Abstract
Queuosine is a hypermodified nucleoside present in the wobble position of tRNAs with a 5′-GUN-3′ sequence in their anticodon (His, Asp, Asn, and Tyr). The 7-deazapurine core of the base is synthesized de novo in prokaryotes from guanosine-5′-triphosphate in a series of eight sequential enzymatic transformations, the final three occurring on tRNA. Epoxyqueuosine reductase (QueG), catalyzes the final step in the pathway, which entails the 2-electron reduction of epoxyqueuosine to form queuosine. Biochemical analyses reveal that this enzyme requires cobalamin and two [4Fe-4S] clusters for catalysis. Spectroscopic studies show that the cobalamin appears to bind in a base-off conformation, whereby the dimethylbenzimidazole moiety of the cofactor is removed from the coordination sphere of the cobalt but not replaced by an imidazole sidechain, which is a hallmark of many cobalamin-dependent enzymes. The bioinformatically-identified residues are shown to have a role in modulating the primary coordination sphere of cobalamin. These studies provide the first demonstration of the cofactor requirements for QueG.
Keywords: queuosine, RNA modification, EPR, cobalamin
RNA is one of the most chemically diverse biological building-blocks with over 110 modifications reported to date1. Modifications can be as complex as the tricyclic ring structure observed in wybutosine2,3, or as simple as acetylation or thiolation of various positions on the purine or pyrimidine core. These modifications contribute to a myriad of functions including, but not limited to, translational efficiency and accuracy, structural stabilization of the tRNA, enhanced recognition between the tRNA and its cognate aminoacyl-tRNA synthetase, and decoding of degenerate codons4,5. However, even simple modifications, such as methylation, are garnering attention as regulators of RNA processes. Only recently has the importance of such modifications come to light through identification of enzymes that reverse this methylation process, in an analogous fashion to DNA, adding a layer of regulation described as RNA epigenetics6,7.
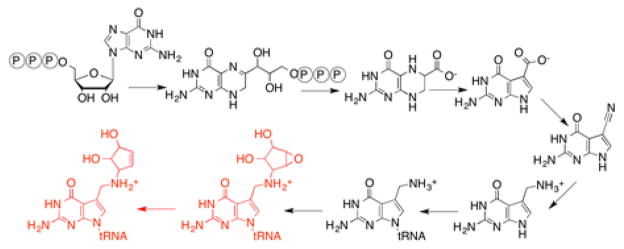
Queuosine is one of the most highly decorated RNA modifications identified to date and has been observed in RNA across all domains of life8. The unique 7-deazapurine core of queuosine, which is also found in a variety of secondary metabolites9, is further adorned with a cyclopentenediol moiety derived from S-adenosyl-L-methionine (SAM)10. Queuosine is present in the wobble position of tRNAs containing a 5′-GUN-3′ anticodon sequence that encodes for Asp, Asn, His, and Tyr amino acids11. In bacteria, it is synthesized from the precursor GTP through an eight step biosynthetic pathway (Fig. 1)10,12–22, which entail biosynthesis of 7-aminomethyl-7-deazaguanine, exchange into the wobble position, followed by two additional steps that occur on tRNA. Higher order organisms obtain queuine, the free base of queuosine, from dietary sources and exchange this base into mature tRNA in place of guanine en masse23. While no discrete function is known for this modification, absence has been correlated with numerous biological phenomena such as cancer pathology24–28, pathogenicity29,30, symbiosis31, and neurological disorders32.
Figure 1.
Biosynthesis of queuosine.
Our understanding of the complete de novo biosynthetic pathway was completed recently with the discovery of epoxyqueuosine reductase (QueG), which catalyzes the final epoxide reduction, converting epoxyqueuosine to queuosine22 (see Fig. 1). QueG is homologous to reductive dehalogenase (RDH) enzymes that are essential to bacteria that utilize halogenated compounds, such as tetrachloroethylene, as their terminal electron acceptors33,34. RDHs are known to contain multiple iron-sulfur clusters and corrinoids as cofactors, and require strong reductants for activity35–39. QueG retains the eight Cys residues that coordinate the two 4Fe-4S clusters in RDHs (Fig. 2). Recent X-ray crystal structures of two RDHs provide views of the active sites of these enzymes40,41. The epoxide reduction reaction catalyzed by QueG is analogous to RDHs in that it is a two-electron reduction requiring a strong reductant. Prior to identification of oQ reductase, bacterial feeding experiments had demonstrated a cobalamin requirement for the conversion of oQ to Q42. In vitro, oQ reduction was shown to be stimulated by addition of exogenous cobalamin22. To date, however, there have been no systematic studies on the role(s) of the cofactors and conserved residues in catalysis in RDHs or QueG.
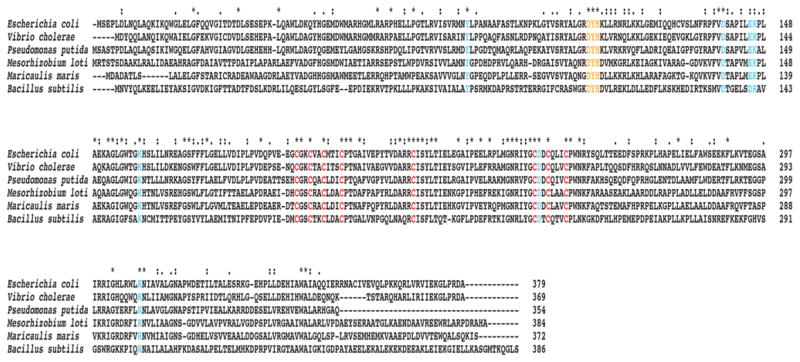
Figure 2. A representative multiple sequence alignment of QueG homologs and in vivo alanine scanning of conserved residues of QueG.
Conserved cysteine residues required for iron-sulfur cluster formation are in red, the conserved DYH motif is shown in orange, and other conserved residues mutated in the in vivo alanine scanning experiment are highlighted in blue. The accession numbers for the sequences are as follows: E. coli (EGI43095), Vibrio cholerae (WP_000386321), Pseudomonas putida (WP_014754621), Mesorhizobium loti (WP_010912587), Maricaulis maris (WP_011642061), and Bacillus subtilis (NP_388772).
Herein we report a biochemical and spectroscopic analysis of the cofactor requirements of QueG. We have established a method to obtain cofactor replete, active recombinant protein in an anaerobic environment allowing the cofactor stoichiometry of the protein to be established unambiguously. The results demonstrate the presence of two iron-sulfur clusters and a cobalamin that are absolutely required for activity. In addition, an in vivo alanine scanning experiment has identified residues that are critical for catalysis. Analysis by electron paramagnetic resonance (EPR) spectroscopy of a subset of conserved residues that are essential for activity has revealed an interesting role for these residues in modulating the coordination environment of the cobalamin cofactor. Taken together, these analyses highlight the unique cofactor requirements necessary to facilitate the novel epoxide reduction catalyzed by QueG on RNA and show that despite the substantially different reaction catalyzed by oQ reductase, it utilizes similar cofactors, suggesting that reductive dehalogenation and epoxide reduction likely follow similar mechanistic pathways.
