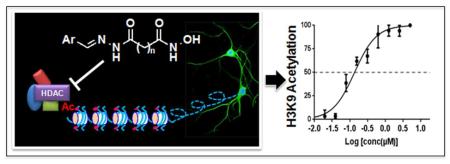
Abstract
Targeting chromatin-mediated epigenetic regulation has emerged as a potential avenue for developing novel therapeutics for a wide range of central nervous system disorders, including cognitive disorders and depression. Histone deacetylase (HDAC) inhibitors have been pursued as cognitive enhancers that impact the regulation of gene expression and other mechanisms integral to neuroplasticity. Through systematic modification of the structure of crebinostat, a previously discovered cognitive enhancer that affects genes critical to memory and enhances synaptogenesis, combined with biochemical and neuronal cell-based screening, we identified a novel hydroxamate-based HDAC inhibitor, here named neurinostat, with increased potency compared to crebinostat in inducing neuronal histone acetylation. In addition, neurinostat was found to have a pharmacokinetic profile in mouse brain modestly improved over that of crebinostat. This discovery of neurinostat and demonstration of its effects on neuronal HDACs adds to the available pharmacological toolkit for dissecting the molecular and cellular mechanisms of neuroepigenetic regulation in health and disease.
Keywords: Cognitive enhancer, nootropic, histone deacetylases, epigenetic, chromatin, acetylation, CREB, neuroepigenetic
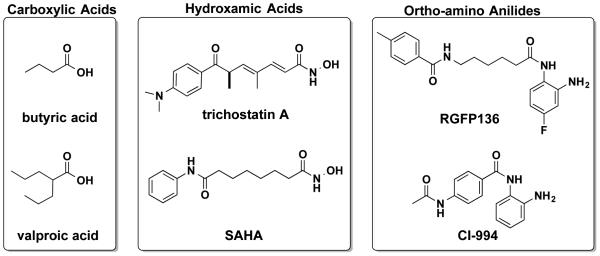
Chromatin-mediated epigenetic regulation of gene expression is an integral part of neuroplasticity [1], dysregulation of which is associated with aging and a broad range of central nervous system (CNS) disorders, including Alzheimer’s disease (AD) [2], and mood disorders [3, 4]. Inhibition of histone deacetylases (HDACs) has emerged as a promising new therapeutic avenue for CNS disorders [5, 6]. As summarized in Figure 1, there are three main chemical structure classes of HDAC inhibitors that have been developed as pharmacological probes with more recent use in the context of CNS disorders: 1) carboxylic acids (e.g., butyric acid, valproic acid); 2) hydroxamic acids (e.g., suberoylanilide hydroxamic acid (SAHA), trichostatin A (TSA)); and 3) ortho-amino anilides (e.g., RGFP136, CI-994). All three classes of HDAC inhibitors have been shown to enhance learning and memory in normal, aged, or age-associated neurodegenerative animal models, and also to increase histone acetylation and change gene expression related to neuroplasticity in relevant brain regions [7-10]. An anti-depressant-like effect has also been shown in preclinical settings in rodent models for several HDAC inhibitors including sodium butyrate [11, 12], SAHA and MS-275 [13], a HDAC1/2-selective inhibitor Cpd-60 [14], and HDAC6-selective inhibitors ACY-738 and ACY-775 [15].
Figure 1.
Three main chemical structure classes of HDAC inhibitors under investigation in the context of central nervous system disorders
While accumulating evidence indicates certain HDACs facilitate, while others play a suppressive role, in cognitive processes [2, 4, 5], the understanding of the precise relationship between HDAC subtype activity, critical epigenetic substrates, the activity of specific HDAC complexes, and therapeutically beneficial effects relevant to neurodegenerative and neuropsychiatric disorders remains incomplete. Thus, the development of potent and subtype-selective HDAC inhibitors that are functionally active toward relevant HDAC complexes and that can achieve sufficient brain exposure to be biochemically and behaviorally active is required [2, 16-18]. Toward this goal, by screening for enhancers of CREB-mediated transcription and modulators of chromatin-mediated neuroplasticity, our previous studies identified a potent hydroxamate HDAC inhibitor, crebinostat, which produced robust enhancement of cognition in mice [19]. Here, through systematic dissection of the structure-activity-relationship (SAR) of crebinostat, we identified an improved analog of crebinostat with enhanced cellular activity in inducing histone acetylation in mouse neuronal cultures and a modestly improved in vivo pharmacokinetic (PK) profile in mice.
