Abstract
The advancements made in tissue culture techniques has made it possible to regenerate various horticultural species in vitro as micropropagation protocols for commercial scale multiplication are available for a wide range of crops. Clonal propagation and preservation of elite genotypes, selected for their superior characteristics, require high degree of genetic uniformity amongst the regenerated plants. However, plant tissue culture may generate genetic variability, i.e., somaclonal variations as a result of gene mutation or changes in epigenetic marks. The occurrence of subtle somaclonal variation is a drawback for both in vitro cloning as well as germplasm preservation. Therefore, it is of immense significance to assure the genetic uniformity of in vitro raised plants at an early stage. Several strategies have been followed to ascertain the genetic fidelity of the in vitro raised progenies comprising morpho-physiological, biochemical, cytological and DNA-based molecular markers approaches. Somaclonal variation can pose a serious problem in any micropropagation program, where it is highly desirable to produce true-to-type plant material. On the other hand, somaclonal variation has provided a new and alternative tool to the breeders for obtaining genetic variability relatively rapidly and without sophisticated technology in horticultural crops, which are either difficult to breed or have narrow genetic base. In the present paper, sources of variations induced during tissue culture cycle and strategies to ascertain and confirm genetic fidelity in a variety of in vitro raised plantlets and potential application of variants in horticultural crop improvement are reviewed.
Keywords: Micropropagation, Somaclones, Oxidative stress, Epignetic variation, Molecular markers, Crop improvement
Introduction
Plant tissue culture techniques proffer a substitute method of vegetative propagation of horticultural crops (Krishna et al. 2005; Alizadeh et al. 2010). Clonal propagation through tissue culture (popularly known as micropropagation) can be realized relatively rapidly within a small space (Krishna et al. 2008; Eftekhari et al. 2012). The uniformity of individual plants within a clone population is a major advantage of clonal cultivars in commercial production (Krishna and Singh 2013). However, genetic variations do occur in undifferentiated cells, isolated protoplasts, calli, tissues and morphological traits of in vitro raised plants (Bairu et al. 2011; Currais et al. 2013). In 1981, Larkin and Scowkraft coined a general term “somaclonal variation” for plant variants derived from any form of cell or tissue cultures.
At present, micropropagated plants, in various crops, such as strawberry, papaya, banana, grapes, pineapple, citrus, tomato, cucumber, watermelon, rhododendron, orchids, etc., are preferred over plants propagated through conventional means. However, ever since the first formal report of morphological variants in sugarcane plants produced in vitro in 1971 (Heinze and Mee 1971), several instances of somaclonal variations have been reported in different horticultural crops. The notable example could be banana in which occurrence of off-types from tissue cultured plantlets ranged from 6 to 38 % in Cavendish cultivars (Sahijram et al. 2003); however, it could be as high as 90 % (Smith 1988). From the point of commercial micropropagation, variation of any kind, in particular, genetic variations may be considered obstructive and worthless; since, such variations may lead to loss of genetic fidelity. However, plant cell and tissue cultures render increased genetic variability comparatively faster and without applying a sophisticated technology. This technology holds ample scope in crop improvement of horticultural crops, which are largely propagated vegetatively, partly, due to reasons like longer juvenile phase as in perennial fruit crops, occasional inbreeding depression, self and cross incompatibility, narrow genetic base especially in ornamentals, etc. Further, somaclonal variations require less space and time for screening of desirable traits in vitro unlike cross seedlings of perennial crops, which require a great deal of land area and time. Somaclones may itself have numerous applications in plant breeding and genetic improvements and recovery of such novel variants can be enhanced by applying suitable in vitro selection pressure (Jain 2001; Lestari 2006).
Sources of variations detected in plant tissue culture
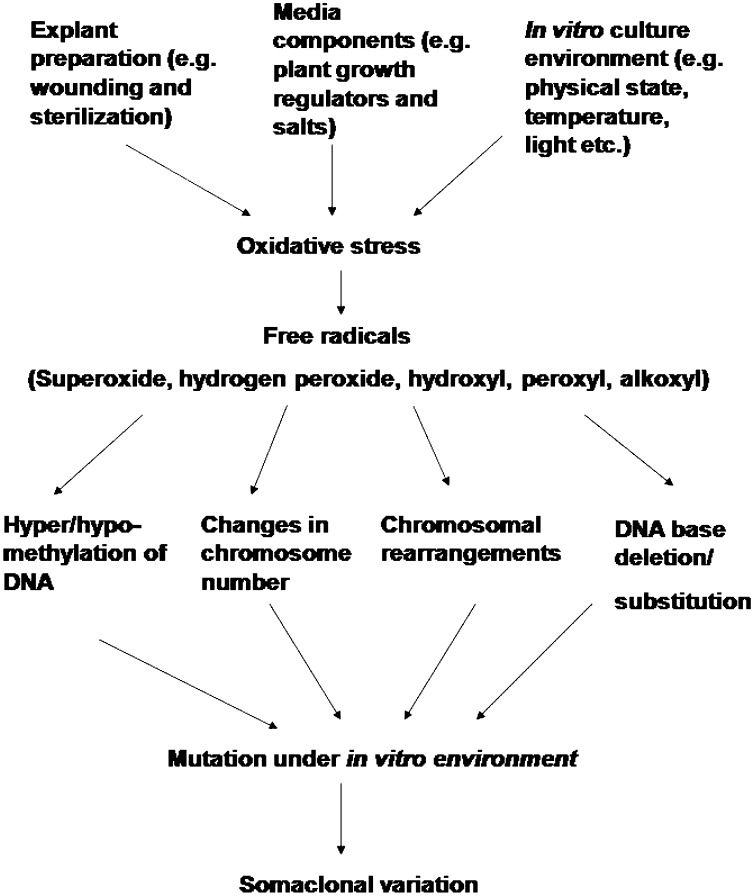
Tissue culture is an efficient method of clonal propagation; however, the resulting regenerants often has a number of somaclonal variations (Larkin and Scowcroft 1981). These somaclonal variations are mainly caused by newly generated mutations arising from tissue culture process (Sato et al. 2011b). The triggers of mutations in tissue culture had been attributed to numerous stress factors, including wounding, exposure to sterilants during sterilization, tissue being incomplete (protoplasts as an extreme example), imbalances of media components such as high concentration of plant growth regulators (auxin and cytokinins), sugar from the nutrient medium as a replacement of photosynthesis in the leaves, lighting conditions, the disturbed relationship between high humidity and transpiration (Joyce et al. 2003; Sato et al. 2011b; Smulders and de Klerk 2011).
Much of the variability expressed in micropropagated plants may be the result of, or related to, oxidative stress damage inflicted upon plant tissues during in vitro culture (Cassells and Curry 2001; Tanurdzic et al. 2008; Nivas and DSouza 2014). Oxidative stress results in elevated levels of pro-oxidants or reactive oxygen species (ROS) such as superoxide, hydrogen peroxide, hydroxyl, peroxyl and alkoxyl radicals. These ROS may involve in altered hyper- and hypo-methylation of DNA (Wacksman 1997); changes in chromosome number from polyploidy to aneuploidy, chromosome strand breakage, chromosome rearrangements, and DNA base deletions and substitutions (Czene and Harms-Ringdahl 1995), which in turn may lead to mutations in plant cells in vitro (Fig. 1). Somaclonal variation shows a similar spectrum of genetic variation to induced mutation as both of them result in qualitatively analogous gamut of DNA changes (Cassells et al. 1998). Different factors affect the frequency of development of somaclones under in vitro conditions.
Fig. 1.
Mechanism of somaclonal variation in micropropagated plants as a result of oxidative burst upon in vitro culture
Explant/explant source
Differences in both the frequency and nature of somaclonal variation may occur when regeneration is achieved from different tissue sources (Sahijram et al. 2003). Highly differentiated tissues such as roots, leaves, and stems generally produce more variations than explants with pre-existing meristems, such as axillary buds and shoot tips (Duncan 1997). In general, the older and/or the more specialized the tissue is used for regeneration, the greater the chances that variation will be recovered in the regenerated plants (Table 1) as under such conditions, adventitious shoot regeneration (shoot organogenesis) takes place from atypical points of origin directly or indirectly through a callus stage (e.g., from leaves, petioles, shoot internodes, root segments, anthers, hypocotyls, cotyledons, etc.; Pijut et al. 2012). Somaclonal variation can also arise from somatic mutations already present in the donor plant, i.e., presence of chimera in explants (Karp 1994).
Table 1 .
Occurrence of somaclonal variations as affected by the choice of explants
| S. no. | Crop species | Explants/explants source | Presence or absence of somaclonal variations (+/−) | References |
|---|---|---|---|---|
| 1 | African violet (Saintpaulia sp.) | Leaf segments | + | Matsuda et al. (2014) |
| 2 | Almond (Prunus dulcis) | Axillary branching | − | Martins et al. (2004) |
| 3 | Chimeric ‘Maricongo’ banana | Vegetative and floral axis tip | + | Krikorian et al. (1993) |
| Cavendish group of bananas (Musa sp.) | Chimeric shoot tip | + | Israeli et al. (1995) | |
| Banana cv. Martaman | Shoot tip | − | Ray et al. (2006) | |
| 4 | Brinjal (Solanum melongena) | Hypocotyl | − | Mallaya and Ravishankar (2013) |
| Callus induction on leaves, nodes and intermodal explants | + | Naseer and Mahmood (2014) | ||
| 5 | Chrysanthemum (Dendranthema grandiflora) | Callus from leaves and internodes | + | Miler and Zalewska (2014) |
| 6 | European violet (Viola uliginosa Besser) | Leaf and petiole fragments | + | Slazak et al. (2015) |
| 7 | Gerbera (Gerbera jamesonii Bolus) | Capitulum | − | Bhatia et al. (2009, 2011) |
| 8 | Gloxinia | Leaf explants | + | Hu and Xu (2010) |
| 9 | Hedychium coronarium Koen. | Axillary bud explants | − | Parida et al. (2013) |
| 10 | Hop (Humulus lupulus L.) | Meristem tissue | − | Patzak (2003) |
| 11 | Kaempferia galanga | Buds of rhizomes | − | Mohanty et al. (2011) |
| 12 | Kiwifruit (Actinidia deliciosa (Chev.) Liang and Ferguson) cv. ‘Tomuri’ | Leaf blades and petioles | + | Prado et al. (2007) |
| 13 | Oil palm (Elaeis guineensis Jacq.) | Mature zygotic embryos | + | Rival et al. (2013) |
| Immature zygotic embryo | + | Sanputawong and Te-chato (2011) | ||
| Immature leaves | + | Lucia et al. (2011) | ||
| 14 | Papaya (Carica papaya L.) | Axillary shoot tips underwent cryopreservation | + | Kaity et al. (2009) |
| 15 | Patchouli (Pogostemon patchouli) | Callus induction on internodal and leaf explants | + | Ravindra et al. (2012) |
| 16 | Potato (Solanum tuberosum) | Callus cultures of stem explant | + | Thieme and Griess (2005) |
| Callus induction via fresh sprouts | + | Munir et al. (2011) | ||
| 17 | Sweet cherry (Prunus avium) | Shoot apical portions | + | Piagnani and Chiozzotto (2010) |
| 18 | Rootstock Mr.S 2/5, selected from a half-sib progeny from Prunus cerasifera Erhr | Leaf | + | Muleo et al. (2006) |
| 19 | Swertia chirayita | Axillary multiplication | − | Joshi and Dhawan (2007) |
| 20 | Turmeric (Curcuma longa L.) | Latent axillary buds of rhizome | − | Nayak et al. (2010) |
| Axillary buds of unsprouted rhizome | − | Panda et al. (2007) | ||
| Callus cultures established from rhizome segments | + | Kar et al. (2014) | ||
| 21 | Vitis spp. | Nodal segment | − | Alizadeh et al. (2008) |
Mode of regeneration
Both culture initiation and subsequent subculture expose explants to oxidative stress (Krishna et al. 2008), which may result in mutations (Cassells and Curry 2001). It seems evident that ‘extreme’ procedures such as protoplast culture and also callus formation impose stress (Smulders and de Klerk 2011). Magnitude of this stress depends on the tissue culture technique. Therefore, the production of plants via axillary branching does not normally result in the production of variants, while cultures that go through a callus phase are the ones that theoretically promote a higher mutation rate (Zayova et al. 2010).
Investigations indicate more chromosome variability in the callus phase than in adventitious shoots (Saravanan et al. 2011), indicating a loss of competence in the more seriously disturbed genomes. This could be explained by the different grade of disturbance with which the cells are confronted. In the first case, cells follow a pattern of division which is the normal one in the developing plant. On the other hand, callus formation implies a dedifferentiation phase followed by uncontrolled cell divisions (Vázquez 2001). Some types of tissue culture mimic, in some aspects, other stressful situations as, for example, protoplast preparation in which cell wall degradation resembles the infective process of some pathogens. Therefore, the type and magnitude of the stress imposed on cultured cells varies according to the technique used. In contrast to popular belief that the growth of unorganized callus is necessary for induction of genetic variation, variability could be noticed in plants regenerated from explants adventitiously (Farahani et al. 2011; Bhojwani and Dantu 2013).
