Abstract
The Arabidopsis stomatal lineage is a microcosm of development; it undergoes selection of precursor cells, asymmetric and stem cell-like divisions, cell commitment and finally, acquisition of terminal cell fates. Recent transcriptomic approaches revealed major shifts in gene expression accompanying each fate transition, and mechanistic analysis of key bHLH transcription factors, along with mathematical modeling, has begun to unravel how these major shifts are coordinated. In addition, stomatal initiation is proving to be a tractable model for defining the genetic and epigenetic basis of stable cell identities and for understanding the integration of environmental responses into developmental programs.
Life on land requires plants to balance acquisition of carbon from the atmosphere with loss of water from internal tissues. More than four hundred million years ago, plants solved this problem by generating stomata in their aerial organs; stomata are small epidermal pores surrounded by guard cells that open and close in response to homeostatic cues and signals from the environment. Today, stomata are present and essential in nearly all land plants, but their structure and distribution display clade-specific patterns and often require multi-step developmental programs. Because of their essential nature, but flexible pattern, stomata are a useful model for understanding a myriad of developmental processes including asymmetric and stem-cell like divisions, cell fate acquisition (and its connection to gene regulation and epigenetics), cell-cell communication, and responses to hormones and environmental cues. A mechanistic understanding of stomatal development has arisen primarily from studies in the dicot Arabidopsis thaliana, and this review will focus on this species, but recent work in Maize has shed light on how cell polarity is established in a multicellular context [1*] and careful morphological assessments in diverse species suggest ways in which control over the orientation of early divisions in the stomatal lineage lead to different leaf patterns [2,3].
Stomatal development in Arabidopsis thaliana proceeds through a stereotyped sequence of cell divisions and cell fate transitions in the epidermis (Figure 1). Stomatal lineage cell types can be defined morphologically and by expression of fate markers, the latter used with great success in combination with extended time-lapse imaging to define the trajectory of individual cells [4–7]. Such studies, along with classical lineage tracing, show that in the young leaf, a subset of protodermal cells in the epidermis, known as meristemoid mother cells, will divide asymmetrically to produce a small, usually triangular, meristemoid and a larger daughter cell known as an SLGC (stomatal lineage ground cell). The SLGC may differentiate directly into a pavement cell, or divide asymmetrically to generate a satellite meristemoid. Each meristemoid continues to divide asymmetrically, typically twice more, before it undergoes a fate and morphological transition into a round guard mother cell (GMC). The GMC is the first committed stage in the stomatal lineage; until they become GMCs, cells are developmentally plastic and may take on other epidermal fates. GMCs will divide a final time, symmetrically, to produce the two guard cells (GCs) of a stoma. Divisions of the meristemoid and SLGC are oriented through cell-cell communication to ensure that that two stomata do not form in direct contact with one another.
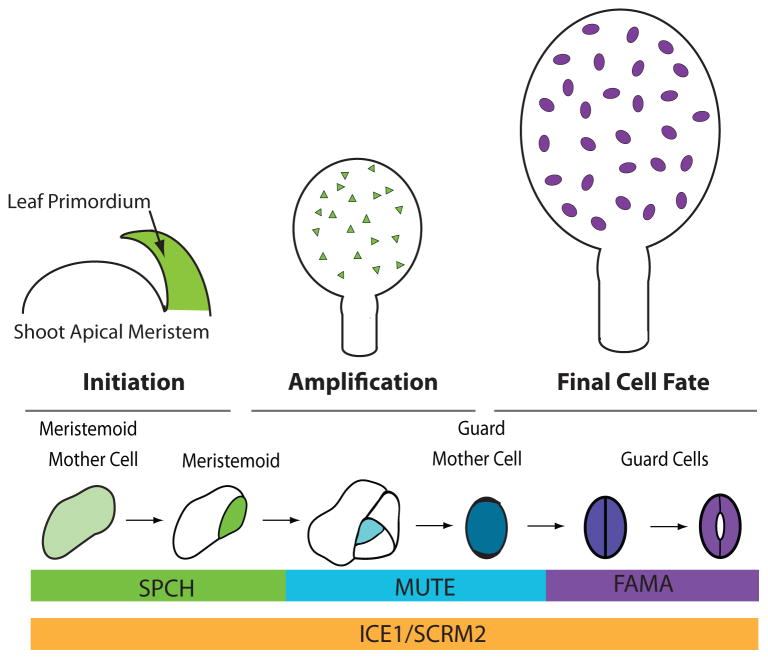
Figure 1. Overview of stomatal development in the context of the developing leaf.
Leaf primordia develop from the shoot apical meristem and the epidermis is already specified at this time. During the initiation phase, meristemoid mother cells (MMCs) are created; these cells in the developing leaf will begin to express SPCH protein and divide asymmetrically to form meristemoids (green). The amplification stage is dominated by cell divisions of MMCs and meristemoids that generate the epidermis and establish proper stomatal patterning. In later phases of leaf development (final cell fate stage) meristemoids transition to guard mother cells (blue) and then to differentiated guard cells (purple) while other cells expand to drive the increase in leaf size. The expression patterns of key transcriptional regulators discussed in this review are indicated at the bottom.
