Abstract
BMP signaling is one of the key pathways regulating craniofacial development. It is involved in the early pattering of the head, the development of cranial neural crest cells, and facial patterning. It regulates development of its mineralized structures, such as cranial bones, maxilla, mandible, palate, and teeth. Targeted mutations in the mouse have been instrumental to delineate the functional involvement of this signaling network in different aspects of craniofacial development. Gene polymorphisms and mutations in BMP pathway genes have been associated with various non-syndromic and syndromic human craniofacial malformations. The identification of intricate cellular interactions and underlying molecular pathways illustrate the importance of local fine-regulation of Bmp signaling to control proliferation, apoptosis, epithelial-mesenchymal interactions, and stem/progenitor differentiation during craniofacial development. Thus, BMP signaling contributes both to shape and functionality of our facial features. BMP signaling also regulates postnatal craniofacial growth and is associated with dental structures life-long. A more detailed understanding of BMP function in growth, homeostasis, and repair of postnatal craniofacial tissues will contribute to our ability to rationally manipulate this signaling network in the context of tissue engineering.
Keywords: Craniofacial development, craniofacial malformations, tooth morphogenesis, cleft palate, BMP signaling, gene targeting, congenital malformations
1. Introduction
Our head is a complex structure, home to several specialized tissues. The majority of congenital malformations involve craniofacial tissues, and BMP signaling is one of the key signaling pathways regulating their development. This review considers BMP signaling in the context of early facial patterning and the subsequent development of several of its mineralized structures. The development of skull and face also occurs in relationship with the brain [1].
Holoprosencephaly (HPE), a common developmental disorder characterized by forebrain midline defects due to failure of its separation into two hemispheres, typically also causes facial midline defects. Gain of Mutations in the BMP signaling pathway can cause HPE [2, 3], reviewed in [4], but will not be considered further.
The first section of this review considers the development of the facial primordium and cranial neural crest (CNC) cells. CNC is a multipotent cell lineage that contributes to all facial tissues, including bone, cartilage, teeth, nerves, and connective tissue, amongst others. This and the section on craniofacial skeletogenesis illustrate how CNC cells are affected by mutations in the BMP pathway. The section on tooth development shows how epithelial-mesenchymal interactions during tooth morphogenesis are regulated by BMP signaling. The involvement of BMP signaling in tooth mineralization and tooth root development is less well understood, though important functions in the development of these structures are emerging.
The fourth section considers BMP signaling in the context of palate morphogenesis. Palate development relies on the coordinated development of several orofacial structures; encompassing palatal shelves themselves, but also tongue and mandible. The development of all these structures is regulated by BMP signaling. Several mutations in the BMP pathway cause cleft palate, a pathology resulting from disturbed palate morphogenesis.
The last sections place BMP signaling into context with common cellular and molecular mechanisms and provide an outlook for future research directions including tissue engineering.
2. BMP signaling in development of the cranial neural crest and facial primordium
Neural crest cells are derived from dorsal midline ectoderm of developing vertebrate embryos [5]. Neural crest cells migrate downwards beside the neural tube and laterally under the surface ectoderm at all axial levels. The cells formed at craniofacial levels migrate into the frontonasal process and pharyngeal arches, and give rise to nearly all tissues of the face and neck. After cranial neural crest cells have completed their migration, facial growth is dominated by regional growth centers and the final differentiation of tissues occurs. Several growth signaling pathways such as BMP, WNT, SHH, and FGF are known to play crucial roles for these events [6-8].
Improper neural crest cell development leads to numerous congenital birth defects, such as Treacher-Collins syndrome (TCS) and DiGeorge syndrome (DGS). TCS is autosomal dominant disorder characterized by abnormal craniofacial development such as micrognathia, hypoplastic zygomatic arches, microtia, and coloboma of the eyelid (OMIM ID:154500) [9-11]. DGS is usually sporadic and results from de novo deletion within chromosome 22. DGS is characterized by hypoplasia of the parathyroid glands, hypoplasia or aplasia of thymus gland, and cardiac malformations (OMIM ID:188400) [12, 13].
2.1 Embryonic craniofacial development
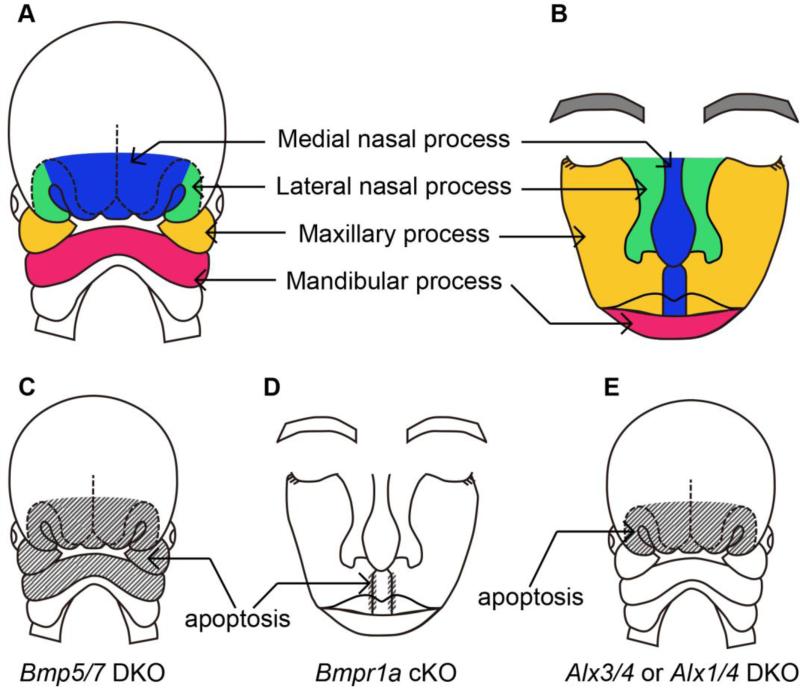
The face is formed by fusion or merger of the five identifiable primordial structures, the frontonasal prominence (FNP) and the paired maxillary and mandibular processes [14]. Fetuses develop the FNP by the end of 5 weeks in humans and at 10.5 days in mice. The frontal portion of the FNP forms the forehead. The FNP splits into the frontonasal process, a pair of the medial nasal process and a pair of the lateral nasal processes due to the invagination of an ectodermal placode (Fig. 1A). The expanding nasomedial limbs merge at the midline to form the primordium, which differentiates into the middle part of nose, philtrum, premaxilla, primary palate, and a portion of the nasal septum (Fig. 1B) [15]. The maxillary and mandibular processes are derived from first branchial arch (Fig. 1A) [16]. The paired maxillary processes expand medially toward each other and the nasomedial processes. This merger between maxillary processes and nasomedial process contributes to formation of the lip (Fig. 1B) [15].
Figure 1.
Schematic representations of fusion of the facial processes. (A) A diagram of human craniofacial structures at 31 day after conception. (B) A diagram of the contribution of the embryonic facial processes to the structures of adult face. The medial nasal process contributes the central part of the nose and the philtrum. The lateral nasal processes form outer parts of the nose. Maxillary processes form lateral part of the upper lip and the cheeks. Lower lip is derived from mandibular processes. (C-E) Schematic representations of affected areas (stripe) and resulted phenotypes caused by mutations in each gene.
2.2 BMP and facial patterning
BMP signaling components are important factors in the growth of facial processes. Bmp2 and Bmp4 are expressed in maxillary and mandibular processes [17]. Bmp5 and Bmp7 are also expressed in branchial arches at the early gastrulation stage [18]. Bmpr1a is nearly ubiquitously expressed in the mouse embryo including craniofacial region [19, 20]. Acvr1 is expressed in the first two pharyngeal arches [21].
Alteration of these BMP signaling components leads to abnormal craniofacial development. Some of the Bmp2 heterozygous mutants exhibit open neural tube defects at E9.5, while all Bmp2 homozygous null mutants die at early embryonic stage [22, 23]. This neural tube phenotype is caused by p53-mediated apoptosis and administration of a p53 inhibitor partially rescues the phenotype in heterozygous mutants. Bmp4 homozygous mutants die between embryonic day (E) 6.5 and 9.5 with abnormal head morphology [24]. Although homozygous Bmp5 null mutants are viable and fertile [25], Bmp5 and Bmp7 double knockout mice have a reduced size of their branchial arches and die at E10.5 [18]. The reduced size of their forebrain and branchial arches is associated with an abnormal pattern of apoptotic cells in the cranial area (Fig. 1C).
Msx1 and Msx2, direct downstream targets of BMP signaling, are also expressed in developing facial processes including FNP, maxillary processes, and mandibular processes in chick embryos [26, 27]. Down-regulation of Msx1 and Msx2 by applying retinoic acid results in inhibition of upper beak outgrowth in chick [28].
2.3 BMP and fusion of facial processes
Cleft lip occurs because of a failure of fusion between the medial and lateral nasal processes and the maxillary processes. The fusion of these processes creates not only the lip but also the alveolar ridge during primary palate formation. Closure of the secondary palate by elevation of the palatal shelves follows that of the primary palate. That means an interference with lip closure can affect formation of the palate. About 60% of patient with cleft lip also have cleft palate (OMIM ID:119530). Cleft palate is explained in a subsequent section and this part of the review focuses on the cleft lip without cleft palate.
In humans, several genetic lesions are known to cause cleft lip such as mutations in MSX1, tumor protein 63 (TP63), interferon regulatory factor 6 (IRF6), and fibroblast growth factor receptor 1 (FGFR1) [29]. Animal studies have dissected the function of BMP signaling in cleft lip etiology. Dental epithelial-specific Bmpr1a knockout mice using Nestin-Cre exhibit elevated apoptosis in the fusing region of the mutant mesial nasal processes and bilateral cleft lips [30] (Fig. 1D).
