Abstract
Over the past decade mutations discovered in genes such as BRCA1, BRCA2, TP53 and PTEN, have emerged as high-penetrance susceptibility genes and are clinically relevant for determination of breast cancer risk. Genetic counseling and subsequent screening for mutations and gene rearrangement has improved patient outcome through early detection and prophylactic interventions in patients with familial breast cancer syndromes. However, these high-penetrance genes only account for a small fraction of the hereditary linked breast cancers. It is currently believed that low-penetrance susceptibility alleles and/or environmental factors may play an important role in the remaining cases. TGFBR1*6A (*6A) is a common hypomorphic variant of the type I TGF-β receptor gene (TGFBR1) that has been associated with risk for several forms of cancer, in particular breast cancer. Several epidemiological studies have suggested that patients who carry the *6A allele have an increased risk of breast cancer. Furthermore, functional analysis suggests that this mutation alters TGF-β signaling and promotes tumorigenesis. Although a decade of research has provided basic information in regards to the prevalence of this mutation in several cancer types and populations the molecular underpinning of its functional effects are poorly understood. A better understanding of the molecular mechanism of TGFBR1 signaling in breast cancer may have an impact on breast cancer risk assessment and breast cancer prevention.
Keywords: TGFBR1, Breast cancer, TGF-β, Cancer genetics, Low-penetrance susceptibility alleles, BRCA1/2, TP53, PTEN, CHEK2, Li Fraumeni syndrome, Cowden’s disease, Hereditary breast and ovarian syndrome
Introduction to breast cancer
Breast cancer is the second leading cause of cancer death for women living in the United States. The American Cancer Society reports that in 2010, there were more than 209,000 new cases of breast cancer and more than 40,000 deaths [1]. Through advances in detection and treatment modalities, the number of breast cancer survivors has steadily increased. The high incidence and prevalence of breast cancer provides a strong rationale for studies into the biology of breast cancer. Screening recommendations, including mammography, MRI in high risk women and self-breast exams, have greatly increased the number of women who are diagnosed at early stage. Breast cancer outcome depends in great part on stage of disease at the time of diagnosis. Additional screening recommendations and new predictive markers of breast cancer risk will likely further decrease breast cancer mortality because of a shift towards diagnosis of early stage breast cancer. While treatments such as surgery, radiation and immunotherapy have proven very effective for localized disease, treatment for metastatic breast cancer remains a major challenge. Of the more than 200,000 cases diagnosed each year, it is estimated that 20%–25% are inherited [2]. Patients that acquire inherited mutations are at a greater risk for development of several cancer types including breast cancer. To date, about 100 genes have been linked to various Mendelian cancer predisposition syndromes [3]. BRCA1, BRCA2, TP53 and PTEN mutations result in significant increase in lifetime risk for breast cancer. Additionally, mutations in several other genes, including STK11/LKB1, CDH1, CHEK2, ATM, MLH1, and MSH2, have also been associated with hereditary breast and/or ovarian tumors.
High-penetrance tumor susceptibility alleles
Mutations in BRCA1 and BRCA2 have been linked to a syndrome known as hereditary breast and ovarian cancer syndrome. BRCA1 mutation is associated with a 65%–81% lifetime risk for breast cancer and a 35%–60% lifetime risk of ovarian or fallopian tube cancer [4–6]. BRCA2 is associated with a 45%–85% lifetime risk for breast cancer and a 10%–27% lifetime risk of ovarian or fallopian tube cancer [4–6]. Both BRCA1 and BRCA2 are associated with an increased risk of male breast cancer [7]. Mutations in these two genes have been associated with ~20% of the familial breast cancer cases diagnosed [8]. Functionally, BRCA1 plays a central role in the repair of double-strand breaks by homologous recombination, with BRCA2 and a number of related proteins in a supportive role for the recognition of such DNA damage [9].
Rare TP53 mutations have been linked to the Li-Fraumeni Syndrome (LS). The TP53 gene encodes for a common tumor suppressor protein TP53, which is found to be mutated in more than 50% of all cancers. TP53 normally acts as a regulator of cell cycle progression in response to DNA damage. In breast cancer, the frequency of TP53 gene mutations is approximately 20% to 30% [10]. Patients with Li-Fraumeni Syndrome also have increased risk to develop several additional forms of cancer including sarcomas, leukemia, and lymphomas. The exact prevalence of Li-Fraumeni is unknown; however, the estimated frequency of the TP53 mutation is 1 in every 20,000 persons [11].
