Abstract
TYW1 catalyzes the formation of 4-demethylwyosine via the condensation of N-methylguanosine (m1G) with carbons 2 and 3 of pyruvate. In this study labeled transfer ribonucleic acid (tRNA) and pyruvate were utilized to determine the site of hydrogen atom abstraction and regiochemistry of the pyruvate addition. tRNA containing a 2H labeled m1G methyl group was used to identify the methyl group of m1G as the site of hydrogen atom abstraction by S-adenosyl-L-methionine. [2-13C1,3,3,3-2H3]-Pyruvate was used to demonstrate retention of all the pyruvate protons indicating that C2 of pyruvate forms the bridging carbon of the imidazoline ring and C3 the methyl.
In the 58 years since the discovery of transfer RNA (tRNA)1 over one hundred modifications of the four canonical RNA bases have been identified that range in complexity from simple methylations of heteroatoms to the hypermodified bases queosine and wybutosine (yW)2.
yW and wyosine derivatives are modifications of a genetically encoded guanosine at position 37 of Phe encoding tRNA in eukaroytes and archaea3. The unique tricyclic core of 4-demethylwyosine (imG-14) is both an intermediate in the biosynthesis of more complex wyosine derivatives in eukaryotes and archaea and a RNA base in archaea4. To date, 8 structural homologs of imG-14 have been identified2.
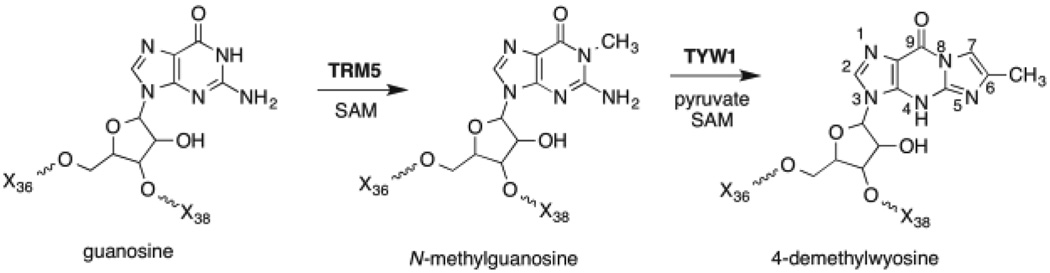
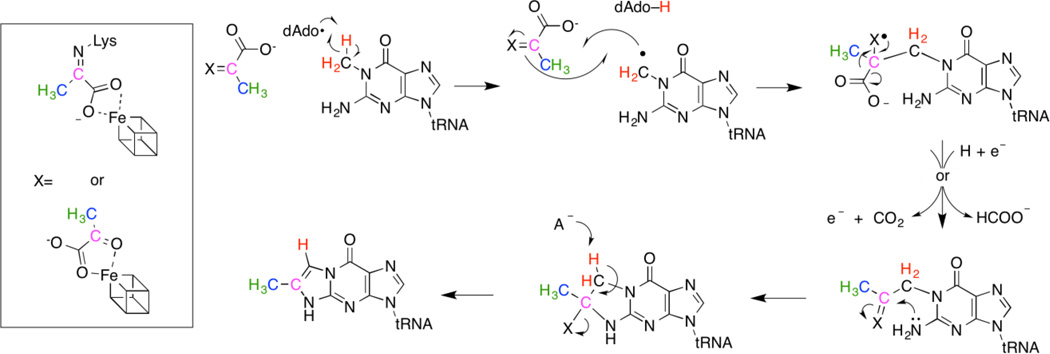
The biosynthesis of imG-14 requires the successive actions of TRM5 and TYW15–7, as shown in Scheme 1. TRM5 is a class I S-adenosyl-l-methionine (SAM) dependent methyl transferase that methylates N1 of guanosine 37 to produce N-methylguanosine (m1G)8,9. The key step involved in the formation of the tricyclic imG-14 core is catalyzed by TYW1, which converts m1G to imG-14 by adding two carbons that are derived from pyruvate, creating the imidazoline ring10. Additional species-specific modifications tailor the core wyosine base to form the known wyosine derivatives4,6,11,12.
Scheme 1.
The biosynthesis of imG-14.
