-error (FWE) corrected p<0.01.
CNR per unit time, which represents the overall sensitivity of the CVR measurement, and coefficient of variation (COV), which represents the reproducibility, were also calculated by:
| [5] |
| [6] |
| [7] |
where μbaseline, and σbaseline are average and standard deviation of baseline BOLD signal respectively. In addition, μinter and σinter represent intersession average and standard deviation, respectively.
To compare data across scan protocols, we performed one-way analysis of variance (ANOVA) tests with repeated measures of scan protocols on mean of CVR, SNR, CNR, and COV. If a scan protocol effect was observed in the ANOVA analysis, we used post-hoc scheffe's tests to further compare the scan protocols in pairs. A multiple-comparison-corrected p value of 0.05 or less was considered significant.
Results
Theoretical study
Figure 1a shows the imaging parameter sets (TR, TE, and the associated FA) that meets the criteria of absence of negative artifact. That is, if the BOLD imaging protocol is chosen to be any TR/TE/FA set on the plot, there should be minimal appearance of negative CVR voxels in the map. It can be seen that not all TR and TE combinations would yield a valid parameter set (e.g. white region in Figure 1a). That is, when TR and TE take on these values, the CSF and blood signal intensities cannot be matched no matter how one chooses the FA.
Figure 1b shows the corresponding CNR per unit time. It can be seen that CNR varies considerably depending on TR and TE. It tends to be that, for a given TR, larger TE yields a greater CNR, but not always the case. Since TR is usually constrained by other considerations (e.g. number of slices, brain coverage) whereas TE is more flexible, we determined, for each TR, the optimal TE that yields the highest CNR. The results are shown in Figure 1c. It is evident from plot that the shorter the TR, the longer the optimal TE. However, all optimal TE are shorter than the typical TE values of 30-50 ms used in typical fMRI experiments. That is, in order to minimize the negative artifacts, the TE in a CVR experiment needs to be shortened. We also examined the dependence of CNR on TR (at the respective optimal TE) (Figure 1d). It can be seen that shorter TR generally yields a greater CNR. Note, however, that TR is often constrained by the number of slices needed to cover the whole brain.
Experiments were therefore performed using two optimal imaging parameter sets suggested by Figure 1c (red symbols). One parameter set, referred to as “Opt1”, used TR/TE=1500 ms/21 ms, FA=89°. A second set, referred to as “Opt2”, used TR/TE=800 ms/22.5 ms, FA=72°. For comparison, we also collected data using a standard BOLD protocol, TR/TE=1500 ms/30 ms, FA=60° (black symbol in Figure 1c). A summary of imaging parameters used in this study is given in Table 2.
Table 2.
Imaging parameters used in the study.
| TR (ms) | TE (ms) | FA (°) | |
|---|---|---|---|
| standard | 1500 | 30 | 60 |
| Opt1 | 1500 | 21 | 89 |
| Opt2 | 800 | 22.5 | 72 |
Experimental study
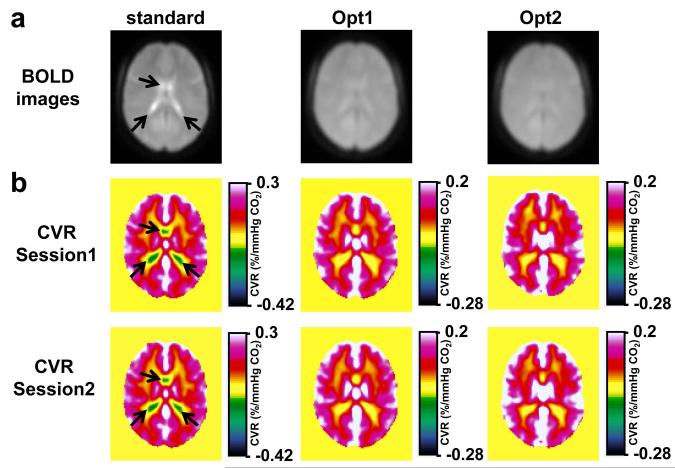
Figure 2a illustrates group-averaged BOLD images for the three protocols tested. It can be seen that, in the standard protocol, the MR signal in the ventricular region is brighter than the rest of the brain, confirming the notion that SCSF is greater than Stissue when typical BOLD protocol is used. In the optimized protocol, on the other hand, the difference between CSF and parenchyma is much less apparent, in accordance with the optimization criteria. Group-averaged CVR maps are shown in Figure 2b. The color scale is selected such that negative CVR is represented in cool colors and positive CVR is represented in warm colors. As expected, CVR map using the standard protocol shows considerable negative CVR voxels in the ventricular region (arrows in Figure 2b). In contrast, negative CVR is minimized when using the two optimized protocols (Figure 2b). Comparing CVR maps between session 1 and session 2 (Figure 2b), it appears that the results show excellent reproducibility.