METHODS
Materials
All materials were purchased commercially (unless otherwise noted) and were of the highest purity. All assays and purification steps were carried out in a Coy anaerobic chamber in an atmosphere of 95% N2, 5% H2. All buffers and materials were deoxygenated in the chamber several days prior to use and were made RNase-free when possible.
Cloning of B. subtilis queG
The codon-optimized gene encoding B. subtilis QueG was obtained from Genscript (Piscataway, NJ). The sequence of the synthetic gene is available in the Supporting Information. The gene was excised from a supplied pUC57 vector by digestion with NdeI and HindIII and ligated into a similarly cut pET28a vector to obtain pZM419. The gene was then amplified from this vector using the primers: 5′-AAAGGATCCATGAATGTTTACCAACTGAAAGAAGAAC-3′ (forward) and 5′-AAAGGATCCGGACAGACCTTGTTTCGTCATACC-3′ (reverse) at an annealing temperature of 43 °C and included flanking BamHI restriction sites. The amplified fragment was digested using BamHI and cloned into a pASK-IBA43plus vector (IBA) that was similarly digested to yield pZM471 for expression of N-terminal His6-tagged and C-terminal Strep-tag II QueG. Mutants of pZM419 and pZM471 were created using Stratagene Mutagenesis Kit according to the manufacturer’s recommended protocol and using the primers listed in Supplementary Table 1.
Expression of B. subtilis QueG
E. coli BL21(DE3) cells containing pZM471 were grown in 12 L of LB containing 0.1 mg/mL ampicillin at 37 °C to an OD600nm of approximately 0.3 at which time ferric chloride was added to a final concentration of 50 μM and the flasks were cooled to 18 °C. The cells were grown further to an OD600nm of approximately 0.6 and protein expression was induced with 20 μL of 10 mg/mL anhydrotetracycline hydrochloride in dimethylformamide (Acros Organics) per 1 L of LB. Cells were grown overnight and harvested the next day by centrifugation (5,000 × g), frozen in liquid N2, and stored at −80 °C.
Purification of QueG
All steps were carried out in an anaerobic chamber as described in the materials. Cells (~20 g) were suspended in buffer containing 100 mM Tris•HCl (pH 8.0), 150 mM NaCl, 10 mM dithiothreitol (DTT), and 1 mM PMSF, and sonified using a Branson digital sonifier (45% amplitude). The lysate was centrifuged for 30 min at 18,000 × g (4 °C) to pellet insoluble material. The cleared lysate was loaded onto a 5 mL StrepTrap HP column (GE Healthcare) that had been equilibrated with buffer containing 100 mM Tris•HCl (pH 8.0) and 150 mM NaCl (buffer A). The column was washed with 25 mL of buffer A and QueG was eluted with 25 mL of buffer A containing 2.5 mM d-desthiobiotin (Sigma-Aldrich). Fractions containing QueG were identified using SDS-PAGE and pooled. The concentration of the pooled protein was determined using the Bradford method with bovine serum albumin (ThermoScientific) as the standard.
Reconstitution of QueG
The reconstitution was carried out on ice and the solution was stirred constantly. First, solid DTT was added to a final concentration of 10 mM and the solution was allowed to mix for 15 min. Ten molar equivalents of ferric chloride dissolved in anaerobic water was added slowly over the course of 5 min and the solution was allowed to mix for an additional 10 min. Ten equivalents of sodium sulfide dissolved in anaerobic water was added slowly over 5 min and the solution was allowed to mix for another 3 h. The protein solution was buffer exchanged using a Bio-Rad DG-10 column into 20 mM Tris•HCl (pH 8.0), 100 mM KCl, and 2 mM DTT (buffer B). The protein was concentrated by centrifugation at 6,000 × g (4 °C) using an Amicon Ultra-4 10 kDa cutoff concentrator to a final volume of less than 1 mL. To the resulting concentrated protein, 100 μL of 20 mM hydroxocobalamin acetate salt (Sigma-Aldrich) was added and the solution was allowed to incubate at room temperature for 5 min. The protein was then loaded onto a Sephacryl S300 HR gel filtration column (16 × 60 cm, GE Healthcare) equilibrated in buffer B, and eluted at a constant flow rate of 1 mL/min. Fractions containing QueG were identified using SDS-PAGE, pooled, and concentrated by centrifugation at 4,000 × g (4 °C) using an Amicon Ultra-15 10 kDa cutoff concentrator. Aliquots were flash frozen in liquid N2 and stored at −80 °C. Protein concentration was determined using the Bradford method with bovine serum albumin (ThermoScientific) as the standard.
Amino acid, iron, labile sulfide, and cobalamin content analyses
All subsequent analyses were completed on at least three independent protein preparations and values are reported as averages.
Amino acid analysis was performed at the Molecular Structure Facility University of California-Davis (Davis, CA). The protein was exchanged by gel filtration using a NICK Sephadex G-50 column (GE Healthcare) into 10 mM NaOH. A Bradford assay was performed on the eluate and 0.3 mL of the remaining protein solution was utilized for the analysis. The amino acid correction factor was used to calculate the resulting stoichiometries during the cofactor content analyses.
Iron analysis was performed at the University of Arizona Department of Hydrology and Water Resources using a Perkin-Elmer Optima 5300 DV ICP-OES. QueG was diluted into 1% (v/v) nitric acid to a concentration of 1 μM for analysis. The concentration of labile sulfide was determined using the Beinert method43.
The cobalamin content of QueG was determined by thermal denaturation of the protein and derivatization of cobalamin to dicyanocob(III)alamin by addition of potassium cyanide as follows. To a solution containing QueG at a known concentration, potassium cyanide was added to a final concentration of 10 mM in a total reaction volume of 0.1 mL. The reaction was allowed to incubate at room temperature in the dark for 30 min and was placed at 99 °C for 15 min. The mixture was placed directly on ice for 5 min and then centrifuged at 21,000 × g for 5 min to pellet the precipitated protein. The concentration of dicyanocob(III)alamin in the supernant was determined by measuring the absorbance at 580 nm using an extinction coefficient of 10.13 mM−1 cm−1 44,45.
Size-exclusion chromatography to determine oligomerization state
Purified QueG (4 mg/mL) was loaded onto a Sephacryl S200 HR column (16 × 60 cm, GE Healthcare) equilibrated in 20 mM Tris•HCl (pH 8.0), 100 mM KCl, and 2 mM DTT. The column was run at a constant flow rate of 1 mL/min and the fractions were analyzed by SDS-PAGE to observe the presence of the standards and QueG. The standards (Bio-Rad) used to determine the molecular weight of QueG were: thyroglobulin (670 kDa), γ-globulin (150 kDa), ovalbumin (44 kDa), myoglobin (17 kDa) and vitamin B12 (1.35 kDa).