Mutations in PTEN have been linked with a syndrome known as Cowden’s disease. PTEN is a tumor suppressor gene that inhibits cell growth during the G1 phase of cell cycle by activating CDKN1B (KIP1) [12]. Individuals with Cowden syndrome are at increased risk for developing breast cancer as well as hamartomas and benign tumors of the skin, thyroid, breast, endometrium, and brain [12]. Although very rare, penetrance is thought to be nearly complete; it approaches 90% by age 20 years. Carcinoma of the breast occurs in 20%–36% of female patients and is one of the most serious consequences of Cowden disease
Although deleterious mutations in high penetrance genes, BRCA1/2, TP53 and PTEN, are causative of a fraction of the familial breast cancer cases it is believed that environmental factors and low-penetrance susceptibility alleles account for the remainder of cases. Examples of genes harboring rare cancer-associated variants include CHEK2, BRIP1 and PALB2 [3]. Inherited deleterious mutations in the cell cycle regulator CHEK2 are associated with a 2-fold increase in breast cancer risk [3, 6]. A recurrent mutation in the CHEK2 gene (1100delC) was reported to be a cause of breast cancer by Meijers-Heijboer et al. [13]. Recent studies suggest that CHEK2 may act as a modifier of risk of other susceptibility genes due to the strong correlation for patients with a family history of breast cancer. Serrano et al. reported a synergistic effect between CHEK2 mutations and a missense variant in BRCA2 [14].
TGFB signaling
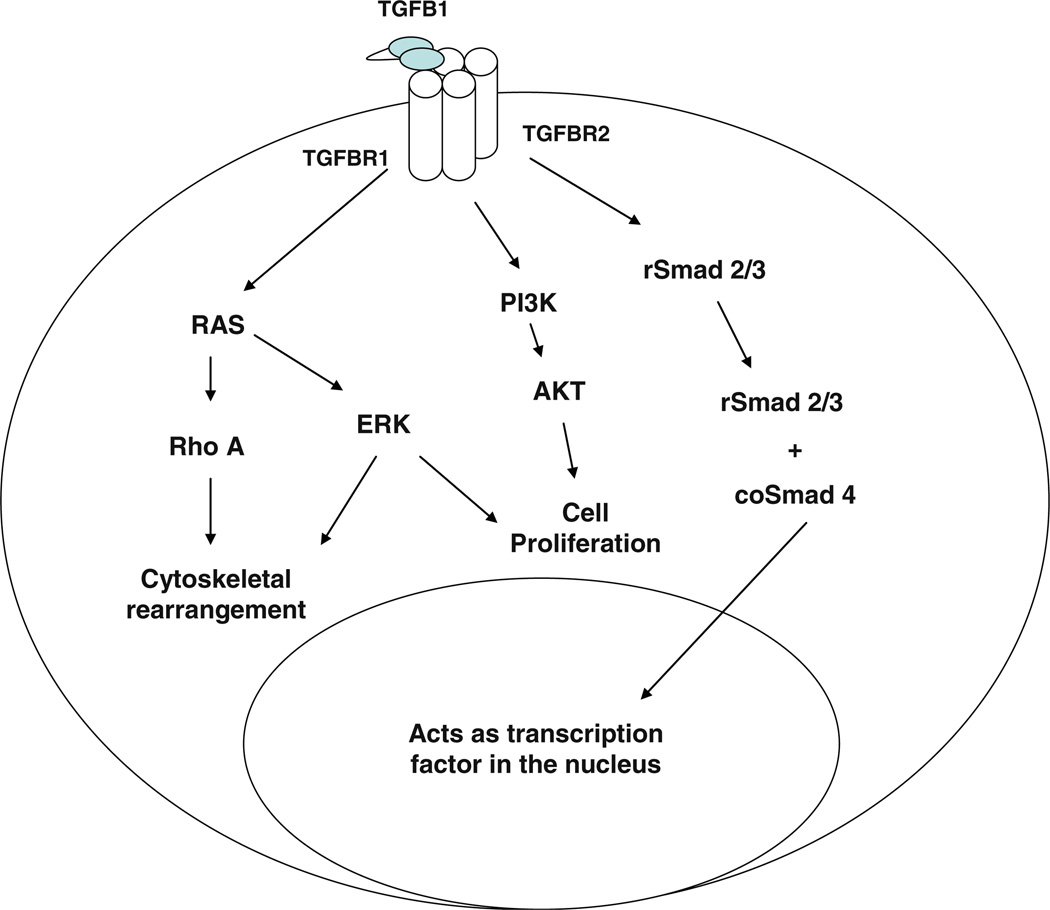
Transforming Growth Factor-β (TGF-β) is one of the most commonly altered cellular signaling pathways in human cancer [15]. TGF-β often has opposing effects on these cellular processes, with its effects being both cell and context specific. A general scheme of TGF-β signaling is depicted in Fig. 1. There are three types of cell surface receptors involved in TGF-β signaling: TGF-β receptor 1, 2 and 3, all of which are dual specificity kinases [16]. TGF-β1 (TGFB1), 2 (TGFB2) and 3 (TGFB3) ligands all bind to the same receptors but with varying affinities. TGFBR3 binds to all three isoforms with very high affinity and may be important in sequestration of TGF-β to the cell surface and for ligand presentation [17]. TGFBR2 is a homodimeric receptor that binds to TGF-β ligand and then forms a heterotetramer with TGFBR1. Autophosphorylation of TGFBR2 causes phosphorylation of TGFBR1 to initiate intracellular signaling. Hence, TGFBR1 is the central propagator of TGF-β signaling.