Sometimes for regeneration under in vitro conditions, somatic embryogenesis is the preferred pathway for generating propagules. It has been suggested that regeneration via embryogenesis has better chance of obtaining genetically uniform plants than through organogenic differentiation (Vázquez 2001). This is so, because DNA in the initial stages of development in somatic embryogenesis contains lower levels of methylation than in the later stages (Sahijram et al. 2003). Variation in in vitro cultures raised through somatic embryogenesis has been reported in several horticultural crops like hazel nut (Diaz-Sala et al. 1995), Citrus paradisi (Hao et al. 2004), oil palm (Jaligot et al. 2004), rose (Xu et al. 2004), potato (Sharma et al. 2007), grapevine (Schellenbaum et al. 2008), coffee (Menéndez-Yuffá et al. 2010), olive (Leva et al. 2012), tamarillo (Currais et al. 2013) and brinjal (Naseer and Mahmood 2014).
Effect of length of culture period and number of subculture cycles
The longer a culture is maintained in vitro, the greater the somaclonal variation is (Kuznetsova et al. 2006; Gao et al. 2010; Farahani et al. 2011; Jevremović et al. 2012; Sun et al. 2013). Variant karyotypes are found to amass with increasing age of callus and as a result the chances of variant plants produced during successive subculture also increases, in general (Zayova et al. 2010). Furthermore, the rapid multiplication of a tissue, during micropropagation, may affect its genetic stability. Khan et al. (2011) reported that after the eighth subculture, the number of somaclonal variants increased with a simultaneous decrease in the multiplication rate of propagules in banana.
Similarly, Clarindo et al. (2012) suggested a limit of less than 4 months storage of coffee cell aggregate suspensions for true-to-type mass propagation as ploidy instability was noticed in long-term in vitro culture. Similarly when Farahani et al. (2011) raised olive cultivars, under in vitro conditions, through internode cuttings, significant difference was observed in morphological characters among the regenerated plants after seventh subculture, which was later confirmed by RAPD analysis. However, C-value analysis showed that no significant change has occurred during subculturing in both olive genotypes. This indicates that the genetic changes accompanied by somaclonal variation could be due to the changes in the nucleotide content of the genome, probably, owing to mutations (insertions/deletions) and not due to quantitative changes.
Not only the number of subculture but their duration also contributes to enhancing the rate of somaclonal variations, especially cell suspension and callus cultures (Bairu et al. 2006; Sun et al. 2013). Studies have shown that somaclonal variation is more apparent in plants regenerated from long-term cultures (Etienne and Bertrand 2003). Rival et al. (2013) noticed that in vitro proliferation induces DNA hypermethylation in a time-dependent fashion and changes in DNA methylation is involved in modulating the expression of embryogenic capacity of oil palm during tissue culture.
Culture environment
External factors like growth regulators, temperature, light, osmolarity and agitation rate of the culture medium are known to influence the cell cycle in vivo in plants, considerably, which indicates that inadequate control of cell cycle in vitro is one of the causes of somaclonal variation (Karp 1994; Nwauzoma and Jaja 2013). Normal cell cycle controls, which prevent cell division before the completion of DNA replication, are presumed to be disrupted by tissue culture, resulting in chromosomal breakage (Phillips et al. 1994). Chromosome breakage and its consequences (deletions, duplications, inversions, and translocations) cause aberrations in vitro (Duncan 1997). Plant growth regulators can affect the rate of somaclonal variation both directly and indirectly by increasing the multiplication rate and inducing adventitious shoots (Gao et al. 2010). According to D’Amato (1985), it cannot be excluded that some plant growth regulators (PGRs) at certain concentrations or in combination with other growth regulators and/or particular constituents of a culture medium, may act as mutagens.
Several growth regulators, such as 2,4-dichlorophenoxy acetic acid (2,4-D), naphthalene acetic acid (NAA) and BAP (6-benzylaminopurine), synthetic phenylurea derivatives (4-CPPU, PBU and 2,3-MDPU) have been most frequently considered to be responsible for genetic variability (Siragusa et al. 2007; Sun et al. 2013; Sales and Butardo 2014).
Prolonged cultivation in medium containing 2,4-D influences higher DNA ploidy levels in callus cells (da Silva and Carvalho 2014). In their experiment with banana, Sales and Butardo (2014) observed that addition of synthetic auxin 2,4-D in culture medium led to high level of methylation events, particularly, cytosine methylation either at the internal or external cytosine end, which largely resulted in variations in tissue cultured plants. Alteration in genomic DNA methylation rate is being attributed for the development of ‘mantled’ somaclonal variant in oil palm (Eeuwens et al. 2002; Jaligot et al. 2011). Similarly, Arnhold-Schmitt (1993) observed that indole-3-acetic acid (IAA) and inositol in the growth medium induced DNA rearrangements and methylation changes in carrot (Daucus carota) callus cultures. Matsuda et al. (2014) observed that percentage of somaclonal variations dramatically increased when PGRs (0.5 ppm BA and 0.1 ppm NAA) were added to the medium inoculated with leaf/leaf segments explants of African violet.
Kinetin has been shown to cause extensive hypomethylation of DNA in proliferating cultures of carrot root explants within 2 weeks (Arnhold-Schmitt 1993), and auxins, including NAA, have the opposite effect and cause hypermethylation (LoSchiavo et al. 1989). Moreover, there is evidence that differential expression in chromatin remodeling genes and histone methylation genes happens during tissue culture, which leads to disruption in the methylation pathway in a non-specific manner and hypo/hypermethylation patterns of DNA induced in tissue culture. This can be stabilized and transmitted to plants regenerated from these cultures (Shearman et al. 2013). Not only the concentration, but also the ratio of different growth regulators affects the occurrence of variations in vitro. Eeuwens et al. (2002) observed that, in general, a relatively high auxin/cytokinin ratio resulted in the lowest incidence of variant ‘mantled’ flowering in oil palm, while using media supplemented with relatively high cytokinins/auxin ratio resulted in a high incidence of mantled flowering. The role of cytokinin was further confirmed by Ooi et al. (2013), who noticed that the mantled inflorescences of oil palm contained higher levels of cytokinins like isopentenyladenine 9-glucoside and lower levels of trans-zeatin 9-glucoside, dihydrozeatin riboside, and dihydrozeatin riboside 5′-monophosphate compared with normal inflorescences.
Genotype and ploidy
Though, the in vitro morphogenesis seems to be highly dependent on plant growth regulators and media used for culture, it is again genotype specific (Alizadeh et al. 2010; Eftekhari et al. 2012). Among factors affecting somaclonal variation, plant genotype is probably the most important determinant of variation (Shen et al. 2007; Tican et al. 2008; Nwauzoma and Jaja 2013). Earlier, Eeuwens et al. (2002) characterized oil palm clones as low/moderate risk and high risk with regard to ‘mantle’ flowering (wherein anther primordia in both male and female flowers turn into fleshy supplementary carpels), on the basis of terminal inflorescence data generated under in vitro conditions. Clones classified as high risk at the outset gave a significantly higher incidence of mantled flowering in the field than low/medium risk clones, confirming that data on terminal inflorescences produced in vitro allows effective screening of material with regard to the risk of mantled flowering. It is likely that this result from a combination of differences in genotype and differences in epigenetically inherited changes are induced during the pre-embryogenic stages of the culture process, i.e., callus initiation and maintenance.
Identification of variation in tissue culture
Both genetic and epigenetic alterations are associated with in vitro propagation, which may have phenotypic consequences, and are collectively called somaclonal variation (Larkin and Scowcroft 1981; Guo et al. 2007). As a result, somaclonal variation is characterized by the intricacy of the changes, which are exhibited at various levels, including phenotypic, cytological, biochemical and genetic/epigenetic (Kaeppler et al. 2000). Therefore, the strategy for the detection of somaclones should be based on such manifestations.
A wide variety of tools are available for the detection and characterization of somaclonal variants which are primarily based on the differences in morphological traits (Pérez et al. 2009, 2011; Nhut et al. 2013), cytogenetical analysis for the determination of numerical and structural variation in the chromosomes (Clarindo et al. 2012; Currais et al. 2013; Abreu et al. 2014), biochemical (Vujovic et al. 2010; Kar et al. 2014), molecular DNA markers (Krishna and Singh 2007; Pathak and Dhawan 2012; Hossain et al. 2013; Bello-Bello et al. 2014) or their combinations (Horáček et al. 2013; Dey et al. 2015; Stanišić et al. 2015). The best test for assessing somaclonal variation is to fruit out the plants and conduct an extensive horticultural evaluation, which is unfortunately a long-term endeavor with woody fruit crops, particularly (Grosser et al. 1996). Every tool has its own advantages and limitations in assessment of the variations (Table 2), which govern their use for restricted or large-scale application. The choice of technique for any given application depends upon the material used and the nature of the question being addressed (Karp 2000).
Table 2.
Strengths and weaknesses of different marker systems for the assessment of clonal fidelity
| Advantages | Disadvantages |
|---|---|
| Morphological traits | |
| Visual differentiation | Sensitive to ontogenic changes and other environmental factors |
| Does not require any laboratory facility | Limited in numbers |
| Suitable for preliminary detection | Time-consuming |
| Cytological markers (flow-cytometry) | |
| Sample preparation and analysis is convenient and rapid in case of in flow-cytometry | Cytosolic compounds may interfere with quantitative DNA staining in flow-cytometry |
| Rapid and efficient method for routine large-scale studies of ploidy level | Absence of a set of internationally agreed DNA reference standards in case of in flow-cytometry |
| Unfailing detection of even the smallest modifications in chromosome number | Time-consuming chromosome counting |
| Isozyme markers | |
| Codominant expression | Sensitive to ontogenic changes and other environmental factors |
| Ease of performance | Limited in numbers Not all of these reagent systems work efficiently with all plant species Tissue-specific expression |
| DNA markers | |
| Codominant expression Any source DNA can be used for the analysis Phenotypically neutral Not sensitive to ontogenic changes and other environmental factors Capability to detect culture-induced variation both at the DNA sequence and methylation pattern levels |
RAPD markers are dominant and do not permit the scoring of heterozygous individuals. Besides, they exclusively identify sequence changes Possible non-homology of similar sized fragments as ISSR is a multilocus technique Disadvantages of AFLPs include the need for purified, high molecular weight DNA, the dominance of alleles and the possible non-homology of comigrating fragments belonging to different loci Involvement of high development costs in SSR markers if adequate primer sequences for the crop species of interest are unavailable. Further, mutations in the primer annealing sites may result in the occurrence of null alleles (no amplification of the intended PCR product), which may lead to errors in scoring |
Molecular basis of somaclonal variation
How a single plant genotype can result in a variety of phenotypic outcomes under the same in vitro culture conditions is still far from being completely understood. Several bases for somaclonal variation have been proposed, which include changes in chromosome number (Mujib et al. 2007; Leva et al. 2012), point mutations (D’Amato 1985; Ngezahayo et al. 2007), somatic crossing over and sister chromatid exchange (Duncan 1997; Bairu et al. 2011), chromosome breakage and rearrangement (Czene and Harms-Ringdahl 1995; Alvarez et al. 2010), somatic gene rearrangement, DNA amplification (Karp 1995; Tiwari et al. 2013), changes in organelle DNA (Cassells and Curry 2001; Bartoszewski et al. 2007), DNA methylation (Guo et al. 2007; Linacero et al. 2011), epigenetic variation (Kaeppler et al. 2000; Guo et al. 2006; Smulders and de Klerk 2011), histone modifications and RNA interference (Miguel and Marum 2011), segregation of pre-existing chimeral tissue (Brar and Jain 1998; Vázquez 2001; Ravindra et al. 2012; Nwauzoma and Jaja 2013) and insertion or excision of transposable elements (Gupta 1998; Sato et al. 2011b). In particular, transposable elements are one of the causes of genetic rearrangements in in vitro culture (Hirochika et al. 1996; Sato et al. 2011a).
Tissue culture is reported to activate silent transposable elements, resulting in somaclonal variations. Insertions of transposable elements and retrotransposons can function as insertional mutagens of plant genomes, whereas widespread activation may result in a wide gamut of chromosomal rearrangements (Tanurdzic et al. 2008). In turn, these rearrangements can lead to misregulation of genes, aneuploidy and new transposon insertions (Smulders and de Klerk 2011).
However, many aspects of the mechanisms, which result in somaclonal variations, remain undefined. It is therefore, inevitable to explore the genome-wide change through sequencing of whole-genome of the concerned crop. Next-generation sequencing technology has enabled the whole-genome sequencing of individual plants (Miyao et al. 2012). A new generation of sequencing technologies, from Illumina/Solexa, ABI/SOLiD, 454/Roche, and Helicos, has provided unprecedented opportunities for high-throughput functional genomic research (Morozova and Marra 2008; Metzker 2010).
Somaclonal variations vis-à-vis crop improvement
Genetic variation is an essential component of any conventional crop breeding program. The typical crop improvement cycle takes 10–15 years to complete and includes germplasm manipulations, genotype selection and stabilization, variety testing, variety increase, proprietary protection and crop production stages. Plant tissue culture is an enabling technology from which many novel tools have been developed to assist plant breeders (Karp 1992; Mathur 2013). Tissue culture-induced somaclonal variation is akin to variations induced with chemical and physical mutagens (Jain 2001) and offers an opportunity to uncover natural variability for their potential exploitation in crop improvement.
