Abstract
Imaging guided techniques have been increasingly employed to investigate the pharmacokinetics (PK) and biodistribution of nanoparticle based drug delivery systems. In most cases, however, the PK profiles of drugs could vary significantly from that of drug delivery carriers upon administration in the blood circulation, which will complicate the interpretation of image findings. Herein we applied a genetically encoded luciferase reporter in conjunction of near infrared (NIR) fluorophores to investigate the respective PK profiles of drug and its carrier in a biodegradable drug delivery system. In this system, a prototype hydrophobic agent, rapamycin (Rapa), was encapsulated into human serum albumin (HSA) to form HSA Rapa nanoparticles, which were then labeled with Cy5 fluorophore to facilitate the fluorescence imaging of HSA carrier. Meanwhile, we employed transgenetic HN12 cells that were modified with a split luciferase reporter, whose bioluminescence function is regulated by Rapa, to reflect the PK profile of the encapsulated agent. It is interesting to uncover that there existed an obvious inconsistency of PK behaviors between HSA carrier and rapamycin in vitro and in vivo through near infrared fluorescence imaging (NIFRI) and bioluminescence imaging (BLI) after treatment with Cy5 labeled HSA Rapa. Nevertheless, HSA Rapa nanoparticles manifested a favorable in vivo PK and tumor suppression efficacy in a follow-up therapeutic study. The developed strategy of combining a molecular reporter and a fluorophore in this study could be extended to other drug delivery systems to provide profound insights for non-invasive real-time evaluation of PK profiles of drug-loaded nanoparticles in pre-clinical studies.
Keywords: Drug delivery, Molecular imaging, Nanoparticles, Pharmacokinetics, Reporter gene
Introduction
With many desirable properties, nanotechnology holds great potential in the areas of cancer diagnosis and therapy1-3. Various nanoparticles have been increasingly investigated for the targeted delivery of drugs especially those are poorly water-soluble yet potent antitumor drugs4-6. The pharmacokinetics (PK) and tissue distribution of nanoparticles play major roles in determining their efficacy and potential toxicity, both of which are fundamental issues needing to be elucidated for the optimal performance of nanoparticles as effective drug carriers in pre-clinical studies7. In most cases, however, the PK profile of drugs could vary significantly from that of drug carriers upon administration in the blood, which may attribute to multiple variations such as shape, size, surface chemistry, and many others8. The dissociation of drugs from their biodegradable carriers after long time circulation in vivo can complicate the interpretation of experimental findings. It is thus imperative to monitor the distribution and disposition of each individual components of a particular drug delivery system to evaluate its in vivo performance and potential side effects.
Currently, various imaging techniques have been applied to evaluate the PK of drug carriers, including magnetic resonance imaging (MRI), positron emission tomography (PET), single-photon emission computed tomography (SPECT), ultrasound and optical imaging, etc.9, 10. Comparing with conventional strategies, such as chromatography analysis, immunoassay, or isotopic tracer determination, in vivo imaging permits the evaluation of intrinsic behavior and tissue biodistribution of these drug loaded nanoparticles with spatiotemporal, quantitative resolution after delivery of different dosages and at the various time of interests11. Among them, optical techniques can provide high detection sensitivity and temporal resolution by tethering fluorophores to drug loaded nanoparticles, and attracted considerable interests. However, this fluorescence imaging modality only represents the PK of drug carriers that are directly conjugated with fluorophores, but reflects little information about the distribution of loaded drugs, especially when the drug released from the carriers. The longitudinal non-invasive optical imaging of the independent behaviors of a therapeutic agent and its delivery carrier after single administration in living subjects remains largely elusive, but is significantly important for the comprehensive understanding of the complicated behavior of drug delivery systems in pre-clinical and clinical studies. Therefore, a high fidelity imaging strategy that can capture the distinctive physical or chemical signals separately from encapsulated drugs and carriers are highly desirable to obtain accurate PK profile of individual component in a drug delivery system.
In this regard, we applied a genetically coded molecular reporter in conjunction of near infrared (NIR) fluorophores to investigate the respective PK profiles of a loaded hydrophobic drug and its carrier in a biodegradable drug delivery system. Rapamycin (Rapa) is a sterling example that is strongly hydrophobic but has been shown to be a potential antitumor agent in a variety of solid tumor models12. Rapamycin exerts its anti-tumor activity by inhibiting the mammalian target of rapamycin (mTOR) pathway, which has a central physiologic role in controlling of eukaryotic cell growth and proliferation13. The inhibition of mTOR by rapamycin is through the formation of a ternary complex comprising rapamycin, FKBP12 (the 12-kDa immunophilin family member FK506-binding and Rapamycin-binding protein), and the FKBP-rapamycin–binding (FRB) domain of mTOR kinase14. However, despite the anti-tumor potency of rapamycin in preclinical studies, clinical development of rapamycin has been impeded due to its poor solubility in water (2.6 μg/ml)15.
Using proteins such as human serum albumin (HSA) as drug carriers has attracted considerable interests for its biodegradability, non-toxicity, non-immunogenicity and preferential uptake in tumor and inflamed tissues, making it an ideal candidate carrier for drug delivery16, 17. Several registered HSA-based nanoparticle formulations such as Albunex™18 and Abraxane™19, 20 were well evaluated in phase I/Π clinical studies. Because of the success of HSA as a robust carrier for hydrophobic drug delivery and the highly potent efficacy of rapamycin in anti-cancer therapy, we are motivated to apply the developed combined imaging system to non-invasively evaluate the PK profiles of individual moieties in the rapamycin loaded HSA nanoparticles (HSA Rapa) delivery system. With the use of split luciferase reporter21, the inducible association of FRB and FKBP12 upon introduction of HSA Rapa enabled conditional activation of the luciferase reporter for bioluminescence imaging (BLI) the PK profile of the encapsulated Rapa, and the visualization of the PK of Cy5 conjugated HSA carrier was realized by near-infrared fluorescence imaging (NIRFI). To the best of our knowledge, it is the first reported study to monitor the respective PK profiles of a loaded drug and its carrier in a nanoparticle-based drug delivery system. This integrated imaging platform provides reliable real-time information on PK profiles and will be of great value for evaluating numerous drug delivery systems in various diseases treatment.