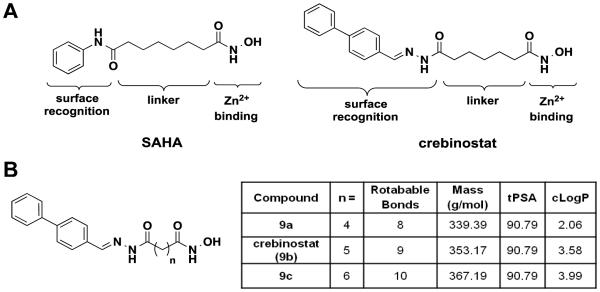
Structural studies of hydroxamate HDAC inhibitors such as SAHA and TSA have revealed three key features: a surface-recognition cap region that interacts with the HDAC enzyme surface adjacent to the catalytic site; a linker region that extends into the catalytic site; and a terminal metal-chelating hydroxamic acid moiety that interacts with the catalytic site zinc ion [20, 21] (Figure 2A). The cognitive enhancer crebinostat, discovered by a high-throughput, cell-based screen of small molecule libraries, shares this “cap-linker-zinc chelator” pharmacophore model [16] (Figure 2A). Structurally, although both are hydroxamic acids, crebinostat differs from SAHA in the cap moiety and length of the aliphatic linker. Functionally, crebinostat demonstrated improved activities over SAHA in in vitro assays and in neuronal cellular assays of histone acetylation [19].
Figure 2. Novel HDAC inhibitors with 4-, 5-, and 6-methylene linkers.
(A) Potent HDAC inhibitors SAHA and crebinostat possess three structural components: surface recognition cap, linker, and a divalent metal (Zn2+) binding element. (B) Novel HDAC inhibitors with 4-, 5-, and 6-methylene linkers and their physicochemical properties. tPSA: topological polar surface area. cLogP: calculated LogP.
Given interest in identifying novel HDAC inhibitors with improved potency, HDAC subtype selectivity and suitable brain PK properties, we synthesized a series of crebinostat derivatives. Crebinostat contains a 5-methylene chain linker while SAHA has a 6-methylene linker. The surface-recognition cap moieties are also different for crebinostat and SAHA. To dissect the structural composition that contributes to the advantageous properties in crebinostat, we first assessed the effect of linker length by synthesizing crebinostat analogs with 4-6 carbon aliphatic chain linkers (Figure 2B). This range of linker lengths was considered appropriate to explore considering the 5- and 6-methylene linker distance in the prototype hydroxamic inhibitors TSA and SAHA (Figure 1), the distance between cap and catalytic site observed in co-crystal structures of HDACs and inhibitors, and based on previous reports in the literature [22, 23]
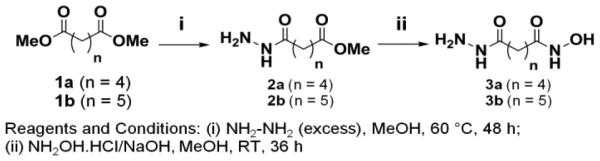
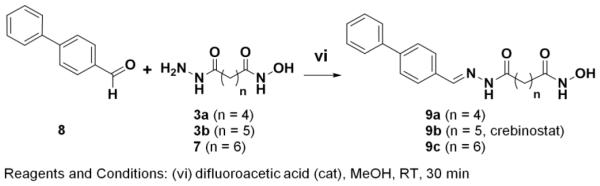
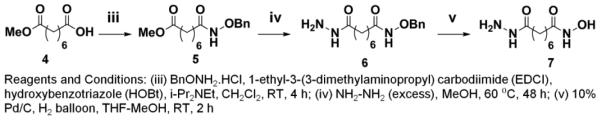
Synthesis of Compounds 9a, 9b and 9c
Chemical Schemes 1-3 describe the procedures used to synthesize three compounds (9a, 9b, 9c) with 4, 5, or 6-methylene linker length respectively, but with the same biphenyl cap and hydroxamate zinc-binding motifs. In order to obtain 9a, 9b, 9c, we synthesized intermediate linker compounds (3a, 3b, 7) terminated by a hydrazide on one side and a hydroxamic acid on the other side (Schemes 1, 2). Acyl hydrazones 9a, 9b and 9c were synthesized according to Scheme 3 by difluoroacetic acid catalyzed condensation of the biphenyl aldehyde (8) with hydrazides 3a, 3b and 7, respectively. Compound 9b corresponds to the previously characterized crebinostat. The physicochemical properties of synthesized compounds are shown in Figure 2B and Supplemental Table S1. Analytical LC-MS spectra for these compounds are included in Supplemental Figure S1.
Scheme 1.
Scheme 3.
Scheme 2.