Each stage in the stomatal lineage requires precise transcriptional control over cell identity and behavior. Five basic helix-loop-helix (bHLH) transcription factors lie at the core of this regulation. Stomatal initiation and subsequent meristemoid self-renewal requires the first of these factors, SPEECHLESS (SPCH). SPCH RNA is broadly expressed in the young leaf, but SPCH protein is restricted to meristemoids and enables their continued asymmetric division [4,8,9]. When meristemoids exit this “SPCH” stage, they begin expressing the transcriptional factor MUTE and exhibit substantial changes in global gene expression [10,11]. MUTE is required for GMC fate, as loss of function mutants get stuck in a continuous self-renewing stage. When overexpressed, MUTE converts all epidermal cells into stomatal precursors [10]. One final transition from GMC to GC requires a symmetric division and is preceded by expression of FAMA [12]. FAMA is necessary for the acquisition of GC identity, but also for continued maintenance of this identity and this latter function is mediated by FAMA in conjunction with RBR (RETINOBLASTOMA RELATED) [13,14]. At each of their specific expression stages, SPCH, MUTE and FAMA act as obligate heterodimers with one of two more broadly expressed, but still stomatal lineage enriched, bHLHs, ICE1/SCREAM (SCRM) or SCRM2 [15]. Loss of both ICE1 and SCRM2 eliminates the stomatal lineage, resembling loss of SPCH, whereas stabilizing mutations (scrm-D) convert all epidermal cells into stomatal guard cells [15]. Because SPCH, MUTE and FAMA expression and activities nicely parallel the different stomatal cell types and transitions, we will use these factors as organizing nodes for the rest of this review.
Limiting stomatal lineage competence to the epidermis
True leaves develop from the shoot apical meristem where tissue layers are already established; the epidermis is derived from anticlinal divisions of the L1 layer, whereas the underlying mesophyll and vascular tissues are derived from deeper L2 and L3 layers. Several HOMEODOMAIN LEUCINE ZIPPER CLASS IV (HD-ZIP IV) proteins are required for epidermal identity beginning in the embryo and loss of two such factors; MERISTEM LAYER 1 (ML1) and PROTODERMAL FACTOR 2 (PDF2), results in plants that lack an epidermis [16]. A third HD-ZIP IV family member, HOMEODOMAIN GLABROUS 2 (HDG2) is expressed in meristemoids, but surprisingly, Peterson, et al. (2013) found that ectopic overexpression of HDG2 (or ML1) can induce stomata to form within the mesophyll. Overexpression of MUTE with the same promoter, however, was not capable of inducing internal guard cells [15] suggesting that epidermal identity is a prerequisite for stomatal lineage fates; consistent with this, overexpression of ML1 and HDG2 leads to the appearance of a SPCHp:GUS reporter in mesophyll cells suggesting a transformation to the earliest stomatal lineage fate [17].
SPCH as master regulator and point of integration for various signals
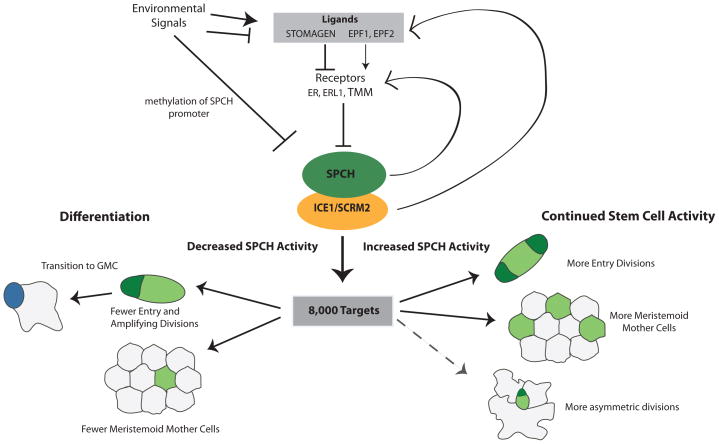
SPCH promotes asymmetric divisions that initiate, amplify and space future stomata in the epidermis and therefore where and how often it is expressed will define stomatal pattern and density [8]. In addition to generating stomata, the stomatal lineage builds most of the epidermis, including pavement cells [18] and possibly trichomes [11]. While protodermal cells that do not express SPCH may divide symmetrically to produce pavement cells, the bulk of the epidermis is derived from cells that have expressed SPCH at some point. Therefore, SPCH represents the logical point for numerous signaling cascades to converge to regulate leaf size, stomatal patterning and stomatal density in response to a variety of signals (Figure 2).
Figure 2. SPCH is a nexus of regulatory inputs and developmental outputs for development of the leaf epidermis.
