Abstract
Dynamic Nuclear Polarization (DNP) has been introduced to overcome the sensitivity limitations of nuclear magnetic resonance (NMR) spectroscopy also of supported lipid bilayers. When investigated by solid-state NMR techniques the approach typically involves doping the samples with biradicals and their investigation at cryo-temperatures. Here we investigated the effects of temperature and membrane hydration on the topology of amphipathic and hydrophobic membrane polypeptides. Although the antimicrobial PGLa peptide in dimyristoyl phospholipids is particularly sensitive to topological alterations, the DNP conditions represent well its membrane alignment also found in bacterial lipids at ambient temperature. With a novel membrane-anchored biradical and purpose-built hardware a 17-fold enhancement in NMR signal intensity is obtained by DNP which is one of the best obtained for a truly static matrix-free system. Furthermore, a membrane anchor sequence encompassing 19 hydrophobic amino acid residues was investigated. Although at cryotemperatures the transmembrane domain adjusts it membrane tilt angle by about 10 degrees, the temperature dependence of two-dimensional separated field spectra show that freezing the motions can have beneficial effects for the structural analysis of this sequence.
Solid-state NMR spectroscopy is unique in providing information at atomic resolution of membrane polypeptides in a native bilayer environment. The technique has been used for investigations of their structure, topology, dynamics and heterogeneous nature in phospholipid bilayers1,2,3,4,5,6,7,8,9. However, NMR suffers from its inherently low signal intensity and the investigation of membrane-associated polypeptides turns out particularly difficult due to the dilution of peptide in lipid (typically around 1-2 mole%) and the increased line width due to inhomogeneous line broadening. Although these limitations are most pronounced when static samples are investigated the large anisotropy of NMR chemical shift, dipolar and quadrupolar interactions has been successively used to determine the structure and/or topology of a number of peptides in oriented lipid bilayers1,2,8,9. These could be refined by additional distance restraints for example from magic angle sample spinning (MAS) solid-state NMR spectra1,2,8,9.
During the last years considerable progress has been made in dynamic nuclear polarization (DNP) solid-state NMR and the technique has been shown to overcome many of the sensitivity limitations of solid-state NMR spectroscopy10,11,12,13. During DNP/solid-state NMR experiments the radicals dispersed in the sample result in a high electron polarization which is transferred to the 1H bath through microwave irradiation. In a next step cross-polarization to the heteronuclei assures enhancements of the NMR signal typically by about two orders of magnitude10,14. Also in the case of membrane samples4,11,15,16 or of membrane proteins in cellular environments considerable improvements have been obtained17.
In order to optimize the magnetization transfer from the unpaired electrons to the nuclei biradicals of suitable geometries have been designed and incorporated into the sample preparation18,19,20,21,22,23. Magnetization transfer through the cross effect24,25,26 requires that the measurements are performed at cryo-temperatures to assure sufficiently long life times of the electron polarization27,28,29,30. Using magic angle oriented sample spinning good DNP signal enhancements were obtained for solid-state NMR measurements of oriented membranes when at the same time it was demonstrated that bilayers made of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) retain an oriented bilayer morphology even at 100K and in the presence of biradicals31.
Following this proof-of-concept study31 a static flat-coil solid-state NMR/DNP probe head has been designed and constructed32. Using this new equipment here we assessed for the first time peptides of biological relevance under DNP conditions. We explore if membrane domains retain the overall structure and topology also under low temperature and DNP conditions, and ask the question if significant DNP efficiencies can be obtained from such systems when reconstituted in supported lipid bilayers. One test sequence that we investigated is the PGLa antimicrobial peptide which is particular sensitive to environmental conditions33. For comparison a transmembrane helical anchor sequence, which was labeled with 15N at five consecutive residues was also studied34.
Antimicrobial peptides of the magainin class have been investigated extensively by biophysical approaches, including oriented solid-state NMR spectroscopy, with good evidence that many of them selectively kill bacteria by disrupting their plasma membranes35,36,37,38. Whereas magainin 2 and related peptides adopt very stable alignments parallel to the membrane surface under all conditions tested so far39,40, the PGLa helical sequence (GMASKAGAIA GKIAKVALKA L-amide) has been shown to be highly affected by the detailed composition of the phospholipid membrane, its peptide-to-lipid ratio, the hydration level or the presence of other peptides33,41. Therefore, this sequence provides a particularly sensitive test case for the potential effects of low temperature conditions used in a variety of physical experiments such as x-ray diffraction or DNP/solid-state NMR4,11,15,16.