Experimental
Preparation of Cy5 labeled rapamycin-loaded HSA nanoparticles
The rapamycin loaded HSA nanoparticles (HSA Rapa) were prepared by two phases emulsion method using high pressure homogenizer (EmulsiFlex-C3, Avestin Inc. Canada). Briefly, 400 mg human serum albumin was dissolved in 15 mL distill water in 50 mL Falcon tube (water phase). 10 mg rapamycin and 60 μl safflower oil were dissolved in 1 mL ethyl acetate (oil phase). The oil phase was added into water phase, followed by vigorous vortex for 5 min to form raw emulsion. The raw emulsion was added into high pressure homogenizer to prepare nanoparticles in ice bath (30 emulsion cycles, 20 K bar pressure). The final milky emulsion was purified by ultracentrifugation at 4000 rpm (Ultracon, 100 K MWCO) three times before nanoparticles were dispersed in PBS. For labeling, Cy5-NHS (GE Healthcare) in DMSO was added into the above nanoparticle solution at a Cy5: HSA ratio 5:1 for incubating 30 min in darkness. The solution was then purified by ultracentrifugation until there is no free dye, which is validated by detecting the fluorescence in the supernatant. Finally, the solvent was exchanged to PBS for further use.
Characterization of HSA Rapa
HSA Rapa nanoparticle size and size distribution measurements were carried out using a Malvern Instrument Zetasizer Nano ZS (Malvern, UK) based on dynamic light scattering. The internal morphology of HSA Rapa was examined by transmission electron microscopic (TEM) using a JEM-2010 (JEOL Ltd., Japan) operated at an accelerating voltage of 200 kv. The loading efficiency and release kinetics of HSA Rapa nanoparticle was examined by HPLC analysis using an Acquity HPLC system (Waters, Milford, MA).
Plasmids construction
The coding sequence of N-terminal fragment (FlucN, aa1-aa398) or the C-terminal fragment (FlucC, aa394-aa550) of firefly luciferase was PCR amplified, digested and inserted into the NheI/BamHI or BamHI/XhoI sites of the pcDNA3.1(+) vector (Invitrogen) to obtain pcFlucN and pcFlucC plasmids. The human FRB or FKBP12 fragments were amplified using the template vector pFRB or pFKBP12 provided by Ariad Pharmaceuticals, Inc. (Cambrige, MA). Then the amplified fragments were cloned downstream of the FlucN or upstream of the FlucC by digesting with corresponding restriction enzymes and finally generated pcFlucN-FRB and pcFKBP12-FlucC plasmids (Fig. S1A). All plasmids contain an in-frame coding sequence for a GGSGGGGSGG linker (colored in red) between the FlucN and FRB or the FKBP12 and FlucC.
Cell culture and stable cell line establishment
Squamous cell carcinoma of the head and neck (HNSCC) cell line HN12, purchased from American Type Culture Collection (Manassas, VA), was grown and maintained in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin solution. To generate stable cell line expressing the luciferase reporter gene, the two plasmids FlucN-FRB and FKBP12-FlucC were cotransfected in HN12 cells with Lipofectamine 2000 (Invitrogen) in accordance with the manufactuer's instructions. Then selection was performed with zeocin (500 μg/ml; Invitrogen) for approximately 3 weeks from the day after transfection. Surviving clones were isolated and subjected to screening for the recovered luciferase activity upon adding rapamycin. One stable cell line (designated HN12 #2), which showed the highest signal-to-noise ratio, was obtained and used for further study.
Western blotting
Cells were rinsed with PBS and lysed with protein lysis buffer [62.5 mmol/L Tris-HCl (pH 6.8), 10% glycerol, 2% SDS, 50 mmol/L DTT], and then transferred to microcentrifuge tubes and boiled for 5 min. Protein yield was quantified using the bicinchoninic acid assay (Pierce). Equivalent amounts of protein (50 ug) were separated by SDS-PAGE, transferred to PVDF membranes, and immunoblotted with primary antibodies for Akt (1:1000, Cell signaling, MA), phosphorylated Akt (1:500, Cell signaling, MA) and β-actin (1:2000, Cell signaling, MA). Bound primary antibodies were visualized with appropriate horseradish peroxidase–conjugated secondary antibodies (1:5000, Cell signaling, MA) with the use of enhanced chemiluminescence (Amersham Pharmacia, GE Healthcare).
Confocal microscope assay
The HN12 cells were plated in Lab TekΠ 8-well chamber slides (NalgeNunc International, Rochester, NY) with a density of 1× 104 cells/ml and grown to 60-80% confluence. Then the cells were incubated for 4 h with 6.25 nM of Cy5 labeled HSA Rapa. The cells were then washed with phosphate buffered saline (PBS) three times. Images were acquired by an IX81 epifluorescence microscope (Olympus, Hamburg, Germany).
Flow cytometry
The HN12 cells were incubated with 6.25 nM of Cy5 labeled HSA Rapa for different time periods (0.5, 1, 2, 4, 6, 24, 48, 72 h). At the end of the incubation period the cells were then washed and resuspended in PBS. The fluorescences from the cells were measured by Accuri C6 flow cytometer using C Flow Plus software (BD, Ann Arbor, MI). The data were analyzed by FlowJo version 7.6.5 (FlowJo, Ashland, OR).
In vitro bioluminescence imaging of cells
To identify the relationship between cell numbers and bioluminescence signals, a dilution series of HN12#2 cells were seeded in a 24-well plate. After 12 h incubation, 20 nM free rapamycin was added to each well for 6 h. Then D-luciferin (final concentration 150 μg/ml) was added and luciferase activity was measured using a Xenogen Lumina Π system (Caliper Life Sciences). Luminescent signal for each well was measured and plotted as average values. In the other set of experiments, the cells were treated with different concentrations of HSA Rapa or ascomycin. Then the cell-containing plates were imaged and measured as described above.