The failure of the fusion between left and right medial nasal processes most probably leads to mid-facial clefting [31]. In human, mutation in aristaless-like (ALX) gene family is known to cause frontonasal dysplasia (FND) (OMIM ID:613451), characterized by hypertelorism, severely depressed nasal bridge and ridge, bifid nasal tip. Similar phenotypes are also seen in Alx3/Alx4 or Alx1/Alx4 compound mutant mice [31, 32]. These mutants exhibit increased apoptotic cells in the outgrowing front nasal processes [31] (Fig. 1E). These reports suggest that Alx genes play an important role in the morphogenesis of craniofacial midline structures. Interestingly, cytogenetic location of ALX4 is in 11p11.2 and the sequence contains the region homologous to the MSX2 gene [33].
BMP signaling members also contribute to abnormal facial morphogenesis in the midline region. A gain-of function mutation in Msx2 results in midfacial clefting [34]. Further, a gain-of-function mutation in Bmpr1a results in a short nasal septum and increased cell death in the nasal septal primordia [35]. These reports suggest that cell death might be affected by BMP signaling in developing facial processes. Further investigations are necessary to reveal the relationships between BMP signaling and apoptosis in craniofacial development.
3. Craniofacial skeletogenesis
The mammalian craniofacial skeleton consists of more than 20 bones and cartilages. The size and shape of those components are strictly determined. The craniofacial skeleton can be divided into 3 major elements such as calvaria, mandible, and nasomaxillary complex [36]. The majority of the cranial bones and cartilage are derived from cranial neural crest cells [37, 38]. Mandible, nasomaxillary complex, and the anterior cranial bones are derived from neural crest, whereas the posterior part is derived from paraxial mesoderm [39]. BMP signaling components along with other signaling molecules play crucial roles in morphogenesis of these elements [40, 41]. Importantly, BMP signaling components are highly expressed in the migrating cranial neural crest cells and later in cranial cartilage and bone [41]. These reports suggest that BMP signaling regulates skeletal development by organizing neural crest cell proliferation and cell death [40].
3.1 BMP and skull formation
BMP signaling alters the expression of Msx homeobox genes, TGFß family ligands, and growth and differentiation factors (Gdfs), which are all important for normal skull development [17, 42, 43]. A conventional gain-of-function mutation in Msx2 results in skeletal defects such as mandibular hypoplasia and aplasia of the interparietal bone [34]. However, the pathogenetic mechanisms of those phenotypes are unknown. Gdf5, 6 and 7 were firstly identified in a screen for BMP related genes corresponding to brachypodism [44]. The Gdf5 null mutant mouse exhibits a brachypodism phenotype, characterized by reduced long bone length in the limbs [44]. A mutation in Gdf6 leads to absence of the coronal suture [45].
Increased BMP signaling in neural crest cells also leads to craniofacial skeletal defects. Constitutive activation of Bmpr1a in the neural crest cell linage using P0-Cre or Wnt1-Cre leads to increased cell death in skeletal primordia. These mutant mice exhibit bone and cartilage defects of the nasomaxillary complex, such as nasal bone and nasal septum [35, 46, 47].
During skull development, the cranial sutures serve as growth centers. Premature fusion of the sutures results in reduced skull growth leading to increased intracranial pressure that causes skull deformity termed craniosynostosis. BMP signaling plays crucial roles in the regulation of cranial suture morphogenesis [48]. BMP signaling components such as Bmp2, Bmp4, Msx1, and Msx2 are expressed in the sagittal suture during its development [48]. Local application of BMP4 protein into mouse calvaria explants induced expression of Msx genes and led to obliteration of the mid sutural space [48]. Enhanced BMP signaling through a constitutively active form of BMPR1A results in induction of craniosynostosis [46]. Gain-of-function mutations in MSX2 also result in Boston-type craniosynostosis in humans (OMIM ID:604757) [49]. Noggin is present in post-natal sutures and its expression is under negative regulation of FGF signaling. FGF gain-of function mutations in syndromic forms of craniosynostosis might inappropriately reduce Noggin expression, such that the suture loses its patency [50].
3.2. BMP and mandibular development
In the early embryonic stage, the mandible develops adjacent to Meckel's cartilage, a first pharyngeal arch derivative. Meckel's cartilage is a transient structure providing early structural stability to the mandible prior to the development of the mandibular bone. Bmp2 and Bmp7 are expressed at early stages of the developing Meckel's cartilage; Noggin expression is continuous [51]. Noggin-deficient mice show a significantly thicker cartilage along with increased pSmad1/5/8 staining leading to ossification rather than degeneration of Meckel's cartilage [51]. Bmp7-deficient mice show reduced growth of Meckel's cartilage, which fails to form a symphysis at its tip [52]. The subsequent development of the mandible begins as a condensation of mesenchyme just lateral to Meckel's cartilage and proceeds entirely as an intramembranous bone formation. BMP signaling also play roles in mandibular morphogenesis. Bmp7-deficient mice have a shorter maxilla and mandible (micrognathia) [52]. Bmp2, and Bmp4 conditional compound mutants using Wnt1-Cre exhibit mandibular and cranial bone defects in a dose dependent manner [53]. In this study, Bmp2 deletion in the Bmp4 conditional knockout background resulted in worsening of micrognathia and enlarged frontal fontanelle phenotype. These studies conclude that BMP signaling is required for self-renewal of cranial neural crest cells [53]. Similar skeletal phenotypes are seen in Acvr1:Wnt1-Cre conditional mutant embryos [21]. Directed Bmp4 expression in neural crest cells leads to a bony fusion of the mandible with the maxilla, reminiscent of the rare human bony syngnathia birth defect [54].
4. BMP and tooth development
4.1 Tooth Morphogenesis
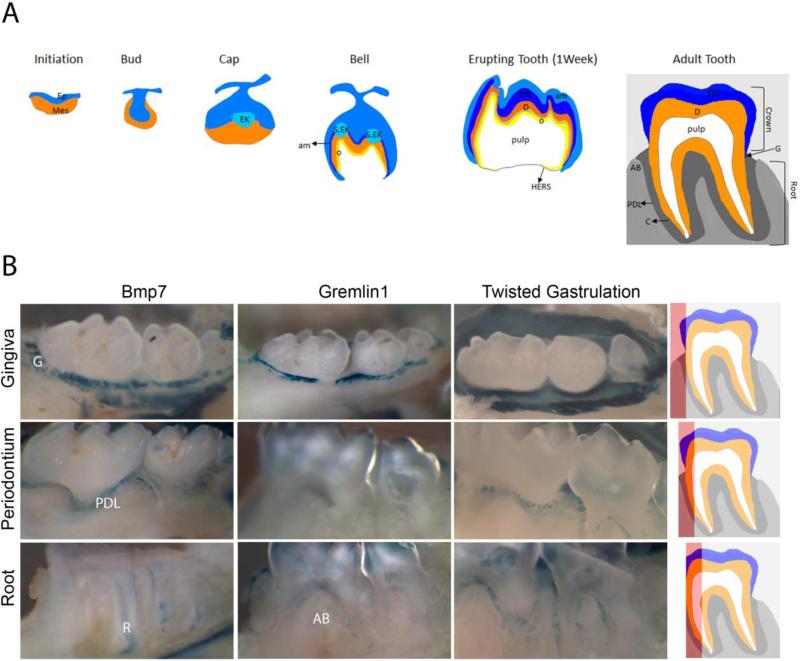
Mammalian tooth development is regulated by sequential and reciprocal epithelial-mesenchymal interactions. As shown in Fig. 2A, it is initiated by thickening of the dental lamina, a band of oral epithelial tissue, to form the dental placode. Tooth morphogenesis starts with its invagination into the underlying neural crest derived mesenchyme, leading to the bud stage. Condensation of the mesenchyme surrounding the bud leads to the cap stage, when the primary enamel knot appears, a signaling center thought to shape tooth morphology, and lateral epithelial protrusions develop. During the bell stage cell differentiation occurs and the first tooth hard tissue is being deposited. Secondary and tertiary enamel knots form to define the crown shape. Tooth mineralization begins late in embryogenesis and continues during root formation towards the end of the first postnatal week. In the mouse, teeth erupt by the second week concomitant with the development of the periodontal tissue that anchors the tooth to the alveolar bone. For a detailed recent review on tooth development refer to [55]. Bmps are expressed in all stages of tooth morphogenesis (dental placode, bud, cap and bell stages), and expression continuous during the secretory stages and root development [56-58], and remains prominent in molar teeth of adult mice (Fig. 2).
Figure 2.
BMP signaling in teeth. (A) Schematic representation of tooth morphogenesis and of an adult tooth. (B) Whole mount lacZ stained molar teeth from 4-6 month old mice. Scheme on the right hand side indicates visible area (red) when viewed from the left. Gingiva = unmanipulated tooth, Periodontium = gingival tissue resected, Root = Periodontium and tooth partly mechanically grounded off. Ep-Epithelium, Mes-Mesenchyme, EK-Enamel Knot, S.EK-Secondary Enamel Knot, o-odontoblasts, EN-Enamel, D-Dentin, am-Ameloblast, AB-Alveolar Bone, PDL-Periodontal Ligament, C-Cementum, G- Gingiva. Fig 2B adapted from [142]
4.2 BMP signaling in tooth initiation and morphogenesis
BMP signaling regulates tooth formation from the earliest stages. Competition between BMP and Noggin regulates the size of the mouse incisor placode, which determines the number of incisors [59], and mice overexpressing Noggin in the oral epithelium lack mandibular molars [60, 61]. BMP4 is an important signal driving tooth morphogenesis from bud to cap transition. In vitro experiments established that Bmp4 regulates expression of Msx1 [62, 63] and Pax9 [64]. Mice lacking Msx1 or Pax9 arrest tooth developmental at the bud stage [64, 65]. Deletion of Bmp4 from dental mesenchyme led altered expression of Msx1 and Pax9, and to arrest of the mandibular molars at the bud stage [66]. Tooth agenesis and hypodontia are the most common tooth malformations in humans. Mutations in MSX1 or PAX9 are known to cause selective tooth agenesis of the second premolar and third molar in humans [67, 68]. Arrest of tooth development at the bud stage has also been observed in mice carrying an epithelial deletion of Bmpr1a [69], indicating the importance of mesenchyme-derived BMP signals to drive development of tooth epithelium. Bmp2, Bmp4 and Bmp7 are expressed in the primary enamel knot, but their roles has not been addressed directly. Loss of the BMP antagonist Ectodin/Usag-1, which is regulated in cultured tooth explants by BMP2 and BM7 [70] results in supernumerary tooth formation [71]. BMP signaling also regulates Sox2-positive dental epithelial stem cells in incisors and molars through Smad-dependent inhibition of the Shh-Gli1 axis [72, 73].