Results and Discussion
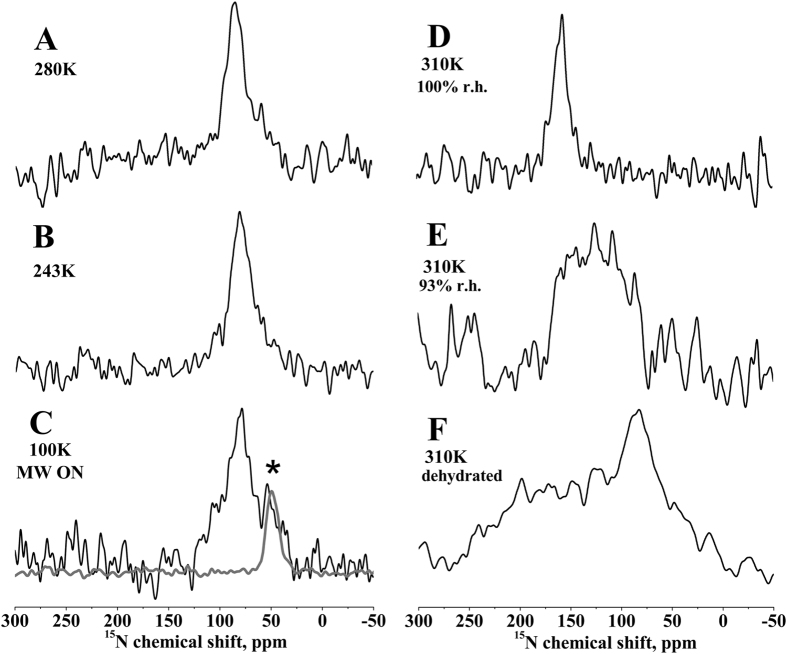
Figure 1 shows the proton-decoupled 15N solid-state NMR spectra of 2 mole% [15N-Ala14]-PGLa reconstituted into uniaxially oriented 1,2-dimyristoyl-sn-3-phosphocholine/1,2-dimyristoyl-sn-3-phospho-(1′-rac-glycerol) (DMPC/DMPG) 3/1 mole/mole bilayers. At 310K and 93% relative humidity (r.h.) a broad signal intensity with a maximum at 125 ppm is observed (Fig. 1E, line width at half height (LWHH) ca. 80 ppm) in agreement with previous investigations where PGLa has been found to adopt a tilt angle of 53°33,42,43. This tilted configuration has been suggested to be due to the formation of homo-dimers42. When the temperature is decreased below the liquid crystalline – gel phase transition temperature of both lipids (Tc = 297K and 296K for DMPC and DMPG, respectively) a more homogeneous topology is observed characterized by a 15N chemical shift maximum of 87 ppm (Fig. 1A,B; LWHH 23 ppm). This chemical shift coincides with the previously published tilt angle of the PGLa helix of 81° in liquid crystalline DMPC, DMPC/DMPG or POPC bilayers at lipid-to-peptide ratios <1 mole%42,44. Importantly the good uniform alignment of PGLa persist at lower temperatures (Fig. 1A–C) and even at sub-gel phase temperatures of 100K and in the presence of the biradical PyPol-C16 (Fig. 1C; LWHH 30 ppm)18, typical conditions of the commercial DNP/solid-state NMR set-up45.
Figure 1. Shows 15N solid-state NMR spectra of 2 mole% [15N-Ala14]-PGLa in uniaxially oriented DMPC/DMPG 3/1 bilayers (membrane normal parallel to Bo) as a function of temperature and hydration.
(A) 280 K, (B) 243 K, (C) 100 K, (D–F): 310 K. (A–C,E) were hydrated at 93% r.h., (D) 100% r.h., and (F) dehydrated during long measurement (likely around 75% r.h.). The spectrum in panel (C) was measured under 7 Watts microwave (MW) irradiation. The gray line in panel (C) shows the spectrum of peptide-free POPC vesicles under microwave irradiation. The position of the PC resonance is labeled with an asterisk.
Notably, the solid-state NMR spectrum of this sample was recorded under the effect of 7W microwave irradiation. The sample contains only 0.7 mg of peptide labeled with 15N at a single site and did not provide any apparent signal intensity without microwave irradiation even after 2 days of acquisition. Therefore, the enhancement factor could not be determined on this sample, however, spectra from related membranes showed 15N signal enhancements of 17-fold under static conditions32. When it is taken into account that spinning of the sample around the magic angle (MAS) increases the DNP efficiency several fold25,26,45 this value is among the best enhancement factors measured so far for matrix-free systems, including oriented membranes30,31,32,46. This favorable property of PyPol-C16 in the presence of membranes was made possible through the addition of a palmitoyl chain, which assures a better distribution in the oriented lipid bilayer samples devoid of bulk water18,22,23.