LC-MS analysis
For quantitative analysis of the concentration of rapamycin, the tumors and major organs as well as the plasma at the indicated time points were harvested and subjected to LC-MS analysis, which employed a Waters LC-MS system (Waters, Milford, MA) including an Acquity UPLC system coupled to the Waters Q-Tof Premier high-resolution mass spectrometer as described previously22. Briefly, An Acquity BEH Shield RP18 column (150 mm × 2.1 mm) was eluted with a two-solution gradient of solution A (2 mM ammonium formate, 0.1% formic acid, and 5% CH3CN) and solution B (2 mM ammonium formate and 0.1% formic acid in CH3CN). The elution profile, at 0.2 mL/min, had the following components: initial condition at 100% (v:v) A and 0% B; gradient 0–40% B over 15 min; isocratic elution at 40% B for an additional 3 min; 40–80% B over 2 min; and re-equilibrated with A for an additional 4 min. For quantitative analysis of the concentration of rapamycin, an LC/MS system consisting of an Agilent 1200 autosampler, Agilent 1200 LC pump and an AB/MDS Sciex 4000 Q-TRAP (AB Sciex, Foster City, CA, USA). Standards were prepared for rapamycin covering the concentration range from 0.001 to 1μM in 1:1 CH3CN-water. Three replicate injections (10 μL) were made for each concentration level.
Animal model
All animal studies were carried out according to NIH approved protocols, in compliance with the Guild for the Care and Use of Laboratory Animals. Athymic nude mice, 4 to 5 weeks of age and weighing 18 to 20 g, were obtained from Harlan laboratories (Frederick, USA). The HNSCC tumor models were generated by subcutaneous injection of 5×106 HN12#2 cells in 100 μl PBS into the right shoulder of nude mice. The mice were used for imaging and therapy when the tumor volume reached ~50 mm3.
In vivo fluorescence and bioluminescence imaging
Mice bearing HN12 xenografted tumors were intravenously injected with 10 mg/kg Cy5 labeled HSA Rapa. At different time points (0.5, 1, 2, 4, 6, 24, 48, 72 h) after injection, fluorescence imaging was performed following by bioluminescence imaging. Fluorescence imaging was carried out with a MaestroΠ optical imaging system (Caliper Life Sciences, Hopkinton, MA). For quantization, regions of interest (ROIs) were drawn over tumors and the average signal (photons per square centimeter per second) for each area was measured. Immediately after fluorescence imaging at each time point, mice was given a single i.p. dose of 150 mg/kg D-luciferin in normal saline. Bioluminescence imaging was accomplished 5 min post-luciferin administration. Signal intensity was quantified as the radiance (photons per second per square centimeter per steradian) within ROIs over the tumor site.
In vivo tumor therapy studies
When the tumor volume reached ~50 mm3, mice bearing HN12 tumor were randomly divided into two groups of 20 animals and injected intravenously with HSA rapa (10 mg/kg) or an equal volume of saline every other day for 5 times. Mice were monitored for up to 3 weeks for tumor growth and body weight. Tumor size was monitored with a digital caliper every 2 days and tumor volume was calculated as vol= LW2/2, where L and W represent the length and the width of the tumor. The body of weight of all mice was measured every 2 days.
TUNEL assay
The excised tumors were collected and immersed into OCT medium. Then the tumors were cut into about 4 μm thick sections and fixed in 4% paraformaldehyde at room temperature for 20 min. After washing with PBS for three times, the sections were stained with In situ Cell Death Detection Kit (Billerica, MA) according to the manufacture's protocols.
Statistical analysis
Quantitative data were expressed as mean ± SD. Means were compared using Student's unpaired test. Data analysis was performed with using GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA). P values <0.05 were considered statistically significant.
Results and discussion
Establishment and validation of stable cell line expressing split luciferase reporter
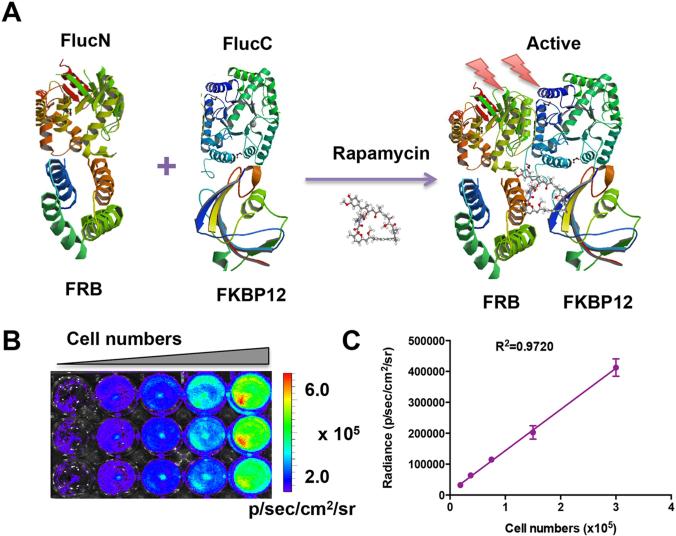
To evaluate the PK profile of HSA Rapa nanoparticles, we employed split firefly luciferase (Fluc) protein fragment-assisted complementation in combination with rapamycin-mediated dimerization of human protein FRB and its interacting partner, FKBP12 (Fig.1A) in cell culture and in living animals by noninvasive imaging. HN12 cancer cells were co-transfected with vector constructs pcFlucN-FRB and pcFKBP12-FlucC under the control of CMV promoter (Fig. S1A). One stable cell line (designated HN12 #2), which showed the highest restored luciferase activity upon addition of rapamycin, was obtained by selection with zeocin and used for further study (Fig. S1B).