Fig. 1.
TGF-β signaling cascade
The canonical TGF-β signaling pathway is mediated through SMAD proteins. The mammalian genome contains eight members of the SMAD family. The receptor-regulated SMAD (rSMADS) are activated through phosphorylation and direct interaction with TGFBR1. Once phosphorylated, these SMADs can complex to form heterodimers, which continue the intracellular signaling cascade. The rSMADs consist of SMAD1, 2, 3, 5 and 8, with SMAD2 and 3 being directly involved in the TGF-β signaling pathway. Co-mediator SMADs (co-SMADs) bind to the rSMAD complex and mediate entry into the nucleus to act as transcription factors. This class of SMADs includes SMAD4 in the TGF-β family signaling cascades. The third group of SMAD proteins inhibits this pathway and these proteins are known as inhibitory-SMADs (i-SMADs). SMAD6 and SMAD7 are members of this class and act to regulate TGF-β signaling and expression levels. While this classical signaling cascade has been shown to mediate many of the physiological effects of TGF-β, studies have shown that both TGF-β receptors and SMADs can interact with other signaling pathways to mediate effects. Studies have shown that signaling through MAPK8, MAPK14, PIK3, MAPK1/3 and RAS pathways may be involved in the physiological and pathological effects of TGF-β signaling [18, 19]. Some of the downstream targets of TGF-β signaling are important cell-cycle checkpoint genes, including CDKN1A, CDKN1B, CDKN2B, and their activation leads to growth arrest in normal cells [20]. Further investigation into the TGF-β signaling through these non- SMAD pathways may help to develop a better understanding of the role of TGF-β in pathological conditions such as cancer and fibrosis.
TGFB signaling and breast cancer
The importance of TGF-β signaling in breast cancer progression is evident through numerous in vitro and in vivo studies in which the pathway is disrupted, as well as naturally occurring mutations are evaluated. In a study of TGF-β modulation of mammary tumor progression, mice transgenic for activated Tgfbr1 and the Erbb2 oncogene displayed reduced mitotic index and proliferation rates but enhanced pulmonary metastasis [21]. MCF-7 breast cancer cells overexpressing dominant negative TGFBR2 or treated with a TGFBR1 inhibitor exhibit increased apoptosis compared to normal breast epithelia [22]. Additionally, mutant TGF-β receptors have been associated with higher tumor grades and increased invasiveness. Studies have shown that overexpressing TGFBR3 in a xenograft breast cancer model decreases angiogenesis in both the primary and metastatic tumor [23]. However, TGF-β upregulation of MMP-9 also induces angiogenesis in a breast cancer cell line [24]. In patients TGF-β ligand expression is elevated in the late stages of breast cancer and can be used as a prognostic marker [25]. Reduced nuclear levels of pSMAD2/3 have been associated with high tumor grade, high architectural grade, larger tumor size, and hormone receptor negativity [26], which suggest that decreased TGF-β signaling is associated with a worse outcome. These previous findings from in vitro, in vivo and patient samples illustrate the important role of TGF-β signaling as both a tumor suppressor and pro-oncogenic mediator of breast carcinogenesis. Newer studies which have focused on changes within the tumor microenvironment have highlighted a role for TGF-β signaling in tumor-stromal interactions. One study demonstrated, for example, that fibroblasts lacking TGFBR2 are able to induce an invasive phenotype in adjacent carcinomas, featuring increased proliferation rate, greater angiogenesis, and reduced apoptosis [27]. Furthermore, TGFBR2-null fibroblasts grafted into the mammary fat pads of wild-type mice induced epithelial changes in the new local microenvironment [28]. Over the past few years, an increasing number of epidemiological studies have pointed to the possibility that genetic variants affecting TGF-β production and/or signaling may be related to the overall risk and survival of breast cancer patients. Several SNPs and/or mutations in TGFB1 and TGFBR1 have been associated with increased breast cancer risk and remain to be thoroughly evaluated. However, for the purposes of this review we will focus on mutations in TGFBR1 and its potential implications for breast cancer risk.