Previously 19F powder pattern line shapes have been observed for PGLa modified with a CF3-carrying Phe-derivative at four different Ala or Ile positions when reconstituted into DMPC/DMPG 3/1 and investigated at 253K or 233K43. In mechanically oriented lipid bilayers that are exposed to intense radiofrequency irradiation during solid-state NMR experiments special consideration has to be given to the exact degree of sample hydration over time within the coil volume (the practical aspects are discussed in reference47). Therefore, spectra of the same samples shown in Fig. 1A–C,E have also been recorded after equilibration at different degrees of hydration. Whereas Fig. 1D shows a defined intensity at 160 ppm at full hydration33, a condition difficult to maintain for oriented lipid bilayers under solid-state NMR conditions47, a broad range of 15N-H vector (i.e. PGLa helix) alignments are observed upon sample dehydration both at 310 K (Fig. 1F) and at 273 K (not shown). Notably, at the same time the phospholipid membrane remains well aligned with an oriented 31P solid-state NMR resonance centered at 35 ppm (not shown).
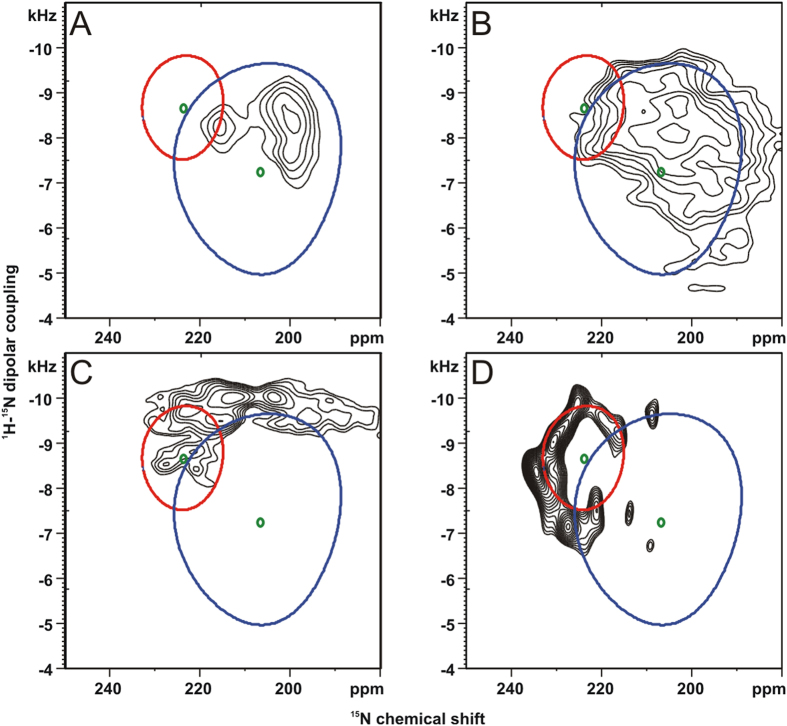
In order to investigate the temperature dependence of a transmembrane sequence upon temperature variation and under DNP conditions the [15N5]-hΦ19W peptide (peptide with a core of 19 hydrophobic residues including a tryptophan; cf. Methods section for details)34 was investigated by two-dimensional separated local field spectroscopy where the resolution in the dipolar dimension is enhanced by phase- and frequency switched Lee-Goldberg decoupling of homonuclear 1H interactions when at the same time cross polarized with the 15N nucleus48,49. The spectrum obtained at 100 K and under DNP conditions is shown in Fig. 2D where a partial helical wheel that arises from the 5 labeled residues is obvious.
Figure 2. PISEMA spectra of [15N5]-hΦ19W carrying five consecutive 15N labels in uniaxially oriented POPC bilayers.
(A) ambient temperature (295K), (B) 253K and (C) 223K. (D) nominal 100K (actual sample temperature due to microwave irradiation ~180K). (A–C) 10 mg of peptide were reconstituted at a peptide-to-lipid ratio (P/L) of 1/35 mole/mole supported by ultra-thin glass plates47. (D) DNP/solid-state NMR spectrum of 8.5mg [15N5]-hΦ19W in the presence of 0.3 mg AMUPol18 at P/L = 1/20 oriented onto films of high-density polyethylene31,32. The simulations show the intensity distributions at static helical tilt angles of 10o (red circle) and 22o (blue circle). Averaging around the helix long axis results in the collapse of the wheel -like pattern into its respective center of gravity (green dots).
