Abstract
Given the difficulty of procuring human brain tissue, a key question in molecular psychiatry concerns the extent to which epigenetic signatures measured in more accessible tissues such as blood can serve as a surrogate marker for the brain. Here, we aimed (1) to investigate the blood-brain correspondence of DNA methylation using a within-subject design and (2) to identify changes in DNA methylation of brain-related biological pathways in schizophrenia.
We obtained paired blood and temporal lobe biopsy samples simultaneously from 12 epilepsy patients during neurosurgical treatment. Using the Infinium 450K methylation array we calculated similarity of blood and brain DNA methylation for each individual separately. We applied our findings by performing gene set enrichment analyses (GSEA) of peripheral blood DNA methylation data (Infinium 27K) of 111 schizophrenia patients and 122 healthy controls and included only Cytosine-phosphate-Guanine (CpG) sites that were significantly correlated across tissues.
Only 7.9% of CpG sites showed a statistically significant, large correlation between blood and brain tissue, a proportion that although small was significantly greater than predicted by chance. GSEA analysis of schizophrenia data revealed altered methylation profiles in pathways related to precursor metabolites and signaling peptides.
Our findings indicate that most DNA methylation markers in peripheral blood do not reliably predict brain DNA methylation status. However, a subset of peripheral data may proxy methylation status of brain tissue. Restricting the analysis to these markers can identify meaningful epigenetic differences in schizophrenia and potentially other brain disorders.
Key words: DNA methylation, cross-tissue, blood, brain, correlation, schizophrenia
Introduction
Despite convincing evidence for strong heritability for psychiatric disorders,1 recently discovered genetic susceptibility loci explain only a small amount of the total variance.1–3 It has therefore been suggested that epigenetic processes might modulate gene expression.4
DNA methylation, one of the key epigenetic processes, involves the addition of a methyl group to the cytosine of a Cytosine-phosphate-Guanine (CpG) dinucleotide and can result in transcriptional silencing.5 DNA methylation has been suggested as the underlying mechanism of crucial developmental processes such as tissue and cellular differentiation. DNA methylation itself is thought to be affected by a wide range of environmental factors including nutritional supplements, chemicals, and parenting behavior.6–9
Recently, groundbreaking studies have shown an association between parenting behavior (eg, licking and grooming in rodents) and DNA methylation alterations in genes, related to hypothalamic-pituitary-adrenal axis activity, underlining the importance of epigenetic mechanisms in (ab)normal development.9–11 Specifically, this discovery spurred the interest of clinician-scientists to focus on differences in DNA methylation related to severe psychiatric disorders such as schizophrenia, eg, in candidate genes12–14 and using methylome-wide approaches.15,16 In all of these studies methylation levels were measured in easily accessible peripheral blood samples. For example, studying 27000 markers in peripheral blood of 22 pairs of monozygotic twins discordant for major psychosis, Dempster et al15 reported altered pathways related to nervous systems development and neurotransmitter receptor signaling. Using a much larger sample of over 700 schizophrenia patients and controls, Aberg et al16 used methyl-CpG binding domain sequencing methods and identified differentially methylated markers linked to hypoxia and infections.
Very few researchers have had the opportunity to investigate DNA methylation in postmortem brain samples of schizophrenia patients. Focusing on candidate genes such as RELN17,18 or MB-COMT19 or applying epigenome-wide approaches,20 these studies showed marked methylation differences between patients and controls.
Studies using blood samples are more feasible but limited because they only allow for indirect conclusions about the associated biological processes in living brain tissue, where epigenetic changes might be of greater relevance with respect to psychiatric disorders.4 Hence, there is a need for studies investigating, how and to what degree methylation patterns obtained from easily accessible blood samples relate to those in living brain tissue.
Several studies analyzed the correlation of DNA methylation patterns across different tissue types—including blood and (postmortem) brain tissue.21–23 Using whole-blood and postmortem brain tissue methylation data sets and a between-subject analysis approach, Horvath et al21 found correlations of around r = 0.9 across the 2 tissues types, while Davies et al23 reported a slightly lower correspondence of mean blood and brain methylation levels using a very similar study design.
