Abstract
Purpose
We present a volumetric sampling method that rotates the spiral interleaves of a Stack-of-Spirals (SOSP) trajectory for reduced aliasing artifacts using parallel imaging with undersampling.
Methods
The aliasing pattern in an undersampled SOSP acquisition was modified by consecutively rotating spiral interleaves in each phase-encoding plane. This allows a sampling scheme with a high reduction factor when using a volumetric multi-receiver array. Phantom and in vivo brain images at a resolution of 1×1×2 mm3 were acquired at 3T using a 32-channel coil. Images reconstructed with a reduction factor of sixteen were compared for aliasing artifacts and g-factor.
Results
Phantom and in vivo brain image results show that the rotated SOSP acquisition with a reduction factor of sixteen produces images with reduced aliasing and lower g-factors than images acquired without rotation.
Conclusion
The proposed rotated SOSP sampling method is a highly efficient way to maximize the encoding power of volumetric receiver arrays in parallel imaging and is applicable to rapid volumetric scanning including susceptibility weighted imaging and functional MRI.
Introduction
The Stack-of-Spirals (SOSP) trajectory (1) is an efficient approach for rapid sampling of a three-dimensional (3D) k-space in MRI. In an SOSP acquisition, each plane is sampled using one or multiple spiral interleaves and the through-plane direction is sampled with multiple phase encoding steps. The SOSP trajectory has the advantages of spiral imaging methods, such as an efficient use of gradient hardware and zero gradient moment nulling, and is useful for volumetric imaging applications such as Quantitative Susceptibility Mapping (QSM) (2), functional MRI (fMRI) (3,4), coronary angiography (5), imaging of short-T2 tissues (6), and 3D spatial excitation (7). The method presented in this paper is based on the multi-shot SOSP parallel imaging method that uses less spiral interleaves than that required by the Nyquist rate. Parallel imaging takes advantage of the spatial sensitivities of the receiver coils to estimate the missing data during image reconstruction and removes aliasing artifacts from undersampling (8). Although theoretically the reduction factor can be as high as the number of the receiver coils, in practice a lower reduction factor (typically two to four in each phase-encoding direction) is used to avoid significant Signal-to-Noise Ratio (SNR) losses and spatially varying noise amplification, which are characterized by a geometry factor (g-factor).
We present a rotated SOSP (R-SOSP) sampling method, in which a reduced number of spiral interleaves are incrementally rotated in the through-plane phase-encoding direction. The concept is similar to the previously proposed methods such as 2D SENSE(9), Diamond-SENSE (10) and 2D CAIPIRINHA (Controlled Aliasing In Parallel Imaging Results IN Higher Acceleration) (11) for sampling a 3D k-space such as Stack-of-Stars (12). The rotation of the spiral interleaves using R-SOSP creates aliasing patterns that vary along the through-plane as well as in-plane phase encoding directions to optimize the encoding power of volumetric multiple-receiver arrays that are now standard on most scanners. From the perspective of the system of linear equations, parallel imaging reconstruction is equivalent to solving an inverse linear problem and R-SOSP improves the condition of the inverse problem such that noise variations cause less perturbation in the reconstructed images. It is demonstrated in the Methods section that R-SOSP has a lower g-factor from in phantom and in vivo experiments.
The undersampled non-Cartesian data can be reconstructed using iterative SENSE (13) or Generalized Auto-calibrating Partially Parallel Acquisition (GRAPPA) (14) for spiral trajectories (15-17). Iterative SENSE is intuitive and simple to apply. However, it's more computationally intensive than GRAPPA even with Non-Uniform Fast Fourier Transform (NUFFT) (18). Among the existing GRAPPA methods for undersampled spiral reconstruction (15-17), the direct reconstruction method (16) is relatively easier to implement compared to the other two methods. In this paper, we present a GRAPPA-based method for undersampled R-SOSP data. This method is an extension of the direct spiral GRAPPA method (16). In the proposed reconstruction the GRAPPA kernels are estimated and the missing data are interpolated using the neighboring data in 3D SOSP k-space. The estimated data are then reconstructed using a gridding method. Finally, to accelerate the gridding procedure further, we use a novel vectorization computing method that exploits the Single Instruction Multiple Data (SIMD) feature on multiple-core Central Processing Units (CPUs).
Theory
The reconstruction of multi-coil SOSP data follows the basic signal equation:
| [1] |
where sn(k) are the data acquired by each coil n, ρ(r) is the image, and cn(r) is the sensitivity of the n-th coil. Equation [1] can also be written in a matrix-vector form and the reconstruction of image ρ seeks the solution of the following linear inverse problem:
| [2] |
where the coefficient matrix A is the product of a matrix that has n encoding matrices e-i2πkir concatenated along the diagonal direction and a matrix vertically concatenated with the diagonal sensitivity matrix for all coils. Here s is the acquired k-space data from all coils. Undersampling k-space reduces the number of equations in Eq. [2], and images from individual coils cannot be reconstructed separately if there are more unknowns than equations. Parallel imaging combines data from all coils. As long as the combined equation has more equations than unknowns, the combined image can be reconstructed using the data from all coils. However, in practice the combined linear equation may be ill-conditioned with a large condition number, which is the ratio between the largest and the smallest singular value of matrix A. The condition number of the matrix can be computed by using singular value decomposition. In an ideal case, all the singular values will have the same value, and fluctuations in data from noise will not cause a significant change in the reconstructed image. In an ill-conditioned matrix A, some singular values are much larger than the others. A small perturbation in the acquired data from noise can lead to a relatively large change in the reconstructed image.