TYW1 was identified as a member of the radical SAM superfamily on the basis of a characteristic CxxxCxxC motif13. The three Cys sidechains of this motif in radical SAM enzymes coordinate three irons of a cubane [4Fe-4S] cluster. SAM is coordinated to the fourth, or unique, iron via its amino and carboxylate groups14,15. Upon reduction of the +2 resting state of the cluster to the +1 state, the cluster reductively cleaves SAM forming the high energy 5′-deoxyadenosyl radical (dAdo•) and methionine. The dAdo• abstracts a hydrogen atom from a substrate to produce an intermediate that undergoes transformations culminating in products16. In some members of the radical SAM superfamily, the cofactor reforms at the end of the catalytic cycle whereas in others SAM is used stoichiometricaly17.
While two X-ray crystal structures of TYW1 are available, neither show electron density for the expected radical SAM [4Fe-4S] cluster18,19. In addition to the three Cys residues in the CxxxCxxC motif, three additional Cys residues are also found in the presumed active site clustered across from the radical SAM cluster binding site. These residues have been proposed to coordinate a second cluster. Electron paramagnetic resonance studies have confirmed the presence of a second [4Fe-4S] cluster20.
In a previous publication, we demonstrated that pyruvate is the source of two carbons in the imidazoline ring, the third being derived from the methyl of m1G10. However, significant gaps in our understanding of the mechanism of TYW1 remain. Herein we utilize isotopically labeled substrates to demonstrate that dAdo• is directly involved in H-atom abstraction from the substrate and that the source of C6 and the C6 methyl of the imidazoline ring are derived from carbons 2 and 3 of pyruvate respectively (see Scheme 1 for numbering).
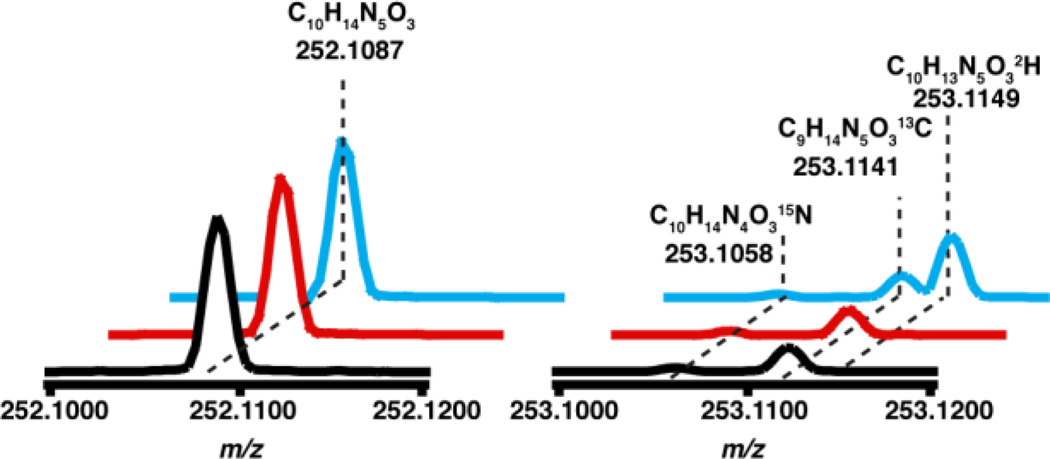
To examine if dAdo•, resulting from the reductive cleavage of SAM, abstracts a H-atom from m1G to initiate the catalytic cycle we synthesized tRNAPhe that contained either unlabeled or deuterated m1G and followed H-transfer to dAdo. The tRNAPhe substrate for these experiments, encoded by the M. jannaschii gene (MJ-t16), was produced by in vitro transcription and treated with TRM5 and SAM to introduce the m1G moiety. Unlabeled (CH3-SAM) or SAM containing a deuterated methyl group (C2H3-SAM) were used to modify the tRNA producing the corresponding protiated or deuterated m1G. The synthetic tRNA substrates were incubated with TYW1 in the presence of pyruvate, and dAdo produced under these conditions was analyzed by LC-MS. The ThermoFisher Orbitrap XL mass spectrometer employed in these studies has mass accuracy of <5 ppm and sufficient resolution to differentiate isotopic content of dAdo. Under the conditions of the experiment, dAdo elutes at 43 min (Fig. S1) and is readily assigned by comparison of retention time and mass spectra to the commercially obtained dAdo. The mass spectrum of dAdo obtained from this analysis is shown in Fig. 1. The peak at m/z 252.1087 corresponds to [M+H+] of dAdo (C10H14N5O3), and the measured mass is within 4 ppm of the theoretical value (m/z 252.1097). The isotope envelope at +1 m/z shows resolved peaks due to 15N, 13C, or 2H, which are present at 1.8, 10.8, and 0.1% of natural abundance relative to the molecular ion. The black trace in Fig. 1 shows the mass spectrum of dAdo standard and shows peaks corresponding to the 15N isotope at m/z 253.1058, the 13C isotope at m/z 253.1141, and a barely discernible 2H isotope peak at m/z 253.1149. Therefore, the mass spectrum will readily allow deuterium transfer to dAdo to be measured sensitively. The obvious advantage of the Orbitrap detector over more traditional mass spectrometry instrumentation is the isotopic resolution, which allows for even small quantities of deuterium transfer to be detected.