Interestingly at room temperature the same peptide exhibits a much smaller wheel where many intensities coincide, indicating that fast motions around the helix long axis occur (Fig. 2A). Furthermore, the center of mass of the spectra obtained at room temperature and at 100 K is shifted by about 20 ppm. This is probably due to one or several phase transitions of the lipid bilayer concomitant with changes in the hydrophobic thickness of the membranes and a smaller tilt angle at the cryo-temperature. At intermediate temperatures where the lipid is in the gel phase broad intensity distributions are obtained that are difficult to analyze (Fig. 2B,C).
Even though PGLa exhibits a number of membrane topologies the peptide retains an oriented alignment even at sub-gel temperatures and is thereby amenable to solid-state/DNP investigations with good signal enhancements. It should be noted that here we have chosen a bilayer lipid composition where PGLa is particularly sensitive to topological transitions33,42,43. Whereas the gel and sub-gel phases exhibit a more homogenous sample orientation with better resolved lines also at 93% r.h. (Fig. 1A–C) and thereby more suitable conditions for a structural analysis, the topological equilibrium is shifted to a more membrane-inserted configuration in the liquid crystalline bilayer (Fig. 1D,E). Interestingly the low-temperature alignment in DMPC/DMPG closely corresponds to topologies observed under more physiological conditions (membranes made from POPC or from E. coli lipid extracts at room temperature)42,44,50. Furthermore, in this work we have characterized hydration as an additional parameter that affects the PGLa topological equilibrium in a sensitive manner. The low temperatures may protect the sample from dehydration when irradiated with strong radio frequency fields and thereby help to maintain a good peptide alignment.
For the transmembrane sequence a 20 ppm shift in the 15N chemical shift position indicates a more upright tilt angle (by about 10o) which is probably due an increased hydrophobic thickness of the bilayer under such conditions. Whereas the peptide undergoes fast motional averaging and exhibits sharp resonances at room temperature the signals are spread over a wider frequency range under DNP conditions which helps to resolve the NMR intensities from individual sites (Fig. 2A,D). Therefore, these studies show that freezing such motions can lead to inhomogeneous line broadening but can also have beneficial effects in spreading the signals over a broader chemical shift/dipolar coupling range.
Notably, investigating oriented membrane systems at cryo-temperatures by DNP/solid-state NMR is only at its beginnings and there remains a large potential of improvement in changing for example the lipid composition51,52. For example in the case of PGLa it has been demonstrated that a stable in-plane alignment such as the one observed at low temperatures including DNP conditions (Fig. 1A–C), is obtained in bilayers made from 1-palmitoyl-2-oleoyl- or in E. coli phospholipids, which represent much better the natural lipid composition than the dimyristoyl-phospholipids33,50. Recently, bicellar lipid mixtures have been presented that orient in the magnetic field of the NMR spectrometer even at temperatures of −15 °C53, a development which encourages further developments to make this alternative approach for oriented membrane preparations also accessible to DNP solid-state NMR technologies. Furthermore, other sequences exhibit a more stable topology and it can be expected that larger proteins have a more densely packed core54,55 that is less sensitive to changes in the lipid macroscopic phase properties than the peptides investigated here which further extends the range of applications of oriented DNP/solid-state NMR spectroscopy.
We estimate that in the experiments presented here it takes several minutes before the final cryo-temperature is obtained. However, methods for rapid freezing have been developed also for membrane-samples, thus that the conformational and topological states that rapidly exchange at room temperature are captured and can be investigated by solid-state NMR or electron paramagnetic resonance spectroscopies56,57. It should therefore be possible to take advantage of the increased sensitivity of DNP solid-state NMR also during investigations of the dynamic features of membrane-associated polypeptides.
Methods
The phospholipids POPC, DMPC and DMPG are from Avanti Polar Lipids (Alabaster, AL). All commercial material was used without further purification.
Peptide sequence and label positions
PGLa (GMASKAGAIA GKIAKVALKA L-amide), labeled with 15N at the underlined position, and the hydrophobic peptide [15N5]- hΦ19W (KKKALLALLALAWALALLALLAKKK) were prepared by solid phase peptide synthesis as described previously34. At five subsequent positions leucine and alanine labeled with 15N were incorporated into the peptide (underlined in the above sequence). The PyPol biradical was prepared as described previously18. The preparation of PyPol-C16, a derivative of PyPol bearing a palmitoyl chain, will be described in detail elsewhere.