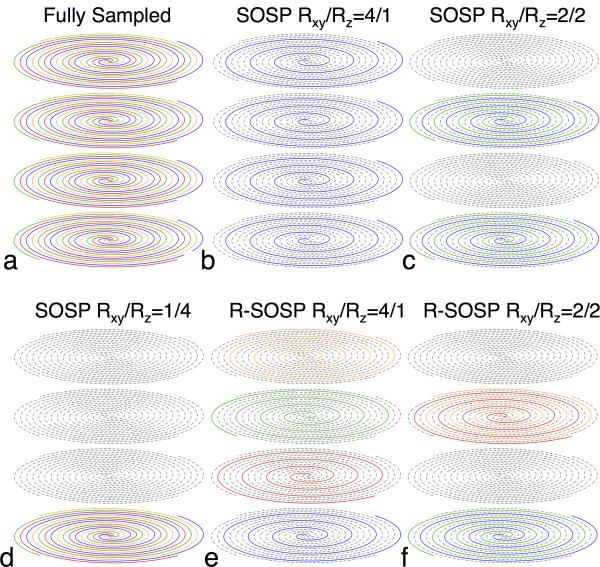
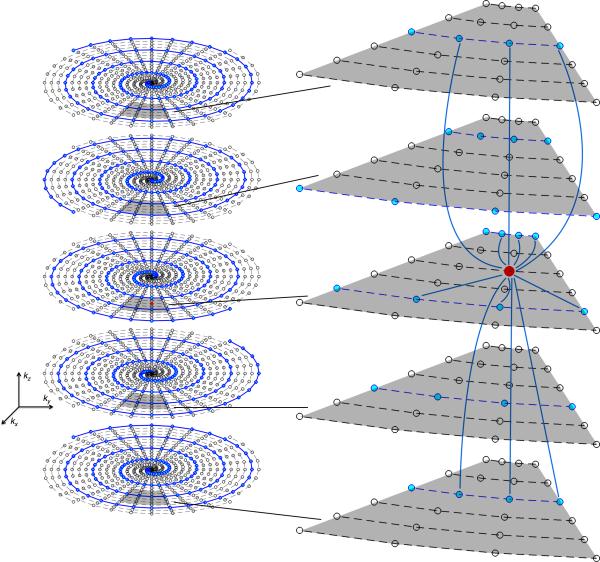
In this paper, we propose a rotated-SOSP (R-SOSP) for improving the condition of the encoding matrix A. To illustrate this concept, Figure 1a shows a schematic plot of an SOSP k-space with four interleaves and four phase-encoding planes. Acceleration of the SOSP acquisition is achieved by using less spiral interleaves or phase encodes than are required by the Nyquist criteria. To reduce the acquisition time by a factor of four (R=4), only 1/4 of the interleaves or phase encodes are used. Figure 1b, 1c, and 1d show examples of three conventional R=4 SOSP sampling strategies. In Figure 1b, each plane is sampled using one interleave (Rxy=4) and all phase encoding planes (Rz=1). In Figure 1c, every other plane (Rz=2) is sampled using two interleaves (Rxy=2). Figure 1d shows that alternatively one out of four (Rz=4) planes can be fully sampled (Rxy=1). For this sampling scheme (Rxy/Rz=1/4), the SOSP sampling is equivalent to R-SOSP. Figure 1e shows an R-SOSP sampling method, in which each plane is sampled using only one interleaf (Rxy=4) that is consecutively rotated by π/2. In this manner, the in-plane aliasing pattern is more evenly distributed in 3D, which will lead to better conditioning of A. Figure 1f shows an alternative R-SOSP sampling pattern, in which every other plane (Rz=2) is partially sampled using two interleaves (Rxy=2).
Figure 1.
(a) A fully sampled SOSP trajectory with four interleaves. Only four through-plane encoding steps are shown. Partial sampling methods with a combined reduction factor of four (R=4) are shown in (b-f). (b) and (e) are SOSP and R-SOSP that has an in-plane reduction factor Rxy=4 and no through plane reduction. (c) and (f) are SOSP and R-SOSP with Rxy/Rz=2/2. (d) is SOSP with a Rxy/Rz=1/4.