Figure 1.
Characterization of the split luciferase reporter. (A) A crystallographic model of FlucN-FRB and FKBP12-FlucC domains in the reporter system. In the absence of rapamycin, there is no luciferase activity. Upon treatment with rapamycin, the dimerization of FRB and FKBP12 will lead to the activation of the luciferase reporter. The Fluc, FRB and FKBP12 images were adapted from RCSB PDB (www.pdb.org) using ID# 1LUC, 1AUE and 2PPN. (B) Bioluminescence imaging analyzed the linearity of recovered luciferase activity with cell numbers after addition of free rapamycin (20 nM) for 6 h. (C) Linear regression analysis between luminescence intensity and cell numbers.
To examine the linear correlation of recovered luciferase activity to the cell number, a series dilution of cells from 1.875 × 104 to 3 × 105 were seeded and the bioluminescence imaging was performed after free rapamycin (20 nM) treatment for 6 hours. As shown in Fig.1B, the bioluminescence signal increased according to the increased cell numbers. An analysis of intensities in the ROI indicated a high correlation (γ2 = 0.9720) between signal intensity and cell numbers (Fig. 1C).
Synthesis and characterization of HSA Rapa
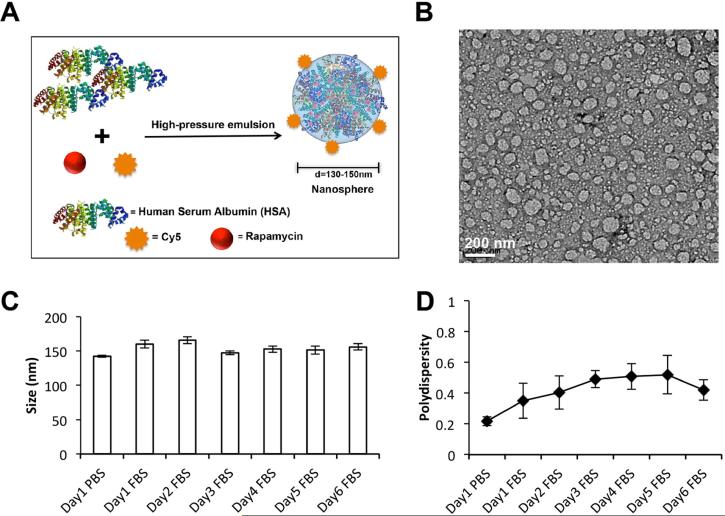
By two phases emulsion method using high-pressure homogenizer, we have successfully prepared Cy5 labeled HSA Rapa, in which Cy5 was conjugated to the surface of HSA carrier (Fig. 2A). The rapamycin was efficiently loaded in HSA nanoparticles, reaching a high encapsulation efficiency of 91.6%. TEM images show that the nanoparticles were discrete with a monodisperse size distribution (Fig. 2B). The nanoparticles were stable when incubated with FBS for 6 days, with an average size of ~150 nm (Fig. 2C) and polydispersity index of ~0.45 (Fig. 2D), as determined by dynamic light scattering. An analysis of the in vitro drug release profile showed approximately 58.3% of encapsulated rapamycin was burst released from HSA Rapa nanoparticles within 5 h, followed by a sustained release (Fig. S2).
Figure 2.
Synthesis and characterization of HSA Rapa nanoparticles. (A) Scheme of the HSA Rapa nanoparticles, which was synthesized by two phases emulsion method using high pressure homogenizer. (B) A representative picture of HSA Rapa nanoparticles obtained by transmission electron microscopy (bar = 200 nm). (C) Mean particle size of HSA Rapa nanoparticles incubated with PBS for 1 day or FBS for 6 days. (D) The polydispersity analysis of the HSA Rapa nanoparticles incubated with PBS or FBS for 6 days.
Concentration response and specificity of HSA Rapa-induced luciferase recovery
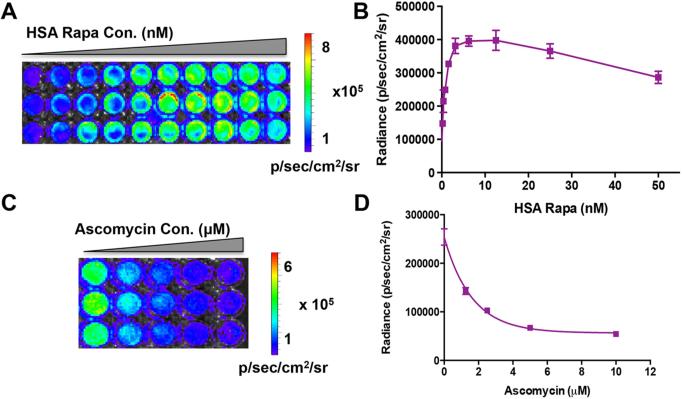
To explore the optimal dose of HSA Rapa for the efficient activation of luciferase reporter, escalating concentrations (from 0.195 nM to 50 nM) of HSA Rapa were added to the HN12 stable cells expressing FRB and FKBP12. Bioluminescence imaging analysis showed that the recovered luciferase activity increased with higher concentrations of HSA Rapa, reached a plateau at 12.5 nM, and then declined at concentrations >12.5 nM (Fig. 3A and 3B).
Figure 3.
Concentration dependent and specificity of HSA Rapa induced luciferase recovery. (A) Bioluminescence imaging and (B) quantification of the recovered luciferase activity in cells on addition of different concentration of HSA Rapa (0.195 nM to 50 nM). The error bars represent the mean ± SD for triplicate experiments. (C) The cells pretreating with 0.78 nM HSA Rapa subsequently received different concentration of ascomycin and bioluminescence imaging were performed. (D) The quantification of bioluminescence imaging data. The error bars represent the mean ± SD for triplicate experiments.