Like any other technology, in vitro induced somaclonal variation has its own merits and demerits, like the two sides of the same coin.
Advantages
The advantages comprise: (1) it is cheaper than other methods of genetic manipulation and does not require ‘containment’ procedures. (2) Tissue culture systems are available for more plant species than can be manipulated by somatic hybridization and transformation at the present time. (3) It is not necessary to have identified the genetic basis of the trait, or indeed, in the case of transformation, to have isolated and cloned it. (4) Novel variants have been reported among somaclones, and evidences indicate that both the frequency and distribution of genetic recombination events can be altered by passage though tissue culture. This implies that variation may be generated from different locations of the genome than those, which are accessible to conventional and mutation breeding (Karp 1992). (5) There is no possibility of obtaining chimeric expression if somaclones are raised through cell culture (Evans 1989).
Somaclonal variation has been most successful in crops with limited genetic systems (e.g., apomicts, vegetative reproducers) and/or narrow genetic bases. In ornamental plants, for instance, the exploitation of in vitro-generated variability has become part of the routine breeding practice of many commercial enterprises.
Disadvantages
One of the serious limitations of somaclonal variation which makes it comparatively difficult to use is that, despite the identification of factors affecting the variation response of a given plant species, it is still not possible to predict the outcome of a somaclonal program (Karp 1992) as it is random and lacks reproducibility. Further, as a large number of genetic changes are based on point mutations or chromosome rearrangements, most R1 segregate. Therefore for quantitative traits such as yield, it is virtually impossible to select individuals with improvements in the R1 generation. Though techniques for selection of somaclones resistant to various biotic and abiotic stresses had been worked out in many horticultural crops, unfortunately, no in vitro selection methods exist for complicated traits such as yield, soluble solids, sweetness, texture or shelf life (Evans 1989).
Somaclonal variation can become a part of plant breeding provided they are heritable and genetically stable. Only a limited numbers of promising varieties so far had been released using somaclonal variations. This is perhaps due to the lack of interaction between plant breeders and tissue culture scientists, and non-predictability of somaclones (Jain 2001). Further, though the new varieties have been produced by somaclonal variation, in a large number of cases improved variants have not been selected due to (1) the variations were all negative; (2) positive changes were also altered in negative ways; (3) the changes were not novel, or (4) the changes were not stable after selfing or crossing (Karp 1992).
Recovery of somaclonal variants
The recovery of variants can be improved by promoting the factors which are responsible for the development of somaclonal variations such as protoplast culture (Kothari et al. 2010) and employing callus and cell suspension culture for several cycles and regeneration of large number of plants from long-term cultures (Barakat and El-Sammak 2011). Indirect organogenesis is an important means of retrieving genetic variation through somaclones with useful traits of agronomic or industrial use. Besides, plant genotype is a major factor, which determines the type and frequency of somaclonal variation. For instances, Solanaceous plants like potato (Sharma et al. 2007) and tomato (Bhatia et al. 2005) produce a gamut of somaclonal variation than many other commercial horticultural crops. However, to be of practical value, the frequency of somaclonal variation should be sufficient enough to select desirable traits, and the selected lines should perform well under multiple environments (Duncan 1997). The efficiency of recovering variants in vitro can further be enhanced by applying selection pressure through screening of desirable traits, e.g., in vitro selection for tolerance against abiotic and biotic stresses (Barakat and El-Sammak 2011). This attains more significance in view of the fact that the selection of desirable traits takes several years and many generations under field conditions. In vitro selection can shorten considerably the time for the selection of desirable traits under in vitro selection pressure with minimal environmental interaction, and can complement field selection (Jain 2001).
The recovery of somaclones can be increased by combining micropropagation with induced mutagenesis in vitro (Afrasiab and Iqbal 2010). Kuksova et al. (1997) noted that somaclonal variation and mutagens can be combined to increase the frequency of induced mutation. Likewise, irradiation followed by adventitious bud regeneration has been reported to have allowed the recovery of mutants with useful agronomic traits in Gypsophila paniculata L. (Barakat and El-Sammak 2011). Yang and Schmidt (1994) treated in vitro leaves of the cherry rootstock ‘209/1’ (Prunus cerasus × P. canescens) with X-rays with LD50 close to 20 Gy. Among plants regenerated from leaves with 20 Gy, one was phenotypically different, and was subsequently isolated and cloned. This somaclone was extremely dwarfed and was stable in both greenhouse and field tests. Employing more than one mutagen results in further improvement in recovery of somaclones in vitro. Murti et al. (2013) exposed the strawberry ‘DNKW001’ to the doses of 0, 30, 80, 130, 180, 230, 280, 300 and 325 Gy and similar doses of gamma rays + EMS 7 µM treatments. Their results showed that Gamma ray irradiation + EMS was more effective to generate more type and magnitude of variants. Purwati and Sudarsono (2007) regenerated four variant lines in abaca banana from (1) embryogenic calli; (2) ethyl methyl sulphonate (EMS)-treated embryogenic calli; (3) EMS-treated embryogenic calli, followed by in vitro selection on Foc (Fusarium oxysporum f.sp. cubense) culture filtrate (EMS + CF line) and (4) EMS-treated embryogenic calli, followed by in vitro selection on fusaric acid. The Foc resistance abaca variants were successfully identified from four tested abaca variant lines, although with different frequencies. However, more Foc resistance abaca plants were identified from EMS + CF line than the others. Earlier, Bidabadi et al. (2012) suggested that the subjecting of shoot tips cultures of banana to EMS (200 mM) treatments could provide an alternative strategy for inducing variants. Recently, Iuliana and Cerasela (2014) suggested irradiation of in vitro raised plants with ultraviolet radiations (UV-C) for induction of somaclones in potato.
Application of somaclonal variations
It is well accepted that somaclonal variations arising out of unique tissue culture environment are very often noticed phenomenon in clonally propagated plants, which can advantageously be utilized as a source of new variation in horticultural crops (Karp 1995). However, suitable tools for detection, evaluation, identification and improvement of resistant clones should be designed in order to realize the benefits of such variations (Sahijram et al. 2003). Crop improvement through somaclonal variation enables breeders to obtain plants tolerant to the biotic or abiotic stress, such as drought, high salinity, high or low soil pH and disease tolerance (Yusnita et al. 2005). A number of cultivars have been developed through somaclonal variation in different horticultural crops for a range of useful traits, which are presented in Table 3.
Table 3.
In vitro selection of desirable traits and development of some commercially exploited varieties through somaclonal variation in different horticultural crops
| S. no. | Horticultural crop | Characteristic of somaclone | References |
|---|---|---|---|
| 1 | Aglaonema | Cultivar ‘Moonlight Bay’ and ‘Diamond Bay’ from ‘Silver Bay,’ and ‘Emerald Bay,’ from ‘Golden Bay’ | Henny et al. (1992, 2003) |
| 2 | Apple (Malus × domestica Borkh.) | Resistance to Erwinia amylovora | Chevreau et al. (1998) |
| 3 | Apple rootstocks M 26 and MM 106 (Malus pumila Mill.) | Resistance to Phytophthora cactorum | Rosati et al. (1990) |
| 4 | Apple rootstock Malling 7 | Resistance to white root rot (Dematophora necatrix) | Modgil et al. (2012) |
| 5 | Anthurium sp. | ‘Orange Hot’ derived from ‘Red Hot’ clone | Henny and Chen (2011) |
| 6 | Banana (Musa acuminata L.) | Semi-dwarf and resistant to Fusarium wilt TC1-229 | Tang et al. (2000) |
| Larger bunch size var. TC2-425; Resistant to Fusarium oxysporum f. sp. cubense (Foc) race 4; bunch 40 % heavier than cv. Formosana | Hwang (2002) | ||
| Fusarium wilt-resistant somaclonal variants of banana cv. Rasthali | Ghag et al. (2014) | ||
| Var. CIEN-BTA-03, resistant to yellow Sigatoka | Giménez et al. (2001) | ||
| 10 somaclones; GCTCV215-1 released for commercial planting | Hwang and Ko (1992, 2004) | ||
| Var. CUDBT-B1, reduced height and early flowering | Martin et al. (2006) | ||
| Var. Tai-Chiao No. 5, superior horticultural traits and resistance to Fusarium wilt | Lee et al. (2011) | ||
| 7 | Begonia (Begonia × elatior) | Plant morphology, number of flowers per plant, and flower size | Jain (1997) |
| 8 | Brinjal (Solanum melongena L.) | Stress-tolerant somaclone selection | Ferdausi et al. (2009) |
| 9 | Blackberry | Thornless var. ‘Lincoln Logan’ | Hall et al. (1986) |
| 10 | Capsicum (Capsicum annuum L.) | Yellow fruited var. Bell sweet | Morrison et al. (1989) |
| 11 | Calthea roseopicta | Developed common cultivars like Angela, Cora, Dottie, Eclipse and Saturn | Chao et al. (2005) |
| 12 | Carrot (Daucus carota L.) | Resistance to leaf spot (Alternaria dauci) | Dugdale et al. (2000) |
| Resistant to drought | Rabiei et al. (2011) | ||
| 13 | Carnation (Dianthus caryoplyllus L.) | Resistant to Fusarium oxysporum f. sp. dianthi | Esmaiel et al. (2012) |
| 14 | Celery (Apium graveolens L.) | Fusarium resistant var. UC-TC | Heath-Pagliuso and Rappaport (1990) |
| Multiple-resistant (insect resistance against Spodoptera exigua and disease resistance against Fusarium yellow) somaclones K-26, K-108 and K-128 | Diawara et al. (1996) | ||
| 15 | Celosia argentea L. | Resistance to nematode | Opabode and Adebooye (2005) |
| 16 | Cereus peruvianus | Shoots with different areoles characteristics | Resende et al. (2010) |
| 17 | Chili pepper (Capsicum annuum L.) | Early flowering and increase of yield components | Hossain et al. (2003) |
| 18 | Chrysanthemum (Dendranthema grandiflora) | Variation in leaf, flower shape and petal size | Ahloowalia (1992) |
| Daisy type chrysanthemum | Jevremović et al. (2012) | ||
| Attractive variants with changed inflorescence colors | Miler and Zalewska (2014) | ||
| 19 | Citrus spp. | Resistant to Phoma tracheiphila | Deng et al. (1995) |
| Salinity tolerance | Ben-Hayyim and Goffer (1989) | ||
| 20 | Cuphea viscosissima Jacq. | Significantly superior over the parents for mean plant height, leaf area, seed yield, per cent caprylic acid and lauric acid contents | Ben-Salah and Roath (1994) |
| 21 | Cymbopogon winterianus Jowitt | Aromatic grass var. CIMAP/Bio-13 with 50–60 % increased oil yield | Mathur et al. (1988) |
| Increased total oil yield and quality with high geraniol content | Nayak et al. (2003) | ||
| Cymbopogon martinii | Increased oil content | Patnaik et al. (1999) | |
