Abstract
Recently, the field of proteomics has rapidly expanded in its application towards clinical research with objectives ranging from elucidating disease pathogenesis to discovering clinical biomarkers. As proteins govern and/or reflect underlying cellular processes, the study of proteomics provides an attractive avenue for research as it allows for the rapid identification of protein profiles in a biological sample. Inflammatory bowel disease (IBD) encompasses several heterogeneous and chronic conditions of the gastrointestinal tract. Proteomic technology provides a powerful means of addressing major challenges in IBD today, especially for identifying biomarkers to improve its diagnosis and management. This review will examine the current state of IBD proteomics research and its use in biomarker research. Furthermore, we also discuss the challenges of translating proteomic research into clinically relevant tools. The potential application of this growing field is enormous and is likely to provide significant insights towards improving our future understanding and management of IBD.
Keywords: Proteomics, Inflammatory bowel disease, Biomarkers, Molecular diagnostic techniques, Mass spectrometry
Core tip: Proteomic methods provide a powerful tool that can be applied to the discovery of disease markers, allowing for rapid identification and quantification of proteins. Inflammatory bowel disease (IBD) currently faces many challenges, ranging from the elucidation of its pathophysiology to the accurate diagnosis in patients. Proteomics has been widely employed in many disease in the search of biomarkers, particularly cancer proteins. It has great potential to improve both our understanding and clinical management of IBD. Our review summarises the current application of proteomics to IBD and discusses challenges relating to translation into clinical practice.
INTRODUCTION
Inflammatory bowel disease (IBD) encompasses a group of conditions characterised by chronic gastrointestinal inflammation, with the two major subtypes being Crohn’s disease (CD) and ulcerative colitis (UC). Differentiating between subtypes of IBD sometimes has a degree of uncertainty due to overlapping clinical and pathological features[1]. Despite clinical evaluation, radiological, endoscopic and histopathological testing by expert physicians, up to 20% of IBD cases are classified as “indeterminate colitis” or “IBD undifferentiated”[2,3]. However, accurate classification of IBD is essential as response to medication, surgical indications and prognosis can vary between UC and CD[4]. The field of proteomics is a rapidly expanding area of research that has been employed in many diseases such as cancer[5,6], exploring everything from understanding disease pathways to discovering diagnostic markers[7-9]. This review examines the current state of biomarkers in IBD, with particular reference to the application of proteomics.
CURRENT BIOMARKERS IN IBD
Biomarkers are measureable substances that can objectively evaluate either physiological processes or therapeutic outcomes[10] and could potentially play a pivotal role in IBD as cheap and non-invasive alternatives to endoscopy[11]. Different biomarkers could be beneficial across all aspects of IBD (illustrated in Figure 1)[12]. The major commercially available biomarkers are summarised below based on their application in Table 1. Whilst some of these biomarkers demonstrate high diagnostic accuracy, they are currently unable to replace endoscopy entirely and limited only to being adjuncts[11,13]. Therefore, there is a prevailing need for the development of additional non-invasive biomarkers that are sufficiently sensitive and specific in the diagnosis and prognosis of IBD.
Figure 1.
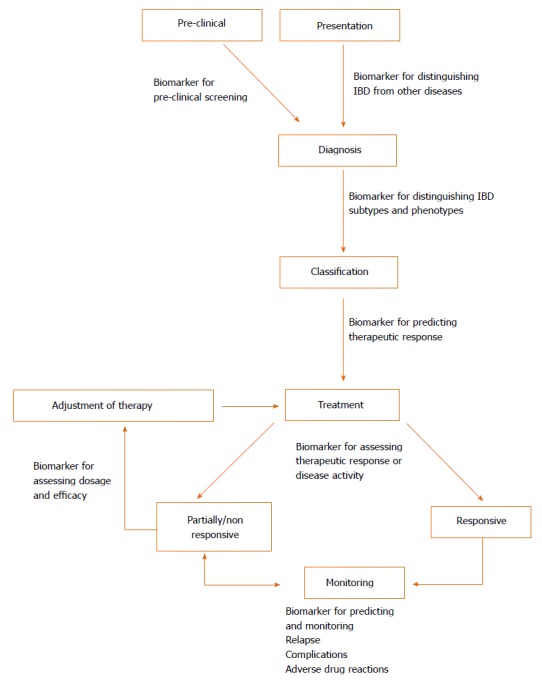
Potential application of biomarkers in inflammatory bowel disease in different stages of clinical management. When presenting clinically, one important use of biomarkers could be in the diagnosis of IBD, as well as differentiating subtypes (e.g., UC vs CD) and phenotypes (e.g., fistulising). Whilst not currently part of management, preclinical screening for IBD may be a possibility. Biomarkers can also be used to predict response to therapy and objectively measure therapeutic response and disease severity. Due to the relapsing and remitting course of IBD, monitoring is necessary for assessing relapse, adverse outcomes and complications (e.g., strictures, fistulas and colorectal cancer). Most of these aspects necessitate endoscopic procedures and would benefit from biomarker substitutes. CD: Crohn’s disease; UC: Ulcerative colitis; IBD: Inflammatory bowel disease.
Table 1.
Current biomarkers and their utility in inflammatory bowel disease management[12]
| Application | Biomarker | Utility |
| Diagnosis of IBD | Fecal calprotectin[69] | Sensitivity: 89%-98%, specificity: 81%-91% |
| Fecal lactoferrin[70] | Sensitivity: 80%, specificity: 82% | |
| Fecal 100A12[71] (differentiating from IBS) | Sensitivity: 86%, specificity: 96% | |
| CRP[72-74] | Sensitivity: Approximately equal 100% in CD, approximately equal 50% in UC poor specificity | |
| Distinguishing UC and CD | ASCA[75] | Sensitivity: 40%-50%, specificity: > 90% in CD |
| pANCA[75] | Sensitivity: 57%, specificity: 92% | |
| Escherichia coli antibodies (Anti-OmpC, Anti-I2, Anti-CBir1)[76] | Sensitivity: 18%-55%, specificity: 76%-93%[76] | |
| Marker of disease activity | Fecal lactoferrin[77,78] | Sensitivity: 66%-80% |
| Specificity: 60%-100% | ||
| Fecal calprotectin[77,78] | Sensitivity: 70%-100% | |
| Specificity: 44%-100% | ||
| CRP[78] | Sensitivity: 48% | |
| Specificity: 91% | ||
| Assessing mucosal healing | Fecal calprotectin | Several studies demonstrate significant reduction in biomarker in the presence of mucosal healing with treatment |
| Predicting disease course | Fecal lactoferrin[77] | May be associated with complications including; structuring or fistulising disease, and small bowel disease pANCA may predict aggressive UC and pouchitis following surgery[79] |
| ASCA | ||
| pANCA Anti-I2, Anti-OmpC[12] | ||
| Predicting Relapse within 12 mo | Fecal calprotectin[80,81] | Sensitivity: 69%-90% |
| Specificity: 69%-82% | ||
| Positive predictive value: 81%/87% (UC/CD) | ||
| Negative predictive value: 90%/43% (UC/CD) | ||
| Fecal lactoferrin[81] | Sensitivity: 62% | |
| Specificity: 65% | ||
| Predicting therapeutic response | pANCA[82] | Conflicting reports, possible lower response rate to infliximab in patients with a positive serology |
| Anti-I2[83] | 94% responded to fecal diversion |
CD: Crohn’s disease; UC: Ulcerative colitis; IBD: Inflammatory bowel disease; ASCA: Anti-Saccharomyces cerevisiae; pANCA: Perinuclear antineutrophil cytoplasmic antibodies; CRP: C-Reactive protein; PF4: Platelet factor 4.
