Abstract
The present article introduces RetFM-J, a semi-automated ImageJ-based module that detects, counts, and collects quantitative data on nuclei of the inner retina from H&E-stained whole-mounted retinas. To illustrate performance, computer-derived outputs were analyzed in inbred C57BL/6J mice. Automated characterization yielded computer-derived outputs that closely matched manual counts. As a method using open-source software that is freely available, inexpensive staining reagents that are robust, and imaging equipment that is routine to most laboratories, RetFM-J could be utilized in a wide variety of experiments benefiting from high-throughput, quantitative, uniform analyses of total cellularity in the inner retina.
Keywords: Retinal whole-mount, retinal ganglion cell layer, image analysis, quantitative analysis, cell counter, ImageJ
1. Introduction
Cells of the inner retina are the subject of many types of research due to their ease of accessibility and importance in blinding eye diseases. The most prominent neurons of the inner retina are retinal ganglion cells (RGCs), which are the only afferent neurons of the retina. Along with them, the inner retina also contains displaced amacrine cells, glia, vascular-associated endothelial cells, pericytes, astrocytes, and a variety of phagocytic cells. Changes in these cells are a feature of many diseases (Levin and Gordon 2002), notably including glaucoma. Accordingly, studies of glaucoma often lead to an experimental need for assessing cellularity using retinal whole-mounts. This is especially common in studies of glaucomatous neurodegeneration or neuroprotection in mice, which often necessitate quantification among relatively large cohorts. While qualitative disease features, such as massive cell loss, presence of vascular proliferation, inflammation, or regional cell loss are often readily apparent from simple visual inspection of retinal whole-mounts, quantification of cellular features is more challenging and can be particularly time-consuming if performed manually.
To expedite cell counting, a handful of software programs have been developed for, or applied to, the analysis of rodent retinal whole-mounts. Among commercial programs, examples of programs that have been applied to automated counting of cells in retinal whole-mounts include MetaMorph (Markand, Saul et al. 2015), StereoInvestigator (Inman, Lambert et al. 2013), and IPlab (Soto, Oglesby et al. 2008). A handful of novel programs built by individual labs have also been reported (Danias, Lee et al. 2003, Salinas-Navarro, Jimenez-Lopez et al. 2009). Open source programs such as ImageJ and CellProfiler also have automated cell counting functionality that can be applied to analysis of retinal whole-mounts (Swanson, Schlieve et al. 2005, Zhong, Bradley et al. 2007), but are largely under-utilized. Among these options, there are recurrent challenges related to limited software accessibility, complicated workflow paths, and a frequent need for specialized equipment or reagents. To address these challenges, we report development of RetFM-J, an open source semi-automated software that detects, counts, and collects quantitative data on features of nuclei in H&E-stained whole-mounted retinas. RetFM-J was developed by reiterative automated and manual analyses of H&E-stained retinal whole-mounts and is packaged in the form of a plugin for Fiji and ImageJ (Abràmoff, Magalhães et al. 2004). As a freely available program that relies on robust, relatively inexpensive staining reagents, and standard light microscopes common to most biological laboratories, the detailed methodology and illustration of its use presented here aim to enable its use in a broad range of potential applications
2. Materials and supplies
2.1 Animal Husbandry
Inbred C57BL/6J mice (The Jackson Laboratory, Bar Harbor, ME) were housed at the University of Iowa Research Animal Facility, where they were maintained on a 4% fat NIH 31 diet provided ad libitum, housed in cages containing dry bedding (Cellu-dri; Shepherd Specialty Papers, Kalamazoo, MI), and kept in a 21°C environment with a 12-hour light:12-hour dark cycle. All experiments included mice of both genders. All mice were treated in accordance with the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research. All experimental protocols were approved by the Animal Care and Use Committee of the University of Iowa.
2.2 Measurement of total retinal area
Whole-mounted retinas stained with H&E were scanned at 40x magnification with a digital whole-slide scanner (ScanScope CS, Aperio, Vista, CA). The total area of each retina was measured by outlining the sample, determining the stained area using the “Color Deconvolution v.9” algorithm, and subtracting the area of the optic nerve. The areas of the central, mid-peripheral, and peripheral zones of eccentricity were calculated using circular regions of interest that divided each petal into approximately thirds. The length of each of the 4 petals (outer edge of optic nerve to edge of retina) was measured with the “Ruler” tool and used to calculate an average petal length. The central zone was defined as the area from the outer edge of the optic nerve to 1/3rd of the average petal length, the mid-peripheral zone as the area from the edge of the central zone to 2/3rds of the average petal length, and the peripheral zone as the area from the edge of the mid-peripheral zone to the edge of the tissue. The peripheral zone was calculated by subtracting the areas of central and mid-peripheral zones from the total retinal area.