Optimal Linker Length for HDAC Inhibitory Activities in Biochemical Assays
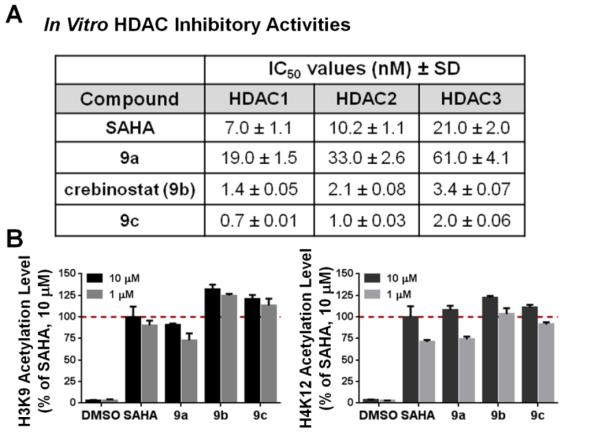
After having synthesized compounds 9a, 9b (crebinostat), and 9c, which share the same cap and zinc-binding elements, and differ in the linker length, we first determined the ability of these compounds to inhibit the enzymatic activity of HDAC1, 2 or 3. The inhibitory activities of compounds 9a, 9b, and 9c were measured in dose responses with the IC50 values reported in Figure 3A. Consistent with our previous report [19], crebinostat (9b) was a more potent inhibitor than SAHA of HDAC1, 2 or 3, with more than a 5-fold reduction in its IC50. 9c exhibited further reduced IC50’s; its IC50 values (0.7, 1.0 and 2.0 nM for HDAC1-3 respectively) are half of those measured for crebinostat (9b). Apparently, the 4-methylene linker conferred a disadvantage to the inhibitory activity; 9a exhibited an approximately 20-fold increase in IC50’s over those of crebinostat (9b). Presumably, the cap to chelator distance produced by the 4-methylene linker precluded correct positioning of the surface-recognition and zinc-interacting elements. These data, though no magnitude difference of IC50 values between 9c and crebinostat (9b), indicate that the 6-methylene linker length is optimal among the three compounds, with ~2 fold improved potency for inhibiting HDAC1-3 activities in the biochemical reaction. Previous work examining linker length of benzothiazole-containing analogs of SAHA reported that a 6-carbon linker showed better inhibition of HDAC3 and HDAC4 than a 4-carbon linker [22]. Studies involving thirty-six acylurea connected aliphatic linker hydroxamates identified close analogs of SAHA with 5-methylene linkers to be more potent than SAHA against HDAC1 or HDAC6 [23].
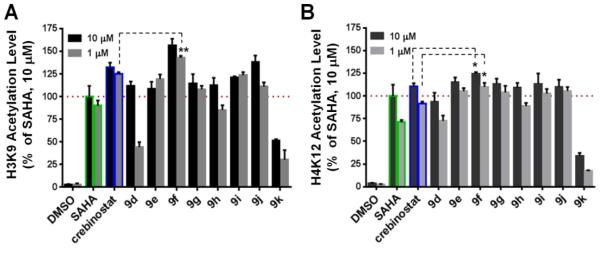
Figure 3. In vitro and ex vivo HDAC inhibitory activities of novel HDAC inhibitors with 4-, 5-, and 6-methylene linkers.
(A) In vitro IC50 (nM) with recombinant human HDAC1-3. (B) Increased acetylation of histone H3K9 and H4K12 in mouse forebrain primary neuronal culture (DIV14) detected at both 10 μM and 1 μM (24 hrs). Cellular activity of SAHA at 10 μM was set to 100% as a reference. N=3. Error bars are SEM.
For all three compounds, we did not observe significant isoform specific activities between HDAC1-3; inhibitory activities for HDAC1-3 were in the same magnitude for each compound albeit inhibitory activities for HDAC1 were about half of inhibitory activities for HDAC2, which were about half of inhibitory activities for HDAC3. Therefore, changing the linker length did not lead to HDAC1, 2 or 3 isoform selective inhibitory activities in this structural series.
Optimal Linker Length for HDAC Inhibitory Activities in Neuronal Assays
Given interest in developing novel HDAC inhibitors as therapeutics of CNS disorders, we examined cellular activities of our novel hydroxamate HDAC inhibitors in induction of neuronal histone acetylation [19, 24, 25]. After 24 hour compound treatments, neuronal cultures were fixed and immunostained for histone acetylation marks H3K9ac and H4K12ac. The acetylation levels of H3K9 and H4K12 have been implicated in cognition enhancement in normal and aged mice [8, 26, 27]. Inhibition of HDAC activities by inhibitors leads to elevated H3K9ac and H4K12ac fluorescence intensities in neuronal nuclei. To assess the HDAC inhibitory activities in neurons, we tested 9a, 9b (crebinostat) and 9c, as well as SAHA, at two concentrations: 1 and 10 μM, and their cellular activities are reported relative to those of SAHA (Figure 3B). Crebinostat (9b) and 9c treatment resulted in greater increases in both H3K9ac and H4K12ac than SAHA at both 1 μM and 10 μM. Despite the higher potencies measured for 9c containing the 6-methylene linker in the in vitro HDAC enzymatic assays described in the previous section, the cellular activities of 9b (crebinostat) were modestly greater than those of 9c, suggesting that the 5-methylene linker may make hydroxamate HDAC inhibitors with biphenyl moieties more accessible to endogenous HDAC complexes.