Methods
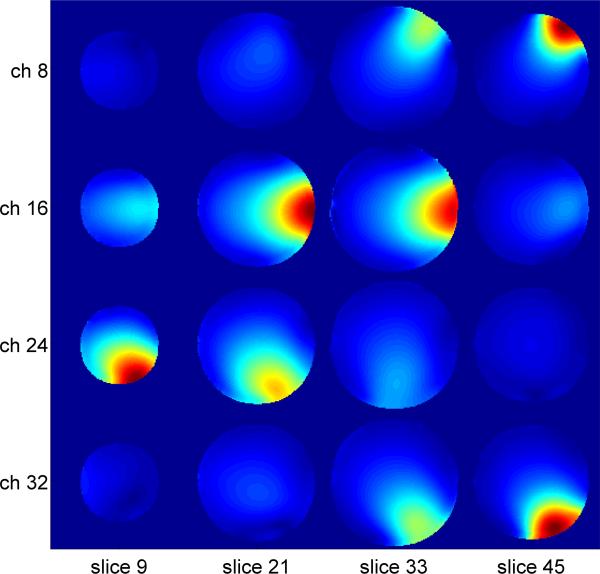
Phantom and in vivo imaging experiments were performed on a Siemens (Erlangen, Germany) 3T Trio Scanner using a Siemens 32-channel helmet-shaped head coil, which has a 12- and 20- coil elements covering the anterior and posterior part of the head, respectively. Figure 2 shows the sensitivities of the 32-channel coil, which are acquired using a fully-sampled SOSP scan. Human subjects were recruited and scanned under informed consent using a protocol approved by the Queens Medical Center and University of Hawaii Internal Review Board. The imaging sequence was a SOSP gradient-echo sequence with following scan parameters: TR=50ms, flip angle=10°, 80 through-plane phase encoding steps, gradient slew rate=125 T/m/sec, gradient amplitude=25 mT/m, FOV=192×192×128 cm3, and resolution=1×1×2 mm3. The echo time was 2ms for phantom experiment, and 30ms for the in vivo scan to show a T2* contrast. The spiral trajectory is a constant velocity Archimedean Spiral (19-21) with the gradient waveform following the equation:
| [5]. |
Figure 2.
The 3D sensitivity map of the 32-channel head coil. Four channels are displayed in rows, and four axial slices are selected for each channel.
Each phase encoding plane was oriented at axial direction and fully sampled using 16 spiral interleaves with 4,096 readout points. The fully sampled data were retrospectively undersampled to simulate the SOSP and R-SOSP undersampling. We used five different sampling methods: SOSP Rxy/Rz=16/1, SOSP Rxy/Rz=1/16, SOSP Rxy/Rz=8/2, R-SOSP Rxy/Rz=16/1, and R-SOSP Rxy/Rz=8/2. The slices are axial. Because the coil sensitivities have less overlap in the in-plane direction than the through-plane direction, it's expected that higher acceleration factors can be used in the in-plane direction than the through-plane direction.
We extended the 2D spiral GRAPPA method proposed in Ref. (16) for reconstruction of the partially sampled SOSP and R-SOSP data. First, data along the constant linear velocity spiral were interpolated to points along a constant angular velocity spiral. This rearranged the spiral sampling points to be along radial lines. The interpolated data were then separated into 64 angular segments and GRAPPA was performed independently for each segment in a spiral plane. Because R-SOSP implicitly uses the 3D sensitivity profile of the coils, the GRAPPA kernel was a 3D kernel in k-space. The 3D GRAPPA kernel is illustrated in Figure 3. The data on the spiral trajectory is shown in blue in the left panel. A magnification of the spiral data is shown in the right panel, where the cyan data points are acquired data points used to estimate the GRAPPA coefficients and the missing data. The point in red is the target point during the GRAPPA calibration and missing data are estimated from the fully sampled data set by summing the twelve neighboring acquired data using the calibrated GRAPPA coefficients. The now fully sampled spiral data were then convolved onto a Cartesian grid. The gridding was the most computationally intensive part of the reconstruction, as it needed to be repeated for every slice and coil. However, because the gridding is the same convolution repeated for multiple data points along the through-plane direction, it is ideal for acceleration through parallel computing. We used a parallel computing model called Single Instruction Multiple Data (SIMD) (22-24) and Advanced Vector Extensions (AVX) (25) to exploit the processing power of a Central Processing Unit (CPU). Discussion of the SIMD AVX parallelization of the gridding process is described in more detail below in Appendix A.
Figure 3.
The schematic of the GRAPPA reconstruction for the rotated SOSP. The pie-shaped gray segment on the left side is zoomed to show more details, shown on the right side. The black circles are un-acquired data points that are estimated from the surrounding acquired data points denoted by solid cyan points. Six in-plane data points and six through-plane data points, connected to the un-acquired data point by blue lines, are used to estimate the un-acquired data point.
The reconstruction of the undersampled data were then analyzed by calculating the g-factor using the pseudo-replica method (26). The g-factor (8) is defined as g = SNRFS/(SNRUS ×R), where SNRFS and SNRUS are signal-to-noise ratio of the fully and under sampled images and R is the reduction factor. A noise covariance matrix required for the simulations was derived from an additional scan using the same scan parameters but without RF applied. To simulate g-factor, Gaussian noises with a mean of zero and a standard deviation of one were used with the noise covariance matrix to generate noise data for multiple coils. This step was repeated 200 times for each under-sampling scenario to calculate SNR and g-factor.