| 22 | Dieffenbachia sp. | Novel and distinct foliar variegation with taller, larger canopy and longer leaves than ‘Camouflage’ parental plants | Shen et al. (2007) |
| 23 | Garlic (Allium sativum L.) | Consistently higher bulb yield than the parental clone | Vidal et al. (1993) |
| Resistance against the pathogenic fungi ‘Sclerotium cepivorum’ | Zhang et al. (2012) | ||
| 24 | Geranium spp. | Vigourous and attractive flower | Skirvin and Janick (1976) |
| Isomenthone-rich somaclonal mutant | Gupta et al. (2001) | ||
| Cv. ‘CIM Pawan, a somaclone of the Bourbon type variety Bipuli, with more herbage and essential oil yield than Bipuli | Saxena et al. (2008) | ||
| 25 | Gerbera (Gerbera jamesonii Bolus) | Novel cultivars | Minerva and Kumar (2013) |
| 26 | Ginger (Zingiber officinale Rosc.) | Tolerant to wilt pathogen (Fusarium oxysporum f.sp. zingiberi Trujillo) | Bhardwaj et al. (2012) |
| 27 | Grapevine (Vitis vinifera L.) | Resistant to Botrytis cinerea and Plasmopara viticola | Kuksova et al. (1997) |
| 28 | Haemerocallis spp. | Dwarf, short flowers, male sterile var. Yellow Tinkerbell | Griesbach (1989) |
| 29 | Hedychium (ornamental ginger) | Ramata, dwarf and variegated cultivar | Sakhanokho et al. (2012) |
| 30 | Java citronella (Cymbopogon winterianus) | Somaclonal variant variety CIMAP/Bio-13, which yields 37 % more oil and 39 % more citronellon than the control variant | Mathur (2010) |
| 31 | Kiwi fruit (Actinidia deliciosa) | 5 somaclones, derived from cv. Tamuri, tolerant to NaCl | Caboni et al. (2003) |
| 32 | Mango (Mangifera indica L.) | Resistant to Colletotrichum gleosporiensis | Litz et al. (1991) |
| 33 | Mint (Mentha arvensis) | Increased herb and oil yield | Kukreja et al. (1991; 2000) |
| 34 | Myrobolan (Prunus cerasifera Erhr) | Water logging-tolerant clone variant (S.4) of myrobolan rootstcock Mr.S 2/5 for peach cv. Sun Crest | Iacona et al. (2013) |
| 35 | Olive (Olive europea) | Bush olive somaclone (BOS), columnar olive somaclone (COS) | Leva et al. (2012) |
| 36 | Patchouli (Pogostemon patchouli) | Higher herb yield and essential oil content | Ravindra et al. (2012) |
| 37 | Pea (Pisum sativum L.) | Resistance to Fusarium solani | Horáček et al. (2013) |
| 38 | Peach (Prunus persica L.) | Somaclones S156 and S122 resistant to leaf spot, moderately resistant to canker in cvs. Sunhigh and Red haven | Hammerschlag and Ognjanov (1990) |
| Resistant to root-knot nematode (Meloidogyne incognita Kofoid and White) | Hashmi et al. (1995) | ||
| Somaclone S 122-1 was found resistant to bacterial canker (Pseudomonas syringae pv. syringae) | Hammerschlag (2000) | ||
| 39 | Pear (Pyrus sp.) | Resistant to Erwinia amylovora | Viseur (1990) |
| Pear rootstock (Pyrus communis L.) ‘Old Home × Farmingdale (OHF 333)’ | Tolerance to the fire blight | Nacheva et al. (2014) | |
| 40 | Philodendron | Cultivars ‘Baby Hope’ from ‘Hope’ | Devanand et al. (2004) |
| 41 | Picrorhiza kurroa | Higher glycoside contents including kutkoside and picroside I in somaclone 14-P derived through Agrobacterium rhizogenes mediated transformed hairy root cultures of P. kurroa | Mondal et al. (2013) |
| 42 | Pineapple (Ananas comosus L., Merr.) | Spineless variant | Jaya et al. (2002) |
| Cvs. P3R5 and Dwarf, variation in fruit color, growth habit, fruit size and length of plant generation cycle | Pérez et al. (2009, 2012) | ||
| 43 | Potato (Solanum tuberosum L.) | Non-browning var. White Baron | Arihara et al. (1995) |
| Somaclones for heat tolerance | Das et al. (2000) | ||
| Somaclones IBP-10, IBP-27 and IBP-30, derived from cultivar Desiree, showed higher resistance to Alternaria solani and Streptomyces scabiei | Veitia-Rodriguez et al. (2002) | ||
| Improved size, shape, appearance, starch content and starch yield | Thieme and Griess (2005) | ||
| Superior processing attributes than cv. ‘Russet Burbank’ | Nassar et al. (2011) | ||
| High-yielding genotype SVP-53 | Hoque and Morshad (2014) | ||
| Increased phytonutrient and antioxidant components over cv. ‘Russet Burbank’ | Nassar et al. (2014) | ||
| 44 | Quince A (Cydonia oblonga) | High soil pH | Dolcet-Sanjuan et al. (1992), Marino et al. (2000) |
| 45 | Stevia rebaudiana | High glycoside contents (steviol, stevioside, and rebaudioside) | Khan et al. (2014) |
| 46 | Strawberry (Fragaria sp.) | Resistant to Fusarium oxysporum f. sp. fragariae | Toyoda et al. (1991) |
| Resistant to Alternaria alternate | Takahashi et al. (1993) | ||
| Resistant to Phytophthora cactorum | Battistini and Rosati (1991) | ||
| Improved horticultural traits | Biswas et al. (2009) | ||
| Resistant to Verticillium dahliae Kleb | Zebrowska (2010) | ||
| ‘Serenity’, a paler skin-colored, late season, resistant to powdery mildew and Verticillium wilt somaclonal variant of the short-day cv. ‘Florence’ | Whitehouse et al. (2014) | ||
| 47 | Sweet potato (Ipomea batatas L. Lam.) | Tolerant to salinity | Anwar et al. (2010) |
| 48 | Sweet orange (Citrus sinensis (L.) Osb.) | Somaclone of OLL (Orie Lee Late) sweet orange; late maturing; suitable for fresh market or processing, exceptional juice quality and flavor | Grosser et al. (2015) |
| 49 | St. Augustine grass [Stenotaphrum secundatum (Walt.) Kuntze] | Freeze-tolerant somaclonal variant SVC3 | Li et al. (2010) |
| 50 | Syngonium podophyllum Schott | 22 cultivars, derived from original ‘White Butterfly’ clone, with distinct and stable foliage characteristics | Henny and Chen (2011) |
| 51 | Tomato (Lycopersicon esculentum L.) | High solid contents var. DNAP9 | Evans (1989) |
| 52 | Tulip (Tulipa sp.) | “Bs6”, selected from among the micropropagated plants of the cultivar ‘Blue Parrot’ with red-violet colored longer flower and stem | Podwyszynska et al. (2010) |
| 53 | Torenia (Torenia fournieri) | Flower color somaclonal variants | Nhut et al. (2013) |
| 54 | Turmeric (Curcuma longa L.) | High essential oil yielding somaclones | Kar et al. (2014) |
| Turmeric somaclone resistant to Fusarium oxysporum f.sp. Zingiberi | Kuanar et al. (2014) | ||
| 55 | Indian ginseng (Withania somnifera (L.) Dunal) | Withanolide (12-deoxywithastramonolide)-rich somaclonal variant | Rana et al. (2012) |
Conclusions
Several strategies have been followed to ascertain the genetic fidelity of the in vitro produced progenies in view of the fact that the commercial viability of micropropagation technology is reliant upon maintenance of genetic fidelity in the regenerated plants. Therefore, a thorough assessment of micropropagated plants becomes very critical, especially, for perennial crops such as fruit species, which have a long pre-bearing growth period. The efficiency and sensitivity of new molecular tools has enabled us to detect somaclonal variation at an early stage. These tools have become very useful for the rapid detection and accurate identification of variants. Nevertheless, the morphological and cytological assays should continue to remain as the primary and essential assay for the sustained success of fidelity tests associated with production of clonal plants. Though, on one hand, tissue culture-induced variations pose a major threat to the genomic integrity of regenerated plants, they provide tools for improvement to plant breeders, particularly for crops with a narrow genetic base, i.e., self pollinated and vegetatively propagated. Irrespective of our goal either for production of true-to-the type planting material or creation of variability, a multidisciplinary approach (involving concerned sciences of horticulture, genetics and plant breeding, physiology, cytology and molecular biology) with all our previous knowledge and experience should be followed to achieve the desideratum.
Acknowledgments
Authors are grateful to the Dr. S.K. Singh, Principal Scientist, Division of Fruits and Horticultural Technology, Indian Agricultural Research Institute, New Delhi 12, India, for his valuable advice during the preparation of the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no potential conflict of interest regarding submission and publication of this manuscript.
References
- Abreu IS, Carvalho CR, Clarindo WR. Massal induction of Carica papaya L. ‘Golden’ somatic embryos and somaclone screening by flow cytometry and cytogenetic analysis. Cytologia. 2014;79(4):475–484. doi: 10.1508/cytologia.79.475. [DOI] [Google Scholar]
- Afrasiab H, Iqbal J. In vitro techniques and mutagenesis for the genetic improvement of potato cvs. Desiree and Diamant. Pak J Bot. 2010;42:1629–1637. [Google Scholar]
- Ahloowalia BS. In vitro radiation induced mutants in Chrysanthemum. Mutat Breed Newsl. 1992;39:6. [Google Scholar]
- Alizadeh M, Singh SK, Jhang T, Sharma TR. Inter simple sequence repeat analysis to confirm genetic stability of micropropagated plantlets in three grape (Vitis spp) rootstock genotypes. J Plant Biochem Biotechnol. 2008;17:77–80. doi: 10.1007/BF03263264. [DOI] [Google Scholar]
- Alizadeh M, Singh SK, Patel VB. Comparative performance of in vitro multiplication in four grape (Vitis spp.) rootstock genotypes. Int J Plant Prod. 2010;4:41–50. [Google Scholar]
- Alvarez ME, Nota F, Cambiagno DA. Epigenetic control of plant immunity. Mol Plant Pathol. 2010;11:563–576. doi: 10.1111/j.1364-3703.2010.00621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar A, Kikuchi A, Watanabe KN. Assessment of somaclonal variation for salinity tolerance in sweet potato regenerated plants. Afr J Biotechnol. 2010;9:7256–7265. [Google Scholar]
- Arihara A, Kita T, Igarashi S, Goto M, Irikura Y. White Baron: a non-browning somaclonal variant of Danshakuimo (Irish cobbler) Am Potato J. 1995;72(11):701–705. doi: 10.1007/BF02849179. [DOI] [Google Scholar]
- Arnhold-Schmitt B. Rapid changes in amplification and methylation pattern of genomic DNA in cultured carrot root explants (Daucus carota L.) Theor Appl Genet. 1993;85:793–800. doi: 10.1007/BF00225021. [DOI] [PubMed] [Google Scholar]
- Bairu MW, Fennell CW, van Staden J. The effect of plant growth regulators on somaclonal variation in Cavendish banana (Musa AAA cv. ‘Zelig’) Sci Hortic. 2006;108:347–351. doi: 10.1016/j.scienta.2006.01.039. [DOI] [Google Scholar]
- Bairu MW, Aremu AO, Staden JV. Somaclonal variation in plants: causes and detection methods. Plant Growth Regul. 2011;63:147–173. doi: 10.1007/s10725-010-9554-x. [DOI] [Google Scholar]
- Barakat MN, El-Sammak H. In vitro mutagenesis, plant regeneration and characterization of mutants via RAPD analysis in baby’s breath ‘Gypsophila paniculata L’. Aust J Crop Sci. 2011;5(2):214–222. [Google Scholar]
- Bartoszewski G, Havey MJ, Ziókowska A, D’ugosz M, Malepszy S. The selection of mosaic (MSC) phenotype after passage of cucumber (Cucumis sativus L.) through cell culture—a method to obtain plant mitochondrial mutants. J Appl Genet. 2007;48:1–9. doi: 10.1007/BF03194652. [DOI] [PubMed] [Google Scholar]
- Battistini C, Rosati P. In vitro evaluation of somaclonal strawberry (Fragaria × ananassa ‘Brighton’) variants for susceptibility to Phytophthora cactorum. In: Dale A, Lubby D, editors. The strawberry into the 21st century. Portland: Timber Press; 1991. pp. 121–123. [Google Scholar]
- Bello-Bello JJ, Iglesias-Andreu LG, Avilés-Viñas SA, Gómez-Uc E, Canto-Flick A, Santana-Buzzy N. Somaclonal variation in habanero pepper (Capsicum chinense Jacq.) as assessed ISSR molecular markers. HortScience. 2014;49(4):481–485. [Google Scholar]
- Ben-Hayyim G, Goffer Y. Plantlet regeneration from NaCl-selected salt-tolerant callus culture of Shamouti oranges (Citrussinesis L. Osbeck) Plant Cell Rep. 1989;7:680–683. doi: 10.1007/BF00272060. [DOI] [PubMed] [Google Scholar]