Despite these very valuable results, limitations of these studies include the study design and the fact that brain tissue was obtained postmortem. First, there is evidence that pre-, peri-, and postmortem factors such as pH, postmortem interval, and preservation methods might have profound effects on DNA methylation (for a review see Pidsley et al24), which can bias study results based on postmortem tissue. Second, comparing methylation levels across tissues, which were obtained from different individuals and/or using mean methylation values averaged across subjects ignores inter-individual variability. It is well possible that—although group-average methylation values show a high level of correlation between blood and brain tissue (BBT)—the correlation within a given subject is weak (see also SM-figure 1). If our goal is to study inter-individual differences in DNA-methylation related to brain-based disorders such as schizophrenia using blood samples as a proxy—data on similarity of DNA-methylation between BBT based on between-subject designs can be misleading (see also SM-figure 2). To date, there are only 2 published reports applying a within-subject design (ie, analyzing paired data from the same subjects) to study blood-brain tissue correlations in DNA methylation in humans. The first study25 focused on a single gene (prodynorphine) while the second one23 included only 2 participants.
The current study had 2 major aims. First, we wanted to explore how DNA methylation signatures in whole blood relate to those in brain tissue of the same individual. Our approach represents the first study to (1) use living brain tissue obtained during neurosurgery, (2) apply a within-subject design in such an analysis and (3) use a large set of methylation markers, including over 450000 CpG sites. The second aim was to utilize the results from the first part of our study to explore brain-relevant DNA methylation differences in schizophrenia using epigenome-wide data obtained in blood samples and gene set enrichment analysis (GSEA). Compared with an epigenome-wide association approach (which tests each individual marker independently), GSEA has the added advantage that results are more reproducible allowing to detect affected pathways even when changes in individual genes are moderate or raw data are noisy.26
Method
Aim 1
Blood-Brain Tissue (BBT) Sample.
Human neocortical tissue samples and blood probes were obtained during neurosurgical treatment of 12 patients with pharmacoresistant temporal lobe epilepsy (mean age = 40.92+/−15.38 years; 4 female; SM-table 1). We only included patients with cortical dysplasia, which is associated with less severe histopathological changes of the brain tissue. In contrast, the more common Ammon’s horn sclerosis (hippocampal sclerosis) includes segmental loss of pyramidal neurons, granule cell dispersion, and reactive gliosis.27 Sections from the temporal lobe were snap-frozen immediately, dissected using a cryomicrotome, histopathologically classified by a trained neuropathologist and stored at −80°C. Blood probes were obtained at the same time and stored together with the brain samples.
Participants were excluded, if they had neurological conditions other than temporal lobe epilepsy, a family history of epilepsy, current substance abuse or dependence, a severe endocrine disorder or an abnormal body mass (<18 or >30), a severe metabolic disorder or incomplete medical records. The institutional review board approved the study and all participants provided written informed consent.
BBT DNA methylation quality control and preprocessing.
Genome-wide DNA methylation of DNA from whole BBT samples was assessed with the Illumina Human Methylation 450 Bead Chip (Illumina Inc, CA, US). No background correction or normalization was performed at this stage. Quality control and further analysis was performed in the statistical programming environment R (version 2.15.2) using various Bioconductor packages (v.2.18.0) including lumi (2.10.0), methylumi (2.4.0), and wateRmelon (0.99.14).28
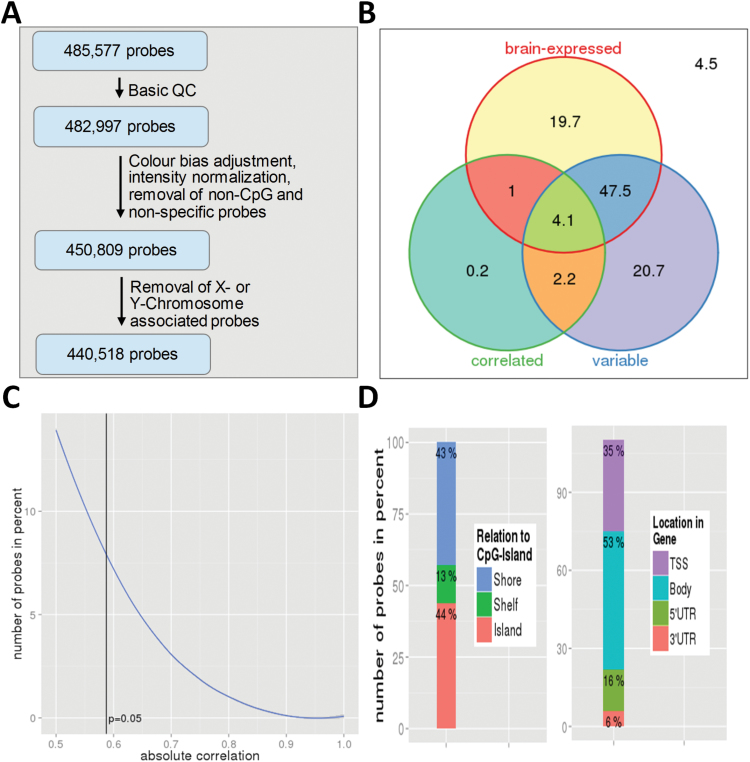