To study the specificity of HSA Rapa-induced luciferase recovery, ascomycin, a competitive inhibitor of rapamycin binding to FKBP12, were added subsequently to the HN12 cells that pretreating with 0.78 nM HSA Rapa. It is showed that the increasing concentrations of ascomycin resulted in a decrease in luciferase activity, indicating that ascomycin hindered the binding of rapamycin to FKBP12 and therefore reduced the dimerization with FRB (Fig. 3C and 3D).
Time-dependent cellular uptake of HSA Rapa
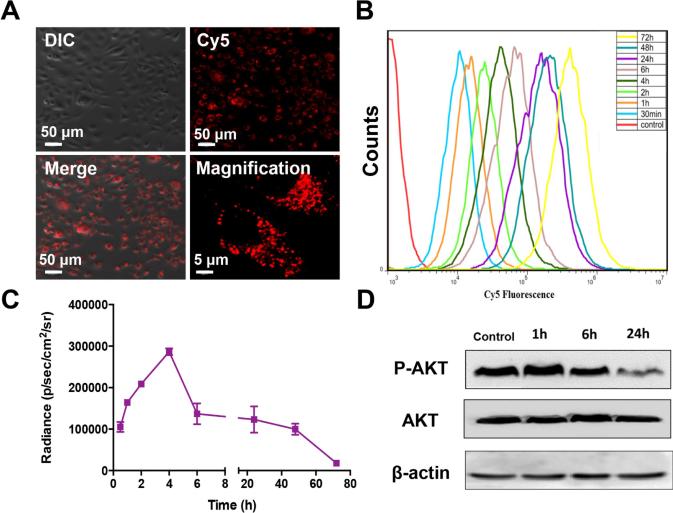
To investigate the cellular uptake of HSA Rapa, the Cy5 labeled HSA Rapa were introduced into cells. Four hours later, the NIFR signal of Cy5 from the cells was observed by confocal microscopy, indicating that HSA Rapa was gradually taken up from extracellular medium into intracellular vesicles (Fig. 4A). The time dependent cellular uptake was further validated by flow cytometry (Fig. 4B). The results demonstrated that the Cy5 fluorescence intensity increased over longer incubation time, indicating the continuous accumulation of HSA Rapa in the cells (Fig. 4A and B).
Figure 4.
Time-dependent cellular uptake of Cy5 labeled HSA Rapa nanoparticles. (A) The Cy5 labeled HSA Rapa nanoparticles were incubated with HN12#2 cells. 4 h later, the cells were investigated with confocal microscopy. The lower right picture is a magnification. (B) The nanoparticles were incubated with cells for different time periods (0.5, 1, 2, 4, 6, 24, 48, 72 h), then the cells were harvested and subjected to flow cytometer analysis. (C) The HN12#2 stable cells received the HSA Rapa for different time periods (0.5, 1, 2, 4, 6, 24, 48, 72 h). At the end of time point, bioluminescence imaging was used to investigate the luciferase activity. The graph is the qualification of the bioluminescence imaging results. The error bars represent the mean ± SD for triplicate experiments. (D) The cells were treated with HSA Rapa for different time periods (1, 6, 24 h), then western blot was performed with primary antibodies for P-AKT, AKT and β-actin.
Then we performed bioluminescence imaging to investigate the luciferase activity from cells receiving HSA Rapa over the same time course as described above. Unexpectedly, we found that the bioluminescence signal reached the peak at 4 h time point and then decreased afterwards. The signal of 72 hours after HSA Rapa treatment was almost undetected (Fig. 4C). To validate the biological effect of rapamycin on mTOR activity, Western blot analysis was performed to monitor the expression of phosphorylated Akt (p-Akt), a key effector of mTOR pathway23, at the designated time points post HSA Rapa treatment in cells (Fig. 4D). When normalized to total Akt levels before treatment, p-AKT was attenuated over time post HSA Rapa exposure, which was in accordance with the bioluminescence imaging results (Fig. 4C).
In vivo imaging and pharmacokinetics of HSA Rapa in HN12 tumor xenografts
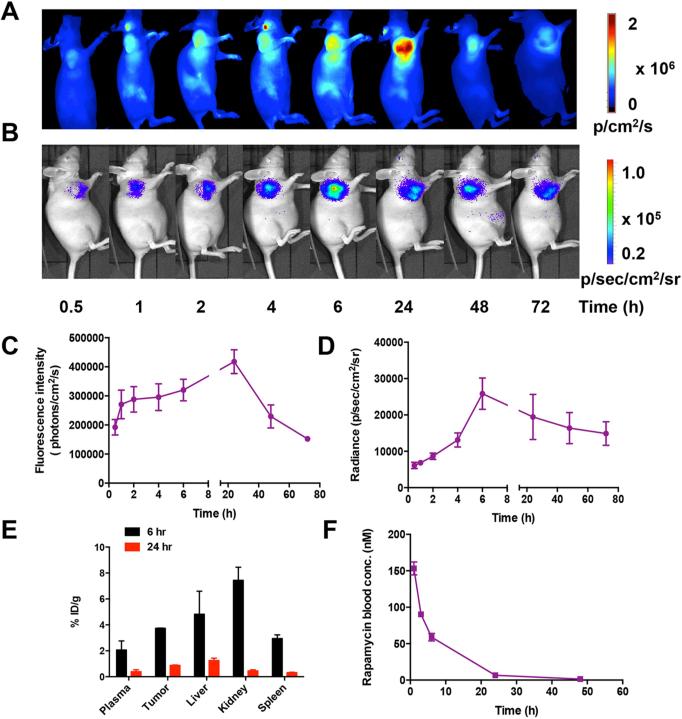
To evaluate the time course of HSA Rapa delivery in vivo, Cy5 labeled HSA Rapa was intravenously injected into the HN12 tumor bearing mice and then NIRFI or BLI was conducted at the indicated time points. For NIRFI results, the fluorescence signal in the tumor area increased over time, reached a plateau at 24 h time point, and then decreased afterwards (Fig. 5A and 5C). In contrast, the bioluminescence signal in vivo was found to be highest at 6 h time point and subsequently declined to a lower level (Fig. 5B and 5D).