Figure 2.
Group-averaged CVR results (N=10). (a) BOLD images in a representative slice (z=46 in MNI template) from the three protocols. Note that CSF-rich ventricles are bright (arrows) in the standard protocol but not so in the optimized protocols (b) CVR maps from the three protocols for both sessions. The color bar is selected such that negative CVR voxels are generally represented in cool colors and positive CVR is represented in warm colors. The center of the color bar is slightly shifted toward the negative range to allow small negative CVR values due to noise, which should be distinguished from the systematic, pronounced negative values as shown in the standard protocol (arrows).
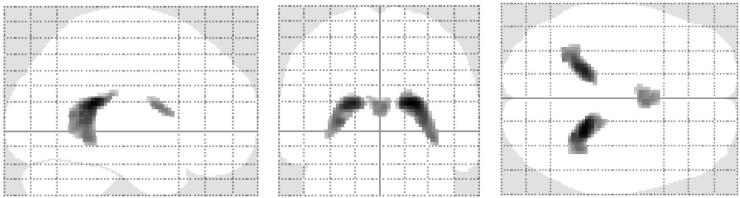
Figure 3 shows the location of the negative-CVR voxels (corrected P<0.01) when using the standard protocol. As expected they are predominantly clustered in ventricular regions. The CVR values in these regions were found to be −0.11±0.02 %BOLD/mmHg CO2. No such clusters were detected when using the Opt1 or Opt2 protocols. We also applied the negative mask derived from the standard protocol data on the Opt1 and Opt2 data. It was found that the CVR values were −0.02±0.01 %BOLD/mmHg CO2 and 0±0.01 %BOLD/mmHg CO2 for Opt1 and Opt2, respectively, suggesting that the negative artifact is substantially mitigated with the optimized protocols. The Opt1 showed a small negative value, which may be due to imperfection in the assumed parameters in the simulation.
Figure 3.
The location of negative CVR voxels when using the standard protocol (FWE corrected p<0.01). Note that the negative CVR voxels are predominantly located in the ventricular region. When applying the same algorithm to Opt1 and Opt2 protocols, no such clusters were detected.
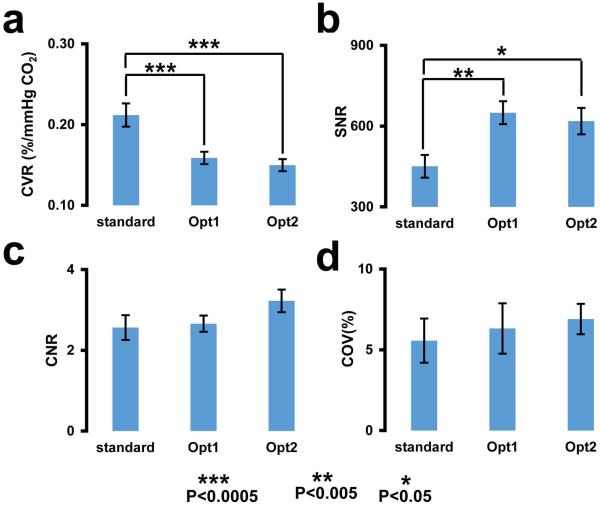
We then performed quantitative analysis of CVR results in the brain parenchyma to compare sensitivity across protocols. The bar plots in Figure 4a depict the whole-brain CVR values for the three protocols. The CVR value for the standard protocol (0.21±0.01 %BOLD/mmHg CO2) was significantly (p<0.001) greater than that of Opt1 (0.16±0.01 %BOLD/mmHg CO2) and Opt2 (0.15±0.01 %BOLD/mmHg CO2). There was no difference between Opt1 and Opt2. This pattern is expected because CVR is related to BOLD percentage signal change, and the BOLD change is approximately proportional to TE, which is longer in the standard protocol. However, the overall sensitivity of the CVR technique is also related to SNR of individual images, which are shown in Figure 4b. The SNR for the standard protocol was found to be significantly lower than both Opt1 (p=0.003) and Opt2 (p=0.013). The lower SNR in the standard protocol is presumably due to the longer TE used. The results for CNR (i.e. overall sensitivity) are shown in Figure 4c. CNR of Opt1 and Opt2 were not different from that of the standard protocol (p=0.1). Another means to evaluate the overall sensitivity of a technique is to examine its COV across repetitions. The COV (i.e. a measure of reproducibility) for the three protocols are shown in Figure 4d. There were no significant differences across the three protocols (p=0.65). We also calculated the COV of individual lobes (frontal, occipital, parietal and temporal) and found no differences across protocols.