4.3 BMP and tooth mineralization
Differentiation of epithelium-derived ameloblasts (enamel forming cells) and mesenchyme-derived odontoblast (dentin forming cells) starts around the bell stage. Bmp2, Bmp4, and Bmp7 have all been shown to strongly induce Ameloblastin expression, a protein associated with enamel mineralization [74]. Loss of Follistatin, a Bmp and Activin antagonist expressed in enamel-free lingual dental epithelium, resulted in ectopic enamel deposition, whereas it's overexpression inhibited ameloblast differentiation [74]. Deletion of Bmp2 in odontoblasts (Osx-Cre) led to deregulated expression of Amel and Enam, two enamel matrix proteins, and Klk4 and Mmp20, two enamel processing proteases [75]. In humans, mutations in MMP20, KLK4 and ENAM are associated with Amelogenesis Imperfecta (AI) [76-81]. In AI enamel fails to form (hypoplastic form) or mature properly (hypocalcified, hypermatured forms) [82]. Bmp2 also regulates the differentiation of odontoblasts and thus dentin formation [83]. Though the molecular mechanism by which mesenchymal BMP2 regulates the maturation of epithelial ameloblasts has not been elucidated, these findings indicate that BMP-dependent epithelial-mesenchymal interactions also regulate the mineralization stage.
4.4 BMP and root formation
Differentiation of the root starts after birth in mice, is regulated by epithelial-mesenchymal interactions, and involves HERS (Hertwig's Epithelial Root Sheath) and the mesenchymal dental follicle and dental papilla cells. Bmp2, Bmp3, Bmp4, and Bmp7 are expressed in the tooth root [57]. Inactivation of Bmp2 in mesenchymal cells affects terminal differentiation of root and periodontal ligament [84]. Inactivation of Smad4 in dental epithelial cells results in arrested molar root development [85]. Deletion of Bmpr1a from dental epithelium at the differentiation stage promoted differentiation of crown epithelium into cementum or root epithelial cells. [86]. Overexpression of Noggin in oral epithelium results in various phenotypic alterations including disrupted root development [60]. Together, these studies indicate that BMP signaling regulates epithelial-mesenchymal interactions during root development. In humans, mutations in the BMP antagonist GREM2 are associated with microdontia, taurodontism and a short tooth root [87]. Grem2 null mice have defects in maxillary and mandibular incisors, but no information on molar root development is available [88].
4.5 BMP in adult teeth
There is comparatively little information available on BMP involvement in development and of cementum and periodontal ligament, though several BMP ligands are expressed in these structures [57]. Inactivation of Bmp2 in mesenchymal cells affects development of the periodontal ligament [84]. BMP signaling has also been associated with orthodontic tooth movement [89, 90]. As shown in Fig. 2B, Bmp7, Gremlin1, and Twisted Gastrulation are expressed in the root and pulp of 4-6 month old mouse teeth. In addition, distinct expression is also observed in several structures of the periodontium (gingiva, periodontal ligament, alveolar bone). Life-long persistence of the Bmp signaling network indicates its potential involvement in alveolar bone remodeling in response to mastication forces and homeostasis of the gingival and periodontal tissues.
5. BMP signaling during palate morphogenesis
5.1 Palatogenesis
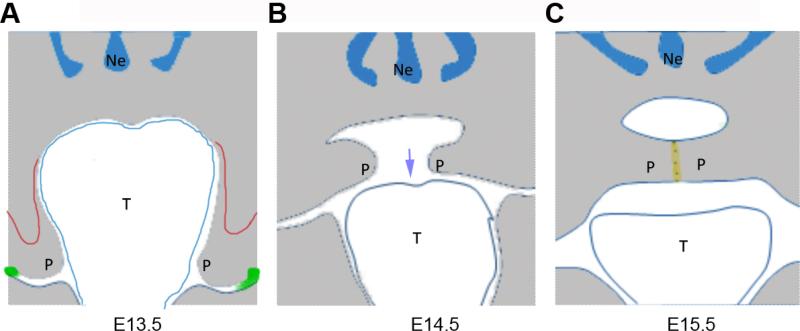
Palatogenesis is a highly regulated multi-step process that starts during the 6th embryonic week in humans and at E11.5 in the mouse, when the initial growth of a pair of palatal shelves at either side of maxillary processes can be observed. Palatal shelves mainly consist of neural crest mesenchyme derived from the first branchial arch encapsulated by a multi-layer of ectodermal epithelial cells. At the initial steps of palatal shelves development, the oral cavity is relatively small and the tongue is interposed between left and right palatal shelves forcing the early palatal shelves to grow downwards. With the expansion of the lower part of oral cavity and downward/forward movement of the tongue the palatal shelves re-orientate, grow in medial direction, and fuse separating oral and nasal cavity [91]. Reorientation, medial growth, and shelve apposition occur rapidly, between E14.0 to E15.0 in the mouse, and shelves fuse by E15.5, as part of which the medial edge epithelium is dissolved. Disturbances to growth, elevation, or fusion of the palatal shelves may result in cleft palate (Fig. 3). Failure of dissolution of the medial edge epithelium can result in sub-mucosal cleft, a situation where the soft tissue has fused, but underlying palatal bone and muscle layer remain separated. The mouse is a well-accepted model to study palate development, as many gene loci associated with cleft palate in humans when deleted result in cleft palate in the mouse [29, 92].
Figure 3.
Schematic representation of palate development and major developmental disturbances. (A) E13.5 palatal shelves. Red = small anterior shelve due to failure to grow, green = ectopic fusion of palate with oral epithelium. (B) E14.5 palatal shelves. Blue arrow indicates tongue contraction. (C) E15.5 palatal shelves. yellow = failure to remove midline epithelial seam cells
5.2 BMP and palatal shelve growth, elevation, and fusion
Early evidence for involvement of BMP signaling in palatal shelves growth came from work showing deregulated BMP expression in the retinoic acid-induced cleft palate model [93, 94] and Msx1-null mice [65]. Msx1 expressed in the anterior mesenchyme during initial palatal shelves growth regulates expression of Bmp4 [95, 96]. Loss of Msx1 results in short palatal shelves associated with impaired cell proliferation, a phenotype that is rescued by transgenic expression of Bmp4 [96]. Bmp4 induces Shh expression, which in turn regulates Bmp2. Bmp2 appears to be the primarily responsible factor regulating cell proliferation [96]. Differences in Bmp2 expression correlate with differences in proliferation also in turtles and birds [97]. Deletion of Bmpr1a in the craniofacial primordium using Nestin-Cre results in cleft palate [30], whereas a neural crest-specific deletion using Wnt1-Cre causes anterior clefting only [98]. This supports the role of BMP signaling in regulating proliferation in the anterior palatal shelve mesenchyme (Fig 3A). Noggin is strongly expressed in palatal shelve epithelium [99] and loss of Noggin also results in cleft palate [100]. Within the anterior palate Noggin negatively affects Bmp2 expression and hence cell proliferation. In the posterior region, loss of Noggin leads to spontaneous fusion of the palate epithelium with epithelia of oral cavity and tongue [100]. Fusion of palatal shelves at their midline is dependent on TGFß3. At the sites of palate-mandible fusion ectopic expression of Tgfß3 is observed [100]. Expression of a constitutively active Bmpr1a in the oral epithelium also causes ectopic epithelial fusions [100]. Together, this implies that repression of Bmp signaling in the oral epithelium is critical to maintain its integrity and prevent premature or ectopic fusion of palatal shelves (Fig. 3A).
5.3 BMP and cleft palate due to other factors
Pierre Robin Sequence stands for a series of developmental events encompassing micrognathia, glossoptosis, and cleft palate. It is thought that micrognathia interferes with the downward movement of the tongue, which in turn disturbs downward movement of the tongue, which is required for palatal shelves reorientation (OMIM ID:261800, [101], Fig 3B). Several loss-of-function mutations in the BMP signaling pathway molecules cause micrognathia [21, 52, 53]. As loss of Bmp7 does neither affect expression of Msx1 or Bmp4, nor does it alter proliferation or apoptosis in the palatal shelves [52], the cleft palate might be caused by a Pierre Robin Sequence. Tak1, a member of Smad-independent BMP pathway also plays crucial roles in craniofacial morphogenesis. Tak1: Wnt1-Cre conditional mutants have a cleft palate, likely the result of micrognathia and malformed tongue rather than changes in the palatal shelves [102, 103]. Bmp11 mutants exhibit abnormal palate, but no detailed information is available [104]. BMP signaling is also involved in regulating maturation and ossification of the fused palate. A constitutively active mutation of Acvr1 in oral epithelium causes a submucosal cleft palate in mice [105]. Enhanced BMP signaling resulted in altered proliferation and apoptosis in the medial edge epithelium leading to its persistence, thereby preventing fusion of palatal mesenchyme and muscle tissue (Fig. 3C).