- Ben-Salah H, Roath WW. Somaclonal variation in Cuphea viscosissima Jacq. for plant improvement. Ind Crop Prod. 1994;2:239–244. doi: 10.1016/0926-6690(94)90041-8. [DOI] [Google Scholar]
- Bhardwaj SV, Thakur T, Sharma R, Sharma P. In vitro selection of resistant mutants of ginger (Zingiberofficinale Rosc.) against wilt pathogen (Fusariumoxysporum f. sp. zingiberi Trujillo) Plant Dis Res. 2012;27:194–199. [Google Scholar]
- Bhatia P, Ashwath N, Senaratna T, Krauss SL. Genetic analysis of cotyledon derived regenerants of tomato using AFLP markers. Curr Sci. 2005;88(2):280–284. [Google Scholar]
- Bhatia R, Singh KP, Jhang T, Sharma TR. Assessment of clonal fidelity of micropropagated gerbera plants by ISSR markers. Sci Hortic. 2009;119:208–211. doi: 10.1016/j.scienta.2008.07.024. [DOI] [Google Scholar]
- Bhatia R, Singh KP, Sharma TR, Jhang T. Evaluation of the genetic fidelity of in vitro-propagated gerbera (Gerbera jamesonii Bolus) using DNA-based markers. Plant Cell Tissue Organ Cult. 2011;104(1):131–135. doi: 10.1007/s11240-010-9806-5. [DOI] [Google Scholar]
- Bhojwani SS, Dantu PK. Plant tissue culture: an introductory text. India: Springer; 2013. [Google Scholar]
- Bidabadi SS, Meon S, Wahab Z, Subramaniam S, Mahmood M. Induced mutations for enhancing variability of banana (Musa spp.) shoot tip cultures using ethyl methanesulphonate (EMS) Aust J Crop Sci. 2012;6:391–401. [Google Scholar]
- Biswas MK, Dutt M, Roy UK, Islam R, Hossain M. Development and evaluation of in vitro somaclonal variation in strawberry for improved horticultural traits. Sci Hortic. 2009;122:409–416. doi: 10.1016/j.scienta.2009.06.002. [DOI] [Google Scholar]
- Brar DS, Jain SM. Somaclonal variation: mechanism and applications in crop improvement. In: Jain SM, Brar DS, Ahloowalia BS, editors. Somaclonal variation and induced mutations in crop improvement. Dordrecht: Kluwer Academic Publishers; 1998. pp. 15–37. [Google Scholar]
- Caboni E, Anselmi S, Donato E, Manes F. In vitro selection of Actinidia deliciosa clones tolerance to NaCl and their molecular and in vivo ecophysiological characterisation. Acta Hortic. 2003;618:77–83. doi: 10.17660/ActaHortic.2003.618.7. [DOI] [Google Scholar]
- Cassells AC, Curry RF. Oxidative stress and physiological, epigenetic and genetic variability in plant tissue culture: implications for micropropagators and genetic engineers. Plant Cell Tissue Organ Cult. 2001;64:145–157. doi: 10.1023/A:1010692104861. [DOI] [Google Scholar]
- Cassells AC, Deadman ML, Brown CA, Griffin E. Field resistance to late blight (Pytophtora infectans (Mont.) De Bary in potato (Solanum tuberosum L.) somaclones associated with instability and pleiotropic effects. Euphytica. 1998;57:157–167. [Google Scholar]
- Chao CCT, Devanand PS, Chen J. AFLP analysis of genetic relationships among Calathea species and cultivars. Plant Sci. 2005;168:1459–1469. doi: 10.1016/j.plantsci.2005.01.012. [DOI] [Google Scholar]
- Chevreau E, Brisset MN, Paulin JP, James DJ. Fire blight resistance and genetic trueness-to-type of four somaclonal variants from the apple cultivar Greensleeves. Euphytica. 1998;104:199–205. doi: 10.1023/A:1018673813980. [DOI] [Google Scholar]
- Clarindo WR, Carvalho CR, Mendonça MAC. Ploidy instability in long-term in vitro cultures of Coffea arabica L. monitored by flow cytometry. Plant Growth Regul. 2012;68(3):533–538. doi: 10.1007/s10725-012-9740-0. [DOI] [Google Scholar]
- Currais L, Loureiro J, Santos C, Canhoto JM. Ploidy stability in embryogenic cultures and regenerated plantlets of tamarillo. Plant Cell Tissue Organ Cult. 2013;114:149–159. doi: 10.1007/s11240-013-0311-5. [DOI] [Google Scholar]
- Czene M, Harms-Ringdahl M. Detection of single-strand breaks and formamidoprymidine-DNA glycosylase-sensitive sites in DNA of cultured human fibroblasts. Mutat Res. 1995;336:235–242. doi: 10.1016/0921-8777(94)00058-E. [DOI] [PubMed] [Google Scholar]
- D’Amato F. Cytogenetics of plant cell and tissue cultures and their regenerants. CRC Crit Rev Plant Sci. 1985;3:73–112. doi: 10.1080/07352688509382204. [DOI] [Google Scholar]
- da Silva TCR, Carvalho CR. Vertical heterogeneity of DNA ploidy level assessed by flow cytometry in calli of Passiflora cincinnata. In Vitro Cell Dev Biol Plant. 2014;50(2):158–165. doi: 10.1007/s11627-013-9582-0. [DOI] [Google Scholar]
- Das A, Gosal SS, Sidhu JS, Dhaliwal HS. Induction of mutations for heat tolerance in potato by using in vitro culture and radiation. Euphytica. 2000;114:205–209. doi: 10.1023/A:1003965724880. [DOI] [Google Scholar]
- Deng ZN, Gentile A, Domina F, Nicolosi E, Tribulato E. Selecting lemon protoplasts for insensitivity to Phoma tracheiphila toxin and regenerating tolerant plants. J Am Soc Hortic Sci. 1995;120:902–905. [Google Scholar]
- Devanand PS, Chen J, Henny RJ, Chao CCT. Assessment of genetic relationships among Philodendron cultivars using AFLP markers. J Am Soc Hortic Sci. 2004;129:690–697. [Google Scholar]
- Dey T, Saha S, Ghosh PD. Somaclonal variation among somatic embryo derived plants—evaluation of agronomically important somaclones and detection of genetic changes by RAPD in Cymbopogon winterianus. S Afr J Bot. 2015;96:112–121. doi: 10.1016/j.sajb.2014.10.010. [DOI] [Google Scholar]
- Diawara MM, Trumble JT, Lacy ML, White KK, Carson WG. Potential of somaclonal celeries for use in integrated pest management. J Econ Entomol. 1996;89:218–223. doi: 10.1093/jee/89.1.218. [DOI] [Google Scholar]
- Diaz-Sala C, Rey M, Boronat A, Besford R, Rodriguez R. Variations in the DNA methylation and polypeptide patterns of adult hazel (Corylus avellana L.) associated with sequential in vitro subcultures. Plant Cell Rep. 1995;15:218–221. doi: 10.1007/BF00193723. [DOI] [PubMed] [Google Scholar]
- Dolcet-Sanjuan R, Mok DWS, Mok MC. Characterization and in vitro selection for iron efficiency in Pyrus and Cydonia. In Vitro Cell Dev Biol Plant. 1992;28:25–29. doi: 10.1007/BF02632188. [DOI] [Google Scholar]
- Dugdale LJ, Mortimer AM, Isaac S, Collin HA. Disease response of carrot and carrot somaclones to Alternaria dauci. Plant Pathol. 2000;49:57–67. doi: 10.1046/j.1365-3059.2000.00389.x. [DOI] [Google Scholar]
- Duncan RR. Tissue culture-induced variation and crop improvement. Adv Agron. 1997;58:201–240. doi: 10.1016/S0065-2113(08)60256-4. [DOI] [Google Scholar]
- Eeuwens CJ, Lord S, Donough CR, Rao V, Vallejo G, Nelson S. Effects of tissue culture conditions during embryoid multiplication on the incidence of “mantled” flowering in clonally propagated oil palm. Plant Cell Tissue Organ Cult. 2002;70:311–323. doi: 10.1023/A:1016543921508. [DOI] [Google Scholar]
- Eftekhari M, Alizadeh M, Mashayekhi K, Asghari HR. In vitro propagation of four Iranian grape varieties: influence of genotype and pretreatment with arbuscular mycorrhiza. Vitis. 2012;51:175–182. [Google Scholar]
- Esmaiel NM, Al-Doss AA, Barakat MN. In vitro selection for resistance to Fusarium oxysporum f. sp. dianthi and detection of genetic polymorphism via RAPD analysis in carnation. J Med Plants Res. 2012;6(23):3997–4004. [Google Scholar]
- Etienne H, Bertrand B. Somaclonal variation in Coffea arabica: effects of genotype and embryogenic cell suspension age on frequency and phenotype of variants. Tree Physiol. 2003;23:419–426. doi: 10.1093/treephys/23.6.419. [DOI] [PubMed] [Google Scholar]
- Evans DA. Somaclonal variation—genetic basis and breeding applications. Trends Genet. 1989;5:46–50. doi: 10.1016/0168-9525(89)90021-8. [DOI] [PubMed] [Google Scholar]
- Farahani F, Yari R, Masoud S. Somaclonal variation in Dezful cultivar of olive (Oleaeuropaea subsp. europaea) Gene Conserve. 2011;10:216–221. [Google Scholar]
- Ferdausi A, Nath UK, Das BL, Alam MS. In vitro regeneration system in brinjal (Solanum melongena L.) for stress tolerant somaclone selection. J Bangladesh Agric Univ. 2009;7(2):253–258. [Google Scholar]
- Gao X, Yang D, Cao D, Ao M, Sui X, Wang Q, Kimatu JN, Wang L. In vitro micropropagation of Freesia hybrid and the assessment of genetic and epigenetic stability in regenerated plantlets. J Plant Growth Regul. 2010;29:257–267. doi: 10.1007/s00344-009-9133-4. [DOI] [Google Scholar]
- Ghag SB, Shekhawat UK, Ganapathi TR. Characterization of Fusarium wilt resistant somaclonal variants of banana cv. Rasthali by cDNA-RAPD. Mol Biol Rep. 2014;41(12):7929–7935. doi: 10.1007/s11033-014-3687-3. [DOI] [PubMed] [Google Scholar]
- Giménez C, Garcia DE, Enrech NX, Blanca I. Somaclonal variation in banana: cytogenetic and molecular characterization of the somaclonal variant CIEN BTA-03. In Vitro Cell Dev Biol Plant. 2001;37:217–222. doi: 10.1007/s11627-001-0038-6. [DOI] [Google Scholar]
- Griesbach RJ. Selection of a dwarf Hemerocallis through tissue culture. HortScience. 1989;24:1027–1028. [Google Scholar]
- Grosser JW, Gmitter FG Jr, Chandler JL (1996) Somaclonal variation in sweet orange—a tool for cultivar improvement. In: Proceedings of International Society of Citriculture VIII Congress, Sun City, South Africa, pp 203–206
- Grosser JW, Gmitter FG, Jr, Dutt M, Calovic M, Ling P, Castle B. Highlights of the University of Florida, Citrus Research and Education Center’s comprehensive citrus breeding and genetics program. Acta Hortic. 2015;1065:405–413. doi: 10.17660/ActaHortic.2015.1065.50. [DOI] [Google Scholar]
- Guo W, Gong L, Ding Z, Li Y, Li F, Zhao S, Liu B. Genomic instability in phenotypically normal regenerants of medicinal plant Codonopsislanceolata Benth. et Hook. f., as revealed by ISSR and RAPD markers. Plant Cell Rep. 2006;25:896–906. doi: 10.1007/s00299-006-0131-8. [DOI] [PubMed] [Google Scholar]
- Guo W, Wu R, Zhang Y, Liu X, Wang H, Gong L, Zhang Z, Liu B. Tissue culture-induced locus-specific alteration in DNA methylation and its correlation with genetic variation in Codonopsislanceolata Benth. et Hook. f. Plant Cell Rep. 2007;26:1297–1307. doi: 10.1007/s00299-007-0320-0. [DOI] [PubMed] [Google Scholar]
- Gupta PK. Chromosomal basis of somaclonal variation in plants. In: Jain SM, Brar DS, Ahloowalia BS, editors. Somaclonal variation and induced mutations in crop improvement. Dordrecht: Kluwer Academic Publishers; 1998. pp. 149–168. [Google Scholar]
- Gupta R, Mallavarapu GR, Banerjee S, Kumar S. Characteristics of an isomenthone-rich somaclonal mutant isolated in a geraniol-rich rose-scented geranium accession of Pelargonium graveolens. Flavour Frag J. 2001;16(5):319–324. doi: 10.1002/ffj.1002. [DOI] [Google Scholar]
- Hall HK, Skirvin RM, Braam WF. Germplasm release of ‘Lincoln Logan’, a tissue culture-derived genetic thornless ‘Loganberry’. Fruit Var J. 1986;40:134–135. [Google Scholar]
- Hammerschlag FA. Resistant responses of peach somaclone 122–1 to Xanthomonas campestris pv. pruni and to Pseudomonassyringae pv. syringae. HortScience. 2000;35:141–143. [Google Scholar]
- Hammerschlag FA, Ognjanov V. Somaclonal variation in peach: screening for resistance to Xanthomonas camperstris pv. pruni and Pseudomonas syringae pv. syringae. Acta Hortic. 1990;280:403–408. doi: 10.17660/ActaHortic.1990.280.65. [DOI] [Google Scholar]
- Hao YJ, Wen XP, Deng XX. Genetic and epigenetic evaluations of citrus calluses recovered from slow-growth culture. J Plant Physiol. 2004;161:479–484. doi: 10.1078/0176-1617-01102. [DOI] [PubMed] [Google Scholar]
- Hashmi G, Hammerschlag FA, Huettel RN, Krusberg LR. Growth, development and response of peach somaclones to the root-knot nematode, Meloidogyneincognita. J Am Soc Hortic Sci. 1995;120:932–937. [Google Scholar]