PROTEOMICS
The term “proteome” was initially defined as the total protein complement encoded by a given genome[14] but now also encompasses any isoforms, post-translational modifications, interactions and effectively anything “post-genomic”[15]. The study of proteomics involves large scale detection, identification and characterisation of proteins, making it highly promising for biomarker discovery across many diseases[16]. The most common method applied is a combination of two-dimensional electrophoresis (2-DE) and mass-spectrometry. 2-DE provides a powerful tool isolating proteins that differ in abundance between cases and controls[17]. Mass spectrometry can then identify proteins utilising techniques such as “surface enhanced laser desorption/ionisation time-of-flight” (SELDI-TOF) or “matrix-assisted laser desorption/ionisation time-time-of-flight” (MALDI-TOF). Both these technique involve fragmentation of proteins into peptides, determining their mass-to-charge ratio based on their “time-of-flight” within an electric field and comparing their peptide mass signatures to a database of known proteins to identify the original protein.
Although mass spectrometry is not inherently quantitative, many methods have been developed to achieve accurate quantitative data[17,18]. The crux of selecting candidate biomarkers in proteomic studies rely detecting differences in abundances between cases and controls; therefore quantitative proteomics is an essential aspect. Multiple reaction monitoring (MRM) is a quantitative technique that achieves absolute quantitation and has a relatively high sensitivity when detecting peptides in low abundance, suiting it towards application in proteomic biomarker studies[19].
APPLYING PROTEOMICS TO IBD
The process leading up to clinical implementation of a novel proteomic biomarker can be divided into three major stages of a pipeline: Discovery, verification and validation, which all vary in both aim and study design (Figure 2)[20]. At present, the application of proteomics in IBD (and many other diseases) remains largely in its infancy in the initial discovery phase. This stage involves the rapid analysis of entire protein profiles within a target sample (e.g., plasma from an IBD patient), to screen for proteins that have relative differences in abundance compared to control samples[21]. The main disadvantage however, is that these discovery experiments do not provide absolute quantification and are labour intensive (and therefore typically have small sample sizes). The “verification” and “validation” stages addresses these issues by confirming the presence of and quantifying candidate markers in larger populations to assess their value in clinical usage.
Figure 2.
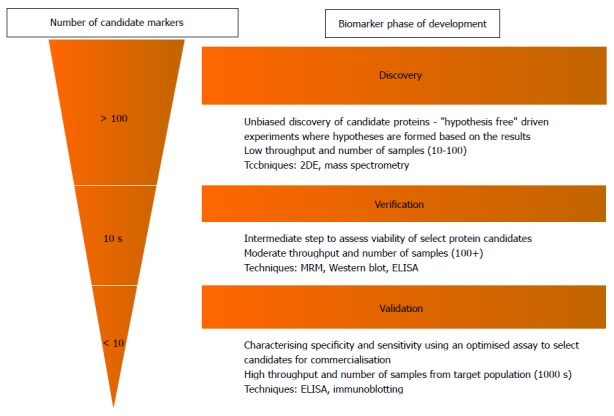
Biomarker “Pipeline” indicating the various stages from biomarker discovery to clinical application[20]. The number of candidate proteins (rough estimate of numbers indicated in figure) is narrowed down significantly in each step, selecting only the best candidates for further assessment and characterisation in a larger sample. The methodology also varies between the different phases. The early discovery phase uses low throughput methods such as 2-DE and mass spectrometry to screen large numbers of proteins in a low number of samples. Verification and validation require much more accurate quantitative methods as candidate proteins are narrowed down from the discovery phase and are assessed for their clinical utility in a large target population. This requires higher throughput methods such as MRM and immunoassays such as ELISA. CD: Crohn’s disease; UC: Ulcerative colitis; IBD: Inflammatory bowel disease; MRM: Multiple reaction monitoring.
Biomarker discovery studies
Proteomic studies involving IBD biomarkers have been divided into those relating to diagnosis and those pertaining to disease characteristics.
The most common approach towards biomarker discovery in proteomics involves assessing relative differences in proteins between cases and controls, for example, identifying which protein is differentially expressed between IBD patients and healthy controls. Furthermore, with the common objective of developing a clinically relevant assay, many groups have analysed plasma/serum for candidate markers (summarised in Table 2).
Table 2.