Figure 4.
Results of quantitative analysis in the whole brain ROI. (a) CVR. (b) SNR per BOLD image. (c) CNR per unit time. (d) Coefficient of variation (COV) across repetitions. The data are presented as mean ± standard error (***: p<0.0005, **: p<0.005 and *: p<0.05).
Discussion
In this study, we conducted theoretical and experimental investigations to optimize BOLD imaging parameters for the removal of artifactually negative CVR signals while preserving the sensitivity of desired signals. We showed that, when the signal intensities in CSF and blood compartment are equated, the artifactually negative CVR can be minimized. We have further shown that CNR and reproducibility of the CVR results when using the optimized protocols are comparable to those using the standard protocol. Overall, we recommend the use of a TE shorter than that typically used in fMRI for CVR mapping.
Negative CVR has been noted in a number of studies (6,28-31). Some of them can be attributed to a vascular stealing phenomenon (31). However, others, especially those in and around posterior horns of the ventricles where the choroid plexus is located, may be due to the artifact described in this study. Therefore, the use of an “artifact-free” BOLD protocol may simplify the interpretation of such results. Comparing BOLD images from the three protocols, the standard protocol showed brighter CSF signal than the brain parenchyma, which is consistent with the previous findings (27). In contrast, this difference is clearly attenuated in the Opt1 and Opt2 images, consistent with the simulation criteria. In accordance with these differences in the raw images, the CVR map using the standard protocol showed clearly discernable negative CVR voxels in the CSF regions, although not all voxels with bright CSF signal showed negative CVR because choroid plexus is known to be absent in certain part (e.g. anterior horns (32)) of the ventricle. The negative voxels are absent in the Opt1 and Opt2 results. These data suggest that the intensity differences between CSF and blood in combination with changes in CSF partial volume is the source of negative CVR, and this artifact could be removed with an imaging protocol that equates CSF and blood signals. Note that this principle also applies to parenchymal voxels, should CSF partial volume in those voxels be replaced by blood partial volume. But it is expected that the size of the effect may be smaller in typical parenchyma voxels.
The CVR values obtained from the Opt1 and Opt2 protocols were considerably lower than that from the standard protocol. This is expected considering their different TE values. Therefore, when comparing reported CVR values across studies or protocols, it is important to evaluate them in the context of the TE used. Shorter TE is known to result in lower CVR values. In terms of sensitivity, the lower CVR values in the Opt protocols appear to be offset by a higher SNR, and consequently the overall CNR was comparable among the protocols tested. Comparing the two optimal protocols with different TR values, theoretical estimation and experimental CNR suggested that the shorter TR one (i.e. Opt2) should yield a higher sensitivity, owing to the greater number of images per unit scan time, which is helpful in boosting the overall CNR (Equations [4] and [5]). However, the CoV result, which is another indicator of sensitivity, revealed that Opt2 is slightly less sensitive, although the difference was not significant due to small sample size. This discrepancy could be because Equations [4] and [5] assumed that the image series are temporally independent, whereas in reality the noise may be partially correlated. Thus, CNR of Opt2 may be slightly over-estimated.
We also evaluated the COV (reproducibility) of the three protocols. The COV values in our experiment were less than 7%. Compared to COV values reported in the literature, e.g. 26% in (33), 8% in (2), and 8.8% in (4), the values in our study for all the protocols were comparatively lower. This suggests that irrespective of BOLD protocol used in this study for measurement of CVR, the experimental design and data analysis used in our experiment is quite reproducible. The COV showed subtle variation across the three protocols tested. However, there were no significant differences among them. We also evaluated COV in the four major lobes (frontal, parietal, occipital, and temporal) of the brain. The findings were similar. Low COV (i.e. high reproducibility) enhances the potential of an imaging tool to find utility in clinical settings.
The clinical importance of the proposed optimal protocol is that it minimizes artifactually negative CVR signals, thereby improving the specificity of pathological indications of negative CVR. Negative CVR has been previously reported in patients with cerebrovascular diseases as evidence of a loss of vascular reserve (6,13). However, the potential of artifactual signal source could cloud the interpretation of such results. Therefore, the optimized BOLD protocol could remove the “volume effect” and ascertain the correct identification of vascular stealing territory in patients with, for example, arterial stenosis.