Membrane samples for DNP
A homogeneous mixture of lipid, peptide, and radical was obtained by co-dissolving the membrane components in trifluoroethanol. To prepare oriented-phospholipid membranes, the solution was spread onto ultra-thin cover glasses for conventional oriented solid-state NMR measurements (8 × 22 mm, thickness 00; Marienfeld, Lauda-Königshofen, Germany), or for DNP on a High-Density PolyEthylene (HDPE) film (3 × 8 mm, Goodfellow, Cambridge, UK), dried first in air and followed by high vacuum overnight31,47. Thereafter, the samples were equilibrated during a day in an atmosphere of 93% relative humidity of D2O/H2O (90/10 by volume). The HDPE film with the membrane layers was carefully folded to fit in the coil and flattened in between two sapphire plates of 3 × 8 mm and 0.5–0.8 mm thickness32.
DNP/solid-state NMR
DNP/solid-state NMR measurements were performed using a Bruker BioSpin wide-bore 9.4 T magnet and an Avance III solid-state NMR spectrometer equipped with a gyrotron producing 263 GHz irradiation, a microwave transmission line, a cooling unit using liquid nitrogen45, and a purpose built static solid-state NMR/DNP probe for oriented samples32. An adiabatic cross polarization (CP) pulse sequence58 was used with a spectral width of 29.8 kHz and acquisition, cross polarization contact and recycle delay times of 8.6 ms, 0.3 ms and 3 s, respectively. The 1H π/2 pulse and SPINAL-64 heteronuclear decoupling field strengths B1 corresponded to a nutation frequency of 50 kHz59. To equilibrate the system before acquisition the sample was exposed to 16 dummy scans. An exponential line-broadening of 100 Hz was applied before Fourier transformation.
The DNP/solid-state NMR Polarization inversion spin exchange at the magic angle (PISEMA) spectrum was recorded on about 9 mg [15N5]-hΦ19W at a nominal temperature of 100K (where the actual sample temperature depends on the micro wave (MW) irradiation (~180K)). A step-by-step protocol for setting up and analyzing the experiment are given in references49,60. The effective B1 field strength during the SEMA pulse train was 50 kHz. During the spin exchange period the amplitude of the 1H B1 field was decreased to 40.9 kHz to maintain the Hartmann– Hahn match condition with an effective field along the magic angle of 50 kHz. 128 t1 increments were recorded by accumulating 64 scans each (total duration 7h; with no significant improvement after 64 t1 increments/3.5 hours).
Conventional (no DNP) CP and PISEMA experiments were performed using a Bruker Avance solid-state NMR (wide-bore 9.4 T magnet) equipped with efree™ static double resonance bioPE™ probe (Bruker Spectrospin, Karlsruhe, Germany). The temperature was controlled using a BCU-Xtreme unit (Bruker Spectrospin, Karlsruhe, Germany). Proton-decoupled 15N solid-state NMR spectra were acquired using an adiabatic CP pulse sequence, a spectral width, acquisition time, CP contact time and recycle delay of 38.5 kHz, 6.7 ms, 0.8 ms and 3 s, respectively. The 1H π/2 pulse and SPINAL-64 heteronuclear decoupling field strengths were 35 kHz59. Typically 60000 scans were accumulated, and the spectra were zero filled to 4096 points. A 100 Hz exponential line- broadening was applied before Fourier transformation. Spectra were externally referenced with 15NH4Cl at 40.0 ppm61.
The field strength during the SEMA pulse train was 60 kHz, 94 t1 increments were recorded by accumulating 2048 scans each (total duration 7 days; with no significant improvement after 64 t1 increments/3 days).
Additional Information
How to cite this article: Salnikov, E. S. et al. Membrane topologies of the PGLa antimicrobial peptide and a transmembrane anchor sequence by Dynamic Nuclear Polarization / solid-state NMR spectroscopy. Sci. Rep. 6, 20895; doi: 10.1038/srep20895 (2016).
Acknowledgments
We are grateful to Melanie Rosay, Werner Maas and Alain Belguise for continuous support in making available DNP time at the Bruker instruments and discussions. We also acknowledge the help by Delphine Hatey during peptide preparation. CNRS and Bruker Biospin, France have conjointly funded the PhD position of H.S. Furthermore the financial contributions of the Agence Nationale de la Recherche (projects ProLipIn 10-BLAN-731, membraneDNP 12-BSV5-0012, MemPepSyn 14-CE34-0001-01 and the LabEx Chemistry of Complex Systems 10-LABX-0026_CSC), the University of Strasbourg, the CNRS, the Région Alsace and the RTRA International Center of Frontier Research in Chemistry are gratefully acknowledged.
Footnotes
Author Contributions E.S.S. and C.A. prepared samples, performed solid-state NMR experiments and prepared the Figures including spectral simulations. The DNP experiments were perfomed with the help of F.A. H.S. helped to construct the prototypic DNP/solid-state NMR probe head under the guidance of F.E. and C.R. and performed preliminary measurements with the help of F.A. O.O. and P.T. provided the biradicals. B.B. designed the experimental strategy and wrote the manuscript.