Environmental signals have been shown to modulate development by increasing or decreasing the levels of EPF ligands, which modulate SPCH protein level through downstream phosphorylation cascades, or by affecting SPCH expression by DNA methylation at the SPCH locus. SPCH, presumably in combination with its binding partners ICE1/SCRM2 binds to over 8,000 targets. This massive reprogramming potential may be needed to create the stem-cell behavior of meristemoids and make them competent to respond to multiple other signals to modulate leaf size and stomatal density in response to the environment. After an initial pulse of SPCH activity, levels of SPCH (low or high) will shift the balance between differentiation and continued renewal, respectively. Solid lines indicate experimentally verified responses to SPCH, dotted lines indicate predicated responses.
Regulating the pattern of stomata: control over initiation and direction
SPCH protein has been shown to be regulated through phosphorylation by MITOGEN ACTIVATED PROTEIN KINASE (MAPKs), GLYCOGEN SYNTHASE 3 KINASE (GSK3) and CYCLIN DEPENDENT KINASE (CDK) families [19–21]. Phosphorylation by MPKs or the GSK3 BIN2 targets SPCH for degradation, and non-phosphorylatable forms of SPCH have increased expression, leading to increased entry divisions and ultimately larger leaves and a higher stomatal density [19,21,22]. MAPKs, GSKs and CDKs upstream of SPCH are broadly expressed in the leaf, so additional information is required to activate these kinase pathways in specific cells to produce the normal pattern of SPCH activity. This information comes, in part, from cell-type specific expression of receptor TOO MANY MOUTHs, some members of the ERECTA (ER) family of receptor kinases and ligands in the EPIDERMAL PATTERNING FACTOR (EPF) family. EPF2, expressed in meristemoids downregulates SPCH, while mesophyll expressed STOMAGEN/EPFL9 leads to an increase in SPCH levels [23–25]. Lee et al. (2015) demonstrated that STOMAGEN’s stomatal-promoting activities are mediated through the same receptors as EPF2 and, through an in vitro binding assay, demonstrate that EPF2 and STOMAGEN compete to bind ER, but only EPF2 can activate the downstream MAP kinase response [25**]; thus STOMAGEN functions as a competitive inhibitor of EPF2. The effect of different EPF ligands on SPCH expression was also tested by reconstituting the signaling pathway in mature Nicotiana benthamiana leaves [27]. In these assays, isolated from confounding changes in cell identities, both EPF1 and EPF2 when co-expressed with their receptors, could decrease SPCH protein levels, while STOMAGEN, when co-expressed with the same receptors increased SPCH protein levels [27].
Modulating stomatal numbers:environmental influences on the early stomatal lineage
EPF family ligands provide not only spatial information about where SPCH should be downregulated to ensure developmental pattern, but can be used to coordinate overall stomatal density with the environment. In response to altered atmospheric carbon dioxide (CO2) levels, CO2 RESPONSE SECRETED PROTEASE (CRSP) levels change and CRSP cleaves the pro-peptide of EPF2 (but not EPF1 or STOMAGEN) to release the active peptide [27*]. Because EPF2 downregulates SPCH [27*], this elegantly links perception of an environmental cue--relative elevation of [CO2]--to a decrease in stomatal lineage initiation and stomatal density [27*]. The identification of CRSP as a specific regulator of EPF2 suggests that multiple environmental signals could converge on the signaling pathways that regulate SPCH and a spate of recent papers underscore this regulatory convergence: ABA (abscisic acid), previously known to induce stomatal closure, was recently shown to also regulate stomatal density through SPCH regulation [29], osmotic stress leads to reduction of leaf size in a SPCH-dependent manner [30], and STOMAGEN expression is activated by light, thereby providing a mechanism for the oft-observed increase of stomatal numbers in high light [31].
Thus far, proteins that directly regulate SPCH transcription (except SPCH itself [31*]) have not been identified, but other forms of gene expression regulation have been found; the locus has high H3K27me3 in samples from seedling tissues suggesting that chromatin marks help repress the locus in mature leaves [33]. In addition, in response to low humidity, DNA methylation at the SPCH locus increases while SPCH expression (and stomatal density) decrease. In a methyltransferase mutant, SPCH expression and stomatal production were insensitive to lowered humidity [34,35].
While much of the regulation of meristemoid activity converges on SPCH, there are other means to regulate divisions in these cells and the overall growth potential of the leaf may have a general effect on individual meristemoids. TIFY transcription factors PEAPOD (PPD) 1 and PPD2 [36] and their associated transcriptional repression complex [36**] appear to negatively regulate continued asymmetric divisions of the meristemoid, directly targeting CYCD3, but not SPCH expression [36**]. An effect on the initial stomatal entry divisions was not tested, but is intriguing to think that the PPD complex controls competence for asymmetric divisions of the meristemoid independent of entry divisions, creating another way for plants to fine tune their final leaf size and epidermal composition.
Downstream of SPCH: Major reprogramming to initiate the stomatal lineage
Due to SPCH’s key role at the start of the stomatal lineage, it is perhaps not surprising to learn that SPCH binds to over 8,000 sites in the genome, and thus can serve as a master regulator of stomatal fate [31*]. SPCH binds to roughly one third of the Arabidopsis genes, a number that while high, is consistent with other lineage establishing transcription factors such as MyoD, a master regulator of mammalian myogenesis [38]. Targets of SPCH include components of the pathway that regulate it including EPF2, TMM and ERL2, and genes encoding its own binding partners ICE1 and SCRM2. Thus SPCH truly sits at the top of the stomatal lineage, first determining entry of protodermal cells into the lineage, and then activating the genes required for downstream regulation, proper patterning, and spacing.