Prior to the calculation of the methylation status of each individual probe several preprocessing and quality control steps were performed (see SM 1.2). Subsequently, we performed a quantile color bias adjustment of the red and green fluorescence channel intensities. After this, intensities were normalized using the Danet function in wateRmelon. We then removed all single nucleotide polymorphism and other non-CpG-associated probes (N = 3145) and 29043 probes, which have been shown to be nonspecific on this chip.29 Last, we removed all CpG sites on the X or Y chromosome (N = 10291), leaving a total of 440518 autosomal CpG sites, which were transformed into beta values for subsequent analyses. For an overview of probe selection during preprocessing see also figure 1A. For additional details regarding data acquisition and preprocessing please see SM 1.2.
Fig. 1.
(A) Probe selection during preprocessing. (B) Percentage of all CpG sites passing quality control being variable (bottom right), correlated between blood and brain tissue (bottom left), or brain-associated (top). 4.5% of all CpG sites were neither variable nor correlated nor brain-associated. (C) Percentage of variable, brain-associated CpG sites, which pass a given Spearman correlation threshold. The vertical line marks the cut-off for significant correlations at P < .05. (D) Characteristics of variable, brain-associated CpG sites (in percent). Due to multiple gene location entries for some CpG sites in the Illumina annotation file, total percentage exceeds 100%.
Statistical Analyses: BBT.
To assess associations between BBT in a within-subject design, we calculated Spearman correlation coefficients and P values for each CpG by correlating paired (ie, same subject) blood and brain data points (see SM-Figure 1, approach B). Correlations were derived using the pspearman package in R, which computes Spearman’s rank tests with a precomputed exact null distribution for N ≤ 22. Furthermore, we created subsets containing only variable CpG sites (defined as those with a SD greater than the maximum of the SD density curve) or CpG sites for which there is evidence that the associated genes are expressed in the human brain (based on results from the Allen Human Brain Atlas, http://www.brain-map.org/). For detailed information see SM 1.3 and SM 1.4.
Because our BBT sample size was moderate making it susceptible to outliers, we used multi-dimensional scaling analysis in R to assess whether the DNA methylation patterns in our BBT dataset were comparable with a much larger dataset of whole blood and brain samples from the superior temporal gyrus. To confirm that the percentage of correlated probes was well above what would have been expected by chance, we carried out a permutation analysis by estimating the proportion distribution of significantly correlated CpG sites under the null hypothesis. For more information see below, SM 2.1 and SM 2.2.
Aim 2
MCIC Study of Schizophrenia.
Cryo-conserved blood samples from participants of the Mind Clinical Imaging Consortium (MCIC) study of schizophrenia from 4 participating sites were used to determine DNA methylation. Patients had a Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) diagnosis of schizophrenia (N = 103), schizophreniform disorder (N = 4) or schizoaffective disorder (N = 3), established using a Structured Clinical Interview for DSM disorders30 and a review of case files by trained clinicians. Severity of positive and negative symptoms was rated using the Scales for Assessment of Positive/Negative Symptoms.31,32 Depression symptoms were assessed using the Calgary Depression Scale.33 Healthy controls were excluded if they had a history of a medical or Axis I psychiatric diagnosis. Patients were excluded if they met criteria for current (within the past month) substance abuse or dependence. All subjects gave written informed consent prior to study enrolment. The human subjects research committees at each of the 4 sites approved the study protocol. For additional details about the participants and clinical measures, see references.34
MCIC DNA Methylation Quality Control and Preprocessing.