Proteomic studies for discovering diagnostic inflammatory bowel disease biomarkers
| Ref. | Bio-sample | Sample size | Proteomic technique | Results |
| Meuwis et al[22] | Serum | CD: 30 | SELDI-TOF | 4 candidate proteins selected for high diagnostic value; PF4, MRP8, FIBA, Hpα 2. PF4 and Hpα 2 were also confirmed and correlated using ELISA and immunoblotting |
| UC: 30 | ||||
| Inflammatory control: 30 | ||||
| Healthy controls: 30 | ||||
| Kanmura et al[23] | Blood | CD: 22 | SELDI-TOF | Plasma concentrations of HNP1, 2 and 3 were significantly higher in active UC compared to inactive UC, CD and control patients |
| UC: 48 | ||||
| Colorectal Cancer: 5 | ||||
| Infectious colitis: 6 | ||||
| Healthy controls: 13 | ||||
| Hatsugai et al[24] | Blood | CD: 13 | 2-DE | Multivariate analysis of peripheral blood mononuclear cells protein profile 58 protein) allowed for accurate discrimination between UC and CD |
| UC: 17 | MALDI-TOF | |||
| Healthy controls: 17 | ||||
| Nanni et al[25] | Blood | Healthy controls: 48 | Liquid chromatography quadrupole-TOF | Exopeptidase activity may distinguish CD from UC. Label free method developed could accurately distinguish synthetic spiked samples of serum |
| CD: 15 | SELDI-TOF | |||
| Sumramanian et al[26] | Serum | CD: 48 | Protein signature of 12 mass: Charge peaks could classify CD with approximately equal 95% sensitivity/specificity | |
| UC: 62 | 4 proteins identified as clinically useful | |||
| Nanni et al[27] | Serum | Healthy controls: 48 | Solid-phase extraction MALDI-TOF | 20 protein signals could be used to accurately classify IBD patients |
| CD: 15 | ||||
| UC: 26 | ||||
| Vaiopoulou et al[28] | Serum | CD: 24 (12 adults, 12 children) | 2-DE | Clusterin was found to be overexpressed in adult CD. Ceruloplasmin and apolipoprotein B-100 was over-expressed in children |
| MALDI-TOF | ||||
| Han et al[34] | Intestinal biopsy | CD: 3 | Liquid chromatography quadrupole-TOF | Increased in UC: TTBK2, SYNE2, SUCLG2, POSTN |
| UC: 4 | Up-regulated in CD: ANXA2, EPX, LAP3, RDX | |||
| Inflammatory polyps: 2 | Up-regulated in IBD: S100A8, MPO, DEFA1B | |||
| Up-regulated in CD (P < 0.05 AND > 2x increase): PRG2, LCP1, PSME1 | ||||
| Normal colon: 3 | ||||
| M’koma et al[35] | Colon samples | CD: 27 | Histology directed MALDI-TOF | 5 m:z peaks were identified and cross-validated for the differentiation of UC and CD |
| UC: 24 | ||||
| Seeley et al[36] | Colon samples | CD: 26 | Histology directed MALDI-TOF | Using a support vector machine and 25 m:z peaks, UC and CD cases were predicted in 93.3% and 60.4% respectively. A lower spectral accuracy cut-off increased sensitivity |
| UC: 36 | ||||
| Wasinger et al[39] | Serum | UC: 27 | MRM | SPP24 differentiated IBD patients from healthy controls |
| CD: 56 | α-1-microglobulin distinguished patients with UC in remission from healthy controls | |||
| Controls: 14 | ||||
| RA controls: 12 |
CD: Crohn’s disease; UC: Ulcerative colitis; IBD: Inflammatory bowel disease; MRM: Multiple reaction monitoring; PF4: Platelet factor 4; MRP8: Myeloid related protein 8; FIBA: Fibrinopeptide A; Hpα2: Haptoglobin α2.
In 2007, Meuwis et al[22] reported a proteomic profile detected with SELDI-TOF MS that could discriminate active UC and CD with a high sensitivity and specificity, performing similarly or better than current ANCA and ASCA serology. From the protein spectra detected, platelet factor 4, myeloid related protein 8, fibrinopeptide A and haptoglobin α2 were considered diagnostically important. Kanmura et al[23] examined UC serum samples using SELDI-TOF MS and identified that human neutrophil peptide (HNP) 1-3 was differentially expressed. HNP 1-3 was confirmed by ELISA to differentiate active UC from inactive UC, all CD cases and controls, but not colorectal cancer. Similar studies using variants of mass spectrometry have yielded similar results where protein profiles could accurately distinguish between selected UC and CD cases[24-27]. A recent study by Vaiopoulou et al[28] sought to investigate pediatric biomarkers for CD by comparing the proteomic profile between adult and pediatric CD patients. 3 proteins (ceruloplasmin, clusterin and apolipoprotein B-100) were shown to be significantly different between the two cohorts. Whilst the plasma proteome is the most comprehensive collection of proteins, potential biomarkers are more difficult to detect as they exist in significantly lower concentrations compared to other proteins such as albumin[20,29,30]. The alternative approach that has also become popular involves sampling “proximal fluid”, as any biological material directly sampled from the site of disease is likely to contain greater concentrations of potential biomarkers relative to plasma[20,30-32]. Employing a similar rationale, direct sampling of diseased tissue in IBD (a far simpler task compared to other diseases due to routine endoscopic biopsies) has been utilised for proteomic experiments (Table 2). Shkoda et al[33] reported the first proteomic study of intestinal tissue, identifying nine statistically significant proteins delineating inflamed IBD tissue from non-inflamed controls. Furthermore, 40 proteins were further detected between inflamed and non-inflamed UC tissue, although only two pairs of patient samples were analysed. Similarly, Han et al[34] identified a large number of differentially expressed proteins (37 relevant for CD, 27 for UC and 11 associated with general IBD) that were seen as candidate biomarkers. M’koma and colleagues conducted two studies that identified spectral peaks representing unknown protein profiles and reported being able to accurately distinguish between the UC and CD using an algorithm[35,36]. These tissue findings however are likely to require validation in plasma samples as the aim involves develop a clinical assay such as a blood test.
A similar demand for objective biomarkers exists across all aspects of IBD patient management, as such these markers have been investigated in a number of studies (summarised in Table 3). Han et al[34] identified 16 additional proteins that were expressed differently between active and inactive CD. Kanmura et al[23] associated a higher level of HNP 1-3 with a positive response following induction of corticosteroid therapy, whilst non-responders had lower HNP 1-3 levels. Meuwis et al[37] published a second report which identified a serum protein profile which correlated with infliximab response. Gazouli et al[38] performed a similar study using MALDI-TOF MS, identifying 15 proteins that were differentially expressed amongst patients that responded differently to infliximab. They were however, unable to confirm the findings by Meuwis et al[37].
Table 3.
Proteomic studies for discovering inflammatory bowel disease management biomarkers
| Ref. | Bio-sample | Sample size | Proteomic technique | Results |
| Disease activity biomarkers | ||||
| Han et al[34] | Intestinal tissue | CD: 3 | LC-QTOF | 16 proteins distinguishing active/inactive CD |
| UC: 4 | 4 proteins distinguishing active/inactive UC | |||
| Inflammatory Polyps: 2 | ||||
| Normal colon: 3 | ||||
| Wasinger et al[39] | Serum | UC: 27 | MRM | SPP24 was able to differentiate active and quiescent disease in both UC and CD |
| CD: 56 | ||||
| Controls: 14 | ||||
| RA controls: 12 | ||||
| Prognostic biomarkers | ||||
| May et al[57] | Intestinal epithelial cells | Non-dysplastic tissue from non-progressors: 5 | High-performance liquid chromatography quadrupole -TOF | 155 candidate proteins were expressed differentially by > 2x between dysplastic/cancerous and non-dysplastic UC tissue. They were identified as mitochondrial, cytoskeletal, apoptotic and RAS superfamily proteins |
| Non-dysplastic tissue from progressors: 5 | ||||
| Highly dysplastic tissue from UC progressors: 5 | ||||
| Response to therapy biomarkers | ||||
| Meuwis et al[37] | Serum | Infliximab responders: 40 | SELDI-TOF | 3 proteins (PF4, sCD40L and IL-6) were identified infliximab non-responders, although PF4 and sCD40L could not be confirmed or correlated with ELISA |
| Infliximab non-responders: 40 | ||||
| Kanmura et al[23] | Blood samples | CD: 22 | SELDI-TOF | Plasma concentration of HNP1, 2 and 3 decreased following successful corticosteroid therapy compared to non-responders |
| UC: 48 | ||||
| Colorectal cancer: 5 | ||||
| Infectious colitis: 6 | ||||
| Healthy controls: 13 | ||||
| Gazouli et al[38] | Serum | Infliximab responders: 6 | 2-DE, MALDI-TOF | 7 proteins were increased in CD patients who did not achieve remission on infliximab. 4 were increased in patients who achieved remission. 3 proteins were lower in remission patients |
| Infliximab non-responders: 6 | ||||
| Infliximab partial responders: 6 | ||||
CD: Crohn’s disease; UC: Ulcerative colitis; IBD: Inflammatory bowel disease; MRM: Multiple reaction monitoring.