2.3 Microscopy and imaging of retinal whole-mounts
Imaging was performed with a light microscope (BX52; Olympus, Tokyo, Japan) equipped with a digital camera (DP72; Olympus, Tokyo, Japan). CellSens Standard imaging software (Olympus, Tokyo, Japan) was used for capturing all color image files (RGB mode, .tif format), using identical software settings. The white balance and 1% spot option of the region tool were used to normalize exposures before capturing images. As needed, the CellSens Extended Focal Imaging module was used to correct for unevenness in the z-plane arising from the variable topography of the whole-mounted retina. For the extraction and counting of nuclei, raw image sets (n=24 images) for each retina were manually reviewed, regions containing artifacts (such as stain precipitate, tears, and debris) were manually excluded from each image, and nuclei counts of each image obtained using RetFM-J. Unless otherwise noted, we report cell densities as mathematical averages of all images analyzed per experiment ±1 SD. The extrapolation to total cells per retina used averages of each zone of eccentricity to derive anatomically weighted averages. For inclusion in the analysis of nuclei counts, each whole-mounted retina was required to pass a uniform set of inclusion criteria. First, 8 images had to be collected from each of the 3 regions (peripheral, mid-peripheral, and central) of the retina, for a total of 24 images. Second, after the removal of artifacts from images, the sum of included areas from all 24 images had to exceed 85% (2.86 of the 3.36 mm2) of the total retinal area sampled.
2.4 Counting and quantitative measurement of nuclei with RetFM-J
Reiterative automated and manual analyses of H&E-stained retinal whole-mounts led to development of a tool, called RetFM-J, a semi-automated imaging tool packaged in the form of a plugin for Fiji and ImageJ, which recognizes, segments, and extracts quantitative features of cell nuclei. For the extraction and counting of nuclei, raw image sets (n=24 images) for each retina were manually reviewed, and regions containing artifacts such as stain precipitate, tears, and debris were manually excluded from each image (Supplemental Figure 1). Analysis and nuclei extraction was performed using a series of sequential automated steps. First, image contrast was adjusted using a contrast limited histogram equalization (CLAHE) method to account for local areas of low contrast (Pizer, Amburn et al. 1987). Second, images were converted to grayscale and binarized using Otsu’s thresholding method (Otsu 1975). Third, nuclei were extracted using a watershed segmentation approach (Vincent and Soille 1991). Fourth, regions not corresponding to nuclei were excluded by applying size-based filters to reject extracted regions smaller than 10 μm2 or larger than 150 μm2. Finally, 41 quantitative features, including measurements of morphology, such as area, as well as color and intensity of each RGB channel, were computed for each extracted nucleus. The complete approach for the extraction and measurement of nuclei is implemented within the ImageJ plugin RetFM-J. Unless otherwise noted, we report cell densities as mathematical averages of all images analyzed per experiment ±1 SD. The extrapolation to total cells per retina used averages of each zone of eccentricity to derive anatomically-weighted averages.
2.5 Tests of imager, analyzer, histologic, and computational variability
To measure the influence that “imager” (person collecting images) had on the output of RetFM-J, three different people independently collected images from the same three retinal whole-mount preparations (n=24 images/retina, 72 images total). All images were subsequently analyzed using RetFM-J by a single person (analyzer). To test the influence that analyzer had on the output of RetFM-J, the same three people performed quantitative analyses on an identical batch of images taken from those described above. To test the potential influence that differences in retinal processing may have on the output of RetFM-J, one person performed analyses on three separate batches of retinas of the same strain and age, but differing with respect to the days on which tissues were collected, mounted, and stained. To test variability of RetFM-J itself, one image from a C57BL/6J retina (post-manual exclusion stage) was analyzed by RetFM-J in ten trials.