In vitro enzymatic assays assessed HDAC inhibitory activities of the reported compounds by direct enzyme engagement whereas the ex vivo, neuron-based functional assays enabled us to evaluate these compounds for additional properties such as cell membrane permeability and target accessibility. The degree of hydrophilicity/hydrophobicity and the size of small molecules can affect their diffusion into intact cells through membrane penetrance as well as their ability to reach target enzymes existing in multi-protein complexes. Crebinostat (9b) and 9c have the same topological polar surface area (tPSA), and crebinostat (9b) has slightly lower calculated LogP than 9c (Figure 2B), the latter of which may give crebinostat (9b) better cell membrane permeability, and therefore better cellular activity. Our results presented a case where 9c, containing a 6-methylene linker, was modestly more potent than crebinostat (9b), containing a 5-methylene linker, in the biochemical assay, but less potent in the cell based assay. Given the necessity for HDAC inhibitors to have strong intracellular activity in order to be effective when used in vivo, and that crebinostat (9b) is well characterized, the 5-methylene linker was chosen for further study of the structure-activity relationship of the crebinostat series.
All of our novel compounds are hydrazone derivatives, which are known to be labile in the cellular environment at lower pH [28]. To ensure that the cellular activities of induction of neuronal histone acetylation that we observed were from the intact molecule, but not from the fragmented linker with zinc-chelating moiety, we also tested the intermediate linker compounds 3a, 3b, and 7 in the neuronal histone acetylation assay (Supplemental Figure S2). No induction of histone acetylation of H3K9 or H4K12 was detected for 3a, 3b, and 7, indicating that the observed functional activities for 9a-c were from the intact synthesized compounds. As these linker-only compounds also contain hydroxamates that may chelate divalent metal ions, this also indicates that the presence of a chelating moiety alone on a linker is insufficient to cause a measurable change in histone acetylation.
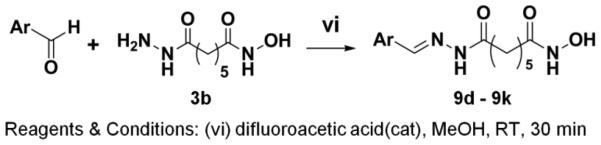
Synthesis of Compounds 9d-9k
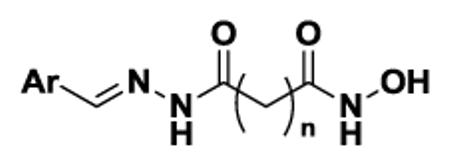
After evaluation of the effect of aliphatic linker length on HDAC inhibitory activities, and having identified the 5-methylene linker as most promising for further SAR studies, we proceeded to varying the surface-recognition caps for potentially improved potency or HDAC subtype selectivity. The biphenyl cap moiety in crebinostat (9b) was replaced with eight different substituted and hetero aromatic moieties (Table 1) that cover a wide range of calculated LogP from 1.26 to 4.06, as LogP is one of the key factors that determine the ability of small molecules to penetrate the cell membrane. Scheme 4 was followed to synthesize eight compounds (9d-9k) using the same strategy as in Scheme 3, in which various aldehydes were coupled with hydrazide 3b. Physicochemical properties of synthesized compounds are reported in Supplemental Tables S1 and S2, and analytical LC-MS spectra for key compounds are shown in Supplemental Figure S1.
Table 1.
In vitro HDAC inhibitory activities of crebinostat analogs.