Figure 5.
In vivo imaging and pharmacokinetics analysis of HSA Rapa. Cy5 labeled HSA Rapa (10 mg/kg) was intravenously injected into the HN12#2 tumor bearing mice (n=6). At different time points (0.5, 1, 2, 4, 6, 24, 48, 72 h) after injection, (A) fluorescence imaging was performed following by (B) bioluminescence imaging. (C) The qualification of the fluorescence imaging results. (D) The qualification of the bioluminescence imaging results. (E) After imaging, the tumors and major organs as well as plasma at 6 h or 24 h time point were harvested and subjected to LC-MS analysis. (F) After intravenous injection of HSA Rapa into nude mice with a dosage of 10 mg/kg (n=6), the concentration of rapamycin in the blood over time was measured by LC-MS. Blood was drawn from the tail vein of mice at different time point, and the content of rapamycin was measured at each time point.
Immediately after imaging, the allograft tumors and major organs were harvested at 6 h or 24 h time point and then subjected to high-performance liquid chromatography-mass spectrometry (LC-MS) analysis (Fig. 5E). The biodistribution results demonstrated that the mean rapamycin concentration from all organs at 6 h was significantly higher than that at 24 h time point. In addition, higher accumulation of rapamycin was observed in the liver and kidney rather than that in the tumor.
To further understand the pharmacokinetics of HSA Rapa, the concentration of rapamycin in the blood at different time intervals was measured by LC-MS after intravenous injection of HSA Rapa into nude mice with a dosage of 10 mg/kg (Fig. 5F). The blood circulation curve showed a blood circulation half-life (T1/2) of 7.44 h for HSA Rapa. Significant plasma concentrations of rapamycin were detectable as early as 1 h after intravenous administration and persisting for 48 h in mice. The pharmacokinetic parameter Cmax value (maximum drug concentration after injection) was 153.20 nM, whereas the corresponding Tmax value (at which Cmax is reached) was 1 h. The mean area under the concentration time curve (AUC0-infinity) was found to be 1344.08 nmol h/L for HSA Rapa.
Furthermore, to prove the stability of HSA Rapa nanoparticles in the blood, we also detected the fluorescence intensity of the blood samples at various time points (1h, 3 h, 6 h, 24 h) and calculated the ratio of fluorescence signal to rapamycin blood concentration obtained by LC-MS. We found that the fluorescence/LC-MS ratio was constant during 24 h, confirming that the HSA Rapa was stable in the blood of mice (Fig. S3).
In vivo antitumor activity of HSA Rapa in HNSCC tumor bearing mice
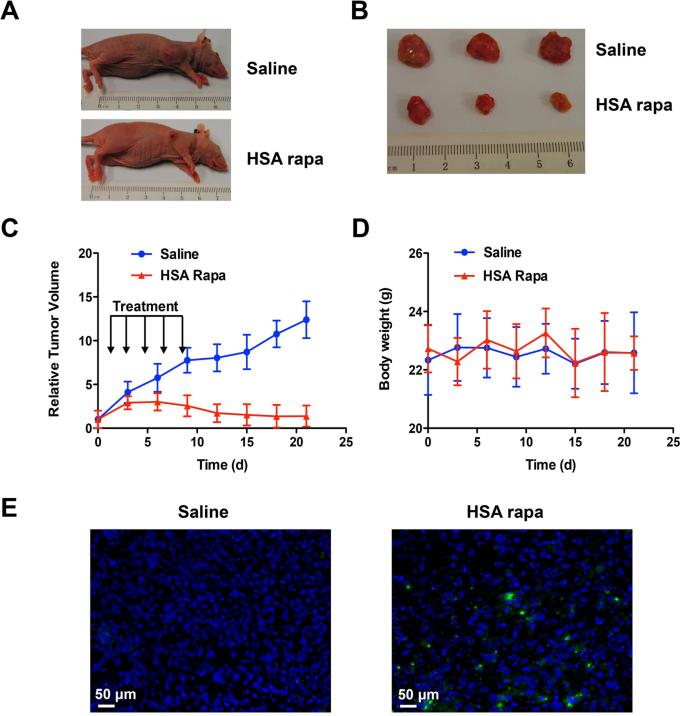
To examine in vivo antitumor efficacy of HSA Rapa, HSA Rapa (10 mg/kg) or saline were injected intravenously every other day into the athymic nude mice bearing human HN12#2 cell xenografts for total of five times injective, when the tumors reached a volume of 50 mm3. The tumor growth was monitored for up to three weeks after the initial treatment. Compared with saline, administration with HSA Rapa resulted in significant growth retardation of xenograft tumors at the end of the treatment (Fig. 6A and 6B). The antitumor effect of HSA Rapa became apparent as early as 3 days after treatment initiation (Fig. 6C), and this effect was sustained over the course of the study. The saline-treated group showed remarkable increase in average tumor volumes, which was 8.9 fold bigger than that in the HSA-Rapa treated group after 21 days. Treatment with HSA-Rapa did not result in any loss of body weight comparing to the control mice during the study (Fig. 6D).
Figure 6.
In vivo evaluation of the antitumor activity of HSA Rapa. HN12 tumor bearing mice were treated i.v. with HSA Rapa (10 mg/kg; n=10) or an equivalent volume of saline (n=10) for five times every other day. (A) An example of tumor regression in HSA Rapa or saline treated animals is showed. (B) A representative of three lesions dissected from HN12 xenograft 21 days after treatment with HSA Rapa or saline control. (C) tumor size from HN12 xenograft in both HSA Rapa and saline treated groups was measured every 2 days as indicated, and tumor volume was calculated as described in Materials and Methods. (D) The body weight of all animals was measured every 2 days for up to 3 weeks as indicated. (E) TUNEL assay in tumor tissues from xenograft model after 5 days of intravenous injection of HSA-Rapa or saline. DAPI was used to stain nuclei of tumor cells. Green: apoptotic cells; blue: DAPI.