Most recently, Wasinger et al[39] reported a panel of protein markers that were progressed into the “validation” stage using MRM. Two proteins [phosphoprotein 24 (SPP24) and α-1 microglobulin], were reported to be able to differentiate IBD patients and health controls whilst guanylin and secretogranin-1 differentiated UC and CD. Furthermore, three of these proteins (secretogranin-1, SPP24 and α-1 microglobulin), were able to distinguish between active and quiescent disease in UC and CD.
An important consideration when investigating IBD biomarkers is that a single protein may not provide the clinical utility desired, but rather a panel of markers governed by a scoring index or algorithm[40]. An existing example is the Brignola score which predicts relapse risk in asymptomatic Crohn’s patients by measures erythrocyte sedimentation rate, white blood cell count, hemoglobin, albumin, alpha 2-globulin, serum iron, C-reactive protein, alpha 1-glycoprotein, and alpha 2-antitrypsin[41]. This has been hinted at in several IBD proteomic studies which differentiated UC and CD using protein profiles rather than discrete markers[22,24]. The role of multiple biomarkers is highlighted by OVA1, the first Food and Drug Administration approved proteomic panel of biomarkers, consisting of 5 markers as a multivariate index assay. This assay combines multiple variables in an algorithm that produces a single diagnostic result[42]. These markers were identified using SELDI[43] and predicts the probability of a malignancy in a woman undergoing surgery for an adnexal mass[44]. Similarly, Plevy et al[45] used a combined panel of 8 serological markers, 4 genetic markers and 5 inflammatory aimed at discriminating CD from UC. The utility of this test however still requires validation in a prospective cohort. Furthermore, as it was a North American cross-sectional study, this warrants additional investigation into its validity when considering factors such as stability of markers over time[45] and ethnical variations[46].
An area that has yet to be addressed relates to the influence of IBD medications on protein abundance levels. Schreiber et al[47] reported the possible link between high dose 5-aminosalicylic acid (5-ASA) and modulated urinary protein concentrations. However, other groups have suggested that these urinary proteins reflect renal extra-intestinal manifestations rather than 5-ASA toxicity[48,49]. Derici et al[50] identified an association between similar urinary proteins and disease activity in UC, however none of these have been conclusive. Similarly, Mishima et al[51] detected elevated plasma levels of osteopontin in IBD patients, whilst Lorenzen et al[52] suggested a possible association between increased urinary osteopontin expression and steroid induced nephrotic syndrome. Whilst the relation between medications and their effect on protein expression is currently unclear, there are a number of implications in the context of biomarker discovery. Depending on the clinical question, the influence of medications would require strict experimental design and patient selection to avoid confounders. Additionally, biomarkers predicting or identifying adverse drug reactions introduces an additional area of research as IBD often requires lifelong medical therapy.
It is clear that proteomics could play a potentially significant role towards improving the clinical management of IBD. Despite this, the value of these studies and their findings remain unknown and require validation in future studies.
FUTURE CONSIDERATIONS FOR IBD PROTEOMICS
Current limitations
Despite significant advancements in discovery-phase technologies and protocols, the rate at which new diagnostic protein assays are being introduced remains static, averaging 1.5/year[29]. The stagnation occurs at the verification stage, effectively obstructing any progression towards the development of a clinical assay[53,54]. This is clearly evident by the inundation of IBD discovery-phase experiments published in the recent decade with little to no candidate proteins undergoing validation.
One common criticism of many proteomic studies is the lack of strict experimental design, resulting in questionable results that cannot be reproduced; in particular, a small sample size and insufficient statistical power biomarker discovery[40,55]. This issue holds true across the aforementioned studies in IBD as out of 19 bio-sample based discovery experiments, 8 studies used ≤ 6 cases and controls[33,34,38,56-59]. In an effort to address this issue, Skates et al[55] designed a statistical model that estimates the statistical power of discovery and verification studies in tissue and plasma. Statistical power is estimated using 5 parameters: Biospecimen used (serum/tissue/proximal fluid), number of candidate proteins selected during discovery, number of cases/controls, percentage of cases where the biomarker is expressed and the difference in standard deviation between the biomarker signal in cases compared to controls. In addition, biomarkers typically occur in low abundance and may randomly exceed machine sensitivity limits, resulting in artificial differences between samples. This combined with inherent biological variations between patient samples further emphasises the importance of achieving sufficient statistical power[60,61]. It has already been noted that the concentration of candidate IBD biomarkers may be more concentrated in the intestinal tissue compared to serum, potentially reducing the chance of false discoveries. This highlights one advantage of analysing tissue samples over serum, although it is unknown which would yield better results[20]. Significant efforts have been made to address such limitations including: Recent requirements on reporting, inclusion of standards, and superior methods. These all aim to improve accuracy and reliability and will all contribute to translatable proteomic markers for disease[62,63].
Hanash et al[30] identified a number of confounding factors that could contribute to variations and false discoveries when identifying potential biomarkers. Patient factors include genetic variations, metabolic state, acute phase reactants and non-specific changes such as cell death. The use of model systems such as cell cultures and animal models, provides an alternative approach that could control for confounding environmental and genetic factors[20,30]. At least 66 different animal models of IBD exist, however these may not accurately reflect the true pathophysiology of IBD. Differences in methodology that could produce artificial differences include: Sample collection and preparation, improper characterisation and randomisation, and sample/statistical analysis. Zhang et al[64] hypothesize that many are likely site specific, suggesting that “multisite sampling” may suffice in the absence of careful prospective sample collection and randomization. This would theoretically reduce the impact of these factors and improve the likelihood of clinically useful candidate biomarkers being detected[64].
The issues highlighted above demonstrate the requirement for standardisation of protocols in large-scale proteomics experiments or at least stringent experimental design to increase the chances of discovering valid biomarkers.
Towards verification and validation
The process of validation differs significantly from the initial discovery stage as candidate proteins are tested in thousands of samples. This phase uses reliable high throughput methods (e.g., immunoassays) in order to evaluate the biomarker’s utility in the target population. Unfortunately this phase requires significant financial investment and produces a major barrier to validating the numerous proteins identified as “candidate biomarkers”[20,53,54]. Consequently, many potential markers are identified in the literature but require further investigation.