Results
Phantom Experiment
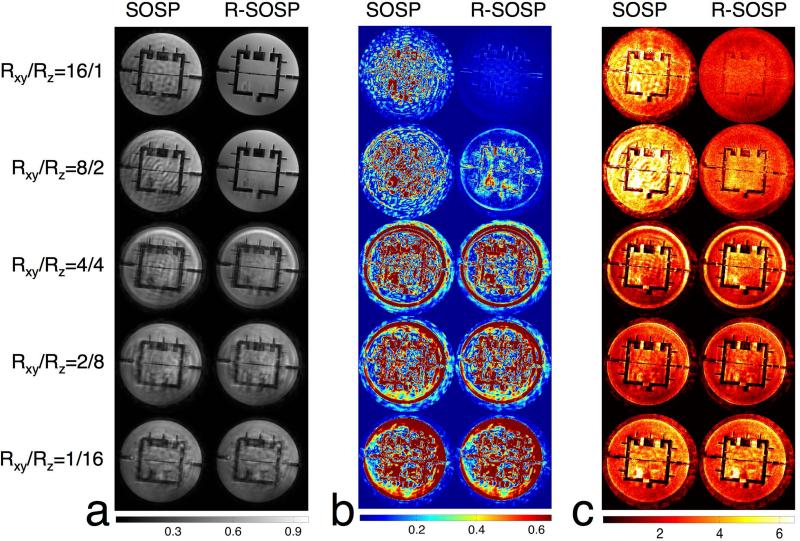
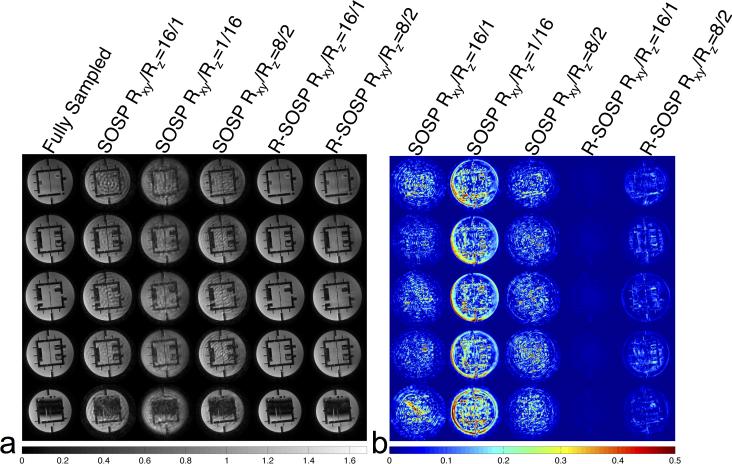
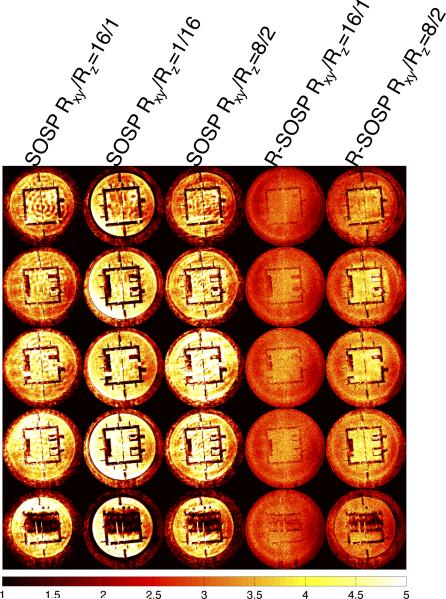
The reconstructed images from fully sampled and retrospectively undersampled data are shown in Figure 4a. The columns in Fig. 4a, from the left to right, are selected images reconstructed from SOSP Rxy/Rz=1/1, SOSP Rxy/Rz=16/1, SOSP Rxy/Rz=1/16, SOSP Rxy/Rz=8/2, R-SOSP Rxy/Rz=16/1, and R-SOSP Rxy/Rz=8/2. The artifacts in the SOSP images are clearly noticeable whereas R-SOSP images have no observable aliasing. This is due to the amplification of artifacts from the high condition number in undersampling. The absolute differences between the fully sampled images and undersampled images are shown in Figure 4b, which shows that the SOSP images differ more from the fully sampled images than the R-SOSP images. In addition, the R-SOSP Rxy/Rz=16/1 images show less difference to the fully sampled images than images from R-SOSP Rxy/Rz=8/2. The g-factors calculated using the pseudo-replica method are shown in Figure 5 in the same order described above. Compared to SOSP, R-SOSP has lower g-factors with R-SOSP Rxy/Rz=16/1 having the lowest. For a comprehensive comparison of different undersampling schemes (Rxy/Rz=16/1, 8/2, 4/4, 2/8, and 1/16) between SOSP and R-SOSP, please refer to Figure A2.
Figure 4.
(a) Magnitude images of a structure phantom acquired with different sampling strategies. From left to right, each column shows five representative axial slices selected from the 3D volume acquired using full sampling, SOSP Rxy/Rz=16/1, SOSP Rxy/Rz=1/16, PPA Rxy/Rz=8/2, R-SOSP Rxy/Rz=16/1, and R-SOSP Rxy/Rz=8/2, respectively. (b) The difference between the fully sampled and partially sampled magnitude images. From left to right, each column shows the magnitude difference between the fully sampled and the undersampled scans. The R-SOSP undersampling shows the best agreement with the full sampled scan.