3. Detailed methods
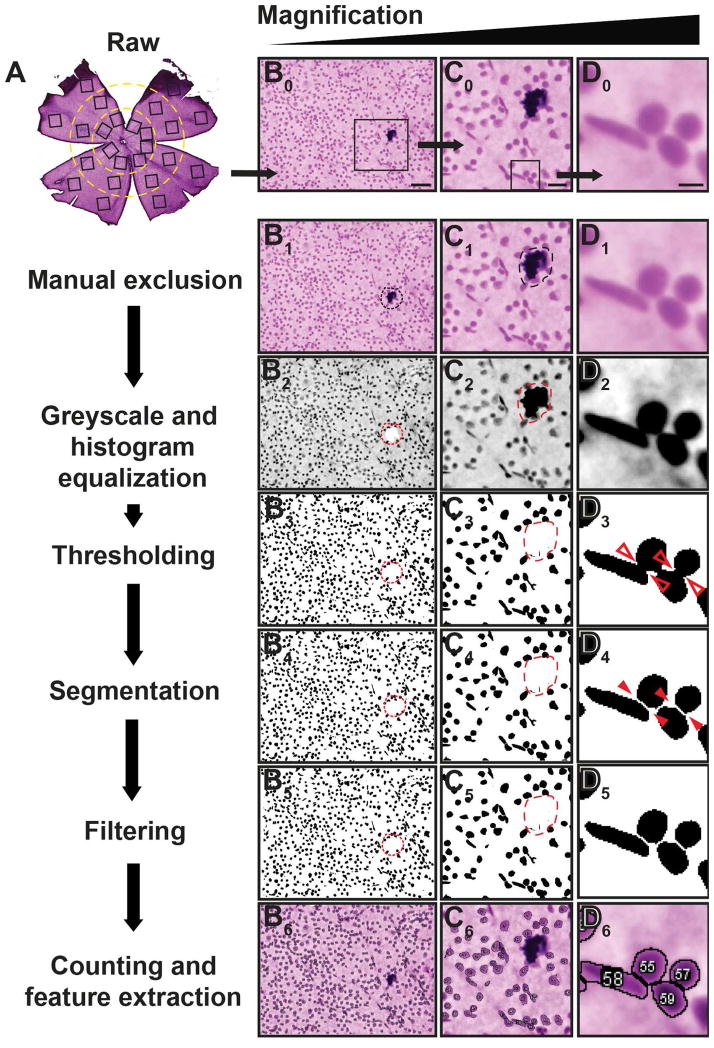
RetFM-J recognizes, segments, and extracts quantitative features of cell nuclei (Figure 1) from H&E-stained whole-mounted retinas. The protocol below has been developed for use in mice:
Figure 1. Processing of images for analysis in RetFM-J.
Flowchart (left, column A) coupled with images of H&E-stained whole-mount retinas at progressively higher magnification (right, columns B-D), illustrating one of the 24 fields (top row) and its subsequent processing (bottom rows). During the manual exclusion stage, artifacts such as debris (dashed circle) are disqualified from quantification for both area and cell count. All following steps, from greyscale conversion to counting and feature extraction, are performed in an automated fashion by RetFM-J. Note that most cells in close proximity to one another (open red arrowheads) are still appropriately segmented (solid red arrowheads). Scale bar = 50 μm (column A), 20 μm (column B), and 5μm (column C).
3.1 Preparation of retinal whole-mounts
The first step to performing nuclei counts with RetFM-J is to prepare H&E stained retinal whole-mounts. The following is our protocol for this procedure, but any method yielding high-quality, flat-mounted, retinas well-stained by H&E would be equally suitable.
All steps are performed at room temperature, unless otherwise noted. Deeply anesthetize mice with 1% ketamine-1% xylazine solution and perfuse transcardially with fresh 4% paraformaldehyde fixative in 1x phosphate-buffered saline (PBS). Enucleate and bisect eyes circumferentially anterior to the limbus, place the posterior cups in 4% paraformaldehyde at 4°C until a total fixation time of 4 hours has been reached, then rinse with and store in PBS at 4°C. Incubate posterior cups in 0.3% Triton X-100 solution overnight at 37°C with agitation, rinse in PBS, and incubate in 3% hydrogen peroxide-1% anhydrous monobasic sodium phosphate solution with agitation for no longer than 1 hour. Use forceps to tease apart the now detached neural retina from the retinal pigmented epithelium, and fine-scissors to help remove the sclera and complete the liberation. Once freed, transfer retinas to fresh 3% hydrogen peroxide:1% anhydrous monobasic sodium phosphate solution, and incubate for an additional 3 hours. Rinse retinas thoroughly in PBS then transfer to glass slides with 4 radial relief cuts to create 4 retinal petals with shallow relief cuts at the periphery of each petal to promote flat-mounting. Dab away excess PBS using filter paper, mount retinas flat with weighted circular coverslips (~10 grams), and place in a humidified chamber (to prevent cracking due to excessive drying) for semi-drying overnight. Gently remove coverslips by prying slip away from slide using a razor blade (coverslips should pop off cleanly and easily), and air dry slides for 15 minutes (min.). Note, retinas should have a translucent appearance with smooth texture, whereas an opaque-white color with brittle texture is indicative of over-drying. Stain slides with hematoxylin and eosin (H&E), under the following conditions: hematoxylin-1 min., rinse in double distilled water, acid-alcohol-3 seconds, rinse in double distilled water, bluing agent-1 min., rinse in double distilled water, 80% ethanol-1 min., eosin-2 seconds, 95% ethanol-30 seconds, 100% ethanol-30 seconds, and xylenes-3 times for 1 min. each. Use MM 24 mounting medium (Leica Biosystems, Buffalo Grove, IL) to apply coverslips, and allow slides to dry overnight. Filtering of hematoxylin prior to each use is recommended. Over time, mounted retinas may begin to develop an epiretinal layer having a “fish scale-like” appearance which can obstruct future imaging, a result of excessive drying and cracking of the mounting media. This issue is remedied by re-mounting; specifically, by immersing slides in 100% xylenes until coverslips passively detach from slides and then applying fresh mounting media and coverslips.