Figure 1.
The mass spectrum of dAdo produced in the presence of deuterated substrate( ), protiated sub-strate(
), protiated sub-strate( ), and standard dAdo (
), and standard dAdo ( ).
).
The mass spectra of dAdo produced when TYW1 was incubated in the presence of either protiated or deuterated tRNA and substoichiometric SAM are shown in Fig. 1, with the traces normalized to the [M+H+] peak at m/z 252.1087. The red and blue traces correspond to the reaction performed with protiated or deuterated tRNA. At m/z 253.1149 there is a large peak present in the dAdo produced in the presence of deuterated tRNA that is not present when protiated tRNA is used or in the standard. dAdo containing one deuterium (C10H13N5O32H) has an expected m/z 253.1159, which corresponds to a difference of 4 ppm to the 253.1149 obtained in the experiment. The dAdo produced in the presence of deuterated tRNA has a 100-fold increase in the peak at m/z 253.1149 relative to that obtained with unlabeled SAM and that in the control sample. This peak corresponds to dAdo containing a single deuterium. In the presence of deuterated m1G, the peak is 28% of the isotope peak. This data is consistent with dAdo• directly abstracting a hydrogen atom from the methyl group of m1G to initiate catalysis.
One would expect that all of the dAdo produced in the presence of deuterated substrate to have a m/z of 253.1149 with no species at m/z 252.1097 corresponding to unlabeled dAdo. The large background of protiated dAdo is due to the abortive cleavage of SAM, wherein dAdo• abstracts a proton from a site other than the substrate. This phenomenon has been observed in nearly all radical SAM enzymes studied to date21. We opted to carry out the experiment with sub-stoichiometric SAM to reduce the background. There is no evidence for transfer of multiple deuteriums to dAdo, as we see no peak at 254.1222.
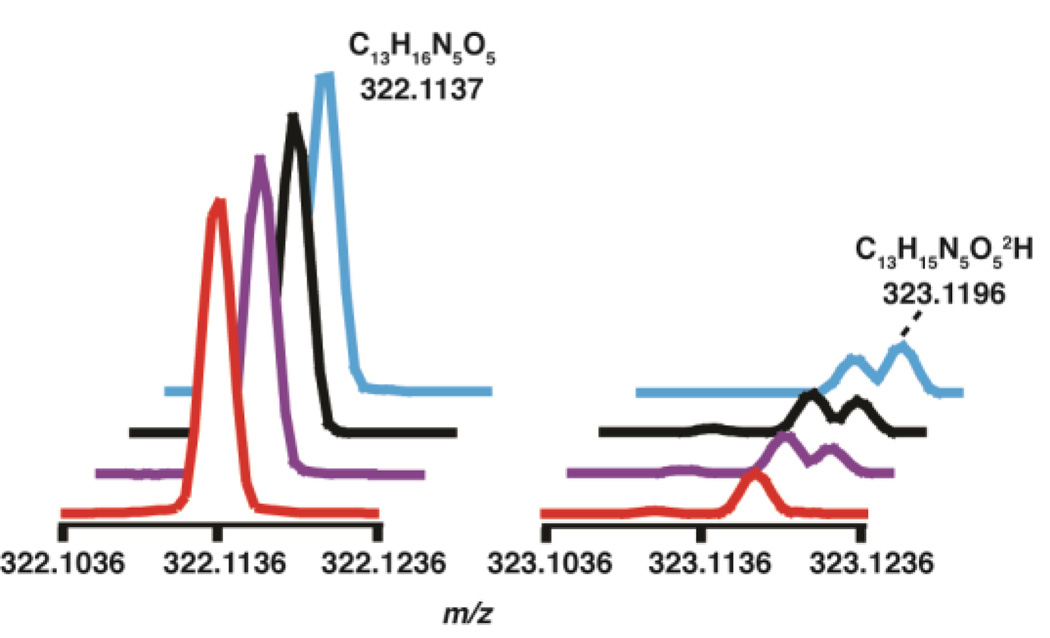
We next probed the fate of the remaining two protons on the methyl group of m1G. In these experiments, the modified tRNA produced in the reaction was digested enzymatically to nucleosides and analyzed by LC-MS. It was expected that one of the hydrogen atoms from the m1G methyl group would be retained in imG-14, surprisingly however the imG-14 produced with the labeled and unlabeled m1G gave rise to MS spectra with a peak at m/z 322.1137, as shown in Fig. 2, indicating that a deuterium was exchanged with a proton during the course of the experiment. To confirm that the starting substrate was fully deuterated, the mass spectra of the m1G digested to the nucleoside level from both the protiated and deuterated tRNA substrates were examined. The MS data clearly show that m1G is fully deuterated (see Fig. S.2–S.3.), suggesting that the absence of m1G deuteriums is either mechanistically relevant, or occurs as part of the workup.