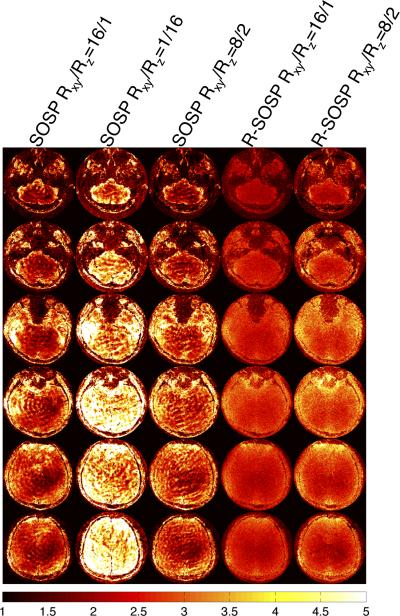
Figure 5.
From left to right, each column shows the g-factor maps generated from the undersampled phantom scans in Fig. 4.
In vivo Experiment
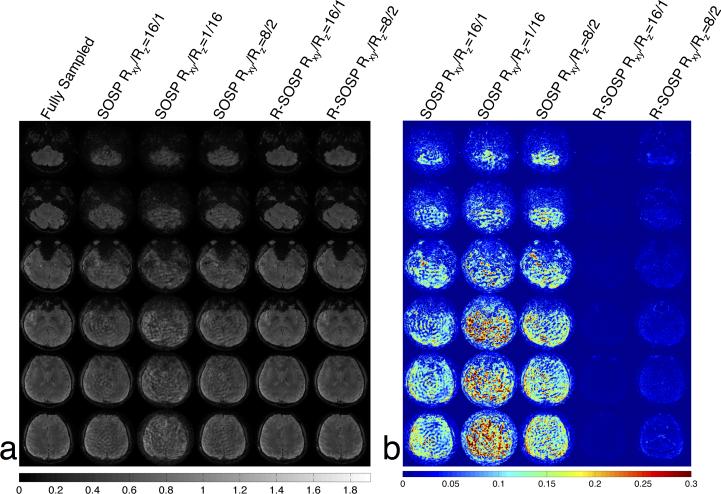
Undersampling methods including SOSP (Rxy/Rz=16/1), SOSP (Rxy/Rz=1/16), SOSP (Rxy/Rz=8/2), R-SOSP (Rxy/Rz=16/1), and R-SOSP (Rxy/Rz=8/2) are used for in vivo experiment. The results from the 3T in vivo experiment on the human brain showed excellent agreement with the results from the phantom experiment. The columns in Figure 6a, shown from the left to right in the same order listed in the paragraph above, are selected axial images from the sampling methods listed in the previous paragraph. Like the phantom images, an excessive level of artifact plague the SOSP images while the R-SOSP images have little observable aliasing. The differences between the fully sampled and the partially sampled images are shown in Figure 6b. The R-SOSP images show very little difference to the fully sampled images while SOSP images show large differences. The g-factors simulated for the in vivo images are shown in Figure 7 and it can be seen that the g-factors from R-SOSP are lower than the ones from SOSP sampling.
Figure 6.
(a) In vivo magnitude images acquired with the undersampled SOSP and R-SOSP trajectories. (b) The difference between the fully sampled magnitude images and the undersampled magnitude images. The R-SOSP undersampling shows the best agreement with the full sampled scan.
Figure 7.
From left to right, each column shows the g-factor maps generated from the undersampled brain scans in Fig. 6.
The square root of the squared differences between the fully sampled images and the undersampled images were calculated and listed in Table 1 for a quantitative comparison. The magnitude of the images was normalized to one. Table 1 also lists the means and standard deviations g-factors of all sampling methods for the in vivo experiment. R-SOSP (Rxy/Rz=16/1) has the lowest means and standard deviations compared to the other under-sampling methods. This indicates that R-SOSP (Rxy/Rz=16/1) has the least noise amplification, and the g-factor pattern is the most uniform throughout the entire volume compared to the other methods.
Table 1.
This table lists the square root of the squared differences between the fully sampled and undersampled images and g-factors for phantom and in vivo experiments.
| SOSP Rxy/Rz=16/1 | SOSP Rxy/Rz=1/16 | SOSP Rxy/Rz=8/2 | R-SOSP Rxy/Rz=16/1 | R-SOSP Rxy/Rz=8/2 | |
|---|---|---|---|---|---|
| Normalized SSE (Phantom) | 72.79 | 148.03 | 65.90 | 14.78 | 37.17 |
| Normalized SSE (in vivo) | 148.03 | 154.24 | 132.92 | 76.40 | 86.79 |
| g-factor (Phantom) | 2.19±1.90 | 2.29±1.78 | 2.27±1.81 | 1.96±1.18 | 2.03±1.37 |
| g-factor (in vivo) | 1.82±1.40 | 2.69±2.27 | 1.96±1.38 | 1.77±1.07 | 1.89±1.24 |
The magnitude of the images is normalized to one. SSE is the square root of the sum of the squared errors.