In vitro activity assays to determine requirements of iron-sulfur clusters and cobalamin
The conversion of oQ to Q by QueG was assayed using a synthetic 17-mer oQ-containing stem loop substrate corresponding to the anticodon loop of Tyr-tRNA and prepared as previously described22. The assays contained 20 mM PIPES•NaOH (pH 7.4), 10 mM sodium dithionite, 0.75 mM methyl viologen, 0.75 mM hydroxocobalamin (if present in assay), and 5 μL of ~0.5 mM oQ stem loop. The reactions were initiated by addition of 5 μM QueG enzyme in a total volume of 0.1 mL. The reaction was allowed to proceed in the dark at room temperature for 5 h and quenched using QIAzol reagent (Qiagen), which contains phenol. RNA from the assay was purified, digested to nucleosides, and analyzed by LC/MS as previously described22.
In vivo alanine scanning complementation experiment
The ΔyjeS E. coli strain from the Keio Collection46 was lysogenized using the λDE3 Lysogenization Kit (Novagen) following the manufacturer’s recommended protocol. The kanamycin resistance cassette, which was inserted in place of the yjeS gene in the Keio Collection, was removed by transformation with the pCP20 plasmid47 following a published method48. Briefly, 30 μL of stationary phase overnight culture of the lysogenized knockout strain was used to inoculate 3 mL of LB containing 34 μg/mL kanamycin. The culture was then left to grow at 37 °C, 200 rpm, to an OD600nm of ~0.6–0.7. The cells were then made electrocompetent by placing 1 mL of the culture in a pre-chilled, sterile 1.5 mL tube and spun at 13,000 × g (4 °C) for 1 min. to pellet the cells. The pellet was resuspended in 1 mL pre-chilled sterile water and was again spun at 13,000 × g (4 °C) for 1 min to pellet the cells. This process was repeated. The pellet from the final spin was resuspended in 50 μL of the residual supernatant and transformed with the pCP20 plasmid, resuspended in 1 mL room temperature SOC media, and left to shake at 30 °C, 200 rpm for 1 h. The culture was then added to 100 mL of LB containing 100 μg/mL ampicillin and 34 μg/mL chloramphenicol, and allowed to shake at 30 °C, 200 rpm overnight. An aliquot (30 μL) of the overnight growth was used to inoculate a 3 mL LB culture and it was allowed to shake at 30 °C, 200 rpm until the OD600nm reached ~0.1, at which time the temperature was shifted to 42 °C and the culture was allowed to grow to an OD600nm of ~0.8–1.0. The culture was then streaked onto an LB/agar plate and left overnight at 37 °C. Colonies were selected that could no longer grow on either kanamycin or ampicillin and verified to be the correct ΔyjeS E. coli strain by colony PCR. Strains verified to be Kan-r and ΔyjeS were made competent and transformed with a pET28a plasmid containing wild-type QueG or QueG variants. Site directed variants were prepared using the primers listed in Table S1. A single colony was used to inoculate 50 mL of LB containing 34 μg/mL kanamycin and was grown at 37 °C, 200 rpm to an OD600nm of ~0.5–0.7. Then, the culture was induced by addition of 5 μL of 1 M IPTG and left to continue growing for 16 h total. The cells were harvested the next day and total RNA was extracted and analyzed on the LC/MS as previously described22. Each variant was analyzed in this manner at least in triplicate.
Electron paramagnetic resonance spectroscopy of wild-type QueG and variants
EPR samples were prepared in anaerobic conditions by mixing concentrated, purified protein with a final concentration of 15% glycerol (v/v) to a total volume of ~0.15 mL and frozen in liquid nitrogen in an EPR tube. Continuous wave (CW) X-band EPR experiments were preformed on a Bruker Biospin EleXsys E500 spectrometer equipped with a cylindrical TE011-mode resonator (SHQE-W), an ESR-900 liquid helium cryostat, and an ITC-5 temperature controller (Oxford Instruments). Spectra were recorded at 40 K at a microwave frequency of 9.323 GHz (WT), 9.333 GHz (D104A), 9.321 GHz (Y105A), 9.373 GHz (H106A), and 9.376 GHz (D134A). The power and modulation amplitude were 0.2 mW and 2 G, respectively. Simulations were carried out in MatLab using the EasySpin suite of programs49.
RESULTS and DISCUSSION
Purification of QueG
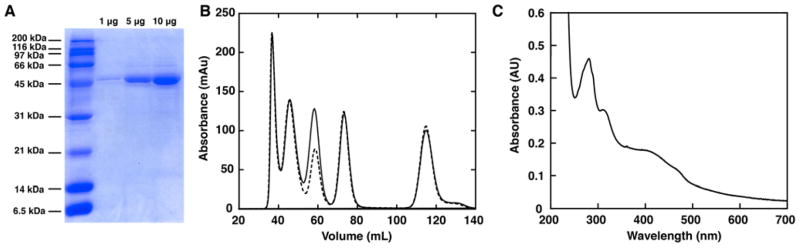
Previous studies on purified QueG confirmed that the enzyme was sufficient for conversion of oQ to Q and demonstrated that addition of exogenous cobalamin stimulated activity22. However, the protein used in those studies was recalcitrant to expression and purification to a more homogenous state, and the observation that cobalamin stimulated activity was presumably due to the lack of cofactor replete protein. Therefore, we obtained a codon-optimized gene encoding the B. subtilis homolog and developed a method of expression in E. coli and purification to obtain >95% pure protein (Fig. 3A). QueG is a monomer as judged by size exclusion chromatography (Fig. 3B). Based on the sequence homology to reductive dehalogenases, we hypothesized that the enzyme would contain iron-sulfur clusters and cobalamin as cofactors. Indeed, the UV-visible spectrum of QueG (Fig. 3C) includes features common to both [4Fe-4S] clusters at ~420 nm50 and cob(II)alamin at ~475 nm51.
Figure 3. Purification, UV/visible spectrum, and quaternary state determination of QueG.
A. An SDS-PAGE gel of purified QueG. Based on the gel, QueG is >95% pure with a molecular weight consistent with the predicted weight of 48.7 kDa. B. A size exclusion chromatogram of protein standards (shown in dotted black line) superimposed with a chromatogram of protein standards including purified QueG enzyme (shown in solid black line). Based on the elution profiles, only the peak pertaining to the ~44 kDa standard changes with inclusion of QueG in the solution mixture. QueG is ~48.7 kDa leading to the increase in only the absorbance of the concurrent standard peak. The standard peaks (from left to right) are: thyroglobulin (670 kDa), γ-globulin (150 kDa), ovalbumin (44 kDa), myoglobin (17 kDa) and vitamin B12 (1350 Da). C. A UV/visible spectrum of the purified enzyme contains features consistent with inclusion of both iron-sulfur clusters (~420 nm) and cob(II)alamin (~475 nm) as cofactors.