5.4 BMP and cleft palate in humans
Cleft lip/cleft palate in humans has been associated with altered Bmp signaling. Mutations in the genes for transcription factor MSX1 [106] and gene polymorphisms in BMP4 and NOGGIN identified by linkage analysis and GWAS have been associated with non-syndromic cleft lip/palate ([107] and references therein, [108-110]. GREMLIN1 has also been associated with cleft palate, which was supported by detailed burden analysis [111]. Microdeletions of 20p12.3 or 14q22-23, which encompass the BMP2 [112] or BMP4 [113] loci respectively have been associated with syndromic forms of cleft palate. Mutations in BMP7 cause a variety of craniofacial malformations including cleft and high-arched palate [114]. The burden is now to demonstrate how these genetic associations, polymorphisms, and mutations functionally alter Bmp signaling in the affected tissues and how this contributes to the observed morbidities.
6. Common mechanism and outlook
6.1 Fine tuning of Bmp signaling
BMP signaling fulfills many diverse functions during craniofacial development. It regulates tissue patterning and organ development, facial growth and morphology. Though many of these structures are mineralized structures, BMP signaling also regulates proliferation and apoptosis in stem and progenitor cells, cell polarization and migration, epithelial-mesenchymal interactions during organ development and differentiation. Craniofacial development is highly coordinated, and many structures develop concomitant and in dependence of each other, as evidenced during palate development. Both an increase and decrease in BMP signaling can affect the development of craniofacial structures, underlining the importance of balanced and locally regulated Bmp signaling. Fine control of production of BMP ligands and BMP antagonists in a precise spatio-temporal manner appears to be critical for this. Several BMPs (e.g. Bmp2, Bmp4, Bmp5) are located in gene deserts that contain a large number of cis-regulatory elements for such tissue- and cell specific gene expression (reviewed in [115]). SNPs in BMP4 associated with non-syndromic cleft palate might relate to changes in tissue-specific enhancers, in contrast to the gene deletion associated with syndromic cleft palate.
It is thus likely that the outcome of BMP signaling is highly context dependent and dosage dependent. Differences in gene dosage of Bmp4 and Noggin affect the shape of the mouse mandible [116]. Gene dosage in Noggin affects susceptibility to arthritis [117], or, beyond mineralized structures, gene dosage in Bmp4 affects vascular remodeling [118, 119]. Juvenile polyposis (OMIM ID: 174900) can be caused by mutations in BMPR1A, which are also associated with dysmorphic facial features. The fact that the craniosynostosis phenotypes caused by expression of the constitutively activated Bmpr1a can be rescued by heterozygous null mutation of Bmpr1a suggests that 50% increase of signaling activity is enough to cause morphological abnormalities [46]. It is thus possible that small differences in BMP signaling contribute to the variability of human craniofacial features as well as variations in tissue physiology or disease susceptibility.
6.2 BMP in the regulation of cell apoptosis
The complexity of BMP signaling is also reflected by the multiple ways it can regulate apoptosis. Apoptosis due to augmented BMP signaling is seen in several craniofacial structures [35, 120, 121]. Smad-dependent BMP signaling prevents degradation of p53, and increased levels of p53 augment downstream targets, such as Bax and caspase-3 to promote apoptosis. Bmp-mediated apoptosis occurs also via interference with the cyclin-dependent kinase inhibitor p21 (WAF1/CIP1). BMP2 and BMP4 enhance p21 expression, which leads to cell cycle arrest and subsequent apoptosis, as shown in the tooth enamel knot [122]. P21 also regulates differentiation of osteoblasts [123], providing a possible link between tissue patterning (proliferation and apoptosis) and tissue differentiation (ossification). In contrast, dissolution of Meckel's cartilage, which in part is mediated via p53-dependent apoptosis [124], requires termination rather than active Bmp signaling [51].
6.3 BMP crosstalk with other signaling pathways
The BMP4-Shh-BMP2 signaling cascade associated with mesenchymal proliferation in the developing anterior palate is well documented [96, 98]. Interaction of BMP with Shh signaling is also seen in the regulation of the epithelial stem cell fate in the developing tooth [73] or the fusion of the facial primordia [125]. Crosstalk between BMP and Wnt/ß-catenin signaling has been demonstrated for early tooth development [61]. Despite the importance of Wnt signaling for craniofacial tissue development and homeostasis [126], this crosstalk has not been widely investigated. For comparison, BMP /Wnt crosstalk regulates stem/progenitor cell proliferation and differentiation in the intestinal crypt (reviewed in [127]) or the developing brain, where BMP signaling promotes Wnt-dependent cell proliferation via stabilization of Lef1 [128]. Bmp signaling regulates cytodifferentiation and epithelial-mesenchymal interactions often in crosstalk with FGF and Notch signaling [129, 130]. The same pathways also regulate osteoblast differentiation and ossification (reviewed in [131]).
6.4 BMP signaling beyond tissue morphology
Most studies investigating BMP signaling in craniofacial development report on morphological features only (e.g. size and shape of mandible, nasal bone, skull bones), but do not investigate if these changes also affect the quality of the mineralized or non-mineralized structures. Bmp signaling controls tooth mineralization [75, 83], bone formation and homeostasis (reviewed in [132-134]). Thus, it is possible to speculate that BMP signaling may regulate quality of minerals such as crystallinity and mineral matrix ratio (MMR). On that basis it might be expected that morphological changes due to altered BMP signaling go along with changes in tissue characteristics such as alterations in collagen crosslinking. In addition to the growth factor signaling, mechanotransduction is another important mechanism by which tissue formation is regulated [135]. How BMP signaling and mechanotransduction pathways interact to regulate bone remodeling is an emerging field [136-138].
7. Summary and Outlook
BMP signaling not only regulates the embryonic development of craniofacial tissues, it also emerges as an important regulator of postnatal craniofacial tissues. BMPs are used in the clinic for bone healing applications, including regeneration of alveolar bone defects. Despite significant progress in the field, our understanding on how BMP signaling regulates tissue homeostasis or repair remains fragmented. Functional genomics studies in the mouse that have been so instrumental for revealing the importance of BMP signaling during embryonic development should therefore embrace adult tissues and their pathologies.
Systematic massive sequencing of human genomes is expected to reveal novel sequence variations in the BMP signaling pathway genes. If and how such differences will affect tissue morphology and function is difficult to predict, not least due to the complexity of the BMP signaling network itself, but also because of its extensive crosstalk to other signaling pathways, such as FGF, HH, WNT, and NOTCH signaling.
Possibly the most important lesson from all these studies is the importance of balanced BMP signaling and its local fine-regulation to organize and maintain tissues. This has important and direct implications for the application of recombinant BMPs or BMP antagonists for tissue engineering in situ, and their current rather general way of application is likely not to achieve the desired results. There are significant efforts under way to develop smart scaffolds and recombinant BMP anchored to specific structures. Their availability should clearly improve the prospects of successful clinical applications. As a last point, BMPs are emerging important immune regulators [139-141] and thus could also play a role in inflammation. Future studies will need to establish whether and how this affects tissue repair, potentially adding further complexity to this pathway.
Table 1.
BMP signaling mouse mutants associated with defects in tooth development: red = mutation in mesenchyme, blue = mutation in epithelia
| Mice | Defects | References | |
|---|---|---|---|
| Bmp2 |
SP7/Osx -Cre Conditional KO SP7/Osx-Cre Conditional KO Collal-Cre Conditional KO |
Root and periodontium development Enamel formation Odontoblast differentiation, delay in amelogenesis |
[84] [75] [83] |
| Bmp4 | Wnt1-Cre Conditional KO | Tooth arrest at bud stage | [66] |
| Bmp7 | Null | Various craniofacial deformities with missing maxillary molars and defective mandible. | [58] |
| Follistatin | K14 transgenic expression | Enamel patterning and ameloblast differentiation | [74] |
| Noggin |
K14 transgenic expression K14 transgenic expression |
Root/crown patterning defects Arrested tooth development at early bud stage. |
[60] [61] |
| Grem2 | Null | Incisors most severely affected with dentin and enamel defects. | [88] |
| Ectodin | Null | Supernumerary teeth | [71] |
| Bmpr1a | K14-Cre Conditional KO (Krt5)-rtTA inducible cond. KO at E14.5 |
Tooth arrest at bud stage Cementoblast like cells in crown of epithelia. Defects in differentiation of crown epithelia | [69] [86] |
Highlights.
BMP signaling is one of the key pathways regulating craniofacial development.
BMP signaling is involved in the early pattering of the head, the development of cranial neural crest cells, and facial patterning.
BMP signaling regulates development of mineralized structures, such as cranial bones, maxilla, mandible, palate, and teeth.
Targeted mutations in the mouse have been instrumental to delineate the functional involvement of BMP signaling in different aspects of craniofacial development.
Gene polymorphisms and mutations in BMP pathway genes have been associated with various non-syndromic and syndromic human craniofacial malformations.
Balanced BMP signaling and its local fine-regulation is critical to organize and maintain craniofacial tissues. This has direct implications for the development of clinical applications using recombinant BMPs or BMP antagonists for tissue engineering in situ.