The gap between the inherent inaccuracies of the discovery phase and the prohibitive cost of validation gave rise to the notion of an intermediate “verification” stage, aimed towards bridging this gap. This is achieved by quantification of selected candidate biomarkers in a larger sample that better represents the target population[55]. Ideally this is performed using reliable and established immunoassays, however, commercial antibodies are unavailable for the majority of protein targets, especially novel candidate markers. Assays must then be developed specifically for testing of the biomarker, an extremely costly endeavour when considering the large numbers of biomarkers[54]. Mass spectrometry can be further utilised here through quantitative techniques. Methods such as MRM have emerged as a viable alternatives towards cost-effectively triaging proteins of interest for further validation[20,53,54,65] and has been published for a number of biomarkers in other diseases[66-68].
CONCLUSION
The field and application of proteomics has expanded greatly in recent years and could have profound implications on the clinical diagnosis and management of IBD through the discovery of novel biomarkers. Many groups have already begun the “discovery” process and have identified many potential candidates. Although the transition into clinical validation is challenging, the tremendous potential of proteomics has garnered great interest and success in other diseases and further investigation into IBD proteomics should certainly be pursued.
Footnotes
Conflict-of-interest statement: No potential conflicts of interest. No financial support.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: September 2, 2015
First decision: September 29, 2015
Article in press: January 4, 2016
P- Reviewer: Bonaz BL, Ciccone MM, Gazouli M, Maric I, Naito Y, Sorrentino D, Tandon R, Vecchi M S- Editor: Qiu S L- Editor: A E- Editor: Jiao XK
References
- 1.Bernstein CN, Fried M, Krabshuis JH, Cohen H, Eliakim R, Fedail S, Gearry R, Goh KL, Hamid S, Khan AG, et al. World Gastroenterology Organization Practice Guidelines for the diagnosis and management of IBD in 2010. Inflamm Bowel Dis. 2010;16:112–124. doi: 10.1002/ibd.21048. [DOI] [PubMed] [Google Scholar]
- 2.Price AB. Overlap in the spectrum of non-specific inflammatory bowel disease--’colitis indeterminate’. J Clin Pathol. 1978;31:567–577. doi: 10.1136/jcp.31.6.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Theodossi A, Spiegelhalter DJ, Jass J, Firth J, Dixon M, Leader M, Levison DA, Lindley R, Filipe I, Price A. Observer variation and discriminatory value of biopsy features in inflammatory bowel disease. Gut. 1994;35:961–968. doi: 10.1136/gut.35.7.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farmer M, Petras RE, Hunt LE, Janosky JE, Galandiuk S. The importance of diagnostic accuracy in colonic inflammatory bowel disease. Am J Gastroenterol. 2000;95:3184–3188. doi: 10.1111/j.1572-0241.2000.03199.x. [DOI] [PubMed] [Google Scholar]
- 5.Chen L, Fang B, Giorgianni F, Gingrich JR, Beranova-Giorgianni S. Investigation of phosphoprotein signatures of archived prostate cancer tissue specimens via proteomic analysis. Electrophoresis. 2011;32:1984–1991. doi: 10.1002/elps.201100101. [DOI] [PubMed] [Google Scholar]
- 6.Glen A, Gan CS, Hamdy FC, Eaton CL, Cross SS, Catto JW, Wright PC, Rehman I. iTRAQ-facilitated proteomic analysis of human prostate cancer cells identifies proteins associated with progression. J Proteome Res. 2008;7:897–907. doi: 10.1021/pr070378x. [DOI] [PubMed] [Google Scholar]
- 7.Alaiya AA, Franzén B, Auer G, Linder S. Cancer proteomics: from identification of novel markers to creation of artifical learning models for tumor classification. Electrophoresis. 2000;21:1210–1217. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1210::AID-ELPS1210>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 8.Celis JE, Gromov P. Proteomics in translational cancer research: toward an integrated approach. Cancer Cell. 2003;3:9–15. doi: 10.1016/s1535-6108(02)00242-8. [DOI] [PubMed] [Google Scholar]
- 9.Jungblut PR, Zimny-Arndt U, Zeindl-Eberhart E, Stulik J, Koupilova K, Pleissner KP, Otto A, Müller EC, Sokolowska-Köhler W, Grabher G, et al. Proteomics in human disease: cancer, heart and infectious diseases. Electrophoresis. 1999;20:2100–2110. doi: 10.1002/(SICI)1522-2683(19990701)20:10<2100::AID-ELPS2100>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 10.Wagner JA, Williams SA, Webster CJ. Biomarkers and surrogate end points for fit-for-purpose development and regulatory evaluation of new drugs. Clin Pharmacol Ther. 2007;81:104–107. doi: 10.1038/sj.clpt.6100017. [DOI] [PubMed] [Google Scholar]
- 11.Vermeire S, Van Assche G, Rutgeerts P. Laboratory markers in IBD: useful, magic, or unnecessary toys? Gut. 2006;55:426–431. doi: 10.1136/gut.2005.069476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis JD. The utility of biomarkers in the diagnosis and therapy of inflammatory bowel disease. Gastroenterology. 2011;140:1817–1826.e2. doi: 10.1053/j.gastro.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langhorst J, Elsenbruch S, Koelzer J, Rueffer A, Michalsen A, Dobos GJ. Noninvasive markers in the assessment of intestinal inflammation in inflammatory bowel diseases: performance of fecal lactoferrin, calprotectin, and PMN-elastase, CRP, and clinical indices. Am J Gastroenterol. 2008;103:162–169. doi: 10.1111/j.1572-0241.2007.01556.x. [DOI] [PubMed] [Google Scholar]
- 14.Wasinger VC, Cordwell SJ, Cerpa-Poljak A, Yan JX, Gooley AA, Wilkins MR, Duncan MW, Harris R, Williams KL, Humphery-Smith I. Progress with gene-product mapping of the Mollicutes: Mycoplasma genitalium. Electrophoresis. 1995;16:1090–1094. doi: 10.1002/elps.11501601185. [DOI] [PubMed] [Google Scholar]
- 15.Ferreri AJ, Illerhaus G, Zucca E, Cavalli F. Flows and flaws in primary central nervous system lymphoma. Nat Rev Clin Oncol. 2010;7:193–197. doi: 10.1038/nrclinonc.2010.9-c1. [DOI] [PubMed] [Google Scholar]