References
- Hong M. & de Grado W. F. Structural basis for proton conduction and inhibition by the influenza M2 protein. Protein Sci 21, 1620–1633 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson M. et al. Allosteric regulation of SERCA by phosphorylation-mediated conformational shift of phospholamban. Proc Natl Acad Sci USA 110, 17338–17343 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker L. A. & Baldus M. Characterization of membrane protein function by solid-state NMR spectroscopy. Curr Opin Struc Biol 27, 48–55 (2014). [DOI] [PubMed] [Google Scholar]
- Ong Y. S., Lakatos A., Becker-Baldus J., Pos K. M. & Glaubitz C. Detecting Substrates Bound to the Secondary Multidrug Efflux Pump EmrE by DNP-Enhanced Solid-State NMR. J Am Chem Soc 135, 15754–15762 (2013). [DOI] [PubMed] [Google Scholar]
- Eddy M. T. et al. Magic angle spinning nuclear magnetic resonance characterization of voltage-dependent anion channel gating in two-dimensional lipid crystalline bilayers. Biochemistry 54, 994–1005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattin Z. et al. Solid-state NMR, electrophysiology and molecular dynamics characterization of human VDAC2. J Biomol NMR 61, 311–320 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke D., Shahid S., Habeck M., Bardiaux B. & van Rossum B. Membrane-protein structure determination by solid-state NMR spectroscopy of microcrystals: The autotransport domain of Yersinia YadA. FASEB J 27, 590:591 (2013). [DOI] [PubMed] [Google Scholar]
- Das N. et al. Structure of CrgA, a cell division structural and regulatory protein from Mycobacterium tuberculosis, in lipid bilayers. Proc Natl Acad Sci USA 112, E119–126 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek M., Salnikov E., Werten S. & Bechinger B. Structure and topology of the huntingtin 1-17 membrane anchor by a combined solution and solid-state NMR approach. . Biophys J 105, 699–710 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Can T. V. et al. Overhauser effects in insulating solids. J Chem Phys 141, 064202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koers E. J. et al. NMR-based structural biology enhanced by dynamic nuclear polarization at high magnetic field. J Biomol NMR 60, 157–168 (2014). [DOI] [PubMed] [Google Scholar]
- Akbey U. et al. Dynamic nuclear polarization of spherical nanoparticles. Phys Chem Chem Phys 15, 20706–20716 (2013). [DOI] [PubMed] [Google Scholar]
- Rossini A. J. et al. DNP Enhanced NMR Spectroscopy for Pharmaceutical Formulations. J Am Chem Soc 136, 2324–2334 (2014). [DOI] [PubMed] [Google Scholar]
- Rossini A. J. et al. Dynamic nuclear polarization surface enhanced NMR spectroscopy. Acc Chem Res 46, 1942–1951 (2013). [DOI] [PubMed] [Google Scholar]
- Su Y., Andreas L. & Griffin R. G. Magic Angle Spinning NMR of Proteins: High-Frequency Dynamic Nuclear Polarization and H Detection. Annu Rev Biochem 84, 465–497 (2015). [DOI] [PubMed] [Google Scholar]
- Salnikov E. S. et al. Developing DNP/Solid-State NMR Spectroscopy of Oriented Membranes. Appl Magn Reson 43, 91–106 (2012). [Google Scholar]
- Yamamoto K., Caporini M. A., Im S. C., Waskell L. & Ramamoorthy A. Cellular solid-state NMR investigation of a membrane protein using dynamic nuclear polarization. Biochim Biophys Acta 1848, 342–349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvee C. et al. Highly Efficient, Water-Soluble Polarizing Agents for Dynamic Nuclear Polarization at High Frequency. Angew Chem Int Ed 52, 10858–10861 (2013). [DOI] [PubMed] [Google Scholar]
- Mao J. F. et al. Host-Guest Complexes as Water-Soluble High-Performance DNP Polarizing Agents. J Am Chem Soc 135, 19275–19281 (2013). [DOI] [PubMed] [Google Scholar]
- Dane E. L. et al. Rigid Orthogonal Bis-TEMPO Biradicals with Improved Solubility for Dynamic Nuclear Polarization. J Org Chem 77, 1789–1797 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicki D. J. et al. Amplifying Dynamic Nuclear Polarization of Frozen Solutions by Incorporating Dielectric Particles. J Am Chem Soc 136, 15711–15718 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. N., Caporini M. A., Fanucci G. E. & Long J. R. A Method for Dynamic Nuclear Polarization Enhancement of Membrane Proteins. Angew Chem Int Ed 54, 1542–1546 (2015). [DOI] [PubMed] [Google Scholar]