SPCH targets include genes that would promote its activity (ICE1/SCRM) as well as those that would ultimately lead to its demise (EPFs, TMM, ERL2) indicating that both feed forward and feedback loops are likely important features of stomatal regulation. These feedback loops were recently included in a formal model by Horst et al. (2015) that demonstrate how such loops can account for maintaining SPCH expression only in the meristemoid. A key aspect of this model is that EPF2 is highly diffusible compared to SPCH/SCRM and this simple model was sufficient to generate an epidermis of SPCH expressing cells, surrounded by cells that do not express SPCH. The inclusion of STOMAGEN and BRASSINOSTEROID signaling as two more regulatory inputs were required to correctly recapitulate additional known signaling mutant phenotypes [8*]. This model is an important device to evaluate the contributions of different factors to pattern, but without biochemical data on ligand movement and receptor behavior, some questions remain, for example meristemoids which express the highest levels of EPF2 and its receptors, do not experience downregulation of SPCH; Horst et al suggest that TMM levels could explain this. Another possibility, however, is that a different target of SPCH, the polarity protein BASL, contributes to this protection. Cortical BASL is polarly localized such that is it always inherited by the SLGC after an asymmetric meristemoid division [39]. BASL interacts with the MAPKKK YODA and could act as a scaffold to segregate MAPK signaling into the SLGC; this would downregulate SPCH protein in this cell following the asymmetric cell division and form a feedback loop that ensures SPCH expression in only the smaller daughter cell [39*].
Transition from meristemoid to guard mother cell fate requires substantial, precisely controlled, transcriptional changes
Early divisions build the leaf, but formation of stomata requires a transition out of the proliferative, meristemoid stage. At the transition to guard mother cell (GMC) identity, the first step in commitment to guard cell fate, the meristemoid stops dividing asymmetrically, stops expressing SPCH and begins expressing MUTE [6]. This transition is accompanied by significant changes in gene expression as seen in several transcriptional profiling experiments; while two of these analyses made use of mutants to enrich for cell types [7], Adrian et al. (2015)** FACS-isolated cells expressing fluorescently tagged with SPCH, MUTE or FAMA (as well as epidermal and mature stomata markers) and performed microarray and RNA-seq analysis on WT cells. The single-cell type transcriptomes represent specific stages, but also defined genes associated with specific bHLH transcription factors. While the SPCH clustering genes include more cell cycle and stem cell maintenance genes, cells that have transitioned from the SPCH to MUTE stage show an enrichment in genes associated with DNA and histone methylation or other modification [11]. This indicates the exciting possibility that it may be possible to uncouple exit from stem-cell like divisions (asymmetric and uncommitted) from wholesale exit from mitotic potential as MUTE expressing cells undergo another round of symmetric division. The complement of cell cycle genes (especially CYCD isoforms) is also different between the SPCH and MUTE stages suggesting another mechanism underlying the switch from asymmetric to symmetric divisions [11].
Does MUTE share SPCH’s role as a nexus of signaling inputs and broad outputs in the GMC fate? Little is know about signals or transcriptional regulators that modulate MUTE expression have been identified (although HDG2 can activate MUTE reporter expression in a transient expression system [Peterson 2015]), and surprisingly, MUTE has been shown to function without its DNA binding residues [6]. While the molecular details of MUTE await future experiments, analysis of the phenotype has led to some interesting ideas about the stage MUTE regulates. In the absence of MUTE, meristemoids fail to transition into GMCs and continue to divide asymmetrically [10]. A version of MUTE with an estrogen responsive promoter expressed in the normal MUTE spatial domain was used to determine how long mute meristemoids were competent to respond to the return of MUTE activity [41*]). When induced early, meristemoids were completely rescued and could progress to make GMCs and stomata. When induced late, the arrested meristemoids could still transition, but neighboring SLGCs also begin to express MUTE and differentiate into guard cells. This suggested that the presence of MUTE might alter the competence of its sister cell to become a GC; one possible explanation for this is that in GMCs, MUTE induces the expression of EPF1, which is secreted and prevents neighboring SLGCs from differentiating into GMCs. In mute, EPF1 levels would be low and so many cells would remain competent to express and respond to MUTE, should it be supplied later. Triviño et al. (2013)* argued against this hypothesis because EPF1 levels recover post MUTE induction; however, this EPF1 expression may be a later consequence of conversion of meristemoids and SLGCs into MUTE-expressing cells.