It was reported that rapamycin could induce cell cycle arrest at G1-S phase and eventually result in apoptosis24-26. Thus, the tumor samples after 5 days of treatment with HSA-Rapa or saline were subjected to in situ TUNEL staining. As shown in Fig. 6E, the HSA-Rapa induced a relative increase of apoptosis in tumor cells compared with saline, further confirming that HSA-Rapa exhibited striking therapeutic efficacy.
An effective reporter in combination with optical imaging, including BLI and NIRFI, enables longitudinally and noninvasively monitoring the effects and metabolisms of candidate drugs in living organisms27. In the present study, we showed the application of BLI and NIRFI to simultaneously monitor the PK profiles of HSA Rapa and intrinsic rapamycin in a noninvasive and continuous manner with the use of a genetically encoded split luciferase reporter. The data presented herein discovered and validated the respective PK behaviors of a model drug and its carrier in a widely used HSA nanoparticle platform. It was found that there existed an obvious inconsistency in PK profiles between the HSA Rapa and rapamycin both in vitro and in vivo after introducing with Cy5 labeled HSA Rapa. The bioluminescence signals from cells treating with HSA Rapa were increased in a concentration-dependent manner up to 12.5 nM, then declined afterwards. This decrease in bioluminescence intensity at a high HSA Rapa concentration could attribute to the cellular activity inhibition by high concentrations of rapamycin, which is an effective antitumor drug. The dose-dependent bioluminescence results were consistent to other reports28. For cellular uptake, the Cy5 fluorescence signal increased persistently over time, indicating HSA nanoparticles translocated cell membrane via endocytosis and subsequently entered into cells. In contrast, the bioluminescence intensity plateaued at the 4 h time point and began to decrease afterwards. Such a discrepancy may have been due to the mechanism of action of rapamycin, which were released from the HSA nanoparticles and then inhibited the mTOR signal pathway, thereby leading to the inhibition of ribosomal protein synthesis and cellular activity over the time frame of experiments29, 30. To further confirm our speculations, western blot was conducted to analysis the expression level of Akt, an upstream protein in mTOR pathway. As anticipated, the phosphorylation of Akt was suppressed over time after exposure to HSA Rapa, further validating the bioluminescence imaging results obtained from cells. Therefore, increased fluorescence intensity simply measures the internalization of particles within cells. The fluorescence intensity does not necessarily indicate the activity of drugs within cells. So only the NIFRI may not accurately represent the PK profile of HSA Rapa and BLI is needed to validate the NIFRI results.
It is worth pointing out that there is also a discrepancy in tumor uptake between the NIRFI observation and that from the BLI study. The fluorescence intensity from the tumor area was shown to increase over time and plateau at 24 h time point, then began to decrease afterwards. In contrast, the BLI reached a highest level of signal induction at 6 h time point after treatment with HSA Rapa and subsequently declined to a lower level. The reason, we believe, is attributable to the fact that rapamycin inhibit the tumor growth via repressing protein translation after exposure to high concentrations of HSA Rapa for 6 h and longer. So we then evaluated the in vivo anti-tumor activity of HSA Rapa in xenografts after administration of HSA Rapa or saline for five times. The results demonstrated that HSA Rapa caused significant tumor growth retardation comparing with saline treated animals. Moreover, to rigorously validate the in vivo imaging findings, LC-MS was performed to analysis the biodistribution and blood circulation of rapamycin in nude mice. Due to the unique sensitivity and mass selectivity, LC-MS has been intensively applied in metabolic profiling and pharmacokinetics studies31, 32. On the basis of LC-MS analysis, we found that the detectable plasma rapamycin could be persisting for 48 h in mice and higher accumulation of rapamycin in the mouse kidney and liver, which are reticuloendothelial systems responsible for the clearance of the intravenously injected nanomaterials by macrophage uptake. Thus, the LC-MS data are consistent with the results of in vivo BLI imaging studies.
Conclusions
In conclusion, we showed the feasibility of a genetically coded molecular reporter in conjunction of near infrared fluorophores to simultaneously monitor the respective PK profiles of a loaded hydrophobic drug and its carrier in a biodegradable drug delivery system by employing BLI and NIRFI. This integrated imaging platform could facilitate the broader application of reporter systems to the pharmacokinetics studies and accelerate the pharmaceutical development process. Furthermore, such a strategy will definitely add significant value to the evaluation of nanoparticle based drug theranostics and bridge the preclinical studies to successful clinical translation.
Supplementary Material
Acknowledgments
This work was supported partly by The National Basic Research and Development Program of China (973 Program) (No.2013CB733803), National Natural Science Foundation of China (No. 81571721, No. 81301214), and the Intramural Research Program, National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH).