Discussion
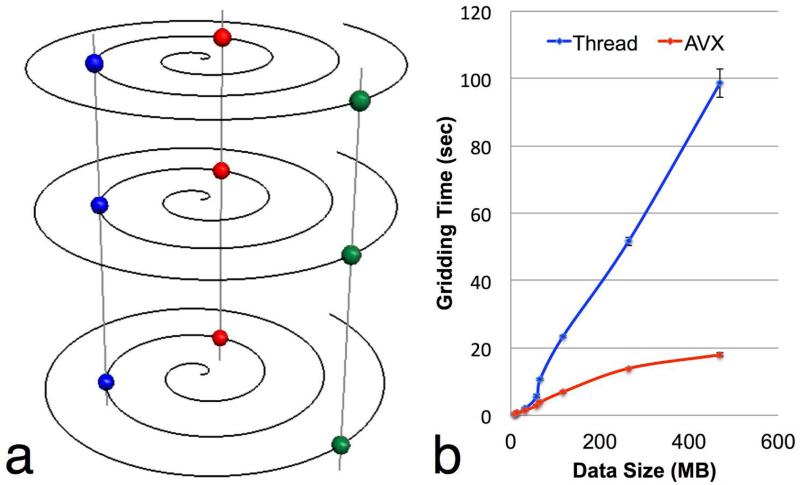
We have presented a rotated stack-of-spirals sampling technique for 3D volumetric imaging. In this method, the spiral interleaves are consecutively rotated along the through-plane direction by an angle dependent on the through-plane k-space location. Therefore, the aliasing artifacts from partial sampling are distributed more evenly to take better advantage of the 3D spatial sensitivities of volumetric multi-receiver arrays. The phantom and in vivo brain experiments showed less aliasing and lower g-factors in R-SOSP than SOSP sampling. G-factors appear different in phantom and in vivo results. This is probably due to different B1 in phantom and in vivo experiment or the B0 field inhomogeneity, which is more prevalent at the air-tissue interfaces in the brains. Intuitively, the B0 inhomogeneity can be approximated using linear terms and be combined with the k-space trajectory in Eq. [1] to modify the vector k. We also presented a GRAPPA method for the reconstruction of the partially sampled SOSP data. The proposed method is computationally faster compared to iterative SENSE, in which the computing time is spent on the computation of the matrix-vector product even with methods such as NUFFT in minimization iterations. In addition, by exploring the parallel structure of SOSP data, the gridding process in the spiral reconstruction can be accelerated using a vectorized SIMD computing. On a desktop quad-core computer with the AVX feature, the computing time was reduced by approximately factor of five (30 seconds using AVX) compared to the multi-threaded method (140 seconds).
The SOSP trajectory is efficient for sampling the 3D k-space and useful for rapid volumetric imaging. The improved image quality at high acceleration factors from the presented method can benefit fMRI experiments and allow images being acquired at faster temperate rate or higher spatial resolution. The R-SOSP approach can also be applied to 3D imaging applications such as QSM acquired at high resolutions, and T1 parameter mapping which usually has long scan times due to the use of the inversion. R-SOSP can benefit fMRI by acquiring a higher spatial resolution without lowering the temporal resolution. Similarly, the resolution can be left unchanged and acquiring more interleaves can shorten readouts. Shorter readouts reduce image blurring and signal dropout in regions such as the orbitofrontal cortex.
A self-calibrated R-SOSP will potentially benefit longer 3D spiral scans including emerging susceptibility tensor imaging (STI) applications that require patients to rotate their heads at different orientations during scans. Self-calibration of R-SOSP can be achieved using a variable-density spiral trajectory that samples the center of the k-space at the Nyquist rate such that each orientation has its own calibration date set for GRAPPA reconstruction. It has been reported that susceptibility maps acquired using spiral with a reduction factor of two are comparable the fully-sampled maps (2). A higher reduction factor can be achieved using the self-calibrated R-SOSP and significantly reduces the scan time in STI.
The GRAPPA reconstruction of the SOSP data was performed in multiple angular segments. As a result, the aliasing pattern and the g-factor estimates are different than ones obtained from SENSE. A comparison between the iterative SENSE and GRAPPA reconstruction was not performed due to the limitation of the scope of the paper. Sixty-four segments were used for SOSP GRAPPA in this paper. Using more segments can improve the image quality. However, the drawback is the increased reconstruction time, and in addition we found the improvement to be marginal from using more segments. The most time-consuming part was actually the estimation of the GRAPPA coefficients that took about fifteen minutes. The combined time used on the estimation of the missing data and the gridding took less than two minutes on a desktop computer with an Intel i7 quad-core processor and 32GB memory for the 32-channel data acquired at an isotropic millimeter resolution. Partitioning the GRAPPA into subtasks and running them in multiple threads can improve the speed.
Conclusion
We have presented an R-SOSP method for rapid volumetric imaging. A reduction factor of sixteen was achieved in phantom and in vivo imaging experiments with a lower g-factor compared to the SOSP undersampling and without penalty of image aliasing. This technique is potentially useful for high-resolution fMRI or reducing long scan time in structural MRI such as SWI.
Acknowledgments
Work supported by the National Institute of Health (R01DA019912, R01EB011517 K02DA020569). Core resources supported by the National Center for Research Resources (G12-RR003061, P20-RR011091) and the Office of National Drug Control Policy.