- Heath-Pagliuso S, Rappaport L. Somaclonal variant UC-T3: the expression of Fusarium wilt resistance in progeny arrays of celery, Apiumgraveolens L. Theor Appl Genet. 1990;80:390–394. doi: 10.1007/BF00210078. [DOI] [PubMed] [Google Scholar]
- Heinze DJ, Mee GWP. Morphologic, cytogenetic and enzymatic variation in Saccharum species hybrid clones derived from callus culture. Am J Bot. 1971;58:257–262. doi: 10.2307/2441162. [DOI] [Google Scholar]
- Henny RJ, Chen J (2011) Tropical foliage plant development: origin of new cultivars. ENH1092. Environmental Horticulture Department, Florida Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida. http://edis.ifas.ufl.edu
- Henny RJ, Poole RT, Conover CA. ‘Silver Bay’ Aglaonema. HortScience. 1992;27:1238. [Google Scholar]
- Henny RJ, Chen J, Norman DJ. ‘Diamond Bay’ and ‘Emerald Bay’Aglaonema. HortScience. 2003;38:1446–1447. [Google Scholar]
- Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M. Retrotransposons of rice involved in mutations induced by tissue culture. Proc Natl Acad Sci USA. 1996;93:7783–7788. doi: 10.1073/pnas.93.15.7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque ME, Morshad MN. Somaclonal variation in potato (Solanum tuberosum L.) using chemical mutagens. Agriculturists. 2014;12(1):15–25. [Google Scholar]
- Horáček J, Švábová L, Šarhanová P, Lebeda A. Variability for resistance to Fusarium solani culture filtrate and fusaric acid among somaclones in pea. Biol Plant. 2013;57(1):133–138. doi: 10.1007/s10535-012-0131-1. [DOI] [Google Scholar]
- Hossain MA, Konisho K, Minami M, Nemoto K. Somaclonal variation of regenerated plants in chili pepper (Capsicum annuum L.) Euphytica. 2003;130:233–239. doi: 10.1023/A:1022856725794. [DOI] [Google Scholar]
- Hossain MM, Kant R, Van PT, Winarto B, Zeng S, Teixeira da Silva JA. The application of biotechnology to orchids. Crit Rev Plant Sci. 2013;32(2):69–139. doi: 10.1080/07352689.2012.715984. [DOI] [Google Scholar]
- Hu X, Xu Q. Morphological Variations of somaclone regenerated from series tissue culture of Gloxinia. Acta Agric Boreal Occident Sin. 2010;5:39. [Google Scholar]
- Hwang S (2002) Somaclonal variational approach to breeding Cavendish banana for resistance to Fusarium wilt race 4. In: Global conference on banana and plantain, Bangalore, India, Abstract 57, October 28–31
- Hwang SC, Ko WH. Breeding for resistance to Fusarium wilt of Cavendish banana by using tissue culture method. Spec Publ Taichung Dist Agric Improv Stn. 1992;29:229–237. [Google Scholar]
- Hwang SC, Ko WH. Cavendish banana cultivars resistant to Fusarium wilt acquired through somaclonal variation in Taiwan. Plant Dis. 2004;88:580–588. doi: 10.1094/PDIS.2004.88.6.580. [DOI] [PubMed] [Google Scholar]
- Iacona C, Cirilli M, Zega A, Frioni A, Silvestri C, Muleo R. A somaclonal myrobalan rootstock increases waterlogging tolerance to peach cultivar in controlled conditions. Sci Hortic. 2013;156:1–8. doi: 10.1016/j.scienta.2013.03.014. [DOI] [Google Scholar]
- Israeli Y, Lahav E, Reuveni O. In vitro culture of bananas. In: Gowen S, editor. Bananas and plantains. London: Chapman and Hall; 1995. pp. 147–178. [Google Scholar]
- Iuliana C, Cerasela P. The effect of the ultraviolet radiation on the somaclonal variability for Solanum tuberosum. Rom Biotechnol Lett. 2014;19(3):9339–9344. [Google Scholar]
- Jain SM. Micropropagation of selected somaclones of Begonia and Saintpaulia. J Biosci. 1997;22:582–592. doi: 10.1007/BF02703396. [DOI] [Google Scholar]
- Jain SM. Tissue culture-derived variation in crop improvement. Euphytica. 2001;118:153–166. doi: 10.1023/A:1004124519479. [DOI] [Google Scholar]
- Jaligot E, Beule T, Baurens FC, Billotte N, Rival A. Search for methylation-sensitive amplification polymorphisms associated with the ‘mantled’ variant phenotype in oil palm (Elaeis guineensis Jacq.) Genome. 2004;47:224–228. doi: 10.1139/g03-085. [DOI] [PubMed] [Google Scholar]
- Jaligot E, Adler S, Debladis É, Beulé T, Richaud F, Ilbert P, Finnegan EJ, Rival A. Epigenetic imbalance and the floral developmental abnormality of the in vitro-regenerated oil palm Elaeisguineensis. Ann Bot. 2011;108:1453–1462. doi: 10.1093/aob/mcq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaya RS, Rao PS, Mhatre M. Suitability of RAPD for analyzing spined and spineless variant regenerants of pineapple (Ananas comosus L., Merr.) Plant Mol Biol Rep. 2002;20:307. doi: 10.1007/BF02782469. [DOI] [Google Scholar]
- Jevremović S, Subotić A, Miljković D, Trifunović M, Petrić M, Cingel A. Clonal fidelity of Chrysanthemum cultivars after long term micropropagation by stem segment culture. Acta Hortic. 2012;961:211–216. doi: 10.17660/ActaHortic.2012.961.25. [DOI] [Google Scholar]
- Joshi P, Dhawan V. Assessment of genetic fidelity of micropropagated Swertia chirayita plantlets by ISSR marker assay. Biol Plant. 2007;51:22–26. doi: 10.1007/s10535-007-0005-0. [DOI] [Google Scholar]
- Joyce SM, Cassells AC, Jain SM. Stress and aberrant phenotypes in vitro culture. Plant Cell Tissue Organ Cult. 2003;74:103–121. doi: 10.1023/A:1023911927116. [DOI] [Google Scholar]
- Kaeppler SM, Kaeppler HF, Rhee Y. Epigenetic aspects of somaclonal variation in plants. Plant Mol Biol. 2000;43:179–188. doi: 10.1023/A:1006423110134. [DOI] [PubMed] [Google Scholar]
- Kaity A, Ashmore SE, Drew RA. Field performance evaluation and genetic integrity assessment of cryopreserved papaya clones. Plant Cell Rep. 2009;28:1421–1430. doi: 10.1007/s00299-009-0742-y. [DOI] [PubMed] [Google Scholar]
- Kar B, Kuanar A, Singh S, Mohanty S, Joshi RK, Subudhi E, Nayak S. In vitro induction, screening and detection of high essential oil yielding somaclones in turmeric (Curcuma longa L.) Plant Growth Regul. 2014;72(1):59–66. doi: 10.1007/s10725-013-9836-1. [DOI] [Google Scholar]
- Karp A. The role of growth regulators in somaclonal variation. Br Soc Plant Growth Regul Annu Bull. 1992;2:1–9. [Google Scholar]
- Karp A. Origins, causes and uses of variation in plant tissue cultures. In: Vasil IK, Thorpe TA, editors. Plant cell and tissue culture. Dordrecht: Kluwer Academic Publishers; 1994. pp. 139–152. [Google Scholar]
- Karp A. Somaclonal variation as a tool for crop improvement. Euphytica. 1995;85:295–302. doi: 10.1007/BF00023959. [DOI] [Google Scholar]
- Karp A. Molecular tools for detecting genetic diversity. Acta Hortic. 2000;530:17–29. doi: 10.17660/ActaHortic.2000.530.1. [DOI] [Google Scholar]
- Khan S, Saeed B, Kauser N. Establishment of genetic fidelity of in vitro raised banana plantlets. Pak J Bot. 2011;43:233–242. [Google Scholar]
- Khan SA, Rahman LU, Shanker K, Singh M. Agrobacteriumtumefaciens-mediated transgenic plant and somaclone production through direct and indirect regeneration from leaves in Stevia rebaudiana with their glycoside profile. Protoplasma. 2014;251(3):661–670. doi: 10.1007/s00709-013-0568-x. [DOI] [PubMed] [Google Scholar]
- Kothari SL, Joshi A, Kachhwaha S, Ochoa-Alejo N. Chilli peppers—a review on tissue culture and transgenesis. Biotechnol Adv. 2010;28(1):35–48. doi: 10.1016/j.biotechadv.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Krikorian AD, Irizarry H, Cronauer-Mitra SS, Rivera E. Clonal fidelity and variation in plantain (Musa AAB) regenerated from vegetative stem and floral axis tips in vitro. Ann Bot. 1993;71:519–535. doi: 10.1006/anbo.1993.1068. [DOI] [Google Scholar]
- Krishna H, Singh SK. Biotechnological advances in mango (Mangifera indica L.) and their future implication in crop improvement—a review. Biotechnol Adv. 2007;25:223–243. doi: 10.1016/j.biotechadv.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Krishna H, Singh D. Micropropagation of lasora (Cordia myxa Roxb.) Indian J Hortic. 2013;70:323–327. [Google Scholar]
- Krishna H, Singh SK, Sharma RR, Khawale RN, Minakshi Patel VB. Biochemical changes in micropropagated grape (Vitis vinifera L.) plantlets due to arbuscular-mycorrhizal fungi (AMF) inoculation during ex vitro acclimatization. Sci Hortic. 2005;106:554–567. doi: 10.1016/j.scienta.2005.05.009. [DOI] [Google Scholar]
- Krishna H, Sairam RK, Singh SK, Patel VB, Sharma RR, Grover M, Nain L, Sachdeva A. Mango explants browning: effect of ontogenic age: mycorrhization and pre-treatments. Sci Hortic. 2008;118:132–138. doi: 10.1016/j.scienta.2008.05.040. [DOI] [Google Scholar]
- Kuanar A, Nayak PK, Subudhi E, Nayak S. In vitro selection of turmeric somaclone resistant to Fusarium oxysporum f. sp. Zingiberi. Proc Natl Acad Sci India Sect B Biol Sci. 2014;84:1077–1082. doi: 10.1007/s40011-013-0295-2. [DOI] [Google Scholar]
- Kukreja AK, Dhawan OP, Mathur AK, Ahuja PS, Mandal S. Screening and evaluation of agronomically useful somaclonal variations in Japanese mint (Mentha arvensis L.) Euphytica. 1991;53:183–191. doi: 10.1007/BF00023270. [DOI] [Google Scholar]
- Kukreja AK, Dhawan OP, Ahuja PS, Sharma S, Kumar S. Yield potential and stability behaviour of in vitro derived somaclones of Japanese mint (Mentha arvensis L.) under different environments. J Genet Breed. 2000;54(2):109–115. [Google Scholar]
- Kuksova VB, Piven NM, Gleba YY. Plant cell variation and in vitro induced mutagenesis in grapevine. Plant Cell Tissue Organ Cult. 1997;49:17–27. doi: 10.1023/A:1005830305206. [DOI] [Google Scholar]
- Kuznetsova OI, Ash OA, Gostimsky SA. The effect of the duration of callus culture on the accumulation of genetic alternation in pea Pisum sativum L. Russ J Genet. 2006;42:555–562. doi: 10.1134/S1022795406050139. [DOI] [Google Scholar]
- Larkin P, Scowcroft W. Somaclonal variation—a novel source of variability from cell cultures for plant improvement. Theor Appl Genet. 1981;60:197–214. doi: 10.1007/BF02342540. [DOI] [PubMed] [Google Scholar]
- Lee SY, Su YU, Chou CS, Liu CC, Chen CC, Chao CP. Selection of a new somaclone cultivar ‘Tai-Chiao No. 5’ (AAA, Cavendish) with resistance to Fusarium wilt of banana in Chinese Taipei. Acta Hortic. 2011;897:391–397. doi: 10.17660/ActaHortic.2011.897.54. [DOI] [Google Scholar]
- Lestari EG. In vitro selection and somaclonal variation for biotic and abiotic stress tolerance. Biodiversitas. 2006;7:297–301. doi: 10.13057/biodiv/d070320. [DOI] [Google Scholar]
- Leva AR, Petruccelli R, Rinaldi LMR. Somaclonal variation in tissue culture: a case study with olive. In: Leva AR, Rinaldi LMR, editors. Recent advances in plant in vitro culture. Croatia: INTECH Open Access Publisher; 2012. pp. 123–150. [Google Scholar]
- Li R, Qu R, Bruneau AH, Livingston DP. Selection for freezing tolerance in St. Augustine grass through somaclonal variation and germplasm evaluation. Plant Breed. 2010;129:417–421. doi: 10.1111/j.1439-0523.2009.01647.x. [DOI] [Google Scholar]
- Linacero R, Rueda J, Esquivel E, Bellido A, Domingo A, Vázquez AM. Genetic and epigenetic relationship in rye, Secale cereale L., somaclonal variation within somatic embryo-derived plants. In Vitro Cell Dev Biol Plant. 2011;47:618–628. doi: 10.1007/s11627-011-9407-y. [DOI] [Google Scholar]
- Litz RE, Mathews WH, Hendrix RC, Yurgalevitch C. Mango somatic cell genetics. Acta Hortic. 1991;291:133–140. doi: 10.17660/ActaHortic.1991.291.15. [DOI] [Google Scholar]
- LoSchiavo F, Pitto L, Giuliano G, Torti G, Nuti-Ronchi V, Marazziti D, Vergara R, Orselli S, Terzi M. DNA methylation of embryogenic carrot cell cultures and its variations as cased by mutation, differentiation, hormones and hypomethylating drugs. Theor Appl Genet. 1989;77:325–331. doi: 10.1007/BF00305823. [DOI] [PubMed] [Google Scholar]