Blood samples were obtained from 234 participants and sent to the Harvard Partners Center for Genetics and Genomics for DNA extraction. All DNA extraction, bisulphite modification, and hybridization steps were done blinded to group assignment, the latter 2 were performed at the Mind Research Network Neurogenetics Core Lab on an Infinium HumanMethylation27 BeadChip using Illumina Infinium Methylation Assay. For more information on MCIC DNA methylation preprocessing, see SM 1.5. The final methylation data set comprised 228 subjects (110 schizophrenia patients and 118 healthy controls) and 27480 CpG sites.
Statistical Analyses: MCIC.
In order to perform GSEA with the goal to identify brain-related biological pathways with altered methylation profiles associated with schizophrenia, we pruned the MCIC dataset for variable, brain-associated and highly correlated CpG sites only (see above and results section). After this step 1218 CpG sites remained in the analysis. Based on the 1454 gene ontology (GO) targets gene sets downloaded from the Molecular Signatures Database (http://www.broadinstitute.org/gsea/msigdb/) containing genes based on the controlled vocabulary of the GO project,35 we performed GSEA using 457 gene sets left in the analysis after filtering for gene set size (min = 5, max = 500) using GSEA software.26 GSEA holds the advantage that pathways can be reliably detected even when effect sizes of individual CpG sites are small or signal-to-noise ratio is low, which is of importance especially for polygenic disorders such as schizophrenia. For further details about GSEA methods, please see SM 2.6 and Subramanian et al.26
Results
Aim 1
BBT Probe Characteristics.
As illustrated in figure 1B, 74.5% of all CpG sites passing quality control were variable. For 72.3% of CpG sites, there was evidence that the associated gene is expressed in the human brain. To ensure biological relevance, the following analyses were restricted to variable CpG sites associated with genes, for which there is evidence of brain-expression (227428, ie, 51.6% of all CpG sites, which passed quality control, figure 1B).
DNA methylation signatures of BBT in our study were similar to those of whole blood and superior temporal gyrus brain tissue (obtained in 2 different samples) of a much larger publicly available dataset of healthy controls (GSE43414; SM-figure 5), suggesting that our results can be generalized (ie, they were not influenced by sample specifics of this study).
Correlation of DNA methylation between BBT.
Figure 1C shows the percentage of CpG sites passing a given correlation threshold. Only 7.9% of a total of 227428 variable, brain-associated CpG sites showed a significant correlation (P < .05, corresponding to rho ≥ 0.59) between BBT. In other words, 4.1% of all CpG sites were variable, brain-associated and significantly correlated (figure 1B). In terms of effects size, 39.2% and 14.0% of all variable, brain-associated CpG sites showed moderate (rho ≥ 0.3) or large (rho ≥ 0.5) correlations, respectively. Permutation analyses (estimating the proportion distribution of significantly correlated CpG sites under the null hypothesis, N = 10000 under rearrangements of subject labels on the observed data points; SM-figure 6) confirmed that the percentage of significantly correlated probes was well above what would have been expected by chance.
Descriptive analyses of these variable, brain-associated, and correlated CpG sites revealed that eighty-seven percent were located either in CpG islands or in CpG shores (defined as within 2kb from CpG islands). Only 13% were associated with CpG shelf areas (between 2–4kb from CpG islands) (figure 1D). With respect to gene location, the majority of correlated CpG sites were located in gene bodies (53%) and in transcription start sites (35%). Comparing CpG sites with highest and lowest correlation values (each N = 100), we found that neither cluster displayed a clear pattern of hypo- or hypermethylation in blood or brain tissue. That is, when looking at the blood-minus-brain nonparametric rank differences in DNA methylation for each of the 200 CpG sites in each of the 12 subjects, we found both CpG sites, with higher and sites, with lower absolute DNA methylation in brain tissue compared with blood, irrespective of correlation strength. However, the variability of rank differences within and across subjects was larger in the low correlation cluster (SM-figure 7A). Compared with correlation results using a within-subject design, averaging probe methylation across subjects (ie, adopting a between-subject design as in previous studies) produced high mean correlation coefficients between BBT for both correlation clusters (SM-figure 7B and 7C). This illustrates that studies using blood as a proxy to study inter-individual differences in brain DNA methylation should be rather based on within-subjects design data. For an illustration of this point, see also SM-figure 1. For a list of the 100 CpG sites with highest within-subject correlation, see SM-table 2.