Cofactor stoichiometry
After establishing the protein purification and cofactor incorporation scheme, we turned our attention to determining whether the iron-sulfur clusters and cobalamin are required for activity. The basic strategy for obtaining replete protein entailed reconstitution of the purified protein in two stages with iron and sulfide first, followed by cobalamin. Excess cofactors were removed by gel filtration prior to activity and stoichiometry measurements.
To determine the cofactor requirements, an aliquot of the reconsitution mixture was assayed for activity following the initial affinity purification steps, after reconstitution with iron and sulfide, and after addition of cobalamin. As shown in Fig. 4, the as isolated protein has no activity whereas protein that has undergone iron, sulfur and cobalamin additions is active. However, reconstitution with iron and sulfide is not sufficient for activity; therefore, oQ reductase activity requires FeS clusters and cobalamin. To determine the stoichiometry of the clusters and cobalamin present, amino acid analysis was carried out on reconstituted QueG after the final size-exclusion step to obtain an accurate protein concentration. Based on the amino acid analysis, the protein concentration obtained using the Bradford method overestimates [QueG] by ~1.7-fold. QueG was subjected to ICP-OES to determine the amount of iron and the Beinert method43 was used to determine the amount of labile sulfide. Finally, a cyanolysis was carried out to quantify cobalamin content44,45. The iron, sulfide and cobalamin stoichiometry derived from the above analyses are summarized in Table 1. Purified and reconstituted QueG contains 6.5 moles of iron, 8 moles of labile sulfide, and one mole of cobalamin per monomer. Recently, two structures of reductive dehalogenase proteins have been reported that are relevant in this discussion40,41. Both structures show that RDHs have two [4Fe-4S] clusters and a cobalamin. While there is limited sequence similarity between QueG and RDHs, the Cys residues that coordinate the two iron-sulfur clusters are conserved in QueG (shown in red in Fig. 2). Intriguingly, oQ reductase has the same complement of cofactors as RDHs, despite the differences in the overall reactions catalyzed by these enzymes.
Figure 4. Iron-sulfur clusters and cobalamin are required for QueG activity in vitro.
Extracted ion chromatograms of digested nucleosides of oQ (m/z = 426) and Q (m/z = 410) from oQ stem loop used as the substrate in an in vitro activity assay of QueG. Protein was assayed for activity before and after reconstitution in the presence or absence of exogenous hydroxocobalamin. The appearance of turnover in only enzyme with reconstituted iron-sulfur clusters and added hydroxocobalamin demonstrates the necessity of both for activity.
Table 1.
Analysis of the cofactor content of QueG.
| Cofactor | Stoichiometry (mol/monomer QueG)a |
|---|---|
| Iron | 6.54 ± 1.61 |
| Labile sulfide | 8.01 ± 1.08 |
| Cobalamin | 1.01 ± 0.11 |
The values are consistent with the presence of two [4Fe-4S] clusters and one cobalamin cofactor per monomer of QueG. The concentration of QueG protein used to obtain the stoichiometric values was calibrated with amino acid analyses as described in Methods.
In vivo screening for essential residues
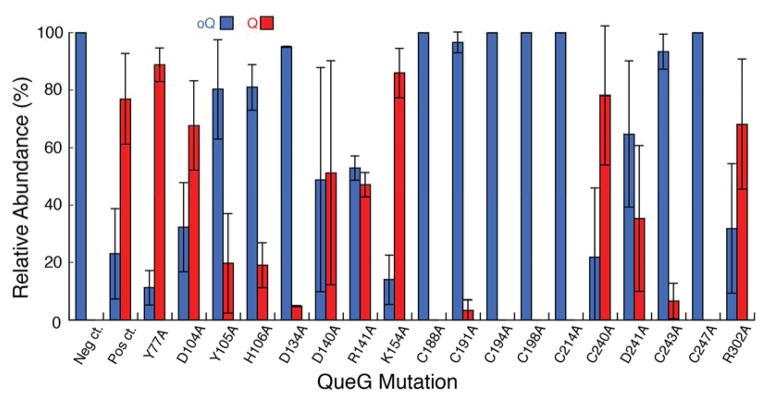
The initial difficulties in expression and purification of QueG and the complicated reconstitution methods that are required to obtain cofactor-replete active protein made it necessary to develop an alternative method to rapidly assess the importance of residues in catalysis. In these experiments we utilized the ΔqueG strain of E. coli from the Keio collection46 and examined the oQ/Q content of the cells upon expression of wild-type and site-directed variants of QueG on a plasmid. Potential catalytic residues were identified from a multiple sequence alignment that was constructed with ~1100 sequences obtained through a protein BLAST search40,41,52,53 of the non-redundant database with the E. coli QueG homolog (YjeS; b4166) as the initial search sequence. The candidates obtained from the BLAST search had 30–80% sequence identity. Analysis of the extensive alignment identified multiple absolutely conserved motifs (Fig. 2). Two of the conserved motifs, CX2CX2CX3C and CX2CX3C with another distally conserved Cys, are likely involved in forming the iron-sulfur clusters as both of these motifs are also the sequence signatures associated with enzymes involved in reductive dehalogenation, where they had been proposed to coordinate two [4Fe-4S] clusters that are necessary for electron transfer and reduction of the cobalamin cofactor35,54–56. Another absolutely conserved sequence was a DYH motif. This DYH motif was of great interest as it was reminiscent of the DxHxxG motif commonly found in cobalamin-dependent enzymes, specifically, enzymes that bind cobalamin in its base-off/His-on conformation, such as methionine synthase57 and glutamate mutase58,59. In this conformation, a histidine side chain displaces the dimethylbenzimidazole moiety of cobalamin, which in solution coordinates the cobalt atom through the N3 atom in the lower axial position. Based on the conserved residues observed in the multiple sequence alignment, mutations were made in a pET28a plasmid harboring the B. subtilis QueG enzyme (pZM419). In all, 18 single mutation variants were generated where the residue of interest was mutated to an alanine. The ΔyjeS E. coli Keio Collection strain was then lysogenized with λDE3 and made electrocompetent to allow for expression of the wild-type or variant queG gene using T7 polymerase after transformation with the corresponding pET28a plasmid. Cells were induced in the log phase of growth and allowed to grow overnight. Total RNA was isolated, digested to nucleosides, and analyzed for the presence of oQ vs Q via LC/MS. The effects of the mutations on the production of both oQ and Q in vivo are shown in Fig. 5. Mutation of almost all of the cysteine residues thought to be involved in cluster formation completely negated the ability to convert oQ to Q. Curiously, the point mutation C240A did not seem to have any effect on activity in comparison to the wild-type protein. However, there have been examples of iron-sulfur cluster containing enzymes where a single mutation in a cysteine residue comprising an iron-sulfur cluster is not sufficient to abolish cluster formation and activity60. Mutations in residues comprising the DYH motif showed differing results. The introduction of Y105A and H106A variants of QueG into the knockout strain reduced the extent of cellular Q relative to oQ. By contrast, the D104A mutation did not alter the oQ/Q ratio relative to wild-type.