Maintenance of terminal cell identities
The final stage of stomatal differentiation is typical in that it results in stable terminal cell identity and the acquisition of physiological function; it is atypical in that the stomatal unit is made of two sister cells, so fate is tied to progression through a single symmetric division. These two requirements—precise cell cycle control and permanent fate, are solved in an elegant way by using transcription factors FAMA and FOUR LIPS (a MYB partially redundant with its paralog MYB88) in combination with RBR and, potentially, POLYCOMB REPRESSIVE COMPLEX 2 (PRC2) activity. FAMA has two distinct roles during guard cell development; first, FAMA activates genes that drive differentiation of the guard cells while simultaneously ending the GMC stage by halting the symmetric divisions typical of this cell [12]. A second, RBR-dependent role, enforces cell fate and prevents ectopic expression of early stomatal lineage genes. If the FAMA-RBR interaction is disrupted by point mutations in FAMA’s RBR binding site [12**] or by blocking the FAMA C-terminus with a YFP tag [42], then SPCH, MUTE, EPFs and other early stomatal lineage genes are reactivated and stomatal lineage division and fate programs are recapitulated, with new GCs formed within an existing GC (Figure 3). This “stoma in stoma” (SIS) phenotype was linked to alterations in H3K27me3 levels at the SPCH and MUTE promoters [14]. In wildtype plants, both SPCH and MUTE have heavy H3K27me3 in mature leaves, but in plants with SIS phenotypes, H3K27me3 levels are low [14]. Overexpression of CURLY LEAF (CLF), a histone methyltransferase of the PRC2 complex, restored high H3K27me3 and suppressed the SIS phenotypes, suggesting the reprogramming of GCs was due to a failure to recruit PRC2 to stomatal lineage genes [14]. Together with previous data that found RBR associated with the SPCH promoter [43], a model was proposed that FAMA recruits RBR (which in turn recruits PRC2 components) to repress the same promoters that SPCH originally activated. Consistent with this model, knocking down RBR expression in mature guard cells can cause them to undergo asymmetric divisions similar to those found in the SIS phenotype [44].
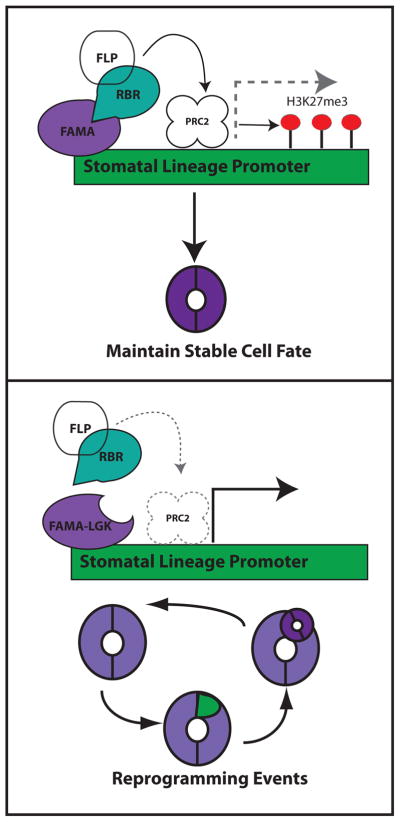
Figure 3. A model for guard cell fate stabilization.
The latest-acting bHLH transcription factor in the stomatal lineage, FAMA, acts in guard cells to bind the promoters of early stomatal lineage genes and to recruit RBR. Based on known interactions between RBR and PRC2 components in other systems, this could lead to establishment of localized deposition of H3K27me3 marks on promoters of stomatal lineage genes and limits to their transcription. When the interaction between FAMA and RBR is disrupted (as in the FAMA-LGK mutant) FAMA does not bring RBR to the early stomatal lineage promoters, PRC2 is not recruited, and early stomatal lineage genes are activated leading to re-capitulation of the entire lineage. The MYB transcription factor FLP may also be involved in this complex, though no functional evidence for it (nor for PRC2) being required for fixing GC fate is currently available and thus this putative components of the model are shown in white.
Chromatin-based mechanisms appear to reinforce cell fate at the end of the stomatal lineage, but their roles in earlier parts of the lineage has not been analyzed. It is possible that an earlier chromatin remodeling occurs when meristemoids transition to GMCs, which is then reinforced by FAMA-RBR in the late stage. The fact that the “reprogrammed” guard cells do not express any embryonic markers, or other markers associated with stages of development prior to the meristemoid, suggests that the meristemoid, while still developmentally plastic, is already restricted to the epidermal fate [12**]. This is consistent with a differentiation model in which stem cells become more fate restricted in a stepwise fashion that leads to a specific mature cell type. In the future, analysis of multiple chromatin modifications in individual stomatal lineage cell types will help address these open questions.
The accessibility of the stomatal lineage to isolation of cell types complements its accessibility to imaging-based studies to follow intact development over time. We predict that future work will develop gene regulatory networks (GRNs) for each cell stage, and begin to link the GRNs between stages and in response to outside perturbation. But as these networks expand, they are likely to still be grounded by the small number of paralogous transcription factors of major effect. Because these factors are conserved throughout the plant kingdom[45–47], analysis of transcriptional regulation in model plants may have wide-ranging implications for optimal plant growth under changing climate conditions.
Highlights.