Aim 2
Application to Schizophrenia (MCIC Sample).
We proceeded to use all variable, brain-expressed and significantly correlated CpG sites, which were also present on the Illumina 27K array (1218 CpG sites, mapping to 1154 genes) in a GSEA of a schizophrenia case-control sample. Patients and controls did not differ in demographic variables such as gender, age or parental socio-economic status but patients had a significantly lower verbal IQ. Clinical variables, eg, mean length of illness was 11.73(±10.26) years, were typical for schizophrenia patients in an in- and outpatient treatment setting (for details see table 1).
Table 1.
Demographics
| Sample | Size | Gender (Female) | Age | WRAT3-RT | Parental SES | Age of Onset | Length of Illness | Negative Symptoms | Positive Symptoms | Calgary Depression Score | Current Antipsychotic Drug Dose | Cumulative Antipsychotic Drug Dose |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | N | Mean (SD) | Mean (SD) | Mean (SD) | Mean (SD) | Mean (SD) | Mean (SD) | Mean (SD) | Mean (SD) | Mean (SD) | ||
| SZ | 110 | 28 | 34.7 (10.59) | 46.17a (6.91) | 2.9 (1.02) | 22.95 (6.30) | 11.73 (10.26) | 8.14 (3.96) | 4.88 (2.86) | 3.42 (4.38) | 131.46 (355.84) | 57.36 (121.87) |
| HC | 118 | 43 | 32.53 (11.11) | 51.09a (3.93) | 2.68 (0.75) | — | — | — | — | — |
Note: Means and standard deviations (SD) are given. SZ, patient with schizophrenia; HC, healthy control, WRAT3-RT, Wide Range Achievement Test 3-Reading Test. Parental SES (socioeconomic status) was classified according to Hollingshead. Severity of positive and negative symptoms was rated using the Scales for Assessment of Positive/Negative Symptoms (SANS),31 and the Scale for Assessment of Positive Symptoms (SAPS).32 Depression symptoms were assessed using the Calgary Depression Scale.33 Cumulative and current antipsychotic exposure was calculated using the chlorpromazine (CPZ) conversion factors Andreasen et al.54
asignificantly different between SZ and HC on basis of Students t test (P < .05).
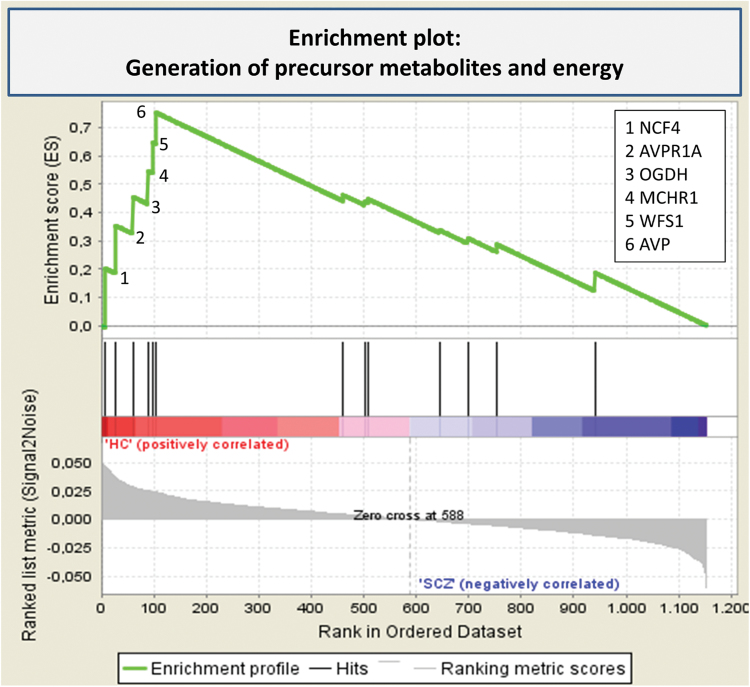
GSEA revealed one GO gene set, related to the generation of precursor metabolites and energy, displaying differential methylation between schizophrenia patients and controls after permutation and false discovery rate (FDR) correction (normalized enrichment score = 2.05, FDR = 0.02; figure 2, SM table 3). Among the genes significantly contributing to the enrichment score, were arginine vasopressin and its receptor (AVPR1A) and arginine vasopressin (AVP). The associated CpG sites were located in the promoter regions close to the transcription start sites of these 2 genes (35bp upstream and 326bp downstream, respectively; see SM-figure 8). The AVP-associated site was also within 100bp of several transcription factor binding sites, such as TGIF, GFI1, and MYC. Other enriched genes in this pathway included MCHR1, WFS1, NCF4, and OGDH. Cumulative antipsychotic drug dose did not correlate with DNA methylation of any of the genes significantly contributing to the enrichment score.