Figure 5. In vivo QueG mutagenesis.
The relative abundances of Q and oQ were calculated from the extracted ion chromatograms of oQ (blue) and Q (red) present in total RNA nucleosides from overnight growths of ΔqueG E. coli overexpressing the indicated variant form of QueG (B. subtilis numbering). All values are normalized to the sum of both oQ and Q and given as a percentage of the total.
Interestingly, the D134A and R141A variants significantly reduced the Q-content of the strain relative to wild-type. The importance of the Cys residues to catalysis in QueG is easy to justify due to the observation that analogous Cys residues in RDH proteins coordinate the two [4Fe-4S] clusters in the protein. The significance of D104, Y105, H106 and D134, however, was not immediately obvious.
EPR spectroscopy of QueG and variants
To further probe the role of residues that appeared to be essential by the in vivo experiments, we carried out electron paramagnetic resonance studies to investigate whether these residues are in the vicinity of the cobalamin cofactor. Cobalamin can exist in three oxidation states with distinct UV/visible spectral signatures. However, in the +2 oxidation state, the cofactor is paramagnetic and may be studied by EPR.
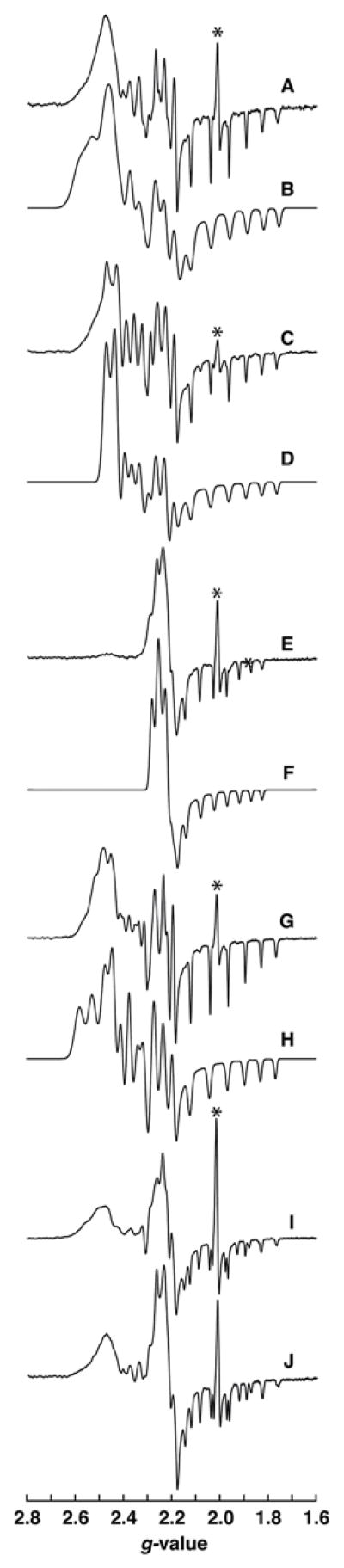
Reconstituted wild-type, D104A, Y105A, H106A, and D134A proteins were analyzed by EPR spectroscopy. The EPR spectrum of the wild-type protein (Fig. 6A) is best simulated by a rhombic g- and A-tensor (Table 2). Interaction of the unpaired spin with cobalt (I=7/2) introduces extensive fine structure, which is best fit with a rhombic A-tensor (Ax=186, Ay=133, Az=340 MHz). We observe no additional strongly coupled nuclei. If a nitrogen (I=1) from the dimethylbenzimidazole or His imidazole sidechain had been present, we would have observed triplet splittings of the gz features, as is observed in other cobalamin-dependent enzymes57,61,62. Therefore, the data clearly show that upon binding of cobalamin to QueG, the dimethylbenzimidazole ligand that coordinates the cobalt in solution is dissociated, but not replaced with a His sidechain. This observation is similar to what has been observed in the structures of the RDH proteins40,41.
Figure 6. EPR of wild-type QueG and variants.
The EPR spectra and simulations of A. wild-type and variants C. D104A, E. Y105A, G. H106A, and I. D134A of QueG in the cob(II)alamin form. Simulations are shown in B, D, F, H, and J for wild-type and variants in the same order as above. The * denotes signal that results from degradation of the iron-sulfur clusters during purification and reconstitution.
Table 2.
EPR simulation parameters for cob(II)alamin in wild-type QueG and site-directed variants.
| Protein | gx | gy | gz | Ax | Ay | Az |
|---|---|---|---|---|---|---|
| (MHz) | ||||||
| wild-type a | 2.437 | 2.355 | 2.061 | 186a | 133 | 340 |
| H106A b | 2.435 | 2.354 | 2.064 | 186 | 130 | 330 |
| D104A | 2.396 | 2.349 | 2.065 | 125 | 125 | 333 |
| Y105A | 2.282 | 2.250 | 2.063 | 9 | 11 | 235 |
Simulation of the wild-type spectrum better matched the experimental when an A-tensor strain of [48.6 17 0] (MHz) was included.
Simulation of the H106A variant was best fit with an A-tensor strain of [30 0 0] (MHz).
To determine if the DYH motif is in the vicinity of the cofactor, the residues in this motif were mutated to Ala, the purified proteins were reconstituted with cobalamin, iron and sulfide and EPR spectra of each variant were recorded (Fig. 6C, E, G). The cobalamin content of the variants was quantified by cyanolysis to be 0.85–1.2 per monomer. The spectra of the wild-type and H106A variant are nearly identical, and can be simulated with very similar sets of parameters (Table 2). The spectrum of the D104A variant, however, shows subtle differences with the wild-type that are best captured in simulations where nearly axial g-and A-tensors are included. By contrast, there is a very dramatic change in the spectrum of the Y105A variant relative to wild-type, whereby there is significantly less g-tensor anisotropy and A-tensors. The differences between the wild-type, D104A, H106A, and Y105A variants are reminiscent of the observations with the H759G variant of cobalamin-dependent methionine synthase (MetH). In MetH, binding of the cobalamin leads to interchange of the dimethylbenzimidazole for the imidazole sidechain of the His759 residue57. Mutation of the His residue reveals two sets of EPR cobalamin spectra, one of which corresponds to a cob(II)alamin interacting with a water molecule (5-coordinate), and another which is essentially 4-coordinate. The EPR spectra of wild-type QueG and all the variants reported here are consistent with 5-coordinate geometry around the cobalamin, but the differences in the g- and A-tensors suggest that in all but the Y105A variant, the interaction between the putative water ligand and the cobalamin is weakened63.