- 16.Vaiopoulou A, Gazouli M, Theodoropoulos G, Zografos G. Current advantages in the application of proteomics in inflammatory bowel disease. Dig Dis Sci. 2012;57:2755–2764. doi: 10.1007/s10620-012-2291-4. [DOI] [PubMed] [Google Scholar]
- 17.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 18.Bachi A, Bonaldi T. Quantitative proteomics as a new piece of the systems biology puzzle. J Proteomics. 2008;71:357–367. doi: 10.1016/j.jprot.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Wasinger VC, Zeng M, Yau Y. Current status and advances in quantitative proteomic mass spectrometry. Int J Proteomics. 2013;2013:180605. doi: 10.1155/2013/180605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 21.Srinivas PR, Verma M, Zhao Y, Srivastava S. Proteomics for cancer biomarker discovery. Clin Chem. 2002;48:1160–1169. [PubMed] [Google Scholar]
- 22.Meuwis MA, Fillet M, Geurts P, de Seny D, Lutteri L, Chapelle JP, Bours V, Wehenkel L, Belaiche J, Malaise M, et al. Biomarker discovery for inflammatory bowel disease, using proteomic serum profiling. Biochem Pharmacol. 2007;73:1422–1433. doi: 10.1016/j.bcp.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 23.Kanmura S, Uto H, Numata M, Hashimoto S, Moriuchi A, Fujita H, Oketani M, Ido A, Kodama M, Ohi H, et al. Human neutrophil peptides 1-3 are useful biomarkers in patients with active ulcerative colitis. Inflamm Bowel Dis. 2009;15:909–917. doi: 10.1002/ibd.20854. [DOI] [PubMed] [Google Scholar]
- 24.Hatsugai M, Kurokawa MS, Kouro T, Nagai K, Arito M, Masuko K, Suematsu N, Okamoto K, Itoh F, Kato T. Protein profiles of peripheral blood mononuclear cells are useful for differential diagnosis of ulcerative colitis and Crohn’s disease. J Gastroenterol. 2010;45:488–500. doi: 10.1007/s00535-009-0183-y. [DOI] [PubMed] [Google Scholar]
- 25.Nanni P, Levander F, Roda G, Caponi A, James P, Roda A. A label-free nano-liquid chromatography-mass spectrometry approach for quantitative serum peptidomics in Crohn’s disease patients. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:3127–3136. doi: 10.1016/j.jchromb.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Subramanian V, Subramanian D, Pollok RC. S1182 Serum Protein Signatures Determined By Mass Spectrometry (SELDI-ToF) Accurately Distinguishes Crohn’s Disease (CD) from Ulcerative Colitis (UC) Gastroenterology. 2008;134:A–196. [Google Scholar]
- 27.Nanni P, Parisi D, Roda G, Casale M, Belluzzi A, Roda E, Mayer L, Roda A. Serum protein profiling in patients with inflammatory bowel diseases using selective solid-phase bulk extraction, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and chemometric data analysis. Rapid Commun Mass Spectrom. 2007;21:4142–4148. doi: 10.1002/rcm.3323. [DOI] [PubMed] [Google Scholar]
- 28.Vaiopoulou A, Gazouli M, Papadopoulou A, Anagnostopoulos AK, Karamanolis G, Theodoropoulos GE, M’Koma A, Tsangaris GT. Serum protein profiling of adults and children with Crohn disease. J Pediatr Gastroenterol Nutr. 2015;60:42–47. doi: 10.1097/MPG.0000000000000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson NL. The clinical plasma proteome: a survey of clinical assays for proteins in plasma and serum. Clin Chem. 2010;56:177–185. doi: 10.1373/clinchem.2009.126706. [DOI] [PubMed] [Google Scholar]
- 30.Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452:571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 31.Kennedy S. Proteomic profiling from human samples: the body fluid alternative. Toxicol Lett. 2001;120:379–384. doi: 10.1016/s0378-4274(01)00269-7. [DOI] [PubMed] [Google Scholar]
- 32.Teng PN, Bateman NW, Hood BL, Conrads TP. Advances in proximal fluid proteomics for disease biomarker discovery. J Proteome Res. 2010;9:6091–6100. doi: 10.1021/pr100904q. [DOI] [PubMed] [Google Scholar]
- 33.Shkoda A, Werner T, Daniel H, Gunckel M, Rogler G, Haller D. Differential protein expression profile in the intestinal epithelium from patients with inflammatory bowel disease. J Proteome Res. 2007;6:1114–1125. doi: 10.1021/pr060433m. [DOI] [PubMed] [Google Scholar]
- 34.Han NY, Choi W, Park JM, Kim EH, Lee H, Hahm KB. Label-free quantification for discovering novel biomarkers in the diagnosis and assessment of disease activity in inflammatory bowel disease. J Dig Dis. 2013;14:166–174. doi: 10.1111/1751-2980.12035. [DOI] [PubMed] [Google Scholar]
- 35.M’Koma AE, Seeley EH, Washington MK, Schwartz DA, Muldoon RL, Herline AJ, Wise PE, Caprioli RM. Proteomic profiling of mucosal and submucosal colonic tissues yields protein signatures that differentiate the inflammatory colitides. Inflamm Bowel Dis. 2011;17:875–883. doi: 10.1002/ibd.21442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seeley EH, Washington MK, Caprioli RM, M’Koma AE. Proteomic patterns of colonic mucosal tissues delineate Crohn’s colitis and ulcerative colitis. Proteomics Clin Appl. 2013;7:541–549. doi: 10.1002/prca.201200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meuwis MA, Fillet M, Lutteri L, Marée R, Geurts P, de Seny D, Malaise M, Chapelle JP, Wehenkel L, Belaiche J, et al. Proteomics for prediction and characterization of response to infliximab in Crohn’s disease: a pilot study. Clin Biochem. 2008;41:960–967. doi: 10.1016/j.clinbiochem.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 38.Gazouli M, Anagnostopoulos AK, Papadopoulou A, Vaiopoulou A, Papamichael K, Mantzaris G, Theodoropoulos GE, Anagnou NP, Tsangaris GT. Serum protein profile of Crohn’s disease treated with infliximab. J Crohns Colitis. 2013;7:e461–e470. doi: 10.1016/j.crohns.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 39.Wasinger VC, Yau Y, Duo X, Zeng M, Campbell B, Shin S, Luber R, Redmond D, Leong RW. Low mass blood peptides discriminative of inflammatory bowel disease (IBD) severity: A quantitative proteomic perspective. Mol Cell Proteomics. 2016;15:256–265. doi: 10.1074/mcp.M115.055095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mischak H, Apweiler R, Banks RE, Conaway M, Coon J, Dominiczak A, Ehrich JH, Fliser D, Girolami M, Hermjakob H, et al. Clinical proteomics: A need to define the field and to begin to set adequate standards. Proteomics Clin Appl. 2007;1:148–156. doi: 10.1002/prca.200600771. [DOI] [PubMed] [Google Scholar]