The primary limitations of this study are the use of only young healthy participants and the relatively small sample size. Our participants have an average age of 29 years thus do not represent typical patients seen in clinical settings. Therefore, a study in elderly individuals with no known pathology would be useful in future investigations. Also, our sample size of 10, is modest. Even though we were able to show significant group-averaged results and an estimation of reproducibility, the lack of significant difference in some comparisons, e.g. CNR across protocol, could be due to insufficient power. Thus, future studies are needed to verify our findings.
In conclusion, an optimized BOLD imaging protocol was devised that allows the removal of negative signal artifacts in CVR mapping. The optimal protocol yields a sensitivity and reproducibility comparable to that of a standard protocol. The availability of such a protocol may improve the interpretability of CVR results in clinical settings.
Acknowledgements
We would like to thank Dr. Peiying Liu for discussion and help with the physiological scans. We are also grateful to Lily Yang for her help with the experimental setup and scanner operation.
Grant sponsors: NIH R01 MH084021, NIH R01 NS067015, NIH R01 AG042753, NIH R21 NS078656
References
- 1.Hare HV, Germuska M, Kelly ME, Bulte DP. Comparison of CO2 in air versus carbogen for the measurement of cerebrovascular reactivity with magnetic resonance imaging. J Cereb Blood Flow Metab. 2013;33(11):1799–1805. doi: 10.1038/jcbfm.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lipp I, Murphy K, Caseras X, Wise RG. Agreement and repeatability of vascular reactivity estimates based on a breath-hold task and a resting state scan. Neuroimage. 2015 doi: 10.1016/j.neuroimage.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tancredi FB, Hoge RD. Comparison of cerebral vascular reactivity measures obtained using breath-holding and CO2 inhalation. J Cereb Blood Flow Metab. 2013;33(7):1066–1074. doi: 10.1038/jcbfm.2013.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tancredi FB, Lajoie I, Hoge RD. Test-retest reliability of cerebral blood flow and blood oxygenation level-dependent responses to hypercapnia and hyperoxia using dual-echo pseudo-continuous arterial spin labeling and step changes in the fractional composition of inspired gases. J Magn Reson Imaging. 2015 doi: 10.1002/jmri.24878. [DOI] [PubMed] [Google Scholar]
- 5.Yezhuvath US, Lewis-Amezcua K, Varghese R, Xiao G, Lu H. On the assessment of cerebrovascular reactivity using hypercapnia BOLD MRI. NMR Biomed. 2009;22(7):779–786. doi: 10.1002/nbm.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mikulis DJ, Krolczyk G, Desal H, et al. Preoperative and postoperative mapping of cerebrovascular reactivity in moyamoya disease by using blood oxygen level-dependent magnetic resonance imaging. Journal of neurosurgery. 2005;103(2):347–355. doi: 10.3171/jns.2005.103.2.0347. [DOI] [PubMed] [Google Scholar]
- 7.Bhogal AA, Siero JC, Fisher JA, et al. Investigating the non-linearity of the BOLD cerebrovascular reactivity response to targeted hypo/hypercapnia at 7T. Neuroimage. 2014;98:296–305. doi: 10.1016/j.neuroimage.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 8.Mark CI, Mazerolle EL, Chen JJ. Metabolic and vascular origins of the BOLD effect: Implications for imaging pathology and resting-state brain function. Journal of magnetic resonance imaging : JMRI. 2015 doi: 10.1002/jmri.24786. [DOI] [PubMed] [Google Scholar]
- 9.Kety SS, Schmidt CF. The Effects of Altered Arterial Tensions of Carbon Dioxide and Oxygen on Cerebral Blood Flow and Cerebral Oxygen Consumption of Normal Young Men. The Journal of clinical investigation. 1948;27(4):484–492. doi: 10.1172/JCI101995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kassner A, Winter JD, Poublanc J, Mikulis DJ, Crawley AP. Blood-oxygen level dependent MRI measures of cerebrovascular reactivity using a controlled respiratory challenge: reproducibility and gender differences. J Magn Reson Imaging. 2010;31(2):298–304. doi: 10.1002/jmri.22044. [DOI] [PubMed] [Google Scholar]