Figure 2.
The mass spectrum of imG-14 produced in the presence of protiated substrate in H2O ( ), deuterated substrate in H2O (
), deuterated substrate in H2O ( ), protiated substrate in D2O (
), protiated substrate in D2O ( ), and deutertated substrate in D2O (
), and deutertated substrate in D2O ( ). The peak due to deuterium incorporation is labeled. The natural abundance 13C peak is visible at m/z 323.1168.
). The peak due to deuterium incorporation is labeled. The natural abundance 13C peak is visible at m/z 323.1168.
To determine if protons in imG-14 were exchanging with solvent, the TYW1 reaction was repeated in D2O. Fig. 2 shows the mass spectrum of imG-14 produced in both D2O and H2O with deuterated and protiated substrate, but worked up in H2O. The predominant peak in all four cases is at m/z 322.1137 corresponding to product with no deuterium. Interestingly, when the reaction is performed with deuterated substrate in D2O, we observe 30-fold enrichment of a species with a single deuterium relative to that observed when protiated tRNA is turned over in H2O. By contrast, when the reaction is performed in D2O using protiated tRNA there is a 22-fold increase in the peak at 323.1196. These observations support the notion that one of the protons from the starting m1G is retained in imG-14, but that there is significant exchange during workup of the reaction. Indeed, in a control experiment, where the reactions were worked up in D2O, the predominant imG-14 species has m/z of 323.1194, which corresponds to C13H15N5O52H (expected m/z 323.1215), indicating substantial proton exchange with solvent (Fig. S4). To pinpoint the site of exchange, we examined the mass spectrum of guanosine from in the same LC-MS runs. The guanosine in the sample also shows elevated monodeutration. However, the exchange does not appear to be to the same extent as with imG-14, as the predominant peak is that of unlabeled guanosine. Our interpretation of this result is that the substantial exchange observed in imG-14 occurs in the imidazoline ring, and not in the guanine-like core of the molecule. Taken together with the data presented in Fig. 2, our data suggest that one of the protons of m1G is retained in the final product.