Appendix
Single Instruction Multiple Data (SIMD) is a parallel computing paradigm in which a single arithmetic operation is simultaneously applied on multiple data sets. This concept is incorporated into the chip design of modern CPUs and GPUs to improve the performance of scalable applications such as computer graphics. It can also be used to accelerate general purpose computing applications such as numerical linear algebra using CPUs or GPUs. Although modern GPUs have hundreds to thousands of lightweight cores and allow simultaneous arithmetic operations on a large data set, their performance is limited by the slow data transfer rate of the PCI-express bus. On the other hand, even though CPUs have fewer cores (two to sixteen) than GPUs, data stored in system memory can be accessed with very low latency.
SIMD on CPUs is implemented into the chip design by two major chip manufactures, Intel and AMD using Advanced Vector Extensions (AVX) to add new registers and instruction sets in addition to the ×86 instruction set. To use AVX, a programmer can either explicitly program AVX registers and instructions in assembly, use libraries supplied by vendors, let compilers implicitly optimize for AVX instructions, or use the Open Computing Language (OpenCL) framework. The drawbacks for the first three approaches are that programming in assembly requires high levels of expertise in instruction set architecture, using libraries from vendors limits the code to the CPU from that specific vendor, and compiler optimization does not guarantee the performance. OpenCL is an open parallel programming standard adopt by most chip vendors. It is easy to use and allows the fine control of the computing device through programming to guarantee the performance.
In this study, the gridding function was programed using OpenCL to run on an Intel quad-core i7 processor, which has a 256-bit wide vector to allow simultaneous operation on eight Single-Precision Floating-Point (SPFP) numbers. Because the CPU has four cores, a total of 32 SPFP numbers can be operated on simultaneously. In OpenCL, a multi-core CPU is treated as one compute device, and the AVX programming on multi-core CPUs is simplified. The computing tasks are partitioned and automatically dispatched to all cores such that the programmers do not have to deal with the thread barrier and synchronization of the multiple threads.
The parallel structure of SOSP data is shown in Fig. A1. Note the data points represented in same color and connected by the vertical lines have same in-plane coordinates. This means the same GRAPPA convolution kernel can be simultaneously applied to all the data points in the through-plane direction for gridding operations. The data points are rearranged such that the data points are stored adjacent to each other in the through-plane direction in memory to improve cache performance. Comparing the vectorized gridding using AVX to the conventional multi-threaded parallel gridding, the gridding computation was accelerated by a factor of five for reconstruction of high-resolution volumetric images in this work. The lower acceleration for smaller data sets is due to the small overhead of OpenCL.
Figure A1.
(a) The data along the through-plane direction have a parallel structure since the same gridding is applied for the same coordinate on each spiral interleaf. (b) An acceleration factor up to five can be achieved using SIMD vectorized gridding compared to using the multi-threaded threading.
Figure A2.
(a) The left and right columns are side-by-side comparisons of the center axial slice of the magnitude image from SOSP and R-SOSP undersampling. From top to bottom are Rxy/Rz=16/1, 8/2, 4/4, 2/8, and 1/16, respectively. The differences between the fully sampled and undersampled iamges are shown in the same order in (b). (c) shows the g-factor maps in the same order.
Bibliography
- 1.Irarrazabal P, Nishimura D. Fast Three Dimensional Magnetic Resonance Imaging. Magn Reson Med. 1995;33(5):656–662. doi: 10.1002/mrm.1910330510. [DOI] [PubMed] [Google Scholar]
- 2.Wu B, Li W, Avram AV, Gho SM, Liu C. Fast and Tissue-Optimized Mapping of Magnetic Susceptibility and T2* with Multi-Echo and Multi-Shot Spirals. NeuroImage. 2012;59(1):297–305. doi: 10.1016/j.neuroimage.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qian Y, Zhao T, Hue YK, Ibrahim TS, Boada FE. High-Resolution Spiral Imaging on a Whole-Body 7T Scanner with Minimized Image Blurring. Magn Reson Med. 2010;63(3):543–552. doi: 10.1002/mrm.22215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu Y, Glover GH. Three-Dimensional Spiral Technique for High-Resolution Functional MRI. Magn Reson Med. 2007;58(5):947–951. doi: 10.1002/mrm.21328. [DOI] [PubMed] [Google Scholar]