In addition to the residues in the conserved DYH motif, we also examined the effect of mutating the absolutely conserved D134 to A on the EPR spectrum of the variant. Interestingly, the EPR spectrum of this variant (Fig. 6I) exhibits features of both the wild-type and Y105A variants. We did not attempt to simulate the complex pattern; however, the spectrum of the protein is reasonably reproduced (Fig. 6J) by mathematically adding experimental spectra of the wild-type and Y105A proteins, suggesting that the cobalamin in this variant exists as a 50:50 mixture of the forms that are present in the wild-type and Y105A proteins.
The molecular mechanisms by which these conserved residues modulate the environment of the cobalamin remain to be established by structural methods, and we cannot exclude the possibility that they are distant from the active site. However, taken in the context of the in vivo analysis of oQ to Q ratios in a ΔqueG knockout strain, it seems likely that these residues are in the active site of the protein.
CONCLUSIONS
The identity of QueG, which catalyzes the final step in the biosynthesis of queuosine, was surprising in that it revealed the protein to be homologous to reductive dehalogenases. In this paper we demonstrate that despite the drastic differences in the reactions catalyzed by these enzymes, QueG and RDHs utilize the same set of cofactors to accomplish their respective chemical transformations. Reconstitution experiments with QueG demonstrate that iron-sulfur clusters and cobalamin are required for function. This data establishes a direct link between cobalamin and RNA modification for the first time since the feeding experiments noted a role for cobalamin in oQ reduction in vivo42. Based on the similarity of the EPR spectroscopic parameters to MetH61–63, the cobalamin cofactor in QueG is bound in a base-off conformation but is 5-coordinate, presumably by interactions with a ligand, such as water, that does not have a nuclear spin. Intriguingly, the conserved DYH motif does not provide an imidazole sidechain to coordinate the cofactor. However, the significant perturbations of the electronic environment of the cobalamin observed in these variants suggest that these residues are in the active site of the protein. Therefore, our findings place QueG in the class of enzymes that displace the dimethylbenzimidazole of the cobalamin cofactor upon binding to the protein. Understanding the mechanism of QueG, the role of the conserved residues in activity, and the correlations between the reactions catalyzed by QueG and RDHs must await high-resolution structures of QueG.
Supplementary Material
Acknowledgments
FUNDING
Research reported in this publication was supported by National Institutes of General Medical Sciences of the National Institutes of Health under award number R01GM72623 (V.B.). Z.D.M. was supported by the Integrative Graduate Education and Research Traineeship Program in Comparative Genomics from the NSF (0654435) and a Biological Chemistry Training grant from the NIH (T32 GM008804). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
ABBREVIATIONS
- EPR
electron paramagnetic resonance
- oQ
epoxyqueuosine
- Q
queuosine
- QueG
epoxyqueuosine reductase
- RDH
reductive dehalogenase
Footnotes
The sequence of the codon optimized gene and the sequences of the primers used for constructing the site-directed variants are reported in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, Helm M, Bujnicki JM, Grosjean H. MODOMICS: a database of RNA modification pathways--2013 update. Nucleic Acids Res. 2012;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Urbonavičius J, Meškys R, Grosjean H. Biosynthesis of wyosine derivatives in tRNA(Phe) of Archaea: role of a remarkable bifunctional tRNA(Phe):m1G/imG2 methyltransferase. RNA. 2014;20:747–753. doi: 10.1261/rna.043315.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young AP, Bandarian V. Radical mediated ring formation in the biosynthesis of the hypermodified tRNA base wybutosine. Curr Opin Chem Biol. 2013;17:613–618. doi: 10.1016/j.cbpa.2013.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El Yacoubi B, Bailly M, de Crécy-Lagard V. Biosynthesis and Function of Posttranscriptional Modifications of Transfer RNAs. Annu Rev Genet. 2012;46:69–95. doi: 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- 5.Jackman JE, Alfonzo JD. Transfer RNA modifications: nature’s combinatorial chemistry playground. WIREs RNA. 2013;4:35–48. doi: 10.1002/wrna.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia G, Fu Y, He C. Reversible RNA adenosine methylation in biological regulation. Trends in Genet. 2013;29:108–115. doi: 10.1016/j.tig.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee M, Kim B, Kim VN. Emerging Roles of RNA Modification: m6A and U-Tail. Cell. 2014;158:980–987. doi: 10.1016/j.cell.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Katze JR, Basile B, McCloskey JA. Queuine, a modified base incorporated posttranscriptionally into eukaryotic transfer RNA: wide distribution in nature. Science. 1982;216:55–56. doi: 10.1126/science.7063869. [DOI] [PubMed] [Google Scholar]
- 9.McCarty RM, Bandarian V. Biosynthesis of pyrrolopyrimidines. Bioorg Chem. 2012;43:15–25. doi: 10.1016/j.bioorg.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slany RK, Bösl M, Crain PF, Kersten H. A New Function of S-Adenosylmethionine: The Ribosyl Moiety of AdoMet Is the Precursor of the Cyclopentenediol Moiety of the tRNA Wobble Base Queuine. Biochemistry. 1993;32:7811–7817. doi: 10.1021/bi00081a028. [DOI] [PubMed] [Google Scholar]
- 11.Nishimura S. Structure, Biosynthesis, and Function of Queuosine in Transfer-Rna. Prog Nucleic Acid Res Mol Biol. 1983;28:49–73. doi: 10.1016/s0079-6603(08)60082-3. [DOI] [PubMed] [Google Scholar]
- 12.Miles ZD, Roberts SA, McCarty RM, Bandarian V. Biochemical and Structural Studies of 6-Carboxy-5,6,7,8-tetrahydropterin Synthase Reveal the Molecular Basis of Catalytic Promiscuity within the Tunnel-fold Superfamily. J Biol Chem. 2014;289:23641–23652. doi: 10.1074/jbc.M114.555680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarty RM, Somogyi A, Bandarian V. Escherichia coli QueD Is a 6-Carboxy-5,6,7,8-tetrahydropterin Synthase. Biochemistry. 2009;48:2301–2303. doi: 10.1021/bi9001437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okada N, Nishimura S. Isolation and characterization of a guanine insertion enzyme, a specific tRNA transglycosylase, from Escherichia coli. J Biol Chem. 1979;254:3061–3066. [PubMed] [Google Scholar]