Characterization of TGFBR1*6A
The receptor serine/threonine kinase family in the human genome comprises 12 members—7 type I and 5 type II receptors—all dedicated to TGF-β signaling [16, 29]. TGFBR1 gene maps to 9q22.33 [30] and is approximately 56 kb in length consisting of 9 exons [31]. Both types of the receptor serine/threonine kinases consist of about 500 amino acids, organized sequentially into an N-terminal extracellular ligand binding domain, a transmembrane region, and a C-terminal serine/threonine kinase domain [16]. Mutations in several components of the TGF-β signaling pathway have been associated with breast cancer, many of which involve TGFBR2 and SMAD alterations. Mutation types such as frameshift and missense mutations in the TGFBRI coding region are present in subsets of ovarian, esophageal, and head and neck cancers [32]. Additionally, decreased expression of TGFBR1 or TGFBR2 in gastric cancers has been linked to methylation of the TGFBR1 promoter suggest epigenetic mechanisms may play a role [32]. Previously there was little evidence that mutations in the TGFBR1 gene occurred in breast cancer, however at study by Chen et al. reported 2 of 31 primary carcinomas and 5 of 12 lymph node metastases carried a C to A transversion mutation resulting in a serine to tyrosine substitution at codon 387 (S387Y) of the TGFBR1. This TGFBR1 mutant has a diminished ability to mediate TGF-β-dependent effects on gene expression as compared with wild-type TGFBR1 [33].
There is growing evidence that a common variant of the type 1 TGF-β receptor, TGFBR1*6A, may account for approximately 5% of all breast cancer cases, a fraction similar to that attributable to BRCA1 and BRCA2 [2]. TGFBR1*6A is a common variant of TGFBR1 which harbors a three alanine deletion within a stretch of nine alanines located in the 3′-end of exon 1 [30, 34, 35]. More than one in seven healthy individuals and one in six patients with cancer is a TGFBR1*6A carrier making this a high frequency, low-penetrance susceptibility allele. In addition to the *6A variant, several deletions/additions within this same region have been identified, including TGFBR1*10A, TGFBR1*8A and TGFBR1*5A. The fact that a significantly higher *6A allelic frequency was found among patients with a diagnosis of cancer than among healthy controls prompted us to postulate that *6A may act functionally as a tumor susceptibility allele [35]. Early studies have shown that somatic acquisition of this allele is uncommon in breast cancer but not in other cancer types [26]. In an early study Baxter et al. investigated the possible influence of the TGFBR1*6A allele on cancer risk in a cases-control study of 248 controls and 355 women with breast cancer occurring under the age of 40 years, bilateral breast cancer, or a family history of breast cancer. The TGFBR1*6A allele was found to be significantly associated with breast cancer (odds ratio, 1.6; 95% confidence interval, 1.1–2.5) [36]. A meta-analysis performed by Colleran et al., found no association (genotypic OR=0.93, 95% CI=0.74–1.19, P=0.57; allelic OR=0.93, 95% CI= 0.74–1.15, P=0.49) for breast cancer risk [37]. However, a larger meta-analysis of 32 case control studies that included 13,662 cases and 14,147 controls showed significantly elevated cancer risk associated with *6A in all genetic models [38]. Overall cancer risk was increased by 11% for *6A heterozygotes and 30% for *6A homozygotes. This meta-analysis included 10,826 breast cancer cases and 12,964 controls and TGFBR1*6A was significantly associated with breast cancer risk: O.R. 1.16, 95% CI 1.01–1.34 [38]. We have recently shown that TGFBR1*6A is associated with constitutively decreased TGFBR1 expression and increased cancer susceptibility [39, 40].