3.2 Microscopy and imaging of retinal whole-mounts for performing nuclei quantification
The second step to cell counting with RetFM-J is to collect digital images. The essential element of this step is to collect non-overlapping color images at 200X total magnification (RGB mode, .tif format) using identical software settings for the digital microscope camera (including options for normalizing exposures). The number and placement of images collected per retina can be varied according to the experimental need. For our purposes, we have sampled two non-overlapping images (1360×1024 px) from each of three zones of eccentricity (central, mid-peripheral, peripheral) in each of the four petals, resulting in a total of 24 images per retina (Supplemental Figure 2). Because each image has an area of 0.14 mm2, this is equivalent to a total sampling coverage of 3.36 mm2 (0.14 mm2 × 24 images) for each retina. This is equal to approximately 23.5% (3.36 mm2/14.4 mm2) of the total measured area of the inner retinal surface.
3.3 Manual exclusion of artifacts from images
In the third step to cell counting with RetFM-J, we advise users to manually exclude regions of artifact from the collected images (Supplemental Figure 1). Raw image files can be opened in Fiji using the drag and drop method, screened for artifacts that may be deemed as regions of exclusion (ROE), and be subsequently excluded from the analysis. ROE are manually traced using the freehand selection tool and left as active selections. Common examples of artifacts that should be excluded include: debris (stain precipitate and pigment granules; Supplemental Figure 1B), artifactual damage (rips and/or holes in the retina incurred during tissue processing; Supplemental Figure 1C), unstained or poorly stained areas providing little, or no, discernible information, and out-of-focus areas occurring by differences in focal plane throughout the sample and present in the final image (Supplemental Figure 1D; “flat-mounts” are seldom flat; careful use of microscope camera tools such as the Extended Focal Imaging module within the CellSens imaging software can greatly improve image quality and allow a greater proportion of each image to be analyzed).
3.4 Nuclei counts using RetFM-J
Automated counting of nuclei from the inner retina of whole-mounted preparations is performed by the ImageJ and Fiji-based plugin, RetFM-J, which is freely available for download from the ImageJ website (http://imagej.nih.gov/ij/). For general instructions on the use and troubleshooting of these image analysis software packages, please refer to the Fiji (http://fiji.sc/Fiji) and ImageJ websites.
In the fourth step to cell counting, RetFM-J is selected to run from the Plugins menu in Fiji or ImageJ. RetFM-J performs counts on all opened image files and excludes ROE (manually traced and active selections) defined during the previous screening step. Two dialogue boxes will prompt the user to: customize a provided parameter set and define the location for which output data files from the analysis should be saved. First, for the parameter box (entitled, cell count setup), define numerical ranges for a series of morphometric parameters, including size and shape, which may be adjusted to customize the inclusion/exclusion criteria for the analysis. Additional options, data filters, and commands appear in the form of a list with check boxes for further customization. For our purposes and all analyses presented in this manuscript, the following values for each parameter were used: size (min-max) = 10–150 μm2; circularity (min-max) = 0–1.0; scale = as appropriate for images; with boxes for Invert current ROI selections, make master output file, and all three color (RGB) channels checked. Second, for the destination box (entitled, output directory), choose a path for saving and storing output text and image files. Once the cell count and directory setup boxes have been completed, the plugin runs on all open image files. During the analysis, RetFM-J performs a series of automated image processing steps on image files to compute size, shape, and color intensity statistics of nuclei detected within input images. Together, these steps segment and identify ROI corresponding to nuclei in images of the inner retina (Figure 1).
In the fifth step to cell counting using RetFM-J, data from the output files are prepared for data extraction. After each analysis, RetFM-J generates three output data files (one image file, .tif and two text files, .txt) that are saved to the indicated directory. The first text file (entitled, image-areas.txt) provides a cumulative list of all image files by name along with the total area sampled within each image. The second text file (title defaults to input image file name.txt) provides a list of every counted nucleus and its corresponding quantitative feature information (from all 41 categories) for each image file analyzed. A master features file (entitled, cell-features.txt), lists all counted nuclei from all images in the analysis within the same file, is generated by checking the make master output file box in the cell count setup box. This master features file takes extra time to generate, and should not be opened until creation of the file is complete (i.e. when the size of the file stops increasing). Once these text files have been created, they are opened and saved as spreadsheet files (.xls) using Microsoft Excel for easier extraction and transfer to other file formats. The third file is a duplicate of each original input image file (entitled, overlayed-input image file name.tif) in which all counted nuclei have been outlined and assigned a unique identification number. This file provides a visual reference for spot-checking what nuclei were counted and not counted.