Fig. 2.
Enrichment plot: Generation of precursor metabolites and energy.
Profile of the running enrichment score and positions of gene set members on the rank ordered list (according to association with a diagnosis of schizophrenia). The leading edge subset is the set of genes that appear in the ranked list prior to the peak. For more information, see SM 2.6.
As a negative control, we then ran a separate GSEA using all variable, brain-expressed CpG sites with low cross-tissue within-subject correlation (defined as (P > .95, corresponding to rho ≤ 0.21,); 923 CpG sites, mapping to 900 genes). No gene set achieved significance after FDR correction (P < .05, see also SM 2.7).
Discussion
Given the difficulty of procuring human brain tissue vs the relative ease of measuring biological markers in blood, it is crucial to understand whether blood samples can be used to ask questions about the epigenetic signatures in the brain. Therefore, we used epigenome-wide markers collected invasively from both blood and living brain tissue of the same subjects at the same time to explore blood-brain tissue correlations of DNA methylation data. Using a within-subject design, we found that only a small proportion of markers (ie, 7.9%) showed a significant, large correlation between both tissue types, ie, most CpG sites are uncorrelated and researchers interested in the epigenetics of brain-related disorders should be careful when interpreting methylome-wide data assessed in peripheral tissues such as blood. The majority of the correlated CpG sites were found in or close to CpG islands and in gene bodies.
The second part of our study shows how knowledge about the within-subject cross-tissue correspondence of DNA methylation markers can be used to identify brain-related epigenetic risk mechanisms of schizophrenia via easily accessible CpG markers assessed in blood cells. By restricting our analysis to probes with a significant correlation with brain DNA methylation markers and using GSEA, we found that alterations in blood DNA methylation in schizophrenia, which correspond to brain tissue DNA methylation, are found in a gene set associated with the generation of precursor metabolites and energy, including peptides such as arginine vasopressin.
Results from previous studies analyzing blood-brain tissue correspondence of methylation profiles were mixed. A few studies reported moderate to high mean correlations across tissue types in humans applying a between-subject design (see also introduction). For example, Fan and Zhang36 analyzed 16000 methylation markers from different human non-CNS tissues types. Averaging each CpG site across all samples, they found high average correlation coefficients ranging between 0.7–0.85. Divergent results can be attributed to the statistical approach selected by the researchers (as demonstrated in SM-Figure 1, 2, and 7) and to the (more common) fact that the different tissue types were obtained from different individuals. There is ample evidence that developmental processes such as cell differentiation are associated with unique methylation profiles for each cell lineage or tissue type, even across species. For example, Hon et al37 reported that methylomes from 17 different mouse tissues clustered according to germ layer. Another recent study described a notable interspecies correspondence of tissue-specific methylation.38 That is, cluster analysis of 3 tissue types in mice and humans perfectly discriminated among tissue types, regardless of the species of origin.37 Similarly, 2 recent studies comparing inter-tissue and inter-subject methylation variability reported greater correspondence of methylation patterns within a tissue across subjects than within a subject across tissues.39,40
Results from our study are in line with these findings and underline the tissue-specificity of methylation profiles. Conflicting results with respect to previous human methylation studies are mainly due to study design. Whereas studies examining mean methylation values—averaged across samples or participants (between-subject design)—claim high tissue correspondence, studies employing cluster analyses or within-subject designs report lower correlations.