- 15.Okada N, Noguchi S, Kasai H, Shindo-Okada N, Ohgi T, Goto T, Nishimura S. Novel mechanism of post-transcriptional modification of tRNA. Insertion of bases of Q precursors into tRNA by a specific tRNA transglycosylase reaction. J Biol Chem. 1979;254:3067–3073. [PubMed] [Google Scholar]
- 16.Phillips G, El Yacoubi B, Lyons B, Alvarez S, Iwata-Reuyl D, de Crécy-Lagard V. Biosynthesis of 7-Deazaguanosine-Modified tRNA Nucleosides: a New Role for GTP Cyclohydrolase I. J Bacteriol. 2008;190:7876–7884. doi: 10.1128/JB.00874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Lanen SG, Reader JS, Swairjo MA, de Crécy-Lagard V, Lee B, Iwata-Reuyl D. From cyclohydrolase to oxidoreductase: Discovery of nitrile reductase activity in a common fold. 2005;102:4264–4269. doi: 10.1073/pnas.0408056102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slany RK, Bösl M, Kersten H. Transfer and isomerization of the ribose moiety of AdoMet during the biosynthesis of queuosine tRNAs, a new unique reaction catalyzed by the QueA protein from Escherichia coli. Biochimie. 1994;76:389–393. doi: 10.1016/0300-9084(94)90113-9. [DOI] [PubMed] [Google Scholar]
- 19.McCarty RM, Bandarian V. Deciphering deazapurine biosynthesis: pathway for pyrrolopyrimidine nucleosides toyocamycin and sangivamycin. Chem Biol. 2008;15:790–798. doi: 10.1016/j.chembiol.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarty RM, Krebs C, Bandarian V. Spectroscopic, steady-state kinetic, and mechanistic characterization of the radical SAM enzyme QueE, which catalyzes a complex cyclization reaction in the biosynthesis of 7-deazapurines. Biochemistry. 2013;52:188–198. doi: 10.1021/bi301156w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reader JS, Metzgar D, Schimmel P, de Crécy-Lagard V. Identification of Four Genes Necessary for Biosynthesis of the Modified Nucleoside Queuosine. J Biol Chem. 2003;279:6280–6285. doi: 10.1074/jbc.M310858200. [DOI] [PubMed] [Google Scholar]
- 22.Miles ZD, McCarty RM, Molnar G, Bandarian V. Discovery of epoxyqueuosine (oQ) reductase reveals parallels between halorespiration and tRNA modification. Proc Natl Acad Sci U S A. 2011;108:7368–7372. doi: 10.1073/pnas.1018636108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reyniers JP, Pleasants JR, Wostmann BS, Katze JR, Farkas WR. Administration of exogenous queuine is essential for the biosynthesis of the queuosine-containing transfer RNAs in the mouse. J Biol Chem. 1981;256:11591–11594. [PubMed] [Google Scholar]
- 24.Aytaç U, Gündüz U. Q-Modification of Transfer-Rnas in Human Brain-Tumors. Cancer Biochem Biophys. 1994;14:93–98. [PubMed] [Google Scholar]
- 25.Baranowski W, Dirheimer G, Jakowicki JA, Keith G. Deficiency of Queuine, a Highly Modified Purine Base, in Transfer RNAs from Primary and Metastatic Ovarian Malignant Tumors in Women. Cancer Res. 1994;54:4468–4471. [PubMed] [Google Scholar]
- 26.Emmerich B, Zubrod E, Weber H, Maubach PA, Kersten H, Kersten W. Relationship of queuine-lacking transfer RNA to the grade of malignancy in human leukemias and lymphomas. Cancer Res. 1985;45:4308–4314. [PubMed] [Google Scholar]
- 27.Huang BS, Wu RT, Chien KY. Relationship of the queuine content of transfer ribonucleic acids to histopathological grading and survival in human lung cancer. Cancer Res. 1992;52:4696–4700. [PubMed] [Google Scholar]
- 28.Katze JR, Beck WT. Administration of queuine to mice relieves modified nucleoside queuosine deficiency in Ehrlich ascites tumor tRNA. Biochem Biophys Res Commun. 1980;96:313–319. doi: 10.1016/0006-291x(80)91216-4. [DOI] [PubMed] [Google Scholar]
- 29.Durand JM, Okada N, Tobe T, Watarai M, Fukuda I, Suzuki T, Nakata N, Komatsu K, Yoshikawa M, Sasakawa C. vacC, a virulence-associated chromosomal locus of Shigella flexneri, is homologous to tgt, a gene encoding tRNA-guanine transglycosylase (Tgt) of Escherichia coli K-12. J Bacteriol. 1994;176:4627–4634. doi: 10.1128/jb.176.15.4627-4634.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Durand JM, Dagberg B, Uhlin BE, Björk GR. Transfer RNA modification, temperature and DNA superhelicity have a common target in the regulatory network of the virulence of Shigella flexneri: the expression of the virF gene. Mol Microbiol. 2000;35:924–935. doi: 10.1046/j.1365-2958.2000.01767.x. [DOI] [PubMed] [Google Scholar]
- 31.Marchetti M, Capela D, Poincloux R, Benmeradi N, Auriac MC, Le Ru A, Maridonneau-Parini I, Batut J, Masson-Boivin C. Queuosine Biosynthesis Is Required for Sinorhizobium meliloti-Induced Cytoskeletal Modifications on HeLa Cells and Symbiosis with Medicago truncatula. PLoS ONE. 2013;8:e56043. doi: 10.1371/journal.pone.0056043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marks T, Farkas WR. Effects of a Diet Deficient in Tyrosine and Queuine on Germfree Mice. Biochem Biophys Res Commun. 1997;230:233–237. doi: 10.1006/bbrc.1996.5768. [DOI] [PubMed] [Google Scholar]
- 33.Holliger C, Wohlfarth G, Diekert G. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol Rev. 1999;22:383–398. [Google Scholar]
- 34.Smidt H, de Vos WM. Anaerobic Microbial Dehalogenation. Annu Rev Microbiol. 2004;58:43–73. doi: 10.1146/annurev.micro.58.030603.123600. [DOI] [PubMed] [Google Scholar]
- 35.Maillard J, Schumacher W, Vazquez F, Regeard C, Hagen WR, Holliger C. Characterization of the Corrinoid Iron-Sulfur Protein Tetrachloroethene Reductive Dehalogenase of Dehalobacter restrictus. Appl Environ Microbiol. 2003;69:4628–4638. doi: 10.1128/AEM.69.8.4628-4638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller E, Wohlfarth G, Diekert G. Purification and characterization of the tetrachloroethene reductive dehalogenase of strain PCE-S. Arch Microbiol. 1998;169:497–502. doi: 10.1007/s002030050602. [DOI] [PubMed] [Google Scholar]