|
|||||||
|---|---|---|---|---|---|---|---|
| IC50 values (nM) ± SD | |||||||
| Compound | Ar | n | tPSA | cLogP | HDAC1 | HDAC2 | HDAC3 |
| SAHA | 7.0 ± 1.10 | 10.2 ± 1.10 | 21.0 ± 2.00 | ||||
| 9b (crebinostat) |
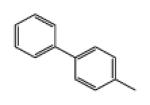
|
5 | 90.79 | 3.58 | 1.4 ± 0.05 | 2.1 ± 0.08 | 3.4 ± 0.07 |
| 9d |
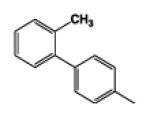
|
5 | 90.79 | 4.06 | 8.0 ± 1.01 | 18.8 ± 2.06 | 5.4 ± 0.07 |
| 9e |
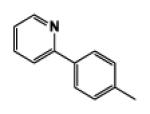
|
5 | 103.15 | 2.66 | 2.3 ± 0.05 | 6.9 ± 1.03 | 1.4 ± 0.03 |
| 9f |
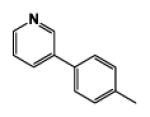
|
5 | 103.15 | 2.24 | 1.1 ± 0.01 | 10.8 ± 2.03 | 0.8 ± 0.01 |
| 9g |
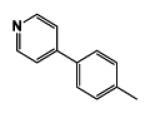
|
5 | 103.15 | 2.24 | 3.1 ± 0.02 | 14.2 ± 1.12 | 1.8 ± 0.02 |
| 9h |
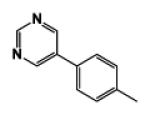
|
5 | 115.51 | 1.64 | 3.2 ± 0.06 | 21.0 ± 3.2 | 2.4 ± 0.04 |
| 9i |
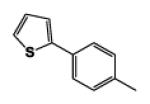
|
5 | 90.79 | 3.56 | 2.1 ± 0.03 | 6.3 ± 0.08 | 1.6 ± 0.02 |
| 9j |
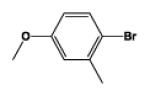
|
5 | 100.02 | 2.06 | 0.6 ± 0.01 | 5.0 ± 0.61 | 1.2 ± 0.03 |
| 9k |
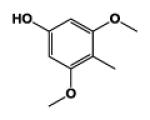
|
5 | 129.48 | 1.26 | 31.4 ± 3.21 | 108.0 ± 8.07 | 25.6 ± 2.33 |
Scheme 4.
Compound 9f Surpasses 9b (crebinostat) in Induction of Neuronal Histone Acetylation
All of the synthesized final analog compounds were tested in the in vitro HDAC1-3 inhibition assays and the results showed that most were potent with low nanomolar IC50 values (Table 1). All analogs except 9k maintained single-digit IC50’s against HDAC1 and 3, around the same magnitude as crebinostat (9b), while four out of seven of these analogs had decreased potency against HDAC2 to double-digit IC50’s. Compounds 9f and 9j surpassed crebinostat (9b) in inhibiting HDAC1 and 3, but both had decreased potency against HDAC2. Changing the terminal phenyl group of the biphenyl moiety of crebinostat structure to a pyridinyl group fine tuned HDAC inhibitory activities (9e, 9f, 9g and 9h). The pyridinyl nitrogen at meta position (9f) improved potency for HDAC1 and 3 inhibition. The result of 9j having improved potency against HDAC1 and 3, and comparable potency against HDAC2 is intriguing in that 9j contains 2-bromo-5-methoxy benzylidene group as a cap element, one of three cap moieties in the series least similar to the biphenyl group of crebinostat. Also interesting is that 9i, which has a thiophenyl phenyl group in the cap region, scored as a potent HDAC inhibitor as well. The least favorable substitution occurred in 9k, with a dimethoxyphenol moiety replacing the biphenyl group, resulting in dropped potency to double-digit IC50’s for HDACs 1 and 3, and a three-digit IC50 for HDAC2.
After evaluating HDAC inhibitory activities of cap-modified compounds in enzymatic assays, we again assessed their cellular activities of induction of neuronal histone acetylation using the ex vivo primary neuronal culture assay. As shown in Figure 4, all compounds except 9k showed cellular activities comparable to SAHA, with several exceeding SAHA’s activities, at both concentrations (with the exception of 9d which had low H3K9ac activity). 9k was the least potent in the neuronal cell assay, consistent with its relative low biochemical activities for HDAC1-3. While all three compounds (crebinostat (9b), 9f, 9j) were more effective in increasing both H3K9ac and H4K12ac than SAHA, 9f was found to be the most potent. The activity differences between 9f and crebinostat (9b) are statistically significant for H3K9ac at 1 μM and for H4K12ac at both 1 and 10 μM (n= 3, unpaired t-test, p<0.05). Based on these neuronal cell-based assay data, demonstrating 9f as the most improved analog of crebinostat (9b) identified in our series, 9f became the focus of additional studies, and we named it neurinostat.
Figure 4. Induction of histone H3K9 and H4K12 acetylation in mouse forebrain primary neuronal cultures by crebinostat analogs.
Neurons (DIV13) were treated at the indicated concentration (n=3) for 24 hrs and then imaged after immunofluorescence-based detection. Error bars are SEM.