Acknowledgements
This review was written with support of funds from the School of Dentistry, University of Alberta (DG), the Swiss National Science Foundation (grant 310030_135530 to DG), the National Institutes of Health (R01DE020843 to YM), the Department of Defense (W81XWH-11-2-0073 to YM), and the Japan Society for the Promotion of Science (15H06423 to SH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hu D, Young NM, Xu Q, Jamniczky H, Green RM, Mio W, et al. Signals from the brain induce variation in avian facial shape. Dev Dyn. 2015 doi: 10.1002/dvdy.24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lana-Elola E, Tylzanowski P, Takatalo M, Alakurtti K, Veistinen L, Mitsiadis TA, et al. Noggin null allele mice exhibit a microform of holoprosencephaly. Hum Mol Genet. 2011;20:4005–15. doi: 10.1093/hmg/ddr329. [DOI] [PubMed] [Google Scholar]
- 3.Petryk A, Anderson RM, Jarcho MP, Leaf I, Carlson CS, Klingensmith J, et al. The mammalian twisted gastrulation gene functions in foregut and craniofacial development. Dev Biol. 2004;267:374–86. doi: 10.1016/j.ydbio.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 4.Petryk A, Graf D, Marcucio R. Holoprosencephaly: signaling interactions between the brain and the face, the environment and the genes, and the phenotypic variability in animal models and humans. Wiley Interdiscip Rev Dev Biol. 2015;4:17–32. doi: 10.1002/wdev.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhatt S, Diaz R, Trainor PA. Signals and switches in Mammalian neural crest cell differentiation. Cold Spring Harb Perspect Biol. 2013:5. doi: 10.1101/cshperspect.a008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Dev Cell. 2004;7:291–9. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Patthey C, Edlund T, Gunhaga L. Wnt-regulated temporal control of BMP exposure directs the choice between neural plate border and epidermal fate. Development. 2009;136:73–83. doi: 10.1242/dev.025890. [DOI] [PubMed] [Google Scholar]
- 8.Stuhlmiller TJ, Garcia-Castro MI. Current perspectives of the signaling pathways directing neural crest induction. Cell Mol Life Sci. 2012;69:3715–37. doi: 10.1007/s00018-012-0991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li C, Mernagh J, Bourgeois J. Novel craniofacial and extracraniofacial findings in a case of Treacher Collins syndrome with a pathogenic mutation and a missense variant in the TCOF1 gene. Clin Dysmorphol. 2009;18:63–6. doi: 10.1097/MCD.0b013e328318c4fb. [DOI] [PubMed] [Google Scholar]
- 10.Lowry RB, Morgan K, Holmes TM, Metcalf PJ, Stauffer GF. Mandibulofacial dysostosis in Hutterite sibs: a possible recessive trait. Am J Med Genet. 1985;22:501–12. doi: 10.1002/ajmg.1320220308. [DOI] [PubMed] [Google Scholar]
- 11.Teber OA, Gillessen-Kaesbach G, Fischer S, Bohringer S, Albrecht B, Albert A, et al. Genotyping in 46 patients with tentative diagnosis of Treacher Collins syndrome revealed unexpected phenotypic variation. Eur J Hum Genet. 2004;12:879–90. doi: 10.1038/sj.ejhg.5201260. [DOI] [PubMed] [Google Scholar]
- 12.Goodship J, Cross I, Scambler P, Burn J. Monozygotic twins with chromosome 22q11 deletion and discordant phenotype. J Med Genet. 1995;32:746–8. doi: 10.1136/jmg.32.9.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vincent MC, Heitz F, Tricoire J, Bourrouillou G, Kuhlein E, Rolland M, et al. 22q11 deletion in DGS/VCFS monozygotic twins with discordant phenotypes. Genet Couns. 1999;10:43–9. [PubMed] [Google Scholar]
- 14.Neskey D, Eloy JA, Casiano RR. Nasal, septal, and turbinate anatomy and embryology. Otolaryngol Clin North Am. 2009;42:193–205, vii. doi: 10.1016/j.otc.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Sulik KK, Johnston MC. Embryonic origin of holoprosencephaly: interrelationship of the developing brain and face. Scan Electron Microsc. 1982:309–22. [PubMed] [Google Scholar]
- 16.Trumpp A, Depew MJ, Rubenstein JL, Bishop JM, Martin GR. Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev. 1999;13:3136–48. doi: 10.1101/gad.13.23.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett JH, Hunt P, Thorogood P. Bone morphogenetic protein-2 and -4 expression during murine orofacial development. Arch Oral Biol. 1995;40:847–54. doi: 10.1016/0003-9969(95)00047-s. [DOI] [PubMed] [Google Scholar]
- 18.Solloway MJ, Robertson EJ. Early embryonic lethality in Bmp5;Bmp7 double mutant mice suggests functional redundancy within the 60A subgroup. Development. 1999;126:1753–68. doi: 10.1242/dev.126.8.1753. [DOI] [PubMed] [Google Scholar]
- 19.Dewulf N, Verschueren K, Lonnoy O, Moren A, Grimsby S, Vande Spiegle K, et al. Distinct spatial and temporal expression patterns of two type I receptors for bone morphogenetic proteins during mouse embryogenesis. Endocrinology. 1995;136:2652–63. doi: 10.1210/endo.136.6.7750489. [DOI] [PubMed] [Google Scholar]
- 20.Mishina Y, Suzuki A, Ueno N, Behringer RR. Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev. 1995;9:3027–37. doi: 10.1101/gad.9.24.3027. [DOI] [PubMed] [Google Scholar]
- 21.Dudas M, Sridurongrit S, Nagy A, Okazaki K, Kaartinen V. Craniofacial defects in mice lacking BMP type I receptor Alk2 in neural crest cells. Mech Dev. 2004;121:173–82. doi: 10.1016/j.mod.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Bradley A. Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development. 1996;122:2977–86. doi: 10.1242/dev.122.10.2977. [DOI] [PubMed] [Google Scholar]
- 23.Castranio T, Mishina Y. Bmp2 is required for cephalic neural tube closure in the mouse. Dev Dyn. 2009;238:110–22. doi: 10.1002/dvdy.21829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–16. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 25.Kingsley DM, Bland AE, Grubber JM, Marker PC, Russell LB, Copeland NG, et al. The mouse short ear skeletal morphogenesis locus is associated with defects in a bone morphogenetic member of the TGF beta superfamily. Cell. 1992;71:399–410. doi: 10.1016/0092-8674(92)90510-j. [DOI] [PubMed] [Google Scholar]
- 26.Brown JM, Wedden SE, Millburn GH, Robson LG, Hill RE, Davidson DR, et al. Experimental analysis of the control of expression of the homeobox-gene Msx-1 in the developing limb and face. Development. 1993;119:41–8. doi: 10.1242/dev.119.Supplement.41. [DOI] [PubMed] [Google Scholar]
- 27.Mina M, Gluhak J, Upholt WB, Kollar EJ, Rogers B. Experimental analysis of Msx-1 and Msx-2 gene expression during chick mandibular morphogenesis. Dev Dyn. 1995;202:195–214. doi: 10.1002/aja.1002020211. [DOI] [PubMed] [Google Scholar]
- 28.Brown JM, Robertson KE, Wedden SE, Tickle C. Alterations in Msx 1 and Msx 2 expression correlate with inhibition of outgrowth of chick facial primordia induced by retinoic acid. Anat Embryol (Berl) 1997;195:203–7. doi: 10.1007/s004290050039. [DOI] [PubMed] [Google Scholar]
- 29.Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 2011;12:167–78. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu W, Sun X, Braut A, Mishina Y, Behringer RR, Mina M, et al. Distinct functions for Bmp signaling in lip and palate fusion in mice. Development. 2005;132:1453–61. doi: 10.1242/dev.01676. [DOI] [PubMed] [Google Scholar]
- 31.Beverdam A, Brouwer A, Reijnen M, Korving J, Meijlink F. Severe nasal clefting and abnormal embryonic apoptosis in Alx3/Alx4 double mutant mice. Development. 2001;128:3975–86. doi: 10.1242/dev.128.20.3975. [DOI] [PubMed] [Google Scholar]
- 32.Qu S, Tucker SC, Zhao Q, deCrombrugghe B, Wisdom R. Physical and genetic interactions between Alx4 and Cart1. Development. 1999;126:359–69. doi: 10.1242/dev.126.2.359. [DOI] [PubMed] [Google Scholar]
- 33.Wu YQ, Badano JL, McCaskill C, Vogel H, Potocki L, Shaffer LG. Haploinsufficiency of ALX4 as a potential cause of parietal foramina in the 11p11.2 contiguous gene-deletion syndrome. Am J Hum Genet. 2000;67:1327–32. doi: 10.1016/s0002-9297(07)62963-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winograd J, Reilly MP, Roe R, Lutz J, Laughner E, Xu X, et al. Perinatal lethality and multiple craniofacial malformations in MSX2 transgenic mice. Hum Mol Genet. 1997;6:369–79. doi: 10.1093/hmg/6.3.369. [DOI] [PubMed] [Google Scholar]
- 35.Hayano S, Komatsu Y, Pan H, Mishina Y. Augmented BMP signaling in the neural crest inhibits nasal cartilage morphogenesis by inducing p53-mediated apoptosis. Development. 2015;142:1357–67. doi: 10.1242/dev.118802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Enlow DH. A morphogenetic analysis of facial growth. Am J Orthod. 1966;52:283–99. doi: 10.1016/0002-9416(66)90169-2. [DOI] [PubMed] [Google Scholar]
- 37.Noden DM. The role of the neural crest in patterning of avian cranial skeletal, connective, and muscle tissues. Dev Biol. 1983;96:144–65. doi: 10.1016/0012-1606(83)90318-4. [DOI] [PubMed] [Google Scholar]