In the sixth and final step to cell counting using RetFM-J, data from these output files is evaluated for inclusion and calculation of cell density. Although we find that the vast majority of samples are of sufficient quality for quantification (>80%), use of a uniform set of inclusion criteria can help identify potentially problematic samples. We have found two criteria to be particularly useful; first, that the correct number of images was able to be collected from the intended areas (n = 24 total, 6 from each petal with 2 from each zone of eccentricity); and second, that after removal of artifacts from images, the sum of included areas from all images must exceed 85% of the total area imaged (3.36 mm2). Once a sample is deemed sufficient for inclusion, cell densities are calculated for each retina. The sampling areas and nuclei counts for each image are summed to calculate the total retinal area sampled and total nuclei counted. These data may be expressed in terms of density for each zone of eccentricity or be extrapolated to total cells for each retina. Mathematical cell density is calculated by dividing the sum of all counted nuclei by the sum of all sampling areas. Anatomically weighted cell density is calculated using average cell densities for each zone of eccentricity, weighted to the area of the total retina that each zone occupies. To convert density to total cell counts for each retina, measure the total area of each whole-mount and each zone of eccentricity, multiply the average density for each zone by its average area, and sum the values for all zones.
4. Potential pitfalls and trouble shooting
The methods described here allow semi-automated counting of nuclei of the inner retina from H&E-stained whole-mounted retinas. The major caveat to this approach is that the inner retina is a diverse cellular environment, which in mice contains approximately equal numbers of RGCs and displaced amacrine cells (Drager and Olsen 1981, Jeon, Strettoi et al. 1998, Schlamp, Montgomery et al. 2013). Thus, data of total cellularity should be interpreted and described accordingly, and in some types of experiments, it is likely that additional experiments would be required to determine cell-type specificity of any observed cell gain or loss. As described in an accompanying manuscript, RetFM-J data can be used in combination with automated classifiers that simultaneously yield cell-type specific quantification to partially overcome this hurdle. In addition to the issue of cell-type specificity, we additionally draw attention to the following issues and potential pitfalls relevant to RetFM-J use:
Because the H&E staining protocol is incompatible with immunofluorescence (Fischer, Jacobson et al. 2008), the approach described above is cumbersome when used in conjunction with immunohistochemistry or the detection of retrograde tracers. Although we have obtained some data by first performing antibody labeling and imaging, then performing H&E staining and imaging of the same areas second, this is a laborious process and cannot easily be adapted for routine comparisons.
Anatomy of the retina creates inherent challenges to counting resulting from the nerve fiber layer, which can partially conceal imaging of stained whole-mounts, and cell clumping, which is particularly common in the central retina. Thus, an experimental approach focusing on cellularity in only the peripheral retina may yield higher quality data with more statistical power for detecting cell loss, though this is obviously a tact that would not be appropriate in all types of experiments.
We have thus far focused our efforts on use of RetFM-J with mice; while the program should be easily adaptable for analysis of human retinas, we point out that the thicker nerve fiber layer of the human retina is likely to be a challenge.
Users should specify whether they are reporting mathematical or anatomically weighted averages of cell densities.
Because total retinal area can change with disease state (Danias, Lee et al. 2003), it should be stated whether corrections for expansion of the total retinal area were considered.