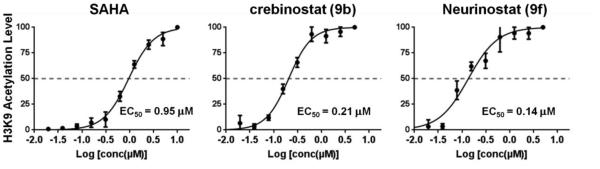
To better evaluate the cellular potency of neurinostat (9f), we determined its EC50 by treating neuronal cultures in a 10-point dose response (1 nM - 5 μM) and measuring the acetylation level of H3K9. Figure 5 shows that neurinostat (9f) was modestly more potent than SAHA and crebinostat (9b) in induction of neuronal histone acetylation; it had an EC50 of 140 nM, whereas crebinostat (9b) and SAHA had EC50s of 210 nM and 950 nM, respectively. The difference in the bioactivities of neurinostat (9f) versus crebinostat (9b) is statistically significant (comparison of 10-point dose curves, exact sum-of-squares F test, p=0.0006). These data indicate that the pyridinyl nitrogen at meta position in the terminal aromatic ring enhances HDAC inhibitory activities in neurons.
Figure 5. Dose response profiles of histone H3K9 acetylation in mouse forebrain primary neuronal cultures.
Neurons (DIV13) were treated at the indicated concentrations (n=2) for 24 hrs and then imaged after immunofluorescence-based detection. Error bars are SEM.
To comprehensively assess inhibitory activities of neurinostat (9f) toward different class categories of HDACs, we tested neurinostat (9f) in HDAC5 (class IIa) and HDAC6 (class IIb) in vitro assays. For both neurinostat (9f) and crebinostat (9b), no activity was observed toward HDAC5, whereas IC50 values were obtained as 0.5 nM and 0.6 nM, respectively, toward HDAC6 (Supplemental Figure S3). Collectively, our in vitro enzymatic assay data suggest that neurinostat (9f) and crebinostat (9b) have similar HDAC subtype selectivity, and that further SAR studies will be needed to yield any subtype selectivity within this chemical series.
Neurinostat (9f) Possesses Modestly Improved Pharmacokinetic Profiles and Brain Penetration
Given our intention to develop neurinostat (9f) as a cognitive enhancer, we proceeded to evaluate its brain PK properties. Neurinostat (9f) was administered intraperitoneally at 25 mg/kg to mice, and its concentration in the brain was monitored over a 6-point time course by collecting brains from three mice at each time point and measuring neurinostat (9f) concentrations in the processed brain samples. As shown in Figure 6 and Supplemental Table S3, neurinostat (9f) reached peak concentration (Cmax) of 79 nM at one hour after systemic administration of the compound. This Cmax exceeds the IC50 concentrations measured for HDAC1-3 in biochemical assays and at the comparable level of the EC50 concentration measured for H3K9ac in cultured mouse neurons. These data indicate that potentially functionally relevant levels of neurinostat (9f) were achieved in the brain after its systemic administration at 25 mg/kg. Though neurinostat’s Cmax was higher than that of crebinostat (9b) administered at the same dose, previously reported to be 60 nM [19], their PK profiles are similar.
Figure 6. Mouse brain pharmacokinetics of compound neurinostat (9f).
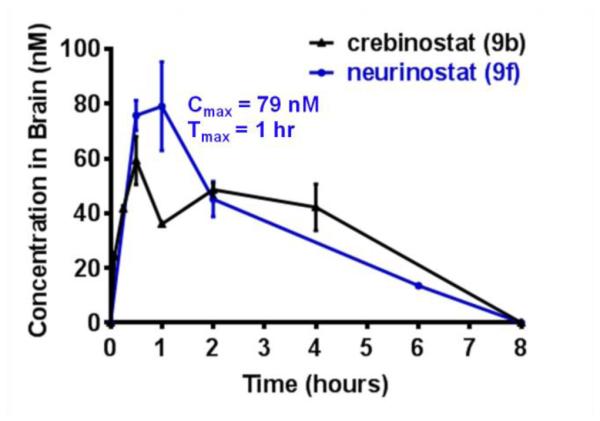
Neurinostat (9f) and Crebinostat (9b) (as published previously [19]) were injected intraperitoneally at 25 mg/kg. N=3 for each time point. Error bars are SEM.
In addition to brain tissue, peripheral blood samples were also collected at each time point and analyzed for neurinostat (9f) concentration (Supplemental Tables S4 and S5). Around one hour after injection, neurinostat (9f) reached a maximum concentration of 1.9 μM with a half life of 1.28 hours. Individual brain-to-plasma concentration ratios were between 0.03-0.07 from 0.5 to 2 hours (Supplemental Table S6). At time points 0.5, 1 and 2 hours, the brain-to-plasma ratios of neurinostat (9f) were about twice of those measured for crebinostat (9b), indicating that neurinostat (9f) was distributed to the brain more effectively than crebinostat (9b). The overall low ratios of brain-to-plasma concentrations suggest brain-penetrant properties of neurinostat (9f) and crebinostat (9b) require further improvement.