The 9-alanine sequence is a component of the signal sequence peptide which is responsible for targeting and membrane insertion of secretory and membrane proteins. The signal sequence is normally cleaved following membrane insertion. Initial characterization of the TGFBR1*6A sequence indicates that the 9-bp deletion in TGFBR1*6A does not interfere with the normal cleavage site [35]. However, a secondary cleavage site, found in normal TGFBR1, is not present in the *6A sequence. An experimental model using dog pancreas microsomes confirmed that neither the 9-bp deletion in the *6A signal sequence nor the 3-bp insertion in the *10A signal sequence measurably affect either targeting to or translocation across the endoplasmic reticulum membrane [35]. TGFBR1*6A has also been shown to mediate TGF-β inhibitory signals less efficiently in reporter assays and growth inhibition assays in mink lung epithelial cells [34]. TGFBR1*10A, another rare variant, transduces TGF-β growth-inhibitory signals as effectively at TGFBR1, which suggests that the 6 Ala repeat of *6A has specific biological properties [34, 35]. In MCF-7 breast cancer cells overexpressing TGFBR1*6A there was an increase in cell proliferation, migration and invasion in response to TGF-β treatment [35, 41]. Furthermore, expression of *6A with abolished kinase activity stimulates MCF-7 cell growth in response to TGF-β providing strong support for the novel notion that *6A biological effects are, at least in part, caused by its shorter signal sequence [35]. An additional colon cancer cell line, DLD-1, which harbors the *6A mutation is also growth stimulated by TGF-β treatment [35]. That *6A was the only somatically acquired TGFBR1 allele in primary and metastatic tumors suggests that, even if instability is present at the TGFBR1 locus, the *6A allele provides selective growth advantage over other TGFBR1 alleles, such as the previously reported *5A, *8A, and *10A alleles [35].
Several additional studies have been performed in a variety of cancer types, which have revealed no association of TGFBR1*6A and cancer risk. Hu et. al recently published a study of *6A risk and osteosarcoma for a Chinese patient population in which no significant association was discovered [42]. Castillejo et al. reported results that suggest that the TGFBR1*6A allele does not confer an increased risk of colorectal cancer in the Spanish population [43]. A small study in lung cancer did not show any significant increase in risk for TGFBR1*6A mutation carriers [44]. However, several TGFBR1 haplotypes were found to be associated with non-small cell lung cancer [45]. A 2007 report by Song et al. reported that in a Swedish population the TGFBR1*6A variant may be associated with an increased risk of low-risk familial breast cancer and might be a marker for poorly differentiated breast cancer [46]. These differences may be attributed to variation in the ethnic populations selected for the various studies or may be due to synergistic effects of TGFBR1*6A and other tumor susceptibility alleles present in some but not all cancer types. However, the most likely explanation is the lack of power of many of these studies given the low penetrance and high prevalence of this allele. Further analysis is warranted to determine the true mechanisms that govern the increased risk associated with TGFBR1*6A in breast cancer patients.
To date, functional analysis of TGFBR1*6A indicates that this mutation may confer a growth advantage to cancer cells by switching TGF-β growth inhibitory signals into growth stimulatory signals [47]. One study has confirmed that negative feedback regulation in terms of SMAD3 expression was attenuated and radiation-induced cell death as a cellular endpoint was enhanced for cells harboring the TGFBR1*6A mutation [47]. We have shown that TGFBR1*6A enhances MCF-7 cell migration and invasion through RHOA and MAPK1 pathway activation and downregulation of ARHGAP5 and FN1 [41]. However, continued analysis using both in vitro and in vivo models will help to elucidate additional mechanisms of action. In addition to the *6A allele, an intronic SNP (Int7G24A, rs334354) in the TGFBR1 gene has also been investigated in relation to breast cancer risk. However, the Int7G24A variant was not associated with breast cancer risk or clinical presentation of the disease including prognosis [48].
Potential clinical applications related to TGFBR1*6A
Clinical options for women at high genetic risk of breast cancer include screening starting at a young age, the use of highly sensitive detection methods, and prophylactic surgeries of the ovaries or breast [6]. Identification of candidates for genetic counseling and genetic testing remains a central priority as only a small fraction of the estimated carriers has been identified to date [8]. The exact role of low-penetrance susceptibility alleles such as *6A, TGFBR1 SNPs associated with constitutively decreased TGFBR1 expression and other SNPs identified through GWA studies remains to be defined. Investigation of low penetrance susceptibility alleles and breast cancer risk are complicated by the fact that the penetrance is highly influenced by other factors such as modifier genes, response to DNA damage, and environmental factors such as exposure to carcinogens, hormonal/reproductive factors, and weight [49].