- Lucia G, Castiglione MR, Turrini A, Ronchi VN, Geri C. Cytogenetic and histological approach for early detection of “mantled” somaclonal variants of oil palm regenerated by somatic embryogenesis: first results on the characterization of regeneration system. Caryologia. 2011;64(2):223–234. doi: 10.1080/00087114.2002.10589787. [DOI] [Google Scholar]
- Mallaya NP, Ravishankar GA. In vitro propagation and genetic fidelity study of plant regenerated from inverted hypocotyl explants of eggplant (Solanum melongena L.) cv. Arka Shirish. 3 Biotech. 2013;3(1):45–52. doi: 10.1007/s13205-012-0068-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Beghelli S, Rombolà AD, Cabrini L. In vitro performance at high culture pH and in vitro responses to Fe-deficiency of leaf-derived quince BA29 (Cydoniaoblonga) somaclones regenerated at variable pH. J Hortic Sci Biotechnol. 2000;75:433–440. doi: 10.1080/14620316.2000.11511264. [DOI] [Google Scholar]
- Martin K, Pachathundikandi S, Zhang C, Slater A, Madassery J. RAPD analysis of a variant of banana (Musa sp.) cv. grande naine and its propagation via shoot tip culture. In Vitro Cell Dev Biol Plant. 2006;42:188–192. doi: 10.1079/IVP2005736. [DOI] [Google Scholar]
- Martins M, Sarmento D, Oliveira MM. Genetic stability of micropropagated almond plantlets, as assessed by RAPD and ISSR markers. Plant Cell Rep. 2004;23(7):492–496. doi: 10.1007/s00299-004-0870-3. [DOI] [PubMed] [Google Scholar]
- Mathur AK. Biotechnological studies in Cymbopogons. In: Akhila A, editor. Essential oil-bearing grasses: the genus Cymbopogons. Boca Raton: CRC Press, Taylor & Francis Group, LLC; 2010. pp. 107–133. [Google Scholar]
- Mathur S. Conservation of biodiversity through tissue culture. Res Rev J Microbiol Biotechnol. 2013;2:1–6. [Google Scholar]
- Mathur AK, Ahuja PS, Pandey B, Kukreja AK, Mathur A (1988) Development of superior strains of an aromatic grass—Cymbopogon winterianus Jowitt—through somaclonal variation. In: International conference on research in plant science and its relevance to future, Abstract, University Press, Delhi, p 160
- Matsuda S, Sato M, Ohno S. Cutting leaves and plant growth regulator application enhance somaclonal variation induced by transposition of VGs1 of Saintpaulia. J Jpn Soc Hortic Sci. 2014;83(4):308–316. doi: 10.2503/jjshs1.MI-009. [DOI] [Google Scholar]
- Menéndez-Yuffá A, Barry-Etienne D, Bertrand B, Georget F, Etienne H. A comparative analysis of the development and quality of nursery plants derived from somatic embryogenesis and from seedlings for large-scale propagation of coffee (Coffea arabica L.) Plant Cell Tissue Organ Cult. 2010;102:297–307. doi: 10.1007/s11240-010-9734-4. [DOI] [Google Scholar]
- Metzker ML. Sequencing technologies—the next generation. Nat Rev. 2010;2010(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Miguel C, Marum L. An epigenetic view of plant cells cultured in vitro somaclonal variation and beyond. J Exp Bot. 2011;62:3713–3725. doi: 10.1093/jxb/err155. [DOI] [PubMed] [Google Scholar]
- Miler N, Zalewska M. Somaclonal variation of chrysanthemum propagated in vitro from different explants types. Acta Sci Pol Hortorum Cultus. 2014;13(2):69–82. [Google Scholar]
- Minerva G, Kumar S (2013) Micropropagation of gerbera (Gerbera jamesonii Bolus). In: Lambardi M, Ozudogru EA, Jain SM (eds) Protocols for micropropagation of selected economically-important horticultural plants. Methods Mol Biol 994:305–316 [DOI] [PubMed]
- Miyao A, Nakagome M, Ohnuma T, Yamagata H, Kanamori H, Katayose Y, Takahashi A, Matsumoto T, Hirochika H. Molecular spectrum of somaclonal variation in regenerated rice revealed by whole-genome sequencing. Plant Cell Physiol. 2012;53:256–264. doi: 10.1093/pcp/pcr172. [DOI] [PubMed] [Google Scholar]
- Modgil M, Guleria N, Ghani M, Sharma JN. Identifying somaclonal variants of the apple rootstock Malling 7 resistant to white root rot. Sci Hortic. 2012;137:148–155. doi: 10.1016/j.scienta.2012.01.006. [DOI] [Google Scholar]
- Mohanty S, Parida R, Singh S, Joshi RK, Subudhi E, Nayak S. Biochemical and molecular profiling of micropropagated and conventionally grown Kaempferia galanga. Plant Cell Tissue Organ Cult. 2011;106:39–46. doi: 10.1007/s11240-010-9891-5. [DOI] [Google Scholar]
- Mondal TK, Bantawa P, Sarkar B, Ghosh P, Chand PK. Cellular differentiation, regeneration, and secondary metabolite production in medicinal Picrorhiza spp. Plant Cell Tissue Organ Cult. 2013;112(2):143–158. doi: 10.1007/s11240-012-0223-9. [DOI] [Google Scholar]
- Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Morrison RA, Loh WHT, Green SK, Griggs TD, McLean BT (1989) Tissue culture of tomato and pepper: new tools for plant breeding. Tomato and pepper production in the tropics. In: Proceedings of international symposium on integrated management practices, Tainen, Taiwan. Shanhua, Taiwan, AVRDC, pp 44–50
- Mujib A, Banerjee S, Dev Ghosh P. Callus induction, somatic embryogenesis and chromosomal instability in tissue culture-raised hippeastrum (Hippeastrumhybridum cv. United Nations) Propag Ornam Plants. 2007;7:169–174. [Google Scholar]
- Muleo R, Iacona C, Pistelli L, Loreti F. A novel Mr. S. 2/5 peach rootstock clone tolerant to flooding stress. Adv Hortic Sci. 2006;20:208–214. [Google Scholar]
- Munir F, Naqvi SMS, Mahmood T. In vitro culturing and assessment of somaclonal variation of Solanum tuberosum var. desiree. Turk J Biochem. 2011;36:296–302. [Google Scholar]
- Murti RH, Kim HY, Yeoung YR. Effectiveness of gamma ray irradiation and ethyl methane sulphonate on in vitro mutagenesis of strawberry. Afr J Biotechnol. 2013;12:4803–4812. doi: 10.5897/AJB12.1386. [DOI] [Google Scholar]
- Nacheva LR, Gercheva PS, Andonova MY, Panayotova DV, Dzhuvinov VT. Somaclonal variation: a useful tool to improve disease resistance of pear rootstock ‘Old Home × Farmingdale’ (OHF 333) (Pyruscommunis L.) Acta Hortic. 2014;1056:253–258. doi: 10.17660/ActaHortic.2014.1056.42. [DOI] [Google Scholar]
- Naseer S, Mahmood T. Tissue culture and genetic analysis of somaclonal variations of Solanum melongena L. cv. Nirrala. Cent Eur J Biol. 2014;9(12):1182–1195. [Google Scholar]
- Nassar AM, Abdulnour J, Leclerc YN, Li XQ, Donnelly DJ. Intraclonal selection for improved processing of NB ‘Russet Burbank’potato. Am J Potato Res. 2011;88(5):387–397. doi: 10.1007/s12230-011-9204-5. [DOI] [Google Scholar]
- Nassar AM, Kubow S, Leclerc YN, Donnelly DJ. Somatic mining for phytonutrient improvement of ‘Russet Burbank’ potato. Am J Potato Res. 2014;91:89–100. doi: 10.1007/s12230-013-9334-z. [DOI] [Google Scholar]
- Nayak S, Debata BK, Srivastava VK, Sangwan NS. Evaluation of agronomically useful somaclonal variants in Jamrosa (a hybrid Cymbopogon) and detection of genetic changes through RAPD. Plant Sci. 2003;164:1029–1035. doi: 10.1016/S0168-9452(03)00090-6. [DOI] [Google Scholar]
- Nayak S, Kaur T, Mohanty S, Ghosh G, Choudhury R, Acharya L, Subudhi E. In vitro and ex vitro evaluation of long-term micropropagated turmeric as analyzed through cytophotometry, phytoconstituents, biochemical and molecular markers. Plant Growth Regul. 2010;64:91–98. doi: 10.1007/s10725-010-9541-2. [DOI] [Google Scholar]
- Ngezahayo F, Dong Y, Liu B. Somaclonal variation at the nucleotide sequence level in rice (Oryzasativa L.) as revealed by RAPD and ISSR markers, and by pairwise sequence analysis. J Appl Genet. 2007;48:329–336. doi: 10.1007/BF03195229. [DOI] [PubMed] [Google Scholar]
- Nhut DT, Hai NT, Thu PTM, Thi NN, Hien TTD, Tuan TT, Nam NB, Huy NP, Chien HX, Jain SM. Protocol for inducing flower color somaclonal variation in torenia (Torenia fournieri Lind.) Methods Mol Biol. 2013;11013:455–462. doi: 10.1007/978-1-62703-074-8_34. [DOI] [PubMed] [Google Scholar]
- Nivas SK, DSouza L. Genetic fidelity in micropropagated plantlets of Anacardium occidentale L. (Cashew) an important fruit tree. Int J Sci Res. 2014;3:2142–2146. [Google Scholar]
- Nwauzoma AB, Jaja ET. A review of somaclonal variation in plantain (Musa spp): mechanisms and applications. J Appl Biosci. 2013;67:5252–5260. doi: 10.4314/jab.v67i0.95046. [DOI] [Google Scholar]
- Ooi SE, Novák O, Doležal K, Ishak Z, Ong-Abdullah M. Cytokinin differences in in vitro cultures and inflorescences from normal and mantled oil palm (Elaeis guineensis Jacq.) J Plant Growth Regul. 2013;32(4):865–874. doi: 10.1007/s00344-013-9352-6. [DOI] [Google Scholar]
- Opabode JT, Adebooye OC. Application of biotechnology for the improvement of Nigerian indigenous leaf vegetables. Afr J Biotechnol. 2005;4(3):138–142. [Google Scholar]
- Panda MK, Mohanty S, Subudhi E, Acharya L, Nayak S. Assessment of genetic stability of micropropagated plants of Curcuma longa L. by cytophotometry and RAPD analysis. Int J Integr Biol. 2007;1(3):189–195. [Google Scholar]
- Parida R, Mohanty S, Nayak S. In vitro propagation of Hedychium coronarium Koen. through axillary bud proliferation. Plant Biosyst. 2013;147(4):905–912. doi: 10.1080/11263504.2012.748102. [DOI] [Google Scholar]
- Pathak H, Dhawan V. ISSR assay for ascertaining genetic fidelity of micropropagated plants of apple rootstock Merton 793. In Vitro Cell Dev Biol Plant. 2012;48:137–143. doi: 10.1007/s11627-011-9385-0. [DOI] [Google Scholar]
- Patnaik J, Sahoo S, Debata BK. Somaclonal variation in cell suspension culture-derived regenerants of Cymbopogonmartinii (Roxb.) Wats var. Motia. Plant Breed. 1999;118:351–354. doi: 10.1046/j.1439-0523.1999.00383.x. [DOI] [Google Scholar]
- Patzak J. Assessment of somaclonal variability in hop (Humulus lupulus L.) in vitro meristem cultures and clones by molecular methods. Euphytica. 2003;131(3):343–350. doi: 10.1023/A:1024096401424. [DOI] [Google Scholar]
- Pérez G, Yanez E, Isidrón M, Lorenzo JC. Phenotypic and AFLP characterization of two new pineapple somaclones derived from in vitro culture. Plant Cell Tissue Organ Cult. 2009;96:113–116. doi: 10.1007/s11240-008-9463-0. [DOI] [Google Scholar]
- Pérez G, Mbogholi A, Sagarra F, Aragón C, González J, Isidrón M, Lorenzo JC. Morphological and physiological characterization of two new pineapple somaclones derived from in vitro culture. In Vitro Cell Dev Biol Plant. 2011;47:428–433. doi: 10.1007/s11627-011-9342-y. [DOI] [Google Scholar]
- Pérez G, Yanez E, Mbogholi A, Valle B, Sagarra F, Yabor L, Aragón C, González J, Isidrón M, Lorenzo JC. New pineapple somaclonal variants: P3R5 and dwarf. Am J Plant Sci. 2012;3:1–11. doi: 10.4236/ajps.2012.31001. [DOI] [Google Scholar]
- Phillips RL, Kappler SM, Olhof R. Genetic instability of plant tissue cultures: breakdown of normal controls. Proc Natl Acad Sci USA. 1994;91:5222–5226. doi: 10.1073/pnas.91.12.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piagnani MC, Chiozzotto R. Shoot regeneration, in vitro performances of regenerated shoots and transient expression in morphogenic explants in Prunus avium cultivar ‘Burlat C1’. Eur J Hortic Sci. 2010;3:132–138. [Google Scholar]
- Pijut PM, Beasley RR, Lawson SS, Palla KJ, Stevens ME, Wang Y. In vitro propagation of tropical hardwood tree species—a review (2001–2011) Propag Ornam Plants. 2012;12(1):25–51. [Google Scholar]
- Podwyszynska M, Kuras A, Korbin M. Somaclonal variation in micropropagated tulips as a source of novel genotypes—field and molecular characteristic. Acta Hortic. 2010;855:225–232. doi: 10.17660/ActaHortic.2010.855.33. [DOI] [Google Scholar]
- Prado MJ, Gonzalez MV, Romo S, Herrera MT. Adventitious plant regeneration on leaf explants from adult male kiwifruit and AFLP analysis of genetic variation. Plant Cell Tissue Organ Cult. 2007;88(1):1–10. doi: 10.1007/s11240-006-9116-0. [DOI] [Google Scholar]