5. Results
5.1 RetFM-J enables uniform quantification of total cellularity in the inner retina
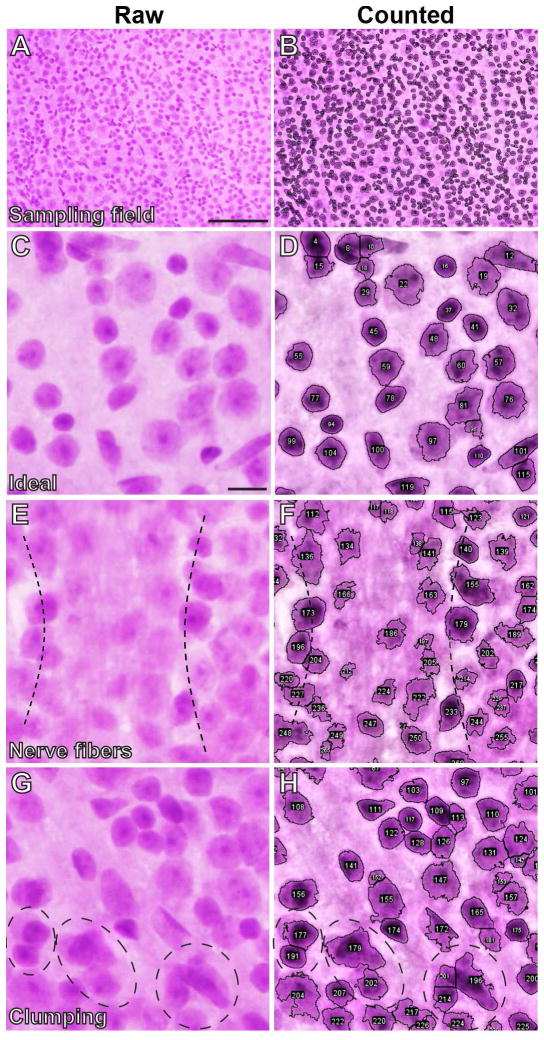
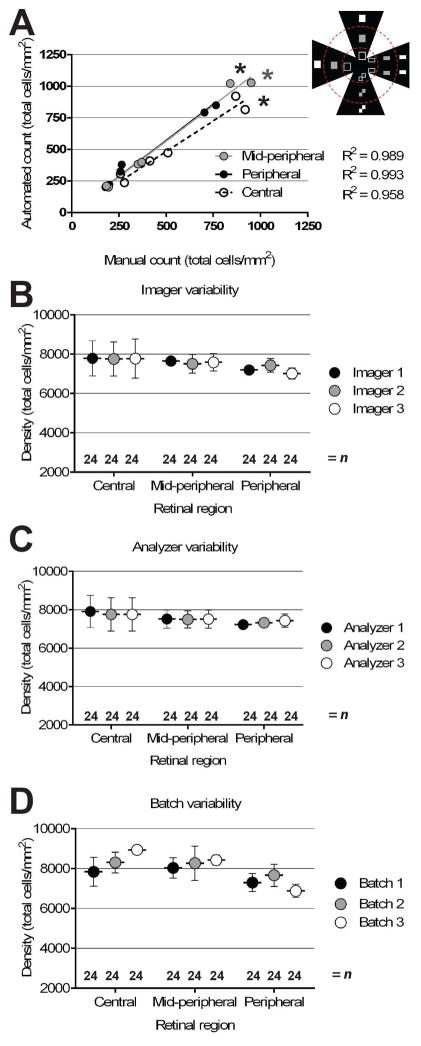
Applying RetFM-J to previously uncharacterized images of whole-mounted retinas from C57BL/6J mice successfully counts nuclei. Qualitatively, comparing images before and after analysis indicated that the vast majority of cells were correctly recognized (Figure 2A–D), with occasional partially concealed views associated with nerve fibers (Figure 2E–F) or cell clumps (Figure 2G–H) typically being associated with instances of less certainty. To test the precision of RetFM-J, we compared manual to automated analyses on a set of images showing incrementally sized fields (quarter-, half-, and full-sized), with equal numbers of images collected from each of three zones of eccentricity (n=18 images; 2 each of the central, mid-peripheral, and peripheral areas) for retinas (n=3). Automated counts of total cellularity were highly correlated with manual counts across all zones (Figure 3A), with highest correlation in the periphery (peripheral R2 = 0.993, p < 1.0E−4; mid-peripheral R2 =0.989, p < 1.0E−4; central R2 = 0.958, p < 7.0E−4). Instances of missed nuclei were often associated with cell clumps or concealment by the nerve fiber layer. In testing the influence that various technical factors have on RetFM-J output, we found that none of the following made a difference in the total cell density measured for each retinal region: imager used (Figure 3B; p=0.77, ANCOVA), analyzer used (Figure 3C; p=0.66, ANCOVA), or histologic batch (Figure 3D; p=0.17, ANCOVA). Moreover, repeat counting of cells with RetFM-J from an individual image resulted in identical cell counts (n=10 repeat trials, 886 ± 0 total cells/mm2).
Figure 2. Qualitative evaluations of RetFM-J in performing automated cell counting.