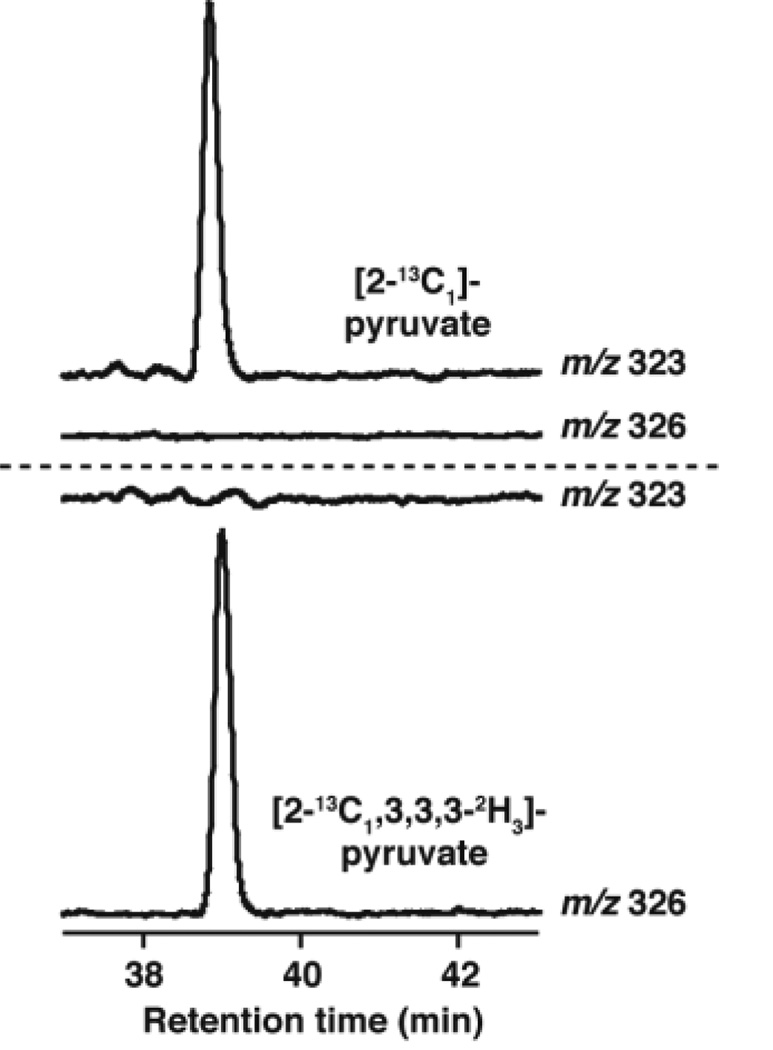
Isotope labeling experiments have demonstrated that C2 and C3 of pyruvate are utilized to form the new carbons that are required to form the imidazoline ring of imG-14. However, these experiments did not provide insights into the regiochemistry of incorporation. To address this we have carried out the TYW1 catalyzed reaction with [2-13C1, 3,3,3-2H3]- and [2-13C1]-pyruvate and analyzed the base produced in the incubation by LC-MS. As shown in Fig. 2, unlabeled imG-14 has a m/z of 322. In the presence of [2-13C1]-pyruvate, the mass of the product shifts to 323, whereas when [2-13C, 3,3,3-2H3]-pyruvate is used, the mass shifts to 326. Fig. 3 shows the extracted ion chromatograms at m/z 323 and 326 in the presence of both substrates. When [2-13C1, 3,3,3-2H3] pyruvate is used as the substrate the prominent peak is at m/z 326, which is consistent with conservation of all of the three deuteriums that are in the starting pyruvate. These data show that the methyl group of the imidazoline ring is formed from carbon 3 of pyruvate and that carbon 2 of pyruvate forms the bridging carbon, closing the imidazoline ring.
Figure 3.
The extracted ion chromatograms showing incorporation of deuterium into imG-14 from pyruvate
A possible mechanism for the transformation of m1G to imG-14 is shown in Scheme 2. The data in this manuscript clearly show that dAdo• abstracts a H-atom from the methyl of m1G. The radical inter mediate combines with pyruvate, which has variously been proposed to be activated by Schiff base formation to an absolutely conserved Lys10 or by interaction with a second [4Fe-4S] cluster, the presence of which has been inferred from spectroscopic measurements20. Oxidative or reductive cleavage of the intermediate and transimination lead to an intermediate, which eliminates the electrophilic center to form products.
Scheme 2.
A proposed mechanism for the formation of imG-14.
In summary this study has identified that hydrogen atom abstraction by dAdo• occurs at the methyl of m1G and established the regiochemistry of the pyruvate incorporation. The role of the iron-sulfur clusters and the fate of carbon 1 of pyruvate remain to be determined.
Supplementary Material
Acknowledgments
Funding Sources
Research reported in this publication was supported the National Institutes of Health under award number R01GM72623. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
ABBREVIATIONS
- tRNA
transfer ribonucleic acid
- yW
wybutosine
- imG-14
4-demethylwyosine
- SAM
S-adenosyl-L-methionine
- m1G
N-methylguanosine
- dAdo•
5′-deoxyadenosyl radical
- dAdo
5′-deoxyadenosine
Footnotes
ASSOCIATED CONTENT
The supporting information contains detailed Materials and Methods and additional figures. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
A.P.Y. and V.B. designed and conducted the experiments and wrote the manuscript.