- Fernandez-de-Alba C. et al. Matrix-free DNP-enhanced NMR spectroscopy of liposomes using a lipid-anchored biradical. Chem Eur J 21, 4512–4517 (2015). [DOI] [PubMed] [Google Scholar]
- Can T. V., Ni Q. Z. & Griffin R. G. Mechanisms of dynamic nuclear polarization in insulating solids. J Magn Reson 253, 23–35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentink-Vigier F. et al. Fast passage dynamic nuclear polarization on rotating solids. J Magn Reson 224, 13–21 (2012). [DOI] [PubMed] [Google Scholar]
- Thurber K. R. & Tycko R. Theory for cross effect dynamic nuclear polarization under magic-angle spinning in solid state nuclear magnetic resonance: the importance of level crossings. J Chem Phys 137, 084508 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber K. R., Potapov A., Yau W. M. & Tycko R. Solid state nuclear magnetic resonance with magic-angle spinning and dynamic nuclear polarization below 25 K. J Magn Reson 226, 100–106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Q. Z. et al. High frequency dynamic nuclear polarization. Acc Chem Res 46, 1933–1941 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbey U., Linden A. H. & Oschkinat H. High-Temperature Dynamic Nuclear Polarization Enhanced Magic-Angle-Spinning NMR. Appl Magn Reson 43, 81–90 (2012). [Google Scholar]
- Jakdetchai O. et al. Dynamic Nuclear Polarization-Enhanced NMR on Aligned Lipid Bilayers at Ambient Temperature. J Am Chem Soc 136, 15533–15536 (2014). [DOI] [PubMed] [Google Scholar]
- Salnikov E. et al. Solid-state NMR spectroscopy of oriented membrane polypeptides at 100 K with signal enhancement by dynamic nuclear polarization. J Am Chem Soc 132, 5940–5941 (2010). [DOI] [PubMed] [Google Scholar]
- Salnikov E. et al. Solid-state NMR/Dynamic Nuclear Polarization of planar supported lipid bilayers. J Phys Chem B 119, 14574–14583 (2015). [DOI] [PubMed] [Google Scholar]
- Salnikov E. & Bechinger B. Lipid-controlled peptide topology and interactions in bilayers: structural insights into the synergistic enhancement of the antimicrobial activities of PGLa and magainin 2. Biophys J 100, 1473–1480 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aisenbrey C. & Bechinger B. Tilt and rotational pitch angles of membrane-inserted polypeptides from combined 15N and 2H solid-state NMR spectroscopy. Biochemistry 43, 10502–10512 (2004). [DOI] [PubMed] [Google Scholar]
- Lee D.-K., Bhunia A., Kotler S. A. & Ramamoorthy A. Detergent-type membrane fragmentation by MSI-78, MSI-367, MSI-594, and MSI-843 antimicrobial peptides and inhibition by cholesterol: a solid-state nuclear magnetic resonance study. Biochemistry 54, 1897–1907 (2015). [DOI] [PubMed] [Google Scholar]
- Bechinger B. Detergent-like properties of magainin antibiotic peptides: A 31P solid-state NMR study. Biochim. Biophys. Acta 1712, 101–108 (2005). [DOI] [PubMed] [Google Scholar]
- Ramamoorthy A. Beyond NMR spectra of antimicrobial peptides: dynamical images at atomic resolution and functional insights. Solid State Nucl Magn Reson. 35, 201–207 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechinger B. The SMART model: Soft Membranes Adapt and Respond, also Transiently, to external stimuli. J Pept Sci 21, 346–355 (2015). [DOI] [PubMed] [Google Scholar]
- Bechinger B. Insights into the mechanisms of action of host defence peptides from biophysical and structural investigations. J Pept Sci 17, 306–314 (2011). [DOI] [PubMed] [Google Scholar]
- Resende J. M. et al. Membrane interactions of Phylloseptin-1, -2, and -3 peptides by oriented solid-state NMR spectroscopy. Biophys J 107, 901–911 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser R. W. et al. Concentration-dependent realignment of the antimicrobial peptide PGLa in lipid membranes observed by solid-state 19F-NMR. Biophys J 88, 3392–3397 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strandberg E., Esteban-Martin S., Salgado J. & Ulrich A. S. Orientation and dynamics of peptides in membranes calculated from 2H-NMR data. Biophys J 96, 3223–3232 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonin S., Grage S. L., Ieronimo M., Wadhwani P. & Ulrich A. S. Temperature-dependent transmembrane insertion of the amphiphilic peptide PGLa in lipid bilayers observed by solid state 19F NMR spectroscopy. J Am Chem Soc 130, 16512–16514 (2008). [DOI] [PubMed] [Google Scholar]