In pursuit of a better understanding of the role of HDACs in chromatin-mediated neuroplasticity and improved CNS disease therapeutics, we systematically dissected the SAR of the hydroxamate-based cognitive enhancer HDAC inhibitor crebinostat (9b), and optimized its linker length and surface recognition cap moieties. These SAR studies identified novel HDAC inhibitors that possess improved potency in HDAC inhibition, especially in functional neurons. The discovery of the neurinostat (9f), which exhibited nanomolar IC50 in HDAC1-3 inhibition, a lower EC50 for induction of neuronal histone acetylation than those of crebinostat (9b) and SAHA, and modestly improved brain PK properties compared to crebinostat (9b) provides a new chemical tool to dissect the role of neuroepigenetic mechanisms in regulating neuroplasticity in health and in disease.
Supplementary Material
Acknowledgements
We would like to thank Dr. Florence F. Wagner for helpful discussions and critical reading of the manuscript. S.J.H. was supported through funding from the NIH (R01DA028301, R01DA030321), and the Tau Consortium. R.M. was supported through funding from the NIH (P50CA086355).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement
R.M. has financial interests in SHAPE Pharmaceuticals and Acetylon Pharmaceuticals. He is also the inventor on IP licensed to these two entities that is unrelated to this study. S.J.H. has financial interest in Rodin Therapeutics and is an inventor on IP licensed to this entity that is unrelated to this study.
REFERENCES
- [1].Guan JS, Xie H, Ding X. The role of epigenetic regulation in learning and memory. Exp Neurol. 2015;268:30–36. doi: 10.1016/j.expneurol.2014.05.006. [DOI] [PubMed] [Google Scholar]
- [2].Nott A, Fass DM, Haggarty SJ, Tsai LH. HDAC Inhibitors as Novel Therapeutics in Aging and Alzheimer’s Disease. Academic Press, USA; Elsevier: 2012. [Google Scholar]
- [3].Nestler EJ. Epigenetic mechanisms in psychiatry. Biol Psychiatry. 2009;65:189–190. doi: 10.1016/j.biopsych.2008.10.030. [DOI] [PubMed] [Google Scholar]
- [4].Fass DM, Schroeder FA, Perlis RH, Haggarty SJ. Epigenetic mechanisms in mood disorders: targeting neuroplasticity. Neuroscience. 2014;264:112–130. doi: 10.1016/j.neuroscience.2013.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Graff J, Tsai LH. The potential of HDAC inhibitors as cognitive enhancers. Annu Rev Pharmacol Toxicol. 2013;53:311–330. doi: 10.1146/annurev-pharmtox-011112-140216. [DOI] [PubMed] [Google Scholar]
- [6].Whittle N, Singewald N. HDAC inhibitors as cognitive enhancers in fear. anxiety and trauma therapy: where do we stand?, Biochem Soc Trans. 2014;42:569–581. doi: 10.1042/BST20130233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dash PK, Orsi SA, Zhang M, Grill RJ, Pati S, Zhao J, Moore AN. Valproate administered after traumatic brain injury provides neuroprotection and improves cognitive function in rats. PLoS One. 2010;5:e11383. doi: 10.1371/journal.pone.0011383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, Bradner JE, DePinho RA, Jaenisch R, Tsai LH. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Graff J, Rei D, Guan JS, Wang WY, Seo J, Hennig KM, Nieland TJ, Fass DM, Kao PF, Kahn M, Su SC, Samiei A, Joseph N, Haggarty SJ, Delalle I, Tsai LH. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GA, Alenghat T, Mullican SE, Jones S, Rusche JR, Lazar MA, Wood MA. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci. 2011;31:764–774. doi: 10.1523/JNEUROSCI.5052-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- [12].Schroeder FA, Lin CL, Crusio WE, Akbarian S. Antidepressant-like effects of the histone deacetylase inhibitor. sodium butyrate, in the mouse, Biol Psychiatry. 2007;62:55–64. doi: 10.1016/j.biopsych.2006.06.036. [DOI] [PubMed] [Google Scholar]