Related bHLH factors are the heart of transcriptional regulation in the stomatal lineage
SPEECHLESS integrates tissue and environmental inputs to modulate cell production
Terminal cell fates appear to require RBR and PRC2-mediated histone modifications
Transcriptional profiles of stomatal lineage cells reveal major reprogramming during fate transitions
Acknowledgments
We thank members of our lab and the vibrant stomatal community for discussions. ARS was supported by NIH training grant 5T32GM007276 and a Donald Kennedy Fellowship from Stanford University. DCB is a Gordon and Betty Moore investigator of the Howard Hughes Medical Institute. We dedicate this review to Prof. Fred Sack, who pioneered the current era of inquiry into stomatal development.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1*.Facette MR, Park Y, Sutimantanapi D, Luo A, Cartwright HN, Yang B, Bennett EJ, Sylvester AW, Smith LG. The SCAR/WAVE complex polarizes PAN receptors and promotes division asymmetry in maize. Nat Plants. 2015;1:1–8. doi: 10.1038/nplants.2014.24. This work upended the expectation that PAN receptors would be the initial polarity cue for Maize subsidiary cell recruitment and instead showed that they are dependent on previous polarization of the actin-organizing SCAR/WAVE complex. [DOI] [PubMed] [Google Scholar]
- 2.Rudall PJ, Knowles EVW. Ultrastructure of stomatal development in early-divergent angiosperms reveals contrasting patterning and pre-patterning. Ann Bot. 2013;112:1031–1043. doi: 10.1093/aob/mct169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudall PJ, Hilton J, Bateman RM. Several developmental and morphogenetic factors govern the evolution of stomatal patterning in land plants. New Phytol. 2013;200:598–614. doi: 10.1111/nph.12406. [DOI] [PubMed] [Google Scholar]
- 4.Robinson S, Barbier de Reuille P, Chan J, Bergmann D, Prusinkiewicz P, Coen E. Generation of spatial patterns through cell polarity switching. Science. 2011;333:1436–40. doi: 10.1126/science.1202185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peterson KM, Torii KU. Long-term, high-resolution confocal time lapse imaging of Arabidopsis cotyledon epidermis during germination. J Vis Exp. 2012 doi: 10.3791/4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies Ka, Bergmann DC. Functional specialization of stomatal bHLHs through modification of DNA-binding and phosphoregulation potential. Proc Natl Acad Sci. 2014;111:15585–15590. doi: 10.1073/pnas.1411766111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pillitteri LJ, Peterson KM, Horst RJ, Torii KU. Molecular profiling of stomatal meristemoids reveals new component of asymmetric cell division and commonalities among stem cell populations in Arabidopsis. Plant Cell. 2011;23:3260–3275. doi: 10.1105/tpc.111.088583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacAlister Ca, Ohashi-Ito K, Bergmann DC. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature. 2007;445:537–40. doi: 10.1038/nature05491. [DOI] [PubMed] [Google Scholar]
- 9*.Horst RJ, Fujita H, Lee JS, Rychel AL, Garrick JM, Kawaguchi M, Peterson KM, Torii KU. Molecular Framework of a Regulatory Circuit Initiating Two-Dimensional Spatial Patterning of Stomatal Lineage. PLOS Genet. 2015;11:e1005374. doi: 10.1371/journal.pgen.1005374. This modeling paper formalizes stomatal patterning as a Turing-type mechanism using known components arranged in feedback and feedforward loops. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU. Termination of asymmetric cell division and differentiation of stomata. Nature. 2007;445:501–5. doi: 10.1038/nature05467. [DOI] [PubMed] [Google Scholar]
- 11**.Adrian J, Chang J, Ballenger CE, Bargmann BOR, Alassimone J, Davies KA, Lau OS, Matos JL, Hachez C, Lanctot A, et al. Transcriptome Dynamics of the Stomatal Lineage: Birth, Amplification, and Termination of a Self-Renewing Population. Dev Cell. 2015;33:107–118. doi: 10.1016/j.devcel.2015.01.025. This study utilized FACS to generate pure populations of cells at different stages within the stomatal lineage, and then characterized them by microarray and RNA-seq. Distinct modules of cells are expressed at different points within the lineage allowing for identification of new stomatal regulators and better understanding of transcriptional regulation at all stages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohashi-Ito K, Bergmann DC. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell. 2006;18:2493–505. doi: 10.1105/tpc.106.046136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13**.Matos JL, Lau OS, Hachez C, Cruz-ramírez A, Scheres B, Dominique C. Irreversible fate commitment in the Arabidopsis stomatal lineage requires a FAMA and RETINOBLASTOMA-RELATED module. Elife. 2014 doi: 10.7554/eLife.03271. [no volume]. This paper demonstrates that when interactions between FAMA and RBR are disrupted, guard cells can be made, but they do not maintain a stable identity. Guard cells inappropriately divide asymmetrically, producing a meristemoid that will recapitulate the entire lineage, ultimately forming a new stoma within the old. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee E, Lucas JR, Goodrich J, Sack FD. Arabidopsis guard cell integrity involves the epigenetic stabilization of the FLP and FAMA transcription factor genes. Plant J. 2014;78:566–77. doi: 10.1111/tpj.12516. [DOI] [PubMed] [Google Scholar]