- 37.Reinhold A, Westermann M, Seifert J, von Bergen M, Schubert T, Diekert G. Impact of Vitamin B12 on Formation of the Tetrachloroethene Reductive Dehalogenase in Desulfitobacterium hafniense Strain Y51. Appl Environ Microbiol. 2012;78:8025–8032. doi: 10.1128/AEM.02173-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schumacher W, Holliger C, Zehnder AJ, Hagen WR. Redox chemistry of cobalamin and iron-sulfur cofactors in the tetrachloroethene reductase of Dehalobacter restrictus. FEBS Lett. 1997;409:421–425. doi: 10.1016/s0014-5793(97)00520-6. [DOI] [PubMed] [Google Scholar]
- 39.van de Pas BA, Smidt H, Hagen WR, van der Oost J, Schraa G, Stams AJM, de Vos WM. Purification and Molecular Characterization of ortho-Chlorophenol Reductive Dehalogenase, a Key Enzyme of Halorespiration in Desulfitobacterium dehalogenans. J Biol Chem. 1999;274:20287–20292. doi: 10.1074/jbc.274.29.20287. [DOI] [PubMed] [Google Scholar]
- 40.Bommer M, Kunze C, Fesseler J, Schubert T, Diekert G, Dobbek H. Structural basis for organohalide respiration. Science. 2014;346:455–458. doi: 10.1126/science.1258118. [DOI] [PubMed] [Google Scholar]
- 41.Payne KAP, Quezada CP, Fisher K, Dunstan MS, Collins FA, Sjuts H, Levy C, Hay S, Rigby SEJ, Leys D. Reductive dehalogenase structure suggests a mechanism for B12-dependent dehalogenation. Nature. 2014;517:513–516. doi: 10.1038/nature13901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frey B, McCloskey JA, Kersten W, Kersten H. New Function of Vitamin B12: Cobamide-Dependent Reduction of Epoxyqueuosine to Queuosine in tRNAs of Escherichia coli and Salmonella typhimurium. J Bacteriol. 1988;170:2078–2082. doi: 10.1128/jb.170.5.2078-2082.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beinert H. Semi-micro Methods for Analysis of Labile Sulfide and of Labile Sulfide plus Sulfane Sulfur in Unusually Stable Iron-Sulfur Proteins. Anal Biochem. 1983;131:373–378. doi: 10.1016/0003-2697(83)90186-0. [DOI] [PubMed] [Google Scholar]
- 44.Ljungdahl LG, LeGall J, Lee JP. Isolation of a protein containing tightly bound 5-methoxybenzimidazolylcobamide (factor IIIm) from Clostridium thermoaceticum. Biochemistry. 1973;12:1802–1808. doi: 10.1021/bi00733a022. [DOI] [PubMed] [Google Scholar]
- 45.Krasotkina J, Walters T, Maruya KA, Ragsdale SW. Characterization of the B12- and Iron-Sulfur-containing Reductive Dehalogenase from Desulfitobacterium chlororespirans. J Biol Chem. 2001;276:40991–40997. doi: 10.1074/jbc.M106217200. [DOI] [PubMed] [Google Scholar]
- 46.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:1–11. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 48.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoll S, Schweiger A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J Magn Reson. 2006;178:42–55. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 50.Grove TL, Radle MI, Krebs C, Booker SJ. Cfr and RlmN Contain a Single [4Fe-4S] Cluster, which Directs Two Distinct Reactivities for S-Adenosylmethionine: Methyl Transfer by SN2 Displacement and Radical Generation. J Am Chem Soc. 2011;133:19586–19589. doi: 10.1021/ja207327v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jarrett JT, Amaratunga M, Drennan CL, Scholten JD, Sands RH, Ludwig ML, Matthews RG. Mutations in the B12-Binding Region of Methionine Synthase: How the Protein Controls Methylcobalamin Reactivity. Biochemistry. 1996;35:2464–2475. doi: 10.1021/bi952389m. [DOI] [PubMed] [Google Scholar]
- 52.Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neumann A, Wohlfarth G, Diekert G. Tetrachloroethene Dehalogenase from Dehalospirillum multivorans: Cloning, Sequencing of the Encoding Genes, and Expression of the pceA Gene in Escherichia coli. J Bacteriol. 1998;180:4140–4145. doi: 10.1128/jb.180.16.4140-4145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suyama A, Yamashita M, Yoshino S, Furukawa K. Molecular Characterization of the PceA Reductive Dehalogenase of Desulfitobacterium sp. Strain Y51. J Bacteriol. 2002;184:3419–3425. doi: 10.1128/JB.184.13.3419-3428.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hölscher T, Krajmalnik-Brown R, Ritalahti KM, von Wintzingerode F, Görisch H, Löffler FE, Adrian L. Multiple Nonidentical Reductive-Dehalogenase-Homologous Genes Are Common in Dehalococcoides. Appl Environ Microbiol. 2004;70:5290–5297. doi: 10.1128/AEM.70.9.5290-5297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drennan CL, Huang S, Drummond JT, Matthews RG, Ludwig ML. How a Protein Binds B12: A 3.0 Å X-ray Structure of B12-Binding Domains of Methionine Synthase. Science. 1994;266:1669–1674. doi: 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- 58.Marsh EN, Holloway DE. Cloning and sequencing of glutamate mutase component S from Clostridium tetanomorphum. Homologies with other cobalamin-dependent enzymes. FEBS Lett. 1992;310:167–170. doi: 10.1016/0014-5793(92)81321-c. [DOI] [PubMed] [Google Scholar]
- 59.Gruber K, Reitzer R, Kratky C. Radical Shuttling in a Protein: Ribose Pseudorotation Controls Alkyl-Radical Transfer in the Coenzyme B12 Dependent Enzyme Glutamate Mutase. Angew Chem Int Ed. 2001;40:3377–3380. doi: 10.1002/1521-3773(20010917)40:18<3377::aid-anie3377>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 60.Kowal AT, Werth MT, Mandori A, Cecchini G, Schroder I, Gunsalus RP, Johnson MK. Effect of Cysteine to Serine Mutations on the Properties of the [4Fe-4S] Center in Escherichia coli Fumarate Reductase. Biochemistry. 1995;34:12284–12293. doi: 10.1021/bi00038a024. [DOI] [PubMed] [Google Scholar]
- 61.Liptak MD, Fleischhacker AS, Matthews RG, Brunold TC. Probing the Role of the Histidine 759 Ligand in Cobalamin-Dependent Methionine Synthase. Biochemistry. 2007;46:8024–8035. doi: 10.1021/bi700341y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liptak MD, Fleischhacker AS, Matthews RG, Telser J, Brunold TC. Spectroscopic and Computational Characterization of the Base-off Forms of Cob(II)alamin. J Phys Chem B. 2009;113:5245–5254. doi: 10.1021/jp810136d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liptak MD, Datta S, Matthews RG, Brunold TC. Spectroscopic Study of the Cobalamin-Dependent Methionine Synthase in the Activation Conformation: Effects of the Y1139 Residue and S-Adenosylmethionine on the B12 Cofactor. J Am Chem Soc. 2008;130:16374–16381. doi: 10.1021/ja8038129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.