- 38.Evans DJ, Noden DM. Spatial relations between avian craniofacial neural crest and paraxial mesoderm cells. Dev Dyn. 2006;235:1310–25. doi: 10.1002/dvdy.20663. [DOI] [PubMed] [Google Scholar]
- 39.Noden DM, Trainor PA. Relations and interactions between cranial mesoderm and neural crest populations. J Anat. 2005;207:575–601. doi: 10.1111/j.1469-7580.2005.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mishina Y, Snider TN. Neural crest cell signaling pathways critical to cranial bone development and pathology. Exp Cell Res. 2014;325:138–47. doi: 10.1016/j.yexcr.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nie X, Luukko K, Kettunen P. BMP signalling in craniofacial development. Int J Dev Biol. 2006;50:511–21. doi: 10.1387/ijdb.052101xn. [DOI] [PubMed] [Google Scholar]
- 42.Barlow AJ, Francis-West PH. Ectopic application of recombinant BMP-2 and BMP-4 can change patterning of developing chick facial primordia. Development. 1997;124:391–8. doi: 10.1242/dev.124.2.391. [DOI] [PubMed] [Google Scholar]
- 43.Chang SC, Hoang B, Thomas JT, Vukicevic S, Luyten FP, Ryba NJ, et al. Cartilage-derived morphogenetic proteins. New members of the transforming growth factor-beta superfamily predominantly expressed in long bones during human embryonic development. J Biol Chem. 1994;269:28227–34. [PubMed] [Google Scholar]
- 44.Storm EE, Huynh TV, Copeland NG, Jenkins NA, Kingsley DM, Lee SJ. Limb alterations in brachypodism mice due to mutations in a new member of the TGF beta-superfamily. Nature. 1994;368:639–43. doi: 10.1038/368639a0. [DOI] [PubMed] [Google Scholar]
- 45.Settle SH, Jr., Rountree RB, Sinha A, Thacker A, Higgins K, Kingsley DM. Multiple joint and skeletal patterning defects caused by single and double mutations in the mouse Gdf6 and Gdf5 genes. Dev Biol. 2003;254:116–30. doi: 10.1016/s0012-1606(02)00022-2. [DOI] [PubMed] [Google Scholar]
- 46.Komatsu Y, Yu PB, Kamiya N, Pan H, Fukuda T, Scott GJ, et al. Augmentation of Smad-dependent BMP signaling in neural crest cells causes craniosynostosis in mice. J Bone Miner Res. 2013;28:1422–33. doi: 10.1002/jbmr.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu S, Wu W, Liu C, Yang L, Sun C, Ye W, et al. BMPRIA mediated signaling is essential for temporomandibular joint development in mice. PLoS One. 2014;9:e101000. doi: 10.1371/journal.pone.0101000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim HJ, Rice DP, Kettunen PJ, Thesleff I. FGF-, BMP- and Shh-mediated signalling pathways in the regulation of cranial suture morphogenesis and calvarial bone development. Development. 1998;125:1241–51. doi: 10.1242/dev.125.7.1241. [DOI] [PubMed] [Google Scholar]
- 49.Jabs EW, Muller U, Li X, Ma L, Luo W, Haworth IS, et al. A mutation in the homeodomain of the human MSX2 gene in a family affected with autosomal dominant craniosynostosis. Cell. 1993;75:443–50. doi: 10.1016/0092-8674(93)90379-5. [DOI] [PubMed] [Google Scholar]
- 50.Warren SM, Brunet LJ, Harland RM, Economides AN, Longaker MT. The BMP antagonist noggin regulates cranial suture fusion. Nature. 2003;422:625–9. doi: 10.1038/nature01545. [DOI] [PubMed] [Google Scholar]
- 51.Wang Y, Zheng Y, Chen D, Chen Y. Enhanced BMP signaling prevents degeneration and leads to endochondral ossification of Meckel's cartilage in mice. Dev Biol. 2013;381:301–11. doi: 10.1016/j.ydbio.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kouskoura T, Kozlova A, Alexiou M, Blumer S, Zouvelou V, Katsaros C, et al. The etiology of cleft palate formation in BMP7-deficient mice. PLoS One. 2013;8:e59463. doi: 10.1371/journal.pone.0059463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bonilla-Claudio M, Wang J, Bai Y, Klysik E, Selever J, Martin JF. Bmp signaling regulates a dose-dependent transcriptional program to control facial skeletal development. Development. 2012;139:709–19. doi: 10.1242/dev.073197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He F, Hu X, Xiong W, Li L, Lin L, Shen B, et al. Directed Bmp4 expression in neural crest cells generates a genetic model for the rare human bony syngnathia birth defect. Dev Biol. 2014;391:170–81. doi: 10.1016/j.ydbio.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jussila M, Thesleff I. Signaling networks regulating tooth organogenesis and regeneration, and the specification of dental mesenchymal and epithelial cell lineages. Cold Spring Harb Perspect Biol. 2012;4:a008425. doi: 10.1101/cshperspect.a008425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aberg T, Wozney J, Thesleff I. Expression patterns of bone morphogenetic proteins (Bmps) in the developing mouse tooth suggest roles in morphogenesis and cell differentiation. Dev Dyn. 1997;210:383–96. doi: 10.1002/(SICI)1097-0177(199712)210:4<383::AID-AJA3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 57.Yamashiro T, Tummers M, Thesleff I. Expression of bone morphogenetic proteins and Msx genes during root formation. J Dent Res. 2003;82:172–6. doi: 10.1177/154405910308200305. [DOI] [PubMed] [Google Scholar]
- 58.Zouvelou V, Luder HU, Mitsiadis TA, Graf D. Deletion of BMP7 affects the development of bones, teeth, and other ectodermal appendages of the orofacial complex. J Exp Zool B Mol Dev Evol. 2009;312B:361–74. doi: 10.1002/jez.b.21262. [DOI] [PubMed] [Google Scholar]
- 59.Munne PM, Felszeghy S, Jussila M, Suomalainen M, Thesleff I, Jernvall J. Splitting placodes: effects of bone morphogenetic protein and Activin on the patterning and identity of mouse incisors. Evol Dev. 2010;12:383–92. doi: 10.1111/j.1525-142X.2010.00425.x. [DOI] [PubMed] [Google Scholar]
- 60.Plikus MV, Zeichner-David M, Mayer JA, Reyna J, Bringas P, Thewissen JG, et al. Morphoregulation of teeth: modulating the number, size, shape and differentiation by tuning Bmp activity. Evol Dev. 2005;7:440–57. doi: 10.1111/j.1525-142X.2005.05048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuan G, Yang G, Zheng Y, Zhu X, Chen Z, Zhang Z, et al. The non-canonical BMP and Wnt/beta-catenin signaling pathways orchestrate early tooth development. Development. 2015;142:128–39. doi: 10.1242/dev.117887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Y, Bei M, Woo I, Satokata I, Maas R. Msx1 controls inductive signaling in mammalian tooth morphogenesis. Development. 1996;122:3035–44. doi: 10.1242/dev.122.10.3035. [DOI] [PubMed] [Google Scholar]
- 63.Vainio S, Karavanova I, Jowett A, Thesleff I. Identification of BMP-4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell. 1993;75:45–58. [PubMed] [Google Scholar]
- 64.Neubuser A, Peters H, Balling R, Martin GR. Antagonistic interactions between FGF and BMP signaling pathways: a mechanism for positioning the sites of tooth formation. Cell. 1997;90:247–55. doi: 10.1016/s0092-8674(00)80333-5. [DOI] [PubMed] [Google Scholar]
- 65.Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6:348–56. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- 66.Jia S, Zhou J, Gao Y, Baek JA, Martin JF, Lan Y, et al. Roles of Bmp4 during tooth morphogenesis and sequential tooth formation. Development. 2013;140:423–32. doi: 10.1242/dev.081927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mostowska A, Kobielak A, Trzeciak WH. Molecular basis of non-syndromic tooth agenesis: mutations of MSX1 and PAX9 reflect their role in patterning human dentition. Eur J Oral Sci. 2003;111:365–70. doi: 10.1034/j.1600-0722.2003.00069.x. [DOI] [PubMed] [Google Scholar]
- 68.Vastardis H, Karimbux N, Guthua SW, Seidman JG, Seidman CE. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat Genet. 1996;13:417–21. doi: 10.1038/ng0896-417. [DOI] [PubMed] [Google Scholar]
- 69.Andl T, Ahn K, Kairo A, Chu EY, Wine-Lee L, Reddy ST, et al. Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development. 2004;131:2257–68. doi: 10.1242/dev.01125. [DOI] [PubMed] [Google Scholar]
- 70.Laurikkala J, Kassai Y, Pakkasjarvi L, Thesleff I, Itoh N. Identification of a secreted BMP antagonist, ectodin, integrating BMP, FGF, and SHH signals from the tooth enamel knot. Dev Biol. 2003;264:91–105. doi: 10.1016/j.ydbio.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 71.Murashima-Suginami A, Takahashi K, Sakata T, Tsukamoto H, Sugai M, Yanagita M, et al. Enhanced BMP signaling results in supernumerary tooth formation in USAG-1 deficient mouse. Biochem Biophys Res Commun. 2008;369:1012–6. doi: 10.1016/j.bbrc.2008.02.135. [DOI] [PubMed] [Google Scholar]