- 15.Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu J-K, Torii KU. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to arabidopsis stomatal differentiation. Plant Cell. 2008;20:1775–85. doi: 10.1105/tpc.108.060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abe M, Katsumata H, Komeda Y, Takahashi T. Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development. 2003;130:635–643. doi: 10.1242/dev.00292. [DOI] [PubMed] [Google Scholar]
- 17.Peterson KM, Shyu C, Burr Ca, Horst RJ, Kanaoka MM, Omae M, Sato Y, Torii KU. Arabidopsis homeodomain-leucine zipper IV proteins promote stomatal development and ectopically induce stomata beyond the epidermis. Development. 2013;140:1924–35. doi: 10.1242/dev.090209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geisler M, Nadeau J, Sack FD. Oriented asymmetric divisions that generate the stomatal spacing pattern in arabidopsis are disrupted by the too many mouths mutation. Plant Cell. 2000;12:2075–2086. doi: 10.1105/tpc.12.11.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gudesblat GE, Schneider-Pizoń J, Betti C, Mayerhofer J, Vanhoutte I, van Dongen W, Boeren S, Zhiponova M, de Vries S, Jonak C, et al. SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat Cell Biol. 2012;14:548–554. doi: 10.1038/ncb2471. [DOI] [PubMed] [Google Scholar]
- 20.Yang K-Z, Jiang M, Wang M, Xue S, Zhu L-L, Wang H-Z, Zou J-J, Lee E-K, Sack F, Le J. Phosphorylation of Serine 186 of bHLH Transcription Factor SPEECHLESS Promotes Stomatal Development in Arabidopsis. Mol Plant. 2014;8:783–795. doi: 10.1016/j.molp.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 21.Lampard G, MacAlister C, Bergmann D. Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science (80- ) 2008;322:1113–1116. doi: 10.1126/science.1162263. [DOI] [PubMed] [Google Scholar]
- 22.Dow GJ, Berry Ja, Bergmann DC. The physiological importance of developmental mechanisms that enforce proper stomatal spacing in Arabidopsis thaliana. New Phytol. 2014;201:1205–1217. doi: 10.1111/nph.12586. [DOI] [PubMed] [Google Scholar]
- 23.Sugano SS, Shimada T, Imai Y, Okawa K, Tamai A, Mori M, Hara-Nishimura I. Stomagen positively regulates stomatal density in Arabidopsis. Nature. 2010;463:241–244. doi: 10.1038/nature08682. [DOI] [PubMed] [Google Scholar]
- 24.Hunt L, Gray JE. The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr Biol. 2009;19:864–9. doi: 10.1016/j.cub.2009.03.069. [DOI] [PubMed] [Google Scholar]
- 25.Hara K, Kajita R, Torii K. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes. 2007 doi: 10.1101/gad.1550707.metric. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26**.Lee JS, Hnilova M, Maes M, Lin Y-CL, Putarjunan A, Han S-K, Avila J, Torii KU. Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature. 2015 doi: 10.1038/nature14561. This study combines careful analysis of combinatorial mutants with in vitro binding assays to determine that EPF2 and STOMAGEN compete for binding with the same set of receptors. They conclude that STOMAGEN increases stomatal number by blocking EPF2 binding to ERECTA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jewaria PK, Hara T, Tanaka H, Kondo T, Betsuyaku S, Sawa S, Sakagami Y, Aimoto S, Kakimoto T. Differential effects of the peptides stomagen, EPF1 and EPF2 on activation of MAP kinase MPK6 and the SPCH protein level. Plant Cell Physiol. 2013;54:1253–1262. doi: 10.1093/pcp/pct076. [DOI] [PubMed] [Google Scholar]
- 28*.Engineer CB, Ghassemian M, Anderson JC, Peck SC, Hu H, Schroeder JI. Carbonic anhydrases, EPF2 and a novel protease mediate CO2 control of stomatal development. Nature. 2014;513:246–250. doi: 10.1038/nature13452. This study determined that stomatal development responds to changes in [CO2] through CRSP (CO2 RESPONSE SECRETED PROTEASE) which cleaves EPF2 to its active form thereby decreasing stomatal density when [CO2] is raised. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka Y, Nose T, Jikumaru Y, Kamiya Y. ABA inhibits entry into stomatal-lineage development in Arabidopsis leaves. Plant J. 2013;74:448–457. doi: 10.1111/tpj.12136. [DOI] [PubMed] [Google Scholar]
- 30.Kumari a, Jewaria PK, Bergmann DC, Kakimoto T. Arabidopsis Reduces Growth Under Osmotic Stress by Decreasing SPEECHLESS Protein. Plant Cell Physiol. 2014;55:2037–2046. doi: 10.1093/pcp/pcu159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hronkova M, Wiesnerova D, Simkova M, Skupa P, Dewitte W, Vrablova M, Zazimalova E, Santrucek J. Light-induced STOMAGEN-mediated stomatal development in Arabidopsis leaves. J Exp Bot. 2015 doi: 10.1093/jxb/erv233. [DOI] [PubMed] [Google Scholar]