REFERENCES
- 1.Hoagland MB, Zamecnik PC, Stephenson ML. BIOCHIMICA ET BIOPHYSICA ACTA. 1957;24:215–216. doi: 10.1016/0006-3002(57)90175-0. [DOI] [PubMed] [Google Scholar]
- 2.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FAP, Fabris D, Agris PF. Nucleic acids research. 2011;39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.RajBhandary UL, Chang SH. The Journal of Biological Chemistry. 1962;243:598–608. [PubMed] [Google Scholar]
- 4.De Crécy-Lagard V, Brochier-Armanet C, Urbonavicius J, Fernandez B, Phillips G, Lyons B, Noma A, Alvarez S, Droogmans L, Armengaud J, Grosjean H. Molecular biology and evolution. 2010;27:2062–2077. doi: 10.1093/molbev/msq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waas WF, Crécy-Lagard V, de, Schimmel P. Journal of Biological Chemistry. 2005;280:37616–37622. doi: 10.1074/jbc.M506939200. [DOI] [PubMed] [Google Scholar]
- 6.Noma A, Kirino Y, Ikeuchi Y, Suzuki T. The EMBO journal. 2006;25:2142–2154. doi: 10.1038/sj.emboj.7601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Droogmans L, Grosjean H. The EMBO journal. 1987;6:477–483. doi: 10.1002/j.1460-2075.1987.tb04778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christian T, Evilia C, Hou Y-M. Biochemsitry. 2006;45:7463–7473. doi: 10.1021/bi0602314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goto-Ito S, Ito T, Ishii R, Muto Y, Bessho Y, Yokoyama S. Proteins: Structure, Function, and Bioinformatics. 2008;72:1274–1289. doi: 10.1002/prot.22019. [DOI] [PubMed] [Google Scholar]
- 10.Young AP, Bandarian V. Biochemistry. 2011;50:10573–10575. doi: 10.1021/bi2015053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young AP, Bandarian V. Current opinion in chemical biology. 2013;17:613–618. doi: 10.1016/j.cbpa.2013.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perche-Letuvée P, Molle T, Forouhar F, Mulliez E, Atta M. RNA Biology. 2014;11:1508–1518. doi: 10.4161/15476286.2014.992271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Nucleic Acids Research. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berkovitch F, Nicolet Y, Wan JT, Jarrett JT, Drennan CL. Science (New York, N.Y.) 2004;303:76–79. doi: 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walsby CJ, Hong W, Broderick WE, Cheek J, Ortillo D, Broderick JB, Hoffman BM. Journal of the American Chemical Society. 2002;124:3143–3151. doi: 10.1021/ja012034s. [DOI] [PubMed] [Google Scholar]
- 16.Frey PA, Hegeman AD, Ruzicka FJ. Critical Reviews in Biochemistry and Molecular Biology. 2008;43:63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 17.Booker SJ. Current Opinion in Chemical Biology. 2009;13:58–73. doi: 10.1016/j.cbpa.2009.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki Y, Noma A, Suzuki T, Senda M, Senda T, Ishitani R, Nureki O. Journal of molecular biology. 2007;372:1204–1214. doi: 10.1016/j.jmb.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 19.Goto-Ito S, Ishii R, Ito T, Shibata R, Fusatomi E, Sekine S, Bessho Y, Yokoyama S. Acta crystallographica Section D, Biological Crystallography. 2007;D63:1059–1068. doi: 10.1107/S0907444907040668. [DOI] [PubMed] [Google Scholar]
- 20.Perche-Letuvée P, Kathirvelu V, Berggren G, Clemancey M, Latour J-M, Maurel V, Douki T, Armengaud J, Mulliez E, Fontecave M, Garcia-Serres R, Gambarelli S, Atta M. The Journal of biological chemistry. 2012;287:41174–41185. doi: 10.1074/jbc.M112.405019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grove TL, Benner JS, Radle MI, Ahlum JH, Landgraf BJ, Krebs C, Booker SJ. Science. 2011;332:604–607. doi: 10.1126/science.1200877. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.