- 5.Thedens DR, Irarrazaval P, Sachs TS, Meyer CH, Nishimura DG. Fast Magnetic Resonance Coronary Angiography with a Three-Dimensional Stack of Spirals Trajectory. Magn Reson Med. 1999;41(6):1170–1179. doi: 10.1002/(sici)1522-2594(199906)41:6<1170::aid-mrm13>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 6.Qian YX, Boada FE. Acquisition-Weighted Stack of Spirals for Fast High-Resolution Three-Dimensional Ultra-Short Echo Time MR Imaging. Magn Reson Med. 2008;60(1):135–145. doi: 10.1002/mrm.21620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stenger VA, Boada FE, Noll DC. Three-Dimensional Tailored RF Pulses for the Reduction of Susceptibility Artifacts in T2*-Weighted Functional MRI. Magn Reson Med. 2000;44(4):525–531. doi: 10.1002/1522-2594(200010)44:4<525::aid-mrm5>3.0.co;2-l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: Sensitivity Encoding for Fast MRI. Magn Reson Med. 1999;42(5):952–962. [PubMed] [Google Scholar]
- 9.Weiger M, Pruessmann KP, Boesiger P. 2D SENSE for Faster 3D MRI. MAGMA. 2002;14(1):10–19. doi: 10.1007/BF02668182. [DOI] [PubMed] [Google Scholar]
- 10.Jurrissen M, Fuderer J, van den Brink J. Diamond-SENSE: Undersampling on a Crystallographic Grid.. Proceedings of the 12th Annual Meeting of ISMRM; Kyoto, Japan. 2004.p. 2643. [Google Scholar]
- 11.Breuer FA, Blaimer M, Mueller MF, Seiberlich N, Heidemann RM, Griswold MA, Jakob PM. Controlled Aliasing in Volumetric Parallel Imaging (2D CAIPIRINHA). Magn Reson Med. 2006;55(3):549–556. doi: 10.1002/mrm.20787. [DOI] [PubMed] [Google Scholar]
- 12.Breuer FA, Bauer S, Blaimer M. Self-Calibrating Stack-of-Stars (SoS)-CAIPIRINHA for Improved Parallel Imaging. Procs Int Soc Magn Reson Med. 2013;21:3824. [Google Scholar]
- 13.Pruessmann KP, Weiger M, Bornert P, Boesiger P. Advances in Sensitivity Encoding with Arbitrary k-Space Trajectories. Magn Reson Med. 2001;46:638–651. doi: 10.1002/mrm.1241. [DOI] [PubMed] [Google Scholar]
- 14.Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized Autocalibrating Partially Parallel Acquisitions (GRAPPA). Magn Reson Med. 2002;47(6):1202–1210. doi: 10.1002/mrm.10171. [DOI] [PubMed] [Google Scholar]
- 15.Lin W, Bornert P, Huang F, Duensing GR, Reykowski A. Generalized GRAPPA Operators for Wider Spiral Bands: Rapid Self-Calibrated Parallel Reconstruction for Variable Density Spiral MRI. Magn Reson Med. 2011;66(4):1067–1078. doi: 10.1002/mrm.22900. [DOI] [PubMed] [Google Scholar]
- 16.Heidemann RM, Griswold MA, Seiberlich N, Kruger G, Kannengiesser SA, Kiefer B, Wiggins G, Wald LL, Jakob PM. Direct Parallel Image Reconstructions for Spiral Trajectories Using GRAPPA. Magn Reson Med. 2006;56(2):317–326. doi: 10.1002/mrm.20951. [DOI] [PubMed] [Google Scholar]
- 17.Seiberlich N, Breuer FA, Ehses P, Moriguchi H, Blaimer M, Jakob PM, Griswold MA. Using the GRAPPA Operator and the Generalized Sampling Theorem to Reconstruct Undersampled Non-Cartesian Data. Magn Reson Med. 2009;61(3):705–715. doi: 10.1002/mrm.21891. [DOI] [PubMed] [Google Scholar]
- 18.Fessler JA, Sutton BP. Nonuniform Fast Fourier Transforms Using Min-Max Interpolation. IEEE Signal Proces. 2003;51(2):560–574. [Google Scholar]
- 19.Meyer CH, Pauly JM, Macovski A. A Rapid, Graphical Method for Optimal Spiral Gradient Design. New York. 1996:392. [Google Scholar]
- 20.King K, Foo T, Crawford C. Optimized Gradient Wavefroms for Spiral Scanning. Magn Reson Med. 1995;34:156–160. doi: 10.1002/mrm.1910340205. [DOI] [PubMed] [Google Scholar]
- 21.Glover GH. Simple Analytical Spiral k-Space Algorithm. Magn Reson Med. 1999;42:412–415. doi: 10.1002/(sici)1522-2594(199908)42:2<412::aid-mrm25>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 22.Manohar M, Tilton JC. Progressive Vector Quantization on a Massively Parallel SIMD Machine with Application to Multispectral Image Data. IEEE Trans Image Process. 1996;5(1):142–147. doi: 10.1109/83.481678. [DOI] [PubMed] [Google Scholar]
- 23.Hong IK, Chung ST, Kim HK, Kim YB, Son YD, Cho ZH. Ultra Fast Symmetry and SIMD-Based Projection-Back projection Algorithm for 3-D PET Image Reconstruction. IEEE Trans Med Imaging. 2007;26(6):789–803. doi: 10.1109/tmi.2007.892644. [DOI] [PubMed] [Google Scholar]
- 24.Agulleiro JI, Garzon EM, Garcia I, Fernandez JJ. Vectorization with SIMD Extensions Speeds up Reconstruction in Electron Tomography. Journal of Structural biology. 2010;170(3):570–575. doi: 10.1016/j.jsb.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 25.Intel Architecture Instruction Set Extensions Programming Reference. Intel; 2014. [Google Scholar]
- 26.Robson PM, Grant AK, Madhuranthakam AJ, Lattanzi R, Sodickson DK, McKenzie CA. Comprehensive Quantification of Signal-to-Noise Ratio and G-Factor for Image-Based and k-Space Based Parallel Imaging Reconstructions. Magn Reson Med. 2008;60(4):895–907. doi: 10.1002/mrm.21728. [DOI] [PMC free article] [PubMed] [Google Scholar]