- 41.Brignola C, Campieri M, Bazzocchi G, Farruggia P, Tragnone A, Lanfranchi GA. A laboratory index for predicting relapse in asymptomatic patients with Crohn’s disease. Gastroenterology. 1986;91:1490–1494. doi: 10.1016/0016-5085(86)90206-4. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Z. An In Vitro Diagnostic Multivariate Index Assay (IVDMIA) for Ovarian Cancer: Harvesting the Power of Multiple Biomarkers. Rev Obstet Gynecol. 2012;5:35–41. [PMC free article] [PubMed] [Google Scholar]
- 43.Rai AJ, Zhang Z, Rosenzweig J, Shih IeM, Pham T, Fung ET, Sokoll LJ, Chan DW. Proteomic approaches to tumor marker discovery. Arch Pathol Lab Med. 2002;126:1518–1526. doi: 10.5858/2002-126-1518-PATTMD. [DOI] [PubMed] [Google Scholar]
- 44.Ueland F, Desimone C, Seamon L, Ware R, Goodrich S, Podzielinski I, Smith A, Santoso J, Van Nagell J, Zhang Z. The OVA1 test improves the preoperative assessment of ovarian tumors. Gynecol Oncol. 2010;116:S23. [Google Scholar]
- 45.Plevy S, Silverberg MS, Lockton S, Stockfisch T, Croner L, Stachelski J, Brown M, Triggs C, Chuang E, Princen F, et al. Combined serological, genetic, and inflammatory markers differentiate non-IBD, Crohn’s disease, and ulcerative colitis patients. Inflamm Bowel Dis. 2013;19:1139–1148. doi: 10.1097/MIB.0b013e318280b19e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prideaux L, De Cruz P, Ng SC, Kamm MA. Serological antibodies in inflammatory bowel disease: a systematic review. Inflamm Bowel Dis. 2012;18:1340–1355. doi: 10.1002/ibd.21903. [DOI] [PubMed] [Google Scholar]
- 47.Schreiber S, Hämling J, Zehnter E, Howaldt S, Daerr W, Raedler A, Kruis W. Renal tubular dysfunction in patients with inflammatory bowel disease treated with aminosalicylate. Gut. 1997;40:761–766. doi: 10.1136/gut.40.6.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fraser JS, Muller AF, Smith DJ, Newman DJ, Lamb EJ. Renal tubular injury is present in acute inflammatory bowel disease prior to the introduction of drug therapy. Aliment Pharmacol Ther. 2001;15:1131–1137. doi: 10.1046/j.1365-2036.2001.01041.x. [DOI] [PubMed] [Google Scholar]
- 49.Mahmud N, O’Toole D, O’Hare N, Freyne PJ, Weir DG, Kelleher D. Evaluation of renal function following treatment with 5-aminosalicylic acid derivatives in patients with ulcerative colitis. Aliment Pharmacol Ther. 2002;16:207–215. doi: 10.1046/j.1365-2036.2002.01155.x. [DOI] [PubMed] [Google Scholar]
- 50.Derici U, Tuncer C, Ebinç FA, Mutluay R, Yakaryilmaz F, Kulaksizoglu S, Soylemezoglu O, Sindel S. Does the urinary excretion of alpha1-microglobulin and albumin predict clinical disease activity in ulcerative colitis? Adv Ther. 2008;25:1342–1352. doi: 10.1007/s12325-008-0109-8. [DOI] [PubMed] [Google Scholar]
- 51.Mishima R, Takeshima F, Sawai T, Ohba K, Ohnita K, Isomoto H, Omagari K, Mizuta Y, Ozono Y, Kohno S. High plasma osteopontin levels in patients with inflammatory bowel disease. J Clin Gastroenterol. 2007;41:167–172. doi: 10.1097/MCG.0b013e31802d6268. [DOI] [PubMed] [Google Scholar]
- 52.Lorenzen J, Shah R, Biser A, Staicu SA, Niranjan T, Garcia AM, Gruenwald A, Thomas DB, Shatat IF, Supe K, et al. The role of osteopontin in the development of albuminuria. J Am Soc Nephrol. 2008;19:884–890. doi: 10.1681/ASN.2007040486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Makawita S, Diamandis EP. The bottleneck in the cancer biomarker pipeline and protein quantification through mass spectrometry-based approaches: current strategies for candidate verification. Clin Chem. 2010;56:212–222. doi: 10.1373/clinchem.2009.127019. [DOI] [PubMed] [Google Scholar]
- 54.Whiteaker JR, Lin C, Kennedy J, Hou L, Trute M, Sokal I, Yan P, Schoenherr RM, Zhao L, Voytovich UJ, et al. A targeted proteomics-based pipeline for verification of biomarkers in plasma. Nat Biotechnol. 2011;29:625–634. doi: 10.1038/nbt.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skates SJ, Gillette MA, LaBaer J, Carr SA, Anderson L, Liebler DC, Ransohoff D, Rifai N, Kondratovich M, Težak Ž, et al. Statistical design for biospecimen cohort size in proteomics-based biomarker discovery and verification studies. J Proteome Res. 2013;12:5383–5394. doi: 10.1021/pr400132j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fogt F, Jian B, Krieg RC, Wellmann A. Proteomic analysis of mucosal preparations from patients with ulcerative colitis. Mol Med Rep. 2008;1:51–54. [PubMed] [Google Scholar]
- 57.May D, Pan S, Crispin DA, Lai K, Bronner MP, Hogan J, Hockenbery DM, McIntosh M, Brentnall TA, Chen R. Investigating neoplastic progression of ulcerative colitis with label-free comparative proteomics. J Proteome Res. 2011;10:200–209. doi: 10.1021/pr100574p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nanni P, Mezzanotte L, Roda G, Caponi A, Levander F, James P, Roda A. Differential proteomic analysis of HT29 Cl.16E and intestinal epithelial cells by LC ESI/QTOF mass spectrometry. J Proteomics. 2009;72:865–873. doi: 10.1016/j.jprot.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 59.Shkoda A, Ruiz PA, Daniel H, Kim SC, Rogler G, Sartor RB, Haller D. Interleukin-10 blocked endoplasmic reticulum stress in intestinal epithelial cells: impact on chronic inflammation. Gastroenterology. 2007;132:190–207. doi: 10.1053/j.gastro.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 60.Alex P, Gucek M, Li X. Applications of proteomics in the study of inflammatory bowel diseases: Current status and future directions with available technologies. Inflamm Bowel Dis. 2009;15:616–629. doi: 10.1002/ibd.20652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Araki K, Mikami T, Yoshida T, Kikuchi M, Sato Y, Oh-ishi M, Kodera Y, Maeda T, Okayasu I. High expression of HSP47 in ulcerative colitis-associated carcinomas: proteomic approach. Br J Cancer. 2009;101:492–497. doi: 10.1038/sj.bjc.6605163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carr SA, Abbatiello SE, Ackermann BL, Borchers C, Domon B, Deutsch EW, Grant RP, Hoofnagle AN, Hüttenhain R, Koomen JM, et al. Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol Cell Proteomics. 