Conclusions
TGF-β signaling plays an important role as both a tumor suppressor in early stage carcinogenesis and pro-metastatic factor in late stage tumorigenesis. It has been known to regulate many aspects of tumor biology including: apoptosis, cell cycle regulation, angiogenesis, immune suppression, migration and invasion. Its numerous roles have made this pathway an excellent candidate for targeted therapeutics. However, a new role for TGF-β signaling components has recently emerged as potential tumor modifiers of cancer susceptibility. Early studies have identified mutations in the receptors, ligands and intermediate signaling components which may be associated with cancer risk. Although some epidemiological studies have been inconclusive it is evident that variants such as TGFBR1*6A play some role in mediating tumorigenic effects. Additional epidemiological and molecular studies of this mutation are warranted to determine its potential as a predictive marker for cancer risk and treatment response.
Acknowledgments
This work was supported by grants CA 137000, CA 112520, and CA 108741 from the National Institutes of Health.
Abbreviations
- ARHGAP5
Rho GTPase-activating protein 5
- ATM
Ataxia telangiectasia mutated
- CDKN
Cyclin-dependent kinase inhibitor
- ERRB2
Human Epidermal growth factor Receptor 2
- FN1
Fibronectin
- GWA
Genome-wide association
- MAPK
Mitogen-activated protein kinase
- PIK3
Phosphatidylinositol 3-kinase
- PTEN
Phosphatase and tensin homologue
- rSMAD
Receptor-regulated SMAD
- coSMAD
Co-mediator SMAD
- iSMAD
Inhibitory SMAD
- STK11
Serine/Threonine kinase
- TGF-β
Transforming growth factor beta
- TGFBR
Transforming growth factor beta receptor
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Rosman DS, Kaklamani V, Pasche B. New insights into breast cancer genetics and impact on patient management. Curr Treat Options Oncol. 2007;8:61–73. doi: 10.1007/s11864-007-0021-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodmer W, Tomlinson I. Rare genetic variants and the risk of cancer. Curr Opin Genet Dev. 2010;20:262–267. doi: 10.1016/j.gde.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 4.Euhus DM. New insights into the prevention and treatment of familial breast cancer. J Surg Oncol. 2011;103:294–298. doi: 10.1002/jso.21664. [DOI] [PubMed] [Google Scholar]
- 5.Walsh CS, Ogawa S, Scoles DR, Miller CW, Kawamata N, Narod SA, et al. Genome-wide loss of heterozygosity and uniparental disomy in BRCA1/2-associated ovarian carcinomas. Clin Cancer Res. 2008;14:7645–7651. doi: 10.1158/1078-0432.CCR-08-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh T, Casadei S, Coats KH, Swisher E, Stray SM, Higgins J, et al. Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA. 2006;295:1379–1388. doi: 10.1001/jama.295.12.1379. [DOI] [PubMed] [Google Scholar]
- 7.Tai YC, Domchek S, Parmigiani G, Chen S. Breast cancer risk among male BRCA1 and BRCA2 mutation carriers. J Natl Cancer Inst. 2007;99:1811–1814. doi: 10.1093/jnci/djm203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasche B. Recent advances in breast cancer genetics. Cancer Treat Res. 2008;141:1–10. doi: 10.1007/978-0-387-73161-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muggia F, Safra T, Dubeau L. BRCA genes: lessons learned from experimental and clinical cancer. Ann Oncol. 2011;22(Suppl 1):i7–i10. doi: 10.1093/annonc/mdq659. [DOI] [PubMed] [Google Scholar]
- 10.Langerod A, Zhao H, Borgan O, Nesland JM, Bukholm IR, Ikdahl T, et al. TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer. Breast Cancer Res. 2007;9:R30. doi: 10.1186/bcr1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemel D, Domchek SM. Breast cancer predisposition syndromes. Hematol Oncol Clin North Am. 2010;24:799–814. doi: 10.1016/j.hoc.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Li DM, Sun H. PTEN/MMAC1/TEP1 suppresses the tumorigenicity and induces G1 cell cycle arrest in human glioblastoma cells. Proc Natl Acad Sci USA. 1998;95:15406–15411. doi: 10.1073/pnas.95.26.15406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meijers-Heijboer H, Wijnen J, Vasen H, Wasielewski M, Wagner A, Hollestelle A, et al. The CHEK2 1100delC mutation identifies families with a hereditary breast and colorectal cancer phenotype. Am J Hum Genet. 2003;72:1308–1314. doi: 10.1086/375121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Serrano-Fernandez P, Debniak T, Gorski B, Bogdanova N, Dork T, Cybulski C, et al. Synergistic interaction of variants in CHEK2 and BRCA2 on breast cancer risk. Breast Cancer Res Treat. 2009;117:161–165. doi: 10.1007/s10549-008-0249-1. [DOI] [PubMed] [Google Scholar]