- [13].Covington HE, I. Maze 3rd, LaPlant QC, Vialou VF, Ohnishi YN, Berton O, Fass DM, Renthal W, Rush AJ, E.Y. Wu 3rd, Ghose S, Krishnan V, Russo SJ, Tamminga C, Haggarty SJ, Nestler EJ. Antidepressant actions of histone deacetylase inhibitors. J Neurosci. 2009;29:11451–11460. doi: 10.1523/JNEUROSCI.1758-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Schroeder FA, Lewis MC, Fass DM, Wagner FF, Zhang YL, Hennig KM, Gale J, Zhao WN, Reis S, Barker DD, Berry-Scott E, Kim SW, Clore EL, Hooker JM, Holson EB, Haggarty SJ, Petryshen TL. A selective HDAC 1/2 inhibitor modulates chromatin and gene expression in brain and alters mouse behavior in two mood-related tests. PLoS One. 2013;8:e71323. doi: 10.1371/journal.pone.0071323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jochems J, Boulden J, Lee BG, Blendy JA, Jarpe M, Mazitschek R, Van Duzer JH, Jones S, Berton O. Antidepressant-like properties of novel HDAC6-selective inhibitors with improved brain bioavailability. Neuropsychopharmacology. 2014;39:389–400. doi: 10.1038/npp.2013.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bieliauskas AV, Pflum MK. Isoform-selective histone deacetylase inhibitors. Chem Soc Rev. 2008;37:1402–1413. doi: 10.1039/b703830p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fischer A, Sananbenesi F, Mungenast A, Tsai LH. Targeting the correct HDAC(s) to treat cognitive disorders. Trends Pharmacol Sci. 2010;31:605–617. doi: 10.1016/j.tips.2010.09.003. [DOI] [PubMed] [Google Scholar]
- [18].Bertrand P. Inside HDAC with HDAC inhibitors. Eur J Med Chem. 2010;45:2095–2116. doi: 10.1016/j.ejmech.2010.02.030. [DOI] [PubMed] [Google Scholar]
- [19].Fass DM, Reis SA, Ghosh B, Hennig KM, Joseph NF, Zhao WN, Nieland TJ, Guan JS, Kuhnle CE, Tang W, Barker DD, Mazitschek R, Schreiber SL, Tsai LH, Haggarty SJ. Crebinostat: a novel cognitive enhancer that inhibits histone deacetylase activity and modulates chromatin-mediated neuroplasticity. Neuropharmacology. 2013;64:81–96. doi: 10.1016/j.neuropharm.2012.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- [21].Vannini A, Volpari C, Filocamo G, Casavola EC, Brunetti M, Renzoni D, Chakravarty P, Paolini C, De Francesco R, Gallinari P, Steinkuhler C, Di Marco S. Crystal structure of a eukaryotic zinc-dependent histone deacetylase. human HDAC8, complexed with a hydroxamic acid inhibitor, Proc Natl Acad Sci U S A. 2004;101:15064–15069. doi: 10.1073/pnas.0404603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Oanh DT, Hai HV, Park SH, Kim HJ, Han BW, Kim HS, Hong JT, Han SB, Hue VT, Nam NH. Benzothiazole-containing hydroxamic acids as histone deacetylase inhibitors and antitumor agents. Bioorg Med Chem Lett. 2011;21:7509–7512. doi: 10.1016/j.bmcl.2011.07.124. [DOI] [PubMed] [Google Scholar]
- [23].Wang H, Lim ZY, Zhou Y, Ng M, Lu T, Lee K, Sangthongpitag K, Goh KC, Wang X, Wu X, Khng HH, Goh SK, Ong WC, Bonday Z, Sun ET. Acylurea connected straight chain hydroxamates as novel histone deacetylase inhibitors: Synthesis. SAR, and in vivo antitumor activity, Bioorg Med Chem Lett. 2010;20:3314–3321. doi: 10.1016/j.bmcl.2010.04.041. [DOI] [PubMed] [Google Scholar]
- [24].Fass DM, Shah R, Ghosh B, Hennig K, Norton S, Zhao WN, Reis SA, Klein PS, Mazitschek R, Maglathlin RL, Lewis TA, Haggarty SJ. Effect of Inhibiting Histone Deacetylase with Short-Chain Carboxylic Acids and Their Hydroxamic Acid Analogs on Vertebrate Development and Neuronal Chromatin. ACS medicinal chemistry letters. 2010;2:39–42. doi: 10.1021/ml1001954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wagner FF, Zhang YL, Fass DM, Joseph N, Gale JP, Weiwer M, McCarren P, Fisher SL, Kaya T, Zhao WN, Reis SA, Hennig KM, Thomas M, Lemercier BC, Lewis MC, Guan JS, Moyer MP, Scolnick E, Haggarty SJ, Tsai LH, Holson EB. Kinetically Selective Inhibitors of Histone Deacetylase 2 (HDAC2) as Cognition Enhancers. Chem Sci. 2015;6:804–815. doi: 10.1039/c4sc02130d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- [27].Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, Salinas-Riester G, Dettenhofer M, Kang H, Farinelli L, Chen W, Fischer A. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- [28].Dong DW, Tong SW, Qi XR. Comparative studies of polyethylenimine-doxorubicin conjugates with pH-sensitive and pH-insensitive linkers. J Biomed Mater Res A. 2013;101:1336–1344. doi: 10.1002/jbm.a.34450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.