- 11.Fierstra J, Conklin J, Krings T, et al. Impaired peri-nidal cerebrovascular reserve in seizure patients with brain arteriovenous malformations. Brain : a journal of neurology. 2011;134:100–109. doi: 10.1093/brain/awq286. Pt 1. [DOI] [PubMed] [Google Scholar]
- 12.Greenberg SM. Small vessels, big problems. The New England journal of medicine. 2006;354(14):1451–1453. doi: 10.1056/NEJMp068043. [DOI] [PubMed] [Google Scholar]
- 13.Fierstra J, Spieth S, Tran L, et al. Severely impaired cerebrovascular reserve in patients with cerebral proliferative angiopathy. Journal of neurosurgery Pediatrics. 2011;8(3):310–315. doi: 10.3171/2011.6.PEDS1170. [DOI] [PubMed] [Google Scholar]
- 14.Donahue MJ, Ayad M, Moore R, et al. Relationships between hypercarbic reactivity, cerebral blood flow, and arterial circulation times in patients with moyamoya disease. J Magn Reson Imaging. 2013;38(5):1129–1139. doi: 10.1002/jmri.24070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall O, Lu H, Brisset JC, et al. Impaired cerebrovascular reactivity in multiple sclerosis. JAMA neurology. 2014;71(10):1275–1281. doi: 10.1001/jamaneurol.2014.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han JS, Mandell DM, Poublanc J, et al. BOLD-MRI cerebrovascular reactivity findings in cocaine-induced cerebral vasculitis. Nature clinical practice Neurology. 2008;4(11):628–632. doi: 10.1038/ncpneuro0918. [DOI] [PubMed] [Google Scholar]
- 17.Hsu YY, Chang CN, Jung SM, et al. Blood oxygenation level-dependent MRI of cerebral gliomas during breath holding. Journal of magnetic resonance imaging : JMRI. 2004;19(2):160–167. doi: 10.1002/jmri.10447. [DOI] [PubMed] [Google Scholar]
- 18.Glodzik L, Randall C, Rusinek H, de Leon MJ. Cerebrovascular reactivity to carbon dioxide in Alzheimer's disease. Journal of Alzheimer's disease : JAD. 2013;35(3):427–440. doi: 10.3233/JAD-122011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yezhuvath US, Uh J, Cheng Y, et al. Forebrain-dominant deficit in cerebrovascular reactivity in Alzheimer's disease. Neurobiology of aging. 2012;33(1):75–82. doi: 10.1016/j.neurobiolaging.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pattinson KT, Rogers R, Mayhew SD, Tracey I, Wise RG. Pharmacological FMRI: measuring opioid effects on the BOLD response to hypercapnia. J Cereb Blood Flow Metab. 2007;27(2):414–423. doi: 10.1038/sj.jcbfm.9600347. [DOI] [PubMed] [Google Scholar]
- 21.Zaca D, Hua J, Pillai JJ. Cerebrovascular reactivity mapping for brain tumor presurgical planning. World journal of clinical oncology. 2011;2(7):289–298. doi: 10.5306/wjco.v2.i7.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kastrup A, Kruger G, Neumann-Haefelin T, Moseley ME. Assessment of cerebrovascular reactivity with functional magnetic resonance imaging: comparison of CO(2) and breath holding. Magn Reson Imaging. 2001;19(1):13–20. doi: 10.1016/s0730-725x(01)00227-2. [DOI] [PubMed] [Google Scholar]
- 23.Rostrup E, Law I, Blinkenberg M, et al. Regional differences in the CBF and BOLD responses to hypercapnia: a combined PET and fMRI study. Neuroimage. 2000;11(2):87–97. doi: 10.1006/nimg.1999.0526. [DOI] [PubMed] [Google Scholar]
- 24.Gauthier CJ, Desjardins-Crepeau L, Madjar C, Bherer L, Hoge RD. Absolute quantification of resting oxygen metabolism and metabolic reactivity during functional activation using QUO2 MRI. Neuroimage. 2012;63(3):1353–1363. doi: 10.1016/j.neuroimage.2012.07.065. [DOI] [PubMed] [Google Scholar]