2014;13:907–917. doi: 10.1074/mcp.M113.036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Percy AJ, Chambers AG, Smith DS, Borchers CH. Standardized protocols for quality control of MRM-based plasma proteomic workflows. J Proteome Res. 2013;12:222–233. doi: 10.1021/pr300893w. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Z, Chan DW. The road from discovery to clinical diagnostics: lessons learned from the first FDA-cleared in vitro diagnostic multivariate index assay of proteomic biomarkers. Cancer Epidemiol Biomarkers Prev. 2010;19:2995–2999. doi: 10.1158/1055-9965.EPI-10-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meng Z, Veenstra TD. Targeted mass spectrometry approaches for protein biomarker verification. J Proteomics. 2011;74:2650–2659. doi: 10.1016/j.jprot.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 66.Cho CK, Drabovich AP, Batruch I, Diamandis EP. Verification of a biomarker discovery approach for detection of Down syndrome in amniotic fluid via multiplex selected reaction monitoring (SRM) assay. J Proteomics. 2011;74:2052–2059. doi: 10.1016/j.jprot.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 67.Drabovich AP, Jarvi K, Diamandis EP. Verification of male infertility biomarkers in seminal plasma by multiplex selected reaction monitoring assay. Mol Cell Proteomics. 2011;10:M110.004127. doi: 10.1074/mcp.M110.004127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim K, Kim SJ, Han D, Jin J, Yu J, Park KS, Yu HG, Kim Y. Verification of multimarkers for detection of early stage diabetic retinopathy using multiple reaction monitoring. J Proteome Res. 2013;12:1078–1089. doi: 10.1021/pr3012073. [DOI] [PubMed] [Google Scholar]
- 69.von Roon AC, Karamountzos L, Purkayastha S, Reese GE, Darzi AW, Teare JP, Paraskeva P, Tekkis PP. Diagnostic precision of fecal calprotectin for inflammatory bowel disease and colorectal malignancy. Am J Gastroenterol. 2007;102:803–813. doi: 10.1111/j.1572-0241.2007.01126.x. [DOI] [PubMed] [Google Scholar]
- 70.Gisbert JP, McNicholl AG, Gomollon F. Questions and answers on the role of fecal lactoferrin as a biological marker in inflammatory bowel disease. Inflamm Bowel Dis. 2009;15:1746–1754. doi: 10.1002/ibd.20920. [DOI] [PubMed] [Google Scholar]
- 71.Kaiser T, Langhorst J, Wittkowski H, Becker K, Friedrich AW, Rueffer A, Dobos GJ, Roth J, Foell D. Faecal S100A12 as a non-invasive marker distinguishing inflammatory bowel disease from irritable bowel syndrome. Gut. 2007;56:1706–1713. doi: 10.1136/gut.2006.113431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beattie RM, Walker-Smith JA, Murch SH. Indications for investigation of chronic gastrointestinal symptoms. Arch Dis Child. 1995;73:354–355. doi: 10.1136/adc.73.4.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Poullis AP, Zar S, Sundaram KK, Moodie SJ, Risley P, Theodossi A, Mendall MA. A new, highly sensitive assay for C-reactive protein can aid the differentiation of inflammatory bowel disorders from constipation- and diarrhoea-predominant functional bowel disorders. Eur J Gastroenterol Hepatol. 2002;14:409–412. doi: 10.1097/00042737-200204000-00013. [DOI] [PubMed] [Google Scholar]
- 74.Shine B, Berghouse L, Jones JE, Landon J. C-reactive protein as an aid in the differentiation of functional and inflammatory bowel disorders. Clin Chim Acta. 1985;148:105–109. doi: 10.1016/0009-8981(85)90219-0. [DOI] [PubMed] [Google Scholar]
- 75.Ruemmele FM, Targan SR, Levy G, Dubinsky M, Braun J, Seidman EG. Diagnostic accuracy of serological assays in pediatric inflammatory bowel disease. Gastroenterology. 1998;115:822–829. doi: 10.1016/s0016-5085(98)70252-5. [DOI] [PubMed] [Google Scholar]
- 76.Peyrin-Biroulet L, Standaert-Vitse A, Branche J, Chamaillard M. IBD serological panels: facts and perspectives. Inflamm Bowel Dis. 2007;13:1561–1566. doi: 10.1002/ibd.20226. [DOI] [PubMed] [Google Scholar]
- 77.Sipponen T, Björkesten CG, Färkkilä M, Nuutinen H, Savilahti E, Kolho KL. Faecal calprotectin and lactoferrin are reliable surrogate markers of endoscopic response during Crohn’s disease treatment. Scand J Gastroenterol. 2010;45:325–331. doi: 10.3109/00365520903483650. [DOI] [PubMed] [Google Scholar]
- 78.Sipponen T, Savilahti E, Kolho KL, Nuutinen H, Turunen U, Färkkilä M. Crohn’s disease activity assessed by fecal calprotectin and lactoferrin: correlation with Crohn’s disease activity index and endoscopic findings. Inflamm Bowel Dis. 2008;14:40–46. doi: 10.1002/ibd.20312. [DOI] [PubMed] [Google Scholar]
- 79.Vecchi M, Gionchetti P, Bianchi MB, Belluzzi A, Meucci G, Campieri M, de Franchis R. p-ANCA and development of pouchitis in ulcerative colitis patients after proctocolectomy and ileoanal pouch anastomosis. Lancet. 1994;344:886–887. doi: 10.1016/s0140-6736(94)92859-2. [DOI] [PubMed] [Google Scholar]
- 80.Costa F, Mumolo MG, Ceccarelli L, Bellini M, Romano MR, Sterpi C, Ricchiuti A, Marchi S, Bottai M. Calprotectin is a stronger predictive marker of relapse in ulcerative colitis than in Crohn’s disease. Gut. 2005;54:364–368. doi: 10.1136/gut.2004.043406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gisbert JP, Bermejo F, Pérez-Calle JL, Taxonera C, Vera I, McNicholl AG, Algaba A, López P, López-Palacios N, Calvo M, et al. Fecal calprotectin and lactoferrin for the prediction of inflammatory bowel disease relapse. Inflamm Bowel Dis. 2009;15:1190–1198. doi: 10.1002/ibd.20933. [DOI] [PubMed] [Google Scholar]
- 82.Dubinsky MC, Mei L, Friedman M, Dhere T, Haritunians T, Hakonarson H, Kim C, Glessner J, Targan SR, McGovern DP, et al. Genome wide association (GWA) predictors of anti-TNFalpha therapeutic responsiveness in pediatric inflammatory bowel disease. Inflamm Bowel Dis. 2010;16:1357–1366. doi: 10.1002/ibd.21174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Spivak J, Landers CJ, Vasiliauskas EA, Abreu MT, Dubinsky MC, Papadakis KA, Ippoliti A, Targan SR, Fleshner PR. Antibodies to I2 predict clinical response to fecal diversion in Crohn’s disease. Inflamm Bowel Dis. 2006;12:1122–1130. doi: 10.1097/01.mib.0000235833.47423.d7. [DOI] [PubMed] [Google Scholar]