- 72.Juuri E, Saito K, Ahtiainen L, Seidel K, Tummers M, Hochedlinger K, et al. Sox2+ stem cells contribute to all epithelial lineages of the tooth via Sfrp5+ progenitors. Dev Cell. 2012;23:317–28. doi: 10.1016/j.devcel.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li J, Feng J, Liu Y, Ho TV, Grimes W, Ho HA, et al. BMP-SHH signaling network controls epithelial stem cell fate via regulation of its niche in the developing tooth. Dev Cell. 2015;33:125–35. doi: 10.1016/j.devcel.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang XP, Suomalainen M, Jorgez CJ, Matzuk MM, Werner S, Thesleff I. Follistatin regulates enamel patterning in mouse incisors by asymmetrically inhibiting BMP signaling and ameloblast differentiation. Dev Cell. 2004;7:719–30. doi: 10.1016/j.devcel.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 75.Guo F, Feng J, Wang F, Li W, Gao Q, Chen Z, et al. Bmp2 deletion causes an amelogenesis imperfecta phenotype via regulating enamel gene expression. J Cell Physiol. 2015;230:1871–82. doi: 10.1002/jcp.24915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hart PS, Hart TC, Michalec MD, Ryu OH, Simmons D, Hong S, et al. Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J Med Genet. 2004;41:545–9. doi: 10.1136/jmg.2003.017657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim JW, Seymen F, Lin BP, Kiziltan B, Gencay K, Simmer JP, et al. ENAM mutations in autosomal-dominant amelogenesis imperfecta. J Dent Res. 2005;84:278–82. doi: 10.1177/154405910508400314. [DOI] [PubMed] [Google Scholar]
- 78.Mardh CK, Backman B, Holmgren G, Hu JC, Simmer JP, Forsman-Semb K. A nonsense mutation in the enamelin gene causes local hypoplastic autosomal dominant amelogenesis imperfecta (AIH2). Hum Mol Genet. 2002;11:1069–74. doi: 10.1093/hmg/11.9.1069. [DOI] [PubMed] [Google Scholar]
- 79.Ozdemir D, Hart PS, Ryu OH, Choi SJ, Ozdemir-Karatas M, Firatli E, et al. MMP20 active-site mutation in hypomaturation amelogenesis imperfecta. J Dent Res. 2005;84:1031–5. doi: 10.1177/154405910508401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rajpar MH, Harley K, Laing C, Davies RM, Dixon MJ. Mutation of the gene encoding the enamel-specific protein, enamelin, causes autosomal-dominant amelogenesis imperfecta. Hum Mol Genet. 2001;10:1673–7. doi: 10.1093/hmg/10.16.1673. [DOI] [PubMed] [Google Scholar]
- 81.Seymen F, Park JC, Lee KE, Lee HK, Lee DS, Koruyucu M, et al. Novel MMP20 and KLK4 Mutations in Amelogenesis Imperfecta. J Dent Res. 2015;94:1063–9. doi: 10.1177/0022034515590569. [DOI] [PubMed] [Google Scholar]
- 82.Witkop CJ., Jr. Amelogenesis imperfecta, dentinogenesis imperfecta and dentin dysplasia revisited: problems in classification. J Oral Pathol. 1988;17:547–53. doi: 10.1111/j.1600-0714.1988.tb01332.x. [DOI] [PubMed] [Google Scholar]
- 83.Yang W, Harris MA, Cui Y, Mishina Y, Harris SE, Gluhak-Heinrich J. Bmp2 is required for odontoblast differentiation and pulp vasculogenesis. J Dent Res. 2012;91:58–64. doi: 10.1177/0022034511424409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rakian A, Yang WC, Gluhak-Heinrich J, Cui Y, Harris MA, Villarreal D, et al. Bone morphogenetic protein-2 gene controls tooth root development in coordination with formation of the periodontium. Int J Oral Sci. 2013;5:75–84. doi: 10.1038/ijos.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang X, Xu X, Bringas P, Jr., Hung YP, Chai Y. Smad4-Shh-Nfic signaling cascade-mediated epithelial-mesenchymal interaction is crucial in regulating tooth root development. J Bone Miner Res. 2010;25:1167–78. doi: 10.1359/jbmr.091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang Z, Hai B, Qin L, Ti X, Shangguan L, Zhao Y, et al. Cessation of epithelial Bmp signaling switches the differentiation of crown epithelia to the root lineage in a beta-catenin-dependent manner. Mol Cell Biol. 2013;33:4732–44. doi: 10.1128/MCB.00456-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kantaputra PN, Kaewgahya M, Hatsadaloi A, Vogel P, Kawasaki K, Ohazama A, et al. GREMLIN 2 Mutations and Dental Anomalies. J Dent Res. 2015 doi: 10.1177/0022034515608168. [DOI] [PubMed] [Google Scholar]
- 88.Vogel P, Liu J, Platt KA, Read RW, Thiel M, Vance RB, et al. Malformation of incisor teeth in Grem2(−)/(−) mice. Vet Pathol. 2015;52:224–9. doi: 10.1177/0300985814528218. [DOI] [PubMed] [Google Scholar]
- 89.Takimoto A, Kawatsu M, Yoshimoto Y, Kawamoto T, Seiryu M, Takano-Yamamoto T, et al. Scleraxis and osterix antagonistically regulate tensile force-responsive remodeling of the periodontal ligament and alveolar bone. Development. 2015;142:787–96. doi: 10.1242/dev.116228. [DOI] [PubMed] [Google Scholar]
- 90.Xue H, Zheng J, Cui Z, Bai X, Li G, Zhang C, et al. Low-intensity pulsed ultrasound accelerates tooth movement via activation of the BMP-2 signaling pathway. PLoS One. 2013;8:e68926. doi: 10.1371/journal.pone.0068926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Enlow DHH. G. H. Essentials of facial growth: W B Saunders Co. 1996 [Google Scholar]
- 92.Kouskoura T, Fragou N, Alexiou M, John N, Sommer L, Graf D, et al. The genetic basis of craniofacial and dental abnormalities. Schweiz Monatsschr Zahnmed. 2011;121:636–46. [PubMed] [Google Scholar]
- 93.Ho CT, Lau TY, Jin Y, Lu HB, Liong E, Leung KM, et al. Overexpression of iNOS and down-regulation of BMPs-2, 4 and 7 in retinoic acid induced cleft palate formation. Histol Histopathol. 2004;19:95–104. doi: 10.14670/HH-19.95. [DOI] [PubMed] [Google Scholar]
- 94.Lu H, Jin Y, Tipoe GL. Alteration in the expression of bone morphogenetic protein-2,3,4,5 mRNA during pathogenesis of cleft palate in BALB/c mice. Arch Oral Biol. 2000;45:133–40. doi: 10.1016/s0003-9969(99)00118-1. [DOI] [PubMed] [Google Scholar]
- 95.Alappat S, Zhang ZY, Chen YP. Msx homeobox gene family and craniofacial development. Cell Res. 2003;13:429–42. doi: 10.1038/sj.cr.7290185. [DOI] [PubMed] [Google Scholar]
- 96.Zhang Z, Song Y, Zhao X, Zhang X, Fermin C, Chen Y. Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development. 2002;129:4135–46. doi: 10.1242/dev.129.17.4135. [DOI] [PubMed] [Google Scholar]
- 97.Abramyan J, Leung KJ, Richman JM. Divergent palate morphology in turtles and birds correlates with differences in proliferation and BMP2 expression during embryonic development. J Exp Zool B Mol Dev Evol. 2014;322:73–85. doi: 10.1002/jez.b.22547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li L, Lin M, Wang Y, Cserjesi P, Chen Z, Chen Y. BmprIa is required in mesenchymal tissue and has limited redundant function with BmprIb in tooth and palate development. Dev Biol. 2011;349:451–61. doi: 10.1016/j.ydbio.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Matsui M, Klingensmith J. Multiple tissue-specific requirements for the BMP antagonist Noggin in development of the mammalian craniofacial skeleton. Dev Biol. 2014;392:168–81. doi: 10.1016/j.ydbio.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.He F, Xiong W, Wang Y, Matsui M, Yu X, Chai Y, et al. Modulation of BMP signaling by Noggin is required for the maintenance of palatal epithelial integrity during palatogenesis. Dev Biol. 2010;347:109–21. doi: 10.1016/j.ydbio.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tan TY, Kilpatrick N, Farlie PG. Developmental and genetic perspectives on Pierre Robin sequence. Am J Med Genet C Semin Med Genet. 2013;163C:295–305. doi: 10.1002/ajmg.c.31374. [DOI] [PubMed] [Google Scholar]
- 102.Yumoto K, Thomas PS, Lane J, Matsuzaki K, Inagaki M, Ninomiya-Tsuji J, et al. TGF-beta-activated kinase 1 (Tak1) mediates agonist-induced Smad activation and linker region phosphorylation in embryonic craniofacial neural crest-derived cells. J Biol Chem. 2013;288:13467–80. doi: 10.1074/jbc.M112.431775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Song Z, Liu C, Iwata J, Gu S, Suzuki A, Sun C, et al. Mice with Tak1 deficiency in neural crest lineage exhibit cleft palate associated with abnormal tongue development. J Biol Chem. 2013;288:10440–50. doi: 10.1074/jbc.M112.432286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McPherron AC, Lawler AM, Lee SJ. Regulation of anterior/posterior patterning of the axial skeleton by growth/differentiation factor 11. Nat Genet. 1999;22:260–4. doi: 10.1038/10320. [DOI] [PubMed] [Google Scholar]
- 105.Noda K, Mishina Y, Komatsu Y. Constitutively active mutation of ACVR1 in oral epithelium causes submucous cleft palate in mice. Dev Biol. 2015 doi: 10.1016/j.ydbio.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.van den Boogaard MJ, Dorland M, Beemer FA, van Amstel HK. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat Genet. 2000;24:342–3. doi: 10.1038/74155. [DOI] [PubMed] [Google Scholar]
- 107.Hu YY, Qin CQ, Deng MH, Niu YM, Long X. Association between BMP4 rs17563 polymorphism and NSCL/P risk: a meta-analysis. Dis Markers. 2015;2015:763090. doi: 10.1155/2015/763090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen Q, Wang H, Schwender H, Zhang T, Hetmanski JB, Chou YH, et al. Joint testing of genotypic and gene-environment interaction identified novel association for BMP4 with non-syndromic CL/P in an Asian population using data from an International Cleft Consortium. PLoS One. 2014;9:e109038. doi: 10.1371/journal.pone.0109038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kempa I, Ambrozaityte L, Stavusis J, Akota I, Barkane B, Krumina A, et al. Association of BMP4 polymorphisms with non-syndromic cleft lip with or without cleft palate and isolated cleft palate in Latvian and Lithuanian populations. Stomatologija. 2014;16:94–101. [PubMed] [Google Scholar]