- 15.Akhurst RJ. TGF beta signaling in health and disease. Nat Genet. 2004;36:790–792. doi: 10.1038/ng0804-790. [DOI] [PubMed] [Google Scholar]
- 16.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 17.Dong M, How T, Kirkbride KC, Gordon KJ, Lee JD, Hempel N, et al. The type III TGF-beta receptor suppresses breast cancer progression. J Clin Invest. 2007;117:206–217. doi: 10.1172/JCI29293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gal A, Sjoblom T, Fedorova L, Imreh S, Beug H, Moustakas A. Sustained TGF beta exposure suppresses Smad and non-Smad signalling in mammary epithelial cells, leading to EMT and inhibition of growth arrest and apoptosis. Oncogene. 2008;27:1218–1230. doi: 10.1038/sj.onc.1210741. [DOI] [PubMed] [Google Scholar]
- 19.Dillon RL, White DE, Muller WJ. The phosphatidyl inositol 3-kinase signaling network: implications for human breast cancer. Oncogene. 2007;26:1338–1345. doi: 10.1038/sj.onc.1210202. [DOI] [PubMed] [Google Scholar]
- 20.Datto MB, Li Y, Panus JF, Howe DJ, Xiong Y, Wang XF. Transforming growth factor beta induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proc Natl Acad Sci USA. 1995;92:5545–5549. doi: 10.1073/pnas.92.12.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siegel PM, Shu W, Cardiff RD, Muller WJ, Massague J. Transforming growth factor beta signaling impairs Neu-induced mammary tumorigenesis while promoting pulmonary metastasis. Proc Natl Acad Sci USA. 2003;100:8430–8435. doi: 10.1073/pnas.0932636100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korpal M, Kang Y. Targeting the transforming growth factor-beta signalling pathway in metastatic cancer. Eur J Cancer. 2010;46:1232–1240. doi: 10.1016/j.ejca.2010.02.040. [DOI] [PubMed] [Google Scholar]
- 23.Gatza CE, Oh SY, Blobe GC. Roles for the type III TGF-beta receptor in human cancer. Cell Signal. 2010;22:1163–1174. doi: 10.1016/j.cellsig.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foda HD, Zucker S. Matrix metalloproteinases in cancer invasion, metastasis and angiogenesis. Drug Discov Today. 2001;6:478–482. doi: 10.1016/s1359-6446(01)01752-4. [DOI] [PubMed] [Google Scholar]
- 25.Levy L, Hill CS. Alterations in components of the TGF-beta superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 2006;17:41–58. doi: 10.1016/j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 26.Zheng W. Genetic polymorphisms in the transforming growth factor-beta signaling pathways and breast cancer risk and survival. Methods Mol Biol. 2009;472:265–277. doi: 10.1007/978-1-60327-492-0_11. [DOI] [PubMed] [Google Scholar]
- 27.Bhowmick NA, Chytil A, Plieth D, Gorska AE, Dumont N, Shappell S, et al. TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004;303:848–851. doi: 10.1126/science.1090922. [DOI] [PubMed] [Google Scholar]
- 28.Bierie B, Moses HL. Tumour microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6:506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- 29.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 30.Pasche B, Luo Y, Rao PH, Nimer SD, Dmitrovsky E, Caron P, et al. Type I transforming growth factor beta receptor maps to 9q22 and exhibits a polymorphism and a rare variant within a polyalanine tract. Cancer Res. 1998;58:2727–2732. [PubMed] [Google Scholar]
- 31.Vellucci VF, Reiss M. Cloning and genomic organization of the human transforming growth factor-beta type i receptor gene. Genomics. 1997;46:278–283. doi: 10.1006/geno.1997.5023. [DOI] [PubMed] [Google Scholar]
- 32.Massague J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen T, Carter D, Garrigue-Antar L, Reiss M. Transforming growth factor beta type I receptor kinase mutant associated with metastatic breast cancer. Cancer Res. 1998;58:4805–4810. [PubMed] [Google Scholar]