Microscopic image pairings collected from the inner retina of H&E-stained whole-mount retinas of the raw (left column) and counted (right column) image to show the performance of automated detection and counting of nuclei by RetFM-J. (A) Image representing a full-sized sampling field from the mid-peripheral zone. (B) Automated nuclei recognition and counting (nuclei outlined and assigned unique identification numbers) of the image shown in panel A by RetFM-J. Magnified image pairs, cropped from full-sized sampling fields, showing one example of an ideal performance by RetFM-J and two examples of inherent biological characteristics that pose challenges to automated detection and counting of nuclei. (C) Representative image from the peripheral retina with features that appear to be relatively discernable and ideal for analysis. (D) Automated nuclei recognition and counting by RetFM-J of the image shown in panel C. (E) Image from the central retina showing concealment of nuclei by overlying nerve fibers (between dotted lines). (F) Automated nuclei recognition and counting by RetFM-J of the image shown in panel E with decreased discrimination of nuclei from non-nuclei in the region concealed by nerve fibers. (G) Image from the mid-peripheral retina showing overlapping or clumped nuclei (circular dotted lines). (H) Automated nuclei recognition and counting by RetFM-J of the image shown in panel G with decreased discrimination of individual nuclei within the clumps. Scale bar = 100 μm (A, B) and 10 μm (C–H).
Figure 3. Quantitative evaluation of RetFM-J in performing automated cell counting.
(A) Automated versus manual counts from an identical set of images captured from differing zones of eccentricity. The image set contained images (n=18 per retina) of various sizes (n=6 for each full-, half-, and quarter-sized) from retinas of three mice, with approximate sizes and locations shown in the schematized inset. Correlations (R2) between manual and automated counts exceeded 0.95 and were strongest in the peripheral and mid-peripheral zones. (B–D) Graphs of cell density versus zone of eccentricity, showing no difference in cell density regardless of which imager (p=0.77, ANCOVA), analyzer (p=0.66, ANCOVA), or histologic batch (p=0.17, ANCOVA) was tested. C57BL/6J mice at 8–9 weeks of age; mean ± 1 SD.
6. Discussion
As a methodology for the quantification of RGCs, the tools and approach presented here could be utilized in a wide variety of experiments. One advantage of RetFM-J-based counting and quantification pertains to uniformity. All manual methods are prone to higher variation than automated methodologies. Among the options for computer-assisted approaches for quantifying cells in retinal whole-mounts, the methodology described here has the advantages of using open-source software and widely available reagents, making it possible for two people to analyze the same retina and obtain the same data in a uniform fashion. Thus, the approach, and its continued evolution, has the potential to be a useful complement to existing methods for assessing retinal whole-mounts.
Supplementary Material
Whole-mounted retina stained with H&E showing: (A) total inner surface area that may be sampled, (B) central, mid-peripheral, and peripheral zones of eccentricity (dotted circles), and (C) approximate size and location of 24 uniform microscopic fields used for sampling each retina (square boxes).
H&E-stained whole-mount retinas showing examples of artifacts that were manually excluded in quantitative analyses using RetFM-J. (A) Representative microscopic field of an ideal area, devoid of artifact. Representative fields highlighting examples of: (B) debris, (C) tears or holes, and (D) out of focus areas.
Highlights and unique aspects of this research include.
RetFM-J counts and quantifies features for all nuclei in the inner retina.
RetFM-J is packaged as a freely available plugin on the open ImageJ-based platform.
Protocols for dissection and H&E staining of mouse retinal whole-mounts are detailed.
Acknowledgments
We thank John Fingert of the Department of Ophthalmology and Visual Sciences at the University of Iowa Hospitals and Clinics for helpful discussions. We thank Katherine Gibson-Corley, Mariah Leidinger, and Allyn Lambertz of the University of Iowa Comparative Pathology Core for technical assistance. In regard to funding, this work was supported by US Dept. of Veterans Affairs, RR&D (C1481-R) and National Institutes of Health, NEI (R01EY017673) grants to MGA. MC and TS are supported by National Institutes of Health, NEI grant (R01023187).
Footnotes
Disclosures
None.