- 110.Suzuki S, Marazita ML, Cooper ME, Miwa N, Hing A, Jugessur A, et al. Mutations in BMP4 are associated with subepithelial, microform, and overt cleft lip. Am J Hum Genet. 2009;84:406–11. doi: 10.1016/j.ajhg.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Al Chawa T, Ludwig KU, Fier H, Potzsch B, Reich RH, Schmidt G, et al. Nonsyndromic cleft lip with or without cleft palate: Increased burden of rare variants within Gremlin-1, a component of the bone morphogenetic protein 4 pathway. Birth defects research Part A, Clinical and molecular teratology. 2014;100:493–8. doi: 10.1002/bdra.23244. [DOI] [PubMed] [Google Scholar]
- 112.Williams ES, Uhas KA, Bunke BP, Garber KB, Martin CL. Cleft palate in a multigenerational family with a microdeletion of 20p12.3 involving BMP2. Am J Med Genet A. 2012;158A:2616–20. doi: 10.1002/ajmg.a.35594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lumaka A, Van Hole C, Casteels I, Ortibus E, De Wolf V, Vermeesch JR, et al. Variability in expression of a familial 2.79 Mb microdeletion in chromosome 14q22.1-22.2. Am J Med Genet A. 2012;158A:1381–7. doi: 10.1002/ajmg.a.35353. [DOI] [PubMed] [Google Scholar]
- 114.Wyatt AW, Osborne RJ, Stewart H, Ragge NK. Bone morphogenetic protein 7 (BMP7) mutations are associated with variable ocular, brain, ear, palate, and skeletal anomalies. Human mutation. 2010;31:781–7. doi: 10.1002/humu.21280. [DOI] [PubMed] [Google Scholar]
- 115.Pregizer S, Mortlock DP. Control of BMP gene expression by long-range regulatory elements. Cytokine Growth Factor Rev. 2009;20:509–15. doi: 10.1016/j.cytogfr.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Boell L, Pallares LF, Brodski C, Chen Y, Christian JL, Kousa YA, et al. Exploring the effects of gene dosage on mandible shape in mice as a model for studying the genetic basis of natural variation. Dev Genes Evol. 2013;223:279–87. doi: 10.1007/s00427-013-0443-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lories RJ, Daans M, Derese I, Matthys P, Kasran A, Tylzanowski P, et al. Noggin haploinsufficiency differentially affects tissue responses in destructive and remodeling arthritis. Arthritis Rheum. 2006;54:1736–46. doi: 10.1002/art.21897. [DOI] [PubMed] [Google Scholar]
- 118.Frank D, Johnson J, de Caestecker M. Bone morphogenetic protein 4 promotes vascular remodeling in hypoxic pulmonary hypertension. Chest. 2005;128:590S–1S. doi: 10.1378/chest.128.6_suppl.590S. [DOI] [PubMed] [Google Scholar]
- 119.Frank DB, Abtahi A, Yamaguchi DJ, Manning S, Shyr Y, Pozzi A, et al. Bone morphogenetic protein 4 promotes pulmonary vascular remodeling in hypoxic pulmonary hypertension. Circ Res. 2005;97:496–504. doi: 10.1161/01.RES.0000181152.65534.07. [DOI] [PubMed] [Google Scholar]
- 120.Billington CJ, Jr., Ng B, Forsman C, Schmidt B, Bagchi A, Symer DE, et al. The molecular and cellular basis of variable craniofacial phenotypes and their genetic rescue in Twisted gastrulation mutant mice. Dev Biol. 2011;355:21–31. doi: 10.1016/j.ydbio.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Saito H, Yamamura K, Suzuki N. Reduced bone morphogenetic protein receptor type 1A signaling in neural-crest-derived cells causes facial dysmorphism. Dis Model Mech. 2012;5:948–55. doi: 10.1242/dmm.009274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jernvall J, Aberg T, Kettunen P, Keranen S, Thesleff I. The life history of an embryonic signaling center: BMP-4 induces p21 and is associated with apoptosis in the mouse tooth enamel knot. Development. 1998;125:161–9. doi: 10.1242/dev.125.2.161. [DOI] [PubMed] [Google Scholar]
- 123.Bellosta P, Masramon L, Mansukhani A, Basilico C. p21(WAF1/CIP1) acts as a brake in osteoblast differentiation. J Bone Miner Res. 2003;18:818–26. doi: 10.1359/jbmr.2003.18.5.818. [DOI] [PubMed] [Google Scholar]
- 124.Trichilis A, Wroblewski J. Expression of p53 and hsp70 in relation to apoptosis during Meckel's cartilage development in the mouse. Anat Embryol (Berl) 1997;196:107–13. doi: 10.1007/s004290050083. [DOI] [PubMed] [Google Scholar]
- 125.Hu D, Young NM, Li X, Xu Y, Hallgrimsson B, Marcucio RS. A dynamic Shh expression pattern, regulated by SHH and BMP signaling, coordinates fusion of primordia in the amniote face. Development. 2015;142:567–74. doi: 10.1242/dev.114835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yin X, Li J, Salmon B, Huang L, Lim WH, Liu B, et al. Wnt Signaling and Its Contribution to Craniofacial Tissue Homeostasis. J Dent Res. 2015;94:1487–94. doi: 10.1177/0022034515599772. [DOI] [PubMed] [Google Scholar]
- 127.Yeung TM, Chia LA, Kosinski CM, Kuo CJ. Regulation of self-renewal and differentiation by the intestinal stem cell niche. Cell Mol Life Sci. 2011;68:2513–23. doi: 10.1007/s00018-011-0687-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Choe Y, Kozlova A, Graf D, Pleasure SJ. Bone morphogenic protein signaling is a major determinant of dentate development. J Neurosci. 2013;33:6766–75. doi: 10.1523/JNEUROSCI.0128-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mitsiadis TA, Graf D, Luder H, Gridley T, Bluteau G. BMPs and FGFs target Notch signalling via jagged 2 to regulate tooth morphogenesis and cytodifferentiation. Development. 2010;137:3025–35. doi: 10.1242/dev.049528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mustonen T, Tummers M, Mikami T, Itoh N, Zhang N, Gridley T, et al. Lunatic fringe, FGF, and BMP regulate the Notch pathway during epithelial morphogenesis of teeth. Dev Biol. 2002;248:281–93. doi: 10.1006/dbio.2002.0734. [DOI] [PubMed] [Google Scholar]
- 131.Lin GL, Hankenson KD. Integration of BMP, Wnt, and notch signaling pathways in osteoblast differentiation. J Cell Biochem. 2011;112:3491–501. doi: 10.1002/jcb.23287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rosen V. BMP and BMP inhibitors in bone. Ann N Y Acad Sci. 2006;1068:19–25. doi: 10.1196/annals.1346.005. [DOI] [PubMed] [Google Scholar]
- 133.Sanchez-Duffhues G, Hiepen C, Knaus P, Ten Dijke P. Bone morphogenetic protein signaling in bone homeostasis. Bone. 2015;80:43–59. doi: 10.1016/j.bone.2015.05.025. [DOI] [PubMed] [Google Scholar]
- 134.Kamiya N, Mishina Y. New insights on the roles of BMP signaling in bone-A review of recent mouse genetic studies. Biofactors. 2011;37:75–82. doi: 10.1002/biof.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Patwari P, Lee RT. Mechanical control of tissue morphogenesis. Circ Res. 2008;103:234–43. doi: 10.1161/CIRCRESAHA.108.175331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Iura A, McNerny EG, Zhang Y, Kamiya N, Tantillo M, Lynch M, et al. Mechanical Loading Synergistically Increases Trabecular Bone Volume and Improves Mechanical Properties in the Mouse when BMP Signaling Is Specifically Ablated in Osteoblasts. PLoS One. 2015;10:e0141345. doi: 10.1371/journal.pone.0141345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kopf J, Petersen A, Duda GN, Knaus P. BMP2 and mechanical loading cooperatively regulate immediate early signalling events in the BMP pathway. BMC Biol. 2012;10:37. doi: 10.1186/1741-7007-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rui YF, Lui PP, Ni M, Chan LS, Lee YW, Chan KM. Mechanical loading increased BMP-2 expression which promoted osteogenic differentiation of tendon-derived stem cells. J Orthop Res. 2011;29:390–6. doi: 10.1002/jor.21218. [DOI] [PubMed] [Google Scholar]
- 139.Martinez VG, Sacedon R, Hidalgo L, Valencia J, Fernandez-Sevilla LM, Hernandez-Lopez C, et al. The BMP Pathway Participates in Human Naive CD4+ T Cell Activation and Homeostasis. PLoS One. 2015;10:e0131453. doi: 10.1371/journal.pone.0131453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tsalavos S, Segklia K, Passa O, Petryk A, O'Connor MB, Graf D. Involvement of twisted gastrulation in T cell-independent plasma cell production. J Immunol. 2011;186:6860–70. doi: 10.4049/jimmunol.1001833. [DOI] [PubMed] [Google Scholar]
- 141.Yasmin N, Bauer T, Modak M, Wagner K, Schuster C, Koffel R, et al. Identification of bone morphogenetic protein 7 (BMP7) as an instructive factor for human epidermal Langerhans cell differentiation. J Exp Med. 2013;210:2597–610. doi: 10.1084/jem.20130275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Graf D, Mitsiadis TA. Den Geheimnissen der Zahnentwicklung auf der Spur. Schweiz Med Forum. 2013;13:168–70. [Google Scholar]