- 25.Lu H, Liu P, Yezhuvath U, Cheng Y, Marshall O, Ge Y. MRI mapping of cerebrovascular reactivity via gas inhalation challenges. Journal of visualized experiments : JoVE. 2014;(94) doi: 10.3791/52306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halani S, Kwinta JB, Golestani AM, Khatamian YB, Chen JJ. Comparing cerebrovascular reactivity measured using BOLD and cerebral blood flow MRI: The effect of basal vascular tension on vasodilatory and vasoconstrictive reactivity. NeuroImage. 2015;110(0):110–123. doi: 10.1016/j.neuroimage.2015.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas BP, Liu P, Aslan S, King KS, van Osch MJ, Lu H. Physiologic underpinnings of negative BOLD cerebrovascular reactivity in brain ventricles. Neuroimage. 2013;83:505–512. doi: 10.1016/j.neuroimage.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandell DM, Han JS, Poublanc J, et al. Selective reduction of blood flow to white matter during hypercapnia corresponds with leukoaraiosis. Stroke; a journal of cerebral circulation. 2008;39(7):1993–1998. doi: 10.1161/STROKEAHA.107.501692. [DOI] [PubMed] [Google Scholar]
- 29.Blockley NP, Driver ID, Francis ST, Fisher JA, Gowland PA. An improved method for acquiring cerebrovascular reactivity maps. Magn Reson Med. 2011;65(5):1278–1286. doi: 10.1002/mrm.22719. [DOI] [PubMed] [Google Scholar]
- 30.Conklin J, Fierstra J, Crawley AP, et al. Impaired cerebrovascular reactivity with steal phenomenon is associated with increased diffusion in white matter of patients with Moyamoya disease. Stroke; a journal of cerebral circulation. 2010;41(8):1610–1616. doi: 10.1161/STROKEAHA.110.579540. [DOI] [PubMed] [Google Scholar]
- 31.Poublanc J, Han JS, Mandell DM, et al. Vascular steal explains early paradoxical blood oxygen level-dependent cerebrovascular response in brain regions with delayed arterial transit times. Cerebrovascular diseases extra. 2013;3(1):55–64. doi: 10.1159/000348841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waxman SG. Ventricles and coverings of the brain. In: Pancotti MWaR., editor. Clinical Neuroanatomy. 26th McGraw-Hill Companies, Inc.; Columbus, OH: 2009. pp. 149–162. [Google Scholar]
- 33.Goode SD, Krishan S, Alexakis C, Mahajan R, Auer DP. Precision of cerebrovascular reactivity assessment with use of different quantification methods for hypercapnia functional MR imaging. AJNR American journal of neuroradiology. 2009;30(5):972–977. doi: 10.3174/ajnr.A1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu H, Golay X, van Zijl PC. Intervoxel heterogeneity of event-related functional magnetic resonance imaging responses as a function of T(1) weighting. Neuroimage. 2002;17(2):943–955. [PubMed] [Google Scholar]
- 35.Whittall KP, MacKay AL, Graeb DA, Nugent RA, Li DK, Paty DW. In vivo measurement of T2 distributions and water contents in normal human brain. Magn Reson Med. 1997;37(1):34–43. doi: 10.1002/mrm.1910370107. [DOI] [PubMed] [Google Scholar]
- 36.Clare S, Jezzard P. Rapid T(1) mapping using multislice echo planar imaging. Magn Reson Med. 2001;45(4):630–634. doi: 10.1002/mrm.1085. [DOI] [PubMed] [Google Scholar]
- 37.Lu H, Clingman C, Golay X, van Zijl PC. Determining the longitudinal relaxation time (T1) of blood at 3.0 Tesla. Magn Reson Med. 2004;52(3):679–682. doi: 10.1002/mrm.20178. [DOI] [PubMed] [Google Scholar]
- 38.Chen L, Bernstein M, Huston J, Fain S. Measurements of T1 relaxation times at 3.0 T: implications for clinical MRA; Proceedings of the 9th Annual Meeting of ISMRM; Glasgow, Scotland. 2001. [Google Scholar]
- 39.Lu H, Golay X, Pekar JJ, Van Zijl PC. Sustained poststimulus elevation in cerebral oxygen utilization after vascular recovery. J Cereb Blood Flow Metab. 2004;24(7):764–770. doi: 10.1097/01.WCB.0000124322.60992.5C. [DOI] [PubMed] [Google Scholar]
- 40.Zhao JM, Clingman CS, Narvainen MJ, Kauppinen RA, van Zijl PC. Oxygenation and hematocrit dependence of transverse relaxation rates of blood at 3T. Magn Reson Med. 2007;58(3):592–597. doi: 10.1002/mrm.21342. [DOI] [PubMed] [Google Scholar]
- 41.Lu H, van Zijl PC. Experimental measurement of extravascular parenchymal BOLD effects and tissue oxygen extraction fractions using multi-echo VASO fMRI at 1.5 and 3.0 T. Magn Reson Med. 2005;53(4):808–816. doi: 10.1002/mrm.20379. [DOI] [PubMed] [Google Scholar]