- Bechinger B., Zasloff M. & Opella S. J. Structure and Dynamics of the Antibiotic Peptide PGLa in Membranes by Multidimensional Solution and Solid-State NMR Spectroscopy. Biophys J 74, 981–987 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosay M. et al. Solid-state dynamic nuclear polarization at 263 GHz: spectrometer design and experimental results. Phys Chem Chem Phys 12, 5850–5860 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Hediger S. & De Paepe G. Matrix-free dynamic nuclear polarization enables solid-state NMR C-13-C-13 correlation spectroscopy of proteins at natural isotopic abundance. Chem Commun 49, 9479–9481 (2013). [DOI] [PubMed] [Google Scholar]
- Aisenbrey C., Bertani P. & Bechinger B. In Antimicrobial Peptides Methods in Molecular Biology eds Guiliani A. & Rinaldi A. C.) Ch. 14, 209–233 (Humana Press, Springer, 2010). [DOI] [PubMed] [Google Scholar]
- Ramamoorthy A., Wei Y. & Lee D. PISEMA Solid-State NMR Spectroscopy. Annual Reports on NMR Spectroscopy 52, 1–52 (2004). [Google Scholar]
- Aisenbrey C., Michalek M., Salnikov E. S. & Bechinger B. In Lipid-Protein Interactions: Methods and Protocols Methods in Molecular Biology (ed Kleinschmidt J. H.) Ch. 16, 357–387 (Springer, 2013). [DOI] [PubMed] [Google Scholar]
- Glattard E., Salnikov E. S., Aisenbrey C. & Bechinger B. Investigations of the synergistic enhancement of antimicrobial activity in mixtures of magainin 2 and PGLa. Biophys Chem, in press S0301–4622(15)30005–3, doi: 10.1016/j.bpc.2015.06.002 (2015). [DOI] [PubMed] [Google Scholar]
- Lee D. K., Kwon B. S. & Ramamoorthy A. Freezing point depression of water in phospholipid membranes: a solid-state NMR study. Langmuir 24, 13598–13604 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikov E. S. et al. Alamethicin topology in phospholipid membranes by oriented solid-state NMR and EPR spectroscopies: A comparison. J Phys Chem.B 113, 3034–3042 (2009). [DOI] [PubMed] [Google Scholar]
- Yamamoto K. et al. Temperature-resistant bicelles for structural studies by solid-state NMR spectroscopy. Langmuir 31, 1496–1504 (2015). [DOI] [PubMed] [Google Scholar]
- Kamihira M. et al. Structural and orientational constraints of bacteriorhodopsin in purple membranes determined by oriented-sample solid-state NMR spectroscopy. J Struct Biol 149, 7–16 (2005). [DOI] [PubMed] [Google Scholar]
- Vogt T. C. B., Schinzel S. & Bechinger B. Biosynthesis of biochemically labelled gramicidins and tyrocidins by Bacillus brevis. J Biomol NMR 26, 1–11 (2003). [DOI] [PubMed] [Google Scholar]
- Lazo N. D., Hu W. & Cross T. A. Low-temperature solid-state 15 N NMR characterization of polypeptide backbone librations. J Magn Reson 107, 43–50 (1995). [DOI] [PubMed] [Google Scholar]
- Dzikovski B. G., Borbat P. P. & Freed J. H. Spin-labeled gramicidin a: channel formation and dissociation. Biophys J 87, 3504–3517 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger S., Meier B. H. & Ernst R. R. Adiabatic passage Hartmann-Hahn cross polarization in NMR under magic angle sample spinning. Chem Phys Lett 240, 449–456 (1995). [Google Scholar]
- Fung B. M., Khitrin A. K. & Ermolaev K. An improved broadband decoupling sequence for liquid crystals and solids. J Magn Reson 142, 97–101 (2000). [DOI] [PubMed] [Google Scholar]
- Salnikov E., Aisenbrey C., Raya J. & Bechinger B. In New Developments in NMR: Advances in Biological Solid-State NMR, Proteins and Membrane-Active Peptides eds Separovic F. & Naito A.) Ch. 12, 214–234 (Royal Society of Chemistry, 2014). [Google Scholar]
- Bertani P., Raya J. & Bechinger B. 15N chemical shift referencing in solid state NMR. Solid-state NMR spec 61–62, 15–18 (2014). [DOI] [PubMed] [Google Scholar]