- 34.Pasche B, Kolachana P, Nafa K, Satagopan J, Chen YG, Lo RS, et al. TbetaR-I(6A) is a candidate tumor susceptibility allele. Cancer Res. 1999;59:5678–5682. [PubMed] [Google Scholar]
- 35.Pasche B, Knobloch TJ, Bian Y, Liu J, Phukan S, Rosman D, et al. Somatic acquisition and signaling of TGFBR1*6A in cancer. JAMA. 2005;294:1634–1646. doi: 10.1001/jama.294.13.1634. [DOI] [PubMed] [Google Scholar]
- 36.Baxter SW, Choong DY, Eccles DM, Campbell IG. Transforming Growth Factor beta Receptor 1 Polyalanine Polymorphism and Exon 5 Mutation Analysis in Breast and Ovarian Cancer. Cancer Epidemiol Biomark Prev. 2002;11:211–214. [PubMed] [Google Scholar]
- 37.Colleran G, McInerney N, Rowan A, Barclay E, Jones AM, Curran C, et al. The TGFBR1*6A/9A polymorphism is not associated with differential risk of breast cancer. Breast Cancer Res Treat. 2010;119:437–442. doi: 10.1007/s10549-009-0395-0. [DOI] [PubMed] [Google Scholar]
- 38.Liao RY, Mao C, Qiu LX, Ding H, Chen Q, Pan HF. TGFBR1*6A/9A polymorphism and cancer risk: a meta-analysis of 13,662 cases and 14,147 controls. Mol Biol Rep. 2010;37:3227–3232. doi: 10.1007/s11033-009-9906-7. [DOI] [PubMed] [Google Scholar]
- 39.Valle L, Serena-Acedo T, Liyanarachchi S, Hampel H, Comeras I, Li Z, et al. Germline allele-specific expression of TGFBR1 confers an increased risk of colorectal cancer. Science. 2008;321:1361–1365. doi: 10.1126/science.1159397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pasche B, Wisinski KB, Sadim M, Kaklamani V, Pennison MJ, Zeng Q, et al. Constitutively decreased TGFBR1 allelic expression is a common finding in colorectal cancer and is associated with three TGFBR1 SNPs. J Exp Clin Cancer Res. 2010;29:57. doi: 10.1186/1756-9966-29-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosman DS, Phukan S, Huang CC, Pasche B. TGFBR1*6A enhances the migration and invasion of MCF-7 breast cancer cells through RhoA activation. Cancer Res. 2008;68:1319–1328. doi: 10.1158/0008-5472.CAN-07-5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu YS, Pan Y, Li WH, Zhang Y, Li J, Ma BA. Association between TGFBR1*6A and osteosarcoma: a Chinese case-control study. BMC Cancer. 2010;10:169. doi: 10.1186/1471-2407-10-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castillejo A, Mata-Balaguer T, Montenegro P, Ochoa E, Lazaro R, Martinez-Canto A, et al. The TGFBR1*6A allele is not associated with susceptibility to colorectal cancer in a Spanish population: a case-control study. BMC Cancer. 2009;9:193. doi: 10.1186/1471-2407-9-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.You W, Liu Z, Zhao J, Zheng M, Zheng SY, Liu X, et al. No association between TGFBR1*6A and lung cancer. J Thorac Oncol. 2007;2:657–659. doi: 10.1097/JTO.0b013e318070ccd7. [DOI] [PubMed] [Google Scholar]
- 45.Lei Z, Liu RY, Zhao J, Liu Z, Jiang X, You W, et al. TGFBR1 Haplotypes and Risk of Non-Small-Cell Lung Cancer. Cancer Res. 2009;69:7046–7052. doi: 10.1158/0008-5472.CAN-08-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song B, Margolin S, Skoglund J, Zhou X, Rantala J, Picelli S, et al. TGFBR1(*)6A and Int7G24A variants of transforming growth factor-beta receptor 1 in Swedish familial and sporadic breast cancer. Br J Cancer. 2007;97:1175–1179. doi: 10.1038/sj.bjc.6603961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schirmer MA, Hoffmann AO, Campean R, Janke JH, Zidek LM, Hoffmann M, et al. Bioinformatic and functional analysis of TGFBR1 polymorphisms. Pharmacogenet Genomics. 2009;19:249–259. doi: 10.1097/FPC.0b013e32831cb5a7. [DOI] [PubMed] [Google Scholar]
- 48.Castillejo A, Mata-Balaguer T, Guarinos C, Castillejo MI, Martinez-Canto A, Barbera VM, et al. The Int7G24A variant of transforming growth factor-beta receptor type I is a risk factor for colorectal cancer in the male Spanish population: a case-control study. BMC Cancer. 2009;9:406. doi: 10.1186/1471-2407-9-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Welcsh PL, King MC. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum Mol Genet. 2001;10:705–713. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]