Roles in the project
A. Hedberg-Buenz developed the methodology, collected experimental data, and spearheaded writing of the manuscript. M. Christopher, T.E. Scheetz, and M.D. Abramoff developed the code for RetFM-J. C. Lewis, K.J. Meyer, D.S. Rudd, and L.M. Dutca contributed to data collection. All authors contributed to revising the final draft of the manuscript. M.G. Anderson oversaw all aspects of this project.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abràmoff MD, Magalhães PJ, Ram SJ. Image processing with ImageJ. Biophotonics international. 2004;11(7):36–43. [Google Scholar]
- Danias J, Lee KC, Zamora MF, Chen B, Shen F, Filippopoulos T, Su Y, Goldblum D, Podos SM, Mittag T. Quantitative analysis of retinal ganglion cell (RGC) loss in aging DBA/2NNia glaucomatous mice: comparison with RGC loss in aging C57/BL6 mice. Invest Ophthalmol Vis Sci. 2003;44(12):5151–5162. doi: 10.1167/iovs.02-1101. [DOI] [PubMed] [Google Scholar]
- Drager UC, Olsen JF. Ganglion cell distribution in the retina of the mouse. Invest Ophthalmol Vis Sci. 1981;20(3):285–293. [PubMed] [Google Scholar]
- Fischer AH, Jacobson KA, Rose J, Zeller R. Hematoxylin and eosin staining of tissue and cell sections. CSH Protoc. 2008;2008 doi: 10.1101/pdb.prot4986. pdb.prot4986. [DOI] [PubMed] [Google Scholar]
- Inman DM, Lambert WS, Calkins DJ, Horner PJ. alpha-Lipoic acid antioxidant treatment limits glaucoma-related retinal ganglion cell death and dysfunction. PLoS One. 2013;8(6):e65389. doi: 10.1371/journal.pone.0065389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon CJ, Strettoi E, Masland RH. The major cell populations of the mouse retina. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1998;18(21):8936–8946. doi: 10.1523/JNEUROSCI.18-21-08936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin LA, Gordon LK. Retinal ganglion cell disorders: types and treatments. Prog Retin Eye Res. 2002;21(5):465–484. doi: 10.1016/s1350-9462(02)00012-5. [DOI] [PubMed] [Google Scholar]
- Markand S, Saul A, Roon P, Prasad P, Martin PM, Rozen R, Ganapathy V, Smith SB. Retinal ganglion cell loss and mild vasculopathy in methylene tetrahydrofolate reductase (Mthfr) deficient mice: a model of mild hyperhomocysteinemia. Invest Ophthalmol Vis Sci. 2015 doi: 10.1167/iovs.14-16190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu N. A threshold selection method from gray-level histograms. Automatica. 1975;11(285–296):23–27. [Google Scholar]
- Pizer SM, Amburn EP, Austin JD, Cromartie R, Geselowitz A, Greer T, ter Haar Romeny B, Zimmerman JB, Zuiderveld K. Adaptive histogram equalization and its variations. Computer Vision, Graphics, and Image Processing. 1987;39(3):355–368. [Google Scholar]
- Salinas-Navarro M, Jimenez-Lopez M, Valiente-Soriano FJ, Alarcon-Martinez L, Aviles-Trigueros M, Mayor S, Holmes T, Lund RD, Villegas-Perez MP, Vidal-Sanz M. Retinal ganglion cell population in adult albino and pigmented mice: a computerized analysis of the entire population and its spatial distribution. Vision Res. 2009;49(6):637–647. doi: 10.1016/j.visres.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Schlamp CL, Montgomery AD, Mac Nair CE, Schuart C, Willmer DJ, Nickells RW. Evaluation of the percentage of ganglion cells in the ganglion cell layer of the rodent retina. Molecular vision. 2013;19:1387–1396. [PMC free article] [PubMed] [Google Scholar]
- Soto I, Oglesby E, Buckingham BP, Son JL, Roberson ED, Steele MR, Inman DM, Vetter ML, Horner PJ, Marsh-Armstrong N. Retinal ganglion cells downregulate gene expression and lose their axons within the optic nerve head in a mouse glaucoma model. J Neurosci. 2008;28(2):548–561. doi: 10.1523/JNEUROSCI.3714-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson KI, Schlieve CR, Lieven CJ, Levin LA. Neuroprotective effect of sulfhydryl reduction in a rat optic nerve crush model. Invest Ophthalmol Vis Sci. 2005;46(10):3737–3741. doi: 10.1167/iovs.05-0155. [DOI] [PubMed] [Google Scholar]
- Vincent L, Soille P. Watersheds in Digital Spaces: An Efficient Algorithm Based on Immersion Simulations. IEEE Trans Pattern Anal Mach Intell. 1991;13(6):583–598. [Google Scholar]
- Zhong L, Bradley J, Schubert W, Ahmed E, Adamis AP, Shima DT, Robinson GS, Ng YS. Erythropoietin promotes survival of retinal ganglion cells in DBA/2J glaucoma mice. Invest Ophthalmol Vis Sci. 2007;48(3):1212–1218. doi: 10.1167/iovs.06-0757. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Whole-mounted retina stained with H&E showing: (A) total inner surface area that may be sampled, (B) central, mid-peripheral, and peripheral zones of eccentricity (dotted circles), and (C) approximate size and location of 24 uniform microscopic fields used for sampling each retina (square boxes).
H&E-stained whole-mount retinas showing examples of artifacts that were manually excluded in quantitative analyses using RetFM-J. (A) Representative microscopic field of an ideal area, devoid of artifact. Representative fields highlighting examples of: (B) debris, (C) tears or holes, and (D) out of focus areas.