Footnotes
Competing interests
The authors declare that they have no competing interests
References
- 1.Farrell D, Alper J, Ptak K, Panaro NJ, Grodzinski P, Barker AD. ACS Nano. 2010;4:589–594. doi: 10.1021/nn100073g. [DOI] [PubMed] [Google Scholar]
- 2.Hong H, Zhang Y, Sun J, Cai W. Nano Today. 2009;4:399–413. doi: 10.1016/j.nantod.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schroeder A, Heller DA, Winslow MM, Dahlman JE, Pratt GW, Langer R, Jacks T, Anderson DG. Nat Rev Cancer. 2012;12:39–50. doi: 10.1038/nrc3180. [DOI] [PubMed] [Google Scholar]
- 4.Luo R, Venkatraman SS, Neu B. Biomacromolecules. 2013 doi: 10.1021/bm4003915. DOI: 10.1021/bm4003915. [DOI] [PubMed] [Google Scholar]
- 5.Namiki Y, Fuchigami T, Tada N, Kawamura R, Matsunuma S, Kitamoto Y, Nakagawa M. Acc Chem Res. 2011;44:1080–1093. doi: 10.1021/ar200011r. [DOI] [PubMed] [Google Scholar]
- 6.Ma Q, Li B, Yu Y, Zhang Y, Wu Y, Ren W, Zheng Y, He J, Xie Y, Song X, He G. Int J Pharm. 2013;445:88–92. doi: 10.1016/j.ijpharm.2013.01.046. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Sui M, Fan W. Curr Drug Metab. 2010;11:129–141. doi: 10.2174/138920010791110827. [DOI] [PubMed] [Google Scholar]
- 8.Moghimi SM, Hunter AC, Andresen TL. Annu Rev Pharmacol Toxicol. 2012;52:481–503. doi: 10.1146/annurev-pharmtox-010611-134623. [DOI] [PubMed] [Google Scholar]
- 9.Ding H, Wu F. Theranostics. 2012;2:1040–1053. doi: 10.7150/thno.4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang F, Wang Z, Hida N, Kiesewetter DO, Ma Y, Yang K, Rong P, Liang J, Tian J, Niu G, Chen X. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:5165–5170. doi: 10.1073/pnas.1321374111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gross S, Piwnica-Worms D. Current opinion in chemical biology. 2006;10:334–342. doi: 10.1016/j.cbpa.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 12.Easton JB, Houghton PJ. Oncogene. 2006;25:6436–6446. doi: 10.1038/sj.onc.1209886. [DOI] [PubMed] [Google Scholar]
- 13.Hidalgo M, Rowinsky EK. Oncogene. 2000;19:6680–6686. doi: 10.1038/sj.onc.1204091. [DOI] [PubMed] [Google Scholar]
- 14.Edwards SR, Wandless TJ. J Biol Chem. 2007;282:13395–13401. doi: 10.1074/jbc.M700498200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simamora P, Alvarez JM, Yalkowsky SH. Int J Pharm. 2001;213:25–29. doi: 10.1016/s0378-5173(00)00617-7. [DOI] [PubMed] [Google Scholar]
- 16.Elzoghby AO, Samy WM, Elgindy NA. J Control Release. 2012;157:168–182. doi: 10.1016/j.jconrel.2011.07.031. [DOI] [PubMed] [Google Scholar]
- 17.Kratz F. J Control Release. 2008;132:171–183. doi: 10.1016/j.jconrel.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 18.Geny B, Mettauer B, Muan B, Bischoff P, Epailly E, Piquard F, Eisenmann B, Haberey P. J Am Coll Cardiol. 1993;22:1193–1198. doi: 10.1016/0735-1097(93)90437-6. [DOI] [PubMed] [Google Scholar]
- 19.Ibrahim NK, Desai N, Legha S, Soon-Shiong P, Theriault RL, Rivera E, Esmaeli B, Ring SE, Bedikian A, Hortobagyi GN, Ellerhorst JA. Clin Cancer Res. 2002;8:1038–1044. [PubMed] [Google Scholar]
- 20.Conlin AK, Seidman AD, Bach A, Lake D, Dickler M, D'Andrea G, Traina T, Danso M, Brufsky AM, Saleh M, Clawson A, Hudis CA. Clin Breast Cancer. 2010;10:281–287. doi: 10.3816/CBC.2010.n.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paulmurugan R, Massoud TF, Huang J, Gambhir SS. Cancer research. 2004;64:2113–2119. doi: 10.1158/0008-5472.can-03-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu D, Ma Y, Niu G, Yan Y, Lang L, Aisa HA, Gao H, Kiesewetter DO, Chen X. Amino acids. 2011;40:669–675. doi: 10.1007/s00726-010-0696-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sekulic A, Hudson CC, Homme JL, Yin P, Otterness DM, Karnitz LM, Abraham RT. Cancer research. 2000;60:3504–3513. [PubMed] [Google Scholar]
- 24.Amornphimoltham P, Patel V, Sodhi A, Nikitakis NG, Sauk JJ, Sausville EA, Molinolo AA, Gutkind JS. Cancer research. 2005;65:9953–9961. doi: 10.1158/0008-5472.CAN-05-0921. [DOI] [PubMed] [Google Scholar]
- 25.Amornphimoltham P, Patel V, Leelahavanichkul K, Abraham RT, Gutkind JS. Cancer research. 2008;68:1144–1153. doi: 10.1158/0008-5472.CAN-07-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amornphimoltham P, Leelahavanichkul K, Molinolo A, Patel V, Gutkind JS. Clin Cancer Res. 2008;14:8094–8101. doi: 10.1158/1078-0432.CCR-08-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stell A, Belcredito S, Ramachandran B, Biserni A, Rando G, Ciana P, Maggi A. The quarterly journal of nuclear medicine and molecular imaging : official publication of the Italian Association of Nuclear Medicine. 2007;51:127–138. [PubMed] [Google Scholar]
- 28.Paulmurugan R, Gambhir SS. Analytical chemistry. 2005;77:1295–1302. doi: 10.1021/ac0484777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaragoza D, Ghavidel A, Heitman J, Schultz MC. Molecular and cellular biology. 1998;18:4463–4470. doi: 10.1128/mcb.18.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cruz MC, Goldstein AL, Blankenship J, Del Poeta M, Perfect JR, McCusker JH, Bennani YL, Cardenas ME, Heitman J. Antimicrobial agents and chemotherapy. 2001;45:3162–3170. doi: 10.1128/AAC.45.11.3162-3170.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui L, Nithipatikom K, Campbell WB. Analytical biochemistry. 2007;369:27–33. doi: 10.1016/j.ab.2007.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edmondson RD, Vondriska TM, Biederman KJ, Zhang J, Jones RC, Zheng Y, Allen DL, Xiu JX, Cardwell EM, Pisano MR, Ping P. Molecular & cellular proteomics : MCP. 2002;1:421–433. doi: 10.1074/mcp.m100036-mcp200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.