- Purwati RD, Sudarsono S. Resistance of Abaca somaclonal variant against Fusarium. Hayati J Biosci. 2007;14:133–139. [Google Scholar]
- Rabiei K, Khodambashi N, Omidi M. Use of somaclonal variation on improvement of drought resistant lines of carrot (Daucuscarota L.) J Hortic Sci (Agric Sci Technol) 2011;25:156–169. [Google Scholar]
- Rana S, Dhar N, Bhat WW, Razdan S, Khan S, Dhar RS, Dutt P, Lattoo SK. A 12-deoxywithastramonolide-rich somaclonal variant in Withania somnifera (L.) Dunal—molecular cytogenetic analysis and significance as a chemotypic resource. In Vitro Cell Dev Biol Plant. 2012;48(5):546–554. doi: 10.1007/s11627-012-9458-8. [DOI] [Google Scholar]
- Ravindra NS, Ramesh SI, Gupta MK, Jhang T, Shukla AK, Darokar MP, Kulkarni RN. Evaluation of somaclonal variation for genetic improvement of patchouli (Pogostemonpatchouli), an exclusively vegetatively propagated aromatic plant. J Crop Sci Biotechnol. 2012;15:33–39. doi: 10.1007/s12892-011-0068-5. [DOI] [Google Scholar]
- Ray T, Dutta I, Saha P, Das S, Roy SC. Genetic stability of three economically important micropropagated banana (Musa spp.) cultivars of lower Indo-Gangetic plains, as assessed by RAPD and ISSR markers. Plant Cell Tissue Organ Cult. 2006;85(1):11–21. doi: 10.1007/s11240-005-9044-4. [DOI] [Google Scholar]
- Resende GA, Mangolin CA, Machado MDF. Somaclonal variation in Cereus peruvianus Mill. (Cactaceae): its potential to generate new varieties and broaden the species’s genetic basis. J Basic Appl Genet. 2010;21:33–42. [Google Scholar]
- Rival A, Ilbert P, Labeyrie A, Torres E, Doulbeau S, Personne A, Dussert S, Beulé T, Durand-Gasselin T, Tregear JW, Jaligot E. Variations in genomic DNA methylation during the long-term in vitro proliferation of oil palm embryogenic suspension cultures. Plant Cell Rep. 2013;32:359–368. doi: 10.1007/s00299-012-1369-y. [DOI] [PubMed] [Google Scholar]
- Rosati P, Mezzetti B, Ancherani M, Foscolo S, Predieri S, Fasolo F. In vitro selection of apple rootstock somaclones with Phytophthora cactorum culture filtrate. Acta Hortic. 1990;280:409–416. doi: 10.17660/ActaHortic.1990.280.66. [DOI] [Google Scholar]
- Sahijram L, Soneji J, Bollamma K. Analyzing somaclonal variation in micropropagated bananas (Musa spp.) In Vitro Cell Dev Biol Plant. 2003;39:551–556. doi: 10.1079/IVP2003467. [DOI] [Google Scholar]
- Sakhanokho HF, Witcher AL, Pounders CT, Spiers JM. ‘Ramata’: a new dwarf variegated Hedychium (ornamental ginger) cultivar. HortSci. 2012;47:803–805. [Google Scholar]
- Sales EK, Butardo NG. Molecular analysis of somaclonal variation in tissue culture derived bananas using MSAP and SSR markers. Int J Biol Vet Agric Food Eng. 2014;8:63–610. [Google Scholar]
- Sanputawong S, Te-Chato S. Analysis of somaclonal variation of callus, somatic embryo and plant regeneration of in vitro oil palm (Elaeis guineensis Jacq.) J Agric Technol. 2011;7(2):531–545. [Google Scholar]
- Saravanan S, Sarvesan R, Vinod MS. Identification of DNA elements involved in somaclonal variants of Rauvolfia serpentina (L.) arising from indirect organogenesis as evaluated by ISSR analysis. Indian J Sci Technol. 2011;4:1241–1245. [Google Scholar]
- Sato M, Kawabe T, Hosokawa M, Tatsuzawam F, Doi M. Tissue culture induced flower-color changes in Saintpaulia caused by excision of the transposon inserted in the flavonoid 39, 59 hydroxylase (F3959H) promoter. Plant Cell Rep. 2011;30:929–939. doi: 10.1007/s00299-011-1016-z. [DOI] [PubMed] [Google Scholar]
- Sato M, Hosokawa M, Doi M. Somaclonal variation is induced de novo via the tissue culture process: a study quantifying mutated cells in Saintpaulia. PLoS ONE. 2011;6:e23541. doi: 10.1371/journal.pone.0023541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena G, Verma PC, Rahman LU, Banerjee S, Kumar S. Field performance of somaclones of rose scented geranium (Pelargonium graveolens L’ Her Ex Ait.) for evolution of their essential oil yield and composition. Ind Crops Prod. 2008;27:86–90. doi: 10.1016/j.indcrop.2007.08.001. [DOI] [Google Scholar]
- Schellenbaum P, Mohler V, Wenzel G, Walter B. Variation in DNA methylation patterns of grapevine somaclones (Vitis vinifera L.) BMC Plant Biol. 2008;8:78. doi: 10.1186/1471-2229-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Bryan G, Winfield M, Millam S. Stability of potato (Solanum tuberosum L.) plants regenerated via somatic embryos, axillary bud proliferated shoots, microtubers and true potato seeds: a comparative phenotypic, cytogenetic and molecular assessment. Planta. 2007;226:1449–1458. doi: 10.1007/s00425-007-0583-2. [DOI] [PubMed] [Google Scholar]
- Shearman JR, Jantasuriyarat C, Sangsrakru D, Yoocha T, Vannavichit A, Tragoonrung S, Tangphatsornruang S. Transcriptome analysis of normal and mantled developing oil palm flower and fruit. Genomics. 2013;101(5):306–312. doi: 10.1016/j.ygeno.2013.02.012. [DOI] [PubMed] [Google Scholar]
- Shen X, Chen J, Kane M, Henny R. Assessment of somaclonal variation in Dieffenbachia plants regenerated through indirect shoot organogenesis. Plant Cell Tissue Organ Cult. 2007;91:21–27. doi: 10.1007/s11240-007-9273-9. [DOI] [Google Scholar]
- Siragusa M, Carra A, Salvia L, Puglia A, De Pasquale F, Carimi F. Genetic instability in calamondin (Citrus madurensis Lour.) plants derived from somatic embryogenesis induced by diphenylurea derivatives. Plant Cell Rep. 2007;26:1289–1296. doi: 10.1007/s00299-007-0326-7. [DOI] [PubMed] [Google Scholar]
- Skirvin RM, Janick J. Tissue culture-induced variation in scented Pelargonium spp. J Am Soc Hortic Sci. 1976;101:281–290. [Google Scholar]
- Slazak B, Sliwinska E, Saługa M, Ronikier M, Bujak J, Słomka A, Göransson U, Kuta E. Micropropagation of Viola uliginosa (Violaceae) for endangered species conservation and for somaclonal variation-enhanced cyclotide biosynthesis. Plant Cell Tissue Organ Cult. 2015;120:179–190. doi: 10.1007/s11240-014-0592-3. [DOI] [Google Scholar]
- Smith MK. A review of factors influencing the genetic stability of micropropagated bananas. Fruits. 1988;43:219–223. [Google Scholar]
- Smulders M, de Klerk G. Epigenetics in plant tissue culture. Plant Growth Regul. 2011;63:137–146. doi: 10.1007/s10725-010-9531-4. [DOI] [Google Scholar]
- Stanišić M, Raspor M, Ninković S, Milošević S, Ćalić D, Bohanec B, Trifunovića M, Petrića M, Subotić A, Jevremović S. Clonal fidelity of Iris sibirica plants regenerated by somatic embryogenesis and organogenesis in leaf-base culture—RAPD and flow cytometer analyses. S Afr J Bot. 2015;96:42–52. doi: 10.1016/j.sajb.2014.10.014. [DOI] [Google Scholar]
- Sun S, Zhong J, Li S, Wang X. Tissue culture-induced somaclonal variation of decreased pollen viability in torenia (Torenia fournieri Lind.) Bot Stud. 2013;54(1):36. doi: 10.1186/1999-3110-54-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Matsumoto T, Takai T. Somaclonal variants from strawberry cv. Morioka-16, M16-AR 1, 2 and 3 resistant to several isolates of Alternaria alternata strawberry pathotype occurring in Tohoku and Hokkaido [Japan] J Jpn Soc Hortic Sci. 1993;61:821–826. doi: 10.2503/jjshs.61.821. [DOI] [Google Scholar]
- Tang CY, Liu CC, Hwang SC. Improvement of the horticultural traits of Cavendish banana (Musa spp., AAA group I). Selection and evaluation of a semi-dwarf clone resistant to Fusarium wilt. J Chin Soc Hortic Sci. 2000;46:173–182. [Google Scholar]
- Tanurdzic M, Vaughn MW, Jiang H, Lee TJ, Slotkin RK, Sosinski B, Thompson WF, Doerge RW, Martienssen RA. Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol. 2008;6(12):2880–2895. doi: 10.1371/journal.pbio.0060302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieme R, Griess H. Somaclonal variation in tuber traits of potato. Potato Res. 2005;48:153–165. doi: 10.1007/BF02742373. [DOI] [Google Scholar]
- Tican A, Câmpeanu G, Chiru N, Ivanovici D. Using of unconventional methods for obtaining somaclonal variations, having as goal making of new potato varieties with resistance at diseases and pests. Rom Biotechnol Lett. 2008;13:3791–3798. [Google Scholar]
- Tiwari JK, Chandel P, Gupta S, Gopal J, Singh BP, Bhardwaj V. Analysis of genetic stability of in vitro propagated potato microtubers using DNA markers. Physiol Mol Biol Plants. 2013;19:587–595. doi: 10.1007/s12298-013-0190-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda H, Horikoshi K, Yamano Y, Ouchi S. Selection of Fusarium wilt disease resistance from regenerant derived from callus of strawberry. Plant Cell Rep. 1991;10:167–170. doi: 10.1007/BF00234287. [DOI] [PubMed] [Google Scholar]
- Vázquez AM. Insight into somaclonal variation. Plant Biosyst. 2001;135:57–62. doi: 10.1080/11263500112331350650. [DOI] [Google Scholar]
- Veitia-Rodriguez N, Francisco-Cardoso J, Perez JN, Garcia-Rodriguez L, Bermudez-Caraballosos I, Garcia-Rodriguez L, Padron-Montesinos Y, Orellana-Perez P, Romero-Quintana C, Hernandez N. Evaluations in field of somaclones of Irish potatoes (Solanum tuberosum Lin.) of the variety Desiree obtained by somaclonal variation and in vitro mutagenesis. Biotechnol Veg. 2002;2:21–26. [Google Scholar]
- Vidal BC, Mello MLS, Illg RD, de Campos Vidal B, Campos De, Vidal B. Chromosome number and DNA content in cells of a biotechnologically selected somaclone of garlic (Allium sativum L) Rev Bras Genet. 1993;16(2):347–356. [Google Scholar]
- Viseur J. Evaluation of fire blight resistance of somaclonal variants obtained from the pear cultivar ‘Durondeau’. Acta Hortic. 1990;273:275–284. doi: 10.17660/ActaHortic.1990.273.41. [DOI] [Google Scholar]
- Vujović T, Ružić Đ, Cerović R, Momirović GŠ. Adventitious regeneration in blackberry (Rubus fruticosus L.) and assessment of genetic stability in regenerants. Plant Growth Regul. 2010;61:265–275. doi: 10.1007/s10725-010-9474-9. [DOI] [Google Scholar]
- Wacksman JT. DNA methylation and the association between genetic and epigenetic changes: relation to carcinogenesis. Mutat Res. 1997;375:1–8. doi: 10.1016/S0027-5107(97)00003-1. [DOI] [PubMed] [Google Scholar]
- Whitehouse AB, Johnson AW, Passey AJ, McLeary KJ, Simpson DW. Serenity’: a paler skin-coloured somaclonal variant of the short-day cultivar ‘Florence. Acta Hortic. 2014;1049:819–821. doi: 10.17660/ActaHortic.2014.1049.130. [DOI] [Google Scholar]
- Xu M, Li X, Korban SS. DNA-methylation alterations and exchanges during in vitro cellular differentiation in rose (Rosa hybrida L.) Theor Appl Genet. 2004;109:899–910. doi: 10.1007/s00122-004-1717-6. [DOI] [PubMed] [Google Scholar]
- Yang H, Schmidt H. Selection of a mutant from adventitious shoots formed in X ray treated cherry leaves and differentiation of standard and mutant with RAPDs. Euphytica. 1994;77:89–92. doi: 10.1007/BF02551467. [DOI] [Google Scholar]
- Yusnita Y, Widodo W, Sudarsono S. In vitro selection of peanut somatic embryos on medium containing culture filtrate of Sclerotium rolfsii and plantlet regeneration. HAYATI J Biosci. 2005;12(2):50–56. [Google Scholar]
- Zayova E, Vassilevska IR, Kraptchev B, Stoeva D. Somaclonal variations through indirect organogenesis in eggplant (Solanum melongena L.) Biol Divers Conserv. 2010;3:1–5. doi: 10.1111/j.1752-4598.2009.00076.x. [DOI] [Google Scholar]
- Zebrowska JI. In vitro selection in resistance breeding of strawberry (Fragaria x ananassa Duch.) Commun Agric Appl Biol Sci. 2010;75:699–704. [PubMed] [Google Scholar]
- Zhang LQ, Cheng ZH, Khan MA, Zhou YL. In vitro selection of resistant mutant garlic lines by using crude pathogen culture filtrate of Sclerotium cepivorum Australas. Plant Pathol. 2012;41:211–217. [Google Scholar]