- 32.Lau O, Davies K, Chang J, Adrian J, Rowe MH, Ballenger CE, Bergmann DC. Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science (80- ) 2014;345:1605–1609. doi: 10.1126/science.1256888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo C, Sidote DJ, Zhang Y, Kerstetter Ra, Michael TP, Lam E. Integrative analysis of chromatin states in Arabidopsis identified potential regulatory mechanisms for natural antisense transcript production. Plant J. 2013;73:77–90. doi: 10.1111/tpj.12017. [DOI] [PubMed] [Google Scholar]
- 34.Tricker PJ, George Gibbings J, Rodríguez López CM, Hadley P, Wilkinson MJ. Low relative humidity triggers RNA-directed de novo DNA methylation and suppression of genes controlling stomatal development. J Exp Bot. 2012;63:3799–3814. doi: 10.1093/jxb/ers076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tricker PJ, López CMR, Gibbings G, Hadley P, Wilkinson MJ. Transgenerational, dynamic methylation of stomata genes in response to low relative humidity. Int J Mol Sci. 2013;14:6674–6689. doi: 10.3390/ijms14046674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White DWR. PEAPOD regulates lamina size and curvature in Arabidopsis. Proc Natl Acad Sci U S A. 2006;103:13238–13243. doi: 10.1073/pnas.0604349103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37**.Gonzalez N, Pauwels L, Baekelandt A, De Milde L, Van Leene J, Besbrugge N, Heyndrickx KS, Pérez AC, Durand AN, De Clercq R, et al. A Repressor Protein Complex Regulates Leaf Growth in Arabidopsis. Plant Cell. 2015 doi: 10.1105/tpc.15.00006. This paper identifies the larger transcriptional repression complex in which PEAPOD (PPD) proteins, which limit asymmetric divisions of meristemoids, play a part. This is the first clear example of transcriptional regulatiors acting in oppostition to SPCH in regulation of the early stomatal lineage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, et al. Genome-wide MyoD Binding in Skeletal Muscle Cells: A Potential for Broad Cellular Reprogramming. Dev Cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong J, MacAlister Ca, Bergmann DC. BASL controls asymmetric cell division in Arabidopsis. Cell. 2009;137:1320–30. doi: 10.1016/j.cell.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40*.Zhang Y, Wang P, Shao W, Zhu J, Dong J. The BASL Polarity Protein Controls a MAPK Signaling Feedback Loop in Asymmetric Cell Division. Dev Cell. 2015;33:1–14. doi: 10.1016/j.devcel.2015.02.022. This study shows that phosphorylated BASL can interact with MAPKKK YODA, and that this interaction aids in polar localization of BASL, as well as segregation of MAPK activity to the larger daughter cell after an asymmetric division. Because SPCH is a target of MAPK activity, segregation of YODA could help reinforce differences in stem-cell behavior between the daughter cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41*.Triviño M, Martín-Trillo M, Ballesteros I, Delgado D, de Marcos A, Desvoyes B, Gutiérrez C, Mena M, Fenoll C. Timely expression of the Arabidopsis stoma-fate master regulator MUTE is required for specification of other epidermal cell types. Plant J. 2013;75:808–22. doi: 10.1111/tpj.12244. This paper rescued mute mutants using an inducible MUTE which expresses the gene in the correct cell types, but only in the presence of estradiol. Late rescue led to stomatal clusters from SLGCs that differentiated into guard mother cells and then guard cells, suggesting a role for MUTE in preventing SLGCs from differentiating into stomata. [DOI] [PubMed] [Google Scholar]
- 42.Desvoyes B, Ramirez-Parra E, Xie Q, Chua N-H, Gutierrez C. Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol. 2006;140:67–80. doi: 10.1104/pp.105.071027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weimer aK, Nowack MK, Bouyer D, Zhao X, Harashima H, Naseer S, De Winter F, Dissmeyer N, Geldner N, Schnittger a. RETINOBLASTOMA RELATED1 Regulates Asymmetric Cell Divisions in Arabidopsis. Plant Cell. 2012;24:4083–4095. doi: 10.1105/tpc.112.104620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee E, Lucas JR, Sack FD. Deep functional redundancy between FAMA and FOUR LIPS in stomatal development. Plant J. 2014;78:555–65. doi: 10.1111/tpj.12489. [DOI] [PubMed] [Google Scholar]
- 45.Ran JH, Shen TT, Liu WJ, Wang XQ. Evolution of the bHLH genes involved in stomatal development: Implications for the expansion of developmental complexity of stomata in land plants. PLoS One. 2013;8 doi: 10.1371/journal.pone.0078997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu T, Ohashi-Ito K, Bergmann DC. Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development. 2009;136:2265–76. doi: 10.1242/dev.032938. [DOI] [PubMed] [Google Scholar]
- 47.MacAlister CA, Bergmann DC. Sequence and function of bHLHs required for stomatal development in Arabidopsis are deeply conserved in land plants. Evol Dev. 2011;13:182–192. doi: 10.1111/j.1525-142X.2011.00468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]