Accordingly, the few existing candidate gene studies applying a within-subject design in rodents reported moderate across tissue correlations in 1 out of 4 CpGs in the Bdnf gene (r = 0.53) and between 2 (out of 7) different CpGs in Fkbp5 (R 2 of 0.23–0.49).41,42 There has been only 1 other human methylome-wide study—apart from ours—that applied a within-subject design. Using postmortem brain tissue and blood obtained pre-mortem from the same 2 individuals, Davies et al23 reported correlation values of 0.66–0.76 between BBT. The even lower correlations described in our study could be a result of a larger sample size (N = 12 vs N = 2), differences in brain tissue retrieval (neurosurgical biopsies vs postmortem tissue) and different methodology (DNA microarray vs methylated DNA immunoprecipitation sequencing).43
Taken together, our results indicate that the common practice of studying DNA methylation in peripheral tissues to study disorders of the brain should be applied with great caution. Although tissue type is only 1 source of variation in DNA methylation data (and future studies should address the effects of age, gender, and cell type, see SM-figure 3 regarding the latter point), we suggest a way forward which includes prioritizing CpG sites that are validated as proxys, ie, with a high correlation in DNA methylation across both tissue types, for future analyses.
Consequently, we excluded all uncorrelated CpG sites for a GSEA of the MCIC data (ie a large blood-based DNA methylation dataset of patients with schizophrenia and healthy controls). Crucial genes of the schizophrenia-linked precursor metabolites and energy pathway identified in GSEA included AVPand AVPR1A. Although we cannot draw conclusions about the functional relevance of DNA methylation changes in these genes and future studies are needed to replicate these patterns in postmortem brain samples as well as investigate the impact on gene expression, it is interesting to note that probes were found in close proximity to the transcription start sites, pointing towards a potential functional role. Furthermore, an association between AVP and schizophrenia has been reported in previous genetic44 and endocrinological studies.45 AVP has an important role in social memory and cognition46–48 – domains that are affected in schizophrenia.49 In line with that, AVP serum concentrations are increased in schizophrenia patients and correlate with positive symptom severity and verbal learning deficits.45 Administration of vasopressin or vasopressin analogues, has been shown to have beneficial effects on social memory47,50,51 and reexpression of AVPR1A in AVPR1A knock-out mice ameliorates social interaction deficits.48
Other highly enriched genes included MCHR1, an important neuroendocrine modulator of appetite that has previously been shown to be associated with schizophrenia,52 as well as WFS1, associated with Wolfram syndrome, which affects central nervous system development.53
Our results have to be seen in the light of the following additional limitations. First, our BBT sample was only of moderate size, although larger than previous studies. Second, brain tissue was obtained from the temporal lobe restricting inference regarding other brain areas. Third, brain tissue was obtained from epilepsy patients with cortical dysplasia, which could bias our results. However, cortical dysplasia in epilepsy is associated with less severe histopathological changes of the brain tissue than the more common Ammon’s horn sclerosis,27 and overall methylation patterns in our temporal lobe samples were highly similar to temporal lobe samples of healthy individuals (SM 2.1). Fourth, we were not able to differentiate between cytosine methylation and hydroxymethylation, which is a common limitation to standard bisulfite-based analysis techniques.
In sum, we explored blood-brain tissue correlations of DNA methylation data and characterized epigenetic risk mechanisms of schizophrenia. As a novel approach, we used living brain tissue, epigenome-wide data and, most importantly, applied a within-subject design. Our correlational findings indicate that only a small proportion of CpG sites show highly similar DNA methylation patterns in BBT within the same subject (as opposed to approaches using group averages). Studies using blood as a proxy to study inter-individual differences in brain DNA methylation should therefore be based on within-subjects design data. Our results can guide future research in psychiatric epigenetics, ie, could help to predict which CpG sites can be studied indirectly by analyzing blood samples. Following this approach, ie, by using easily accessible CpG markers taken from blood samples, for which there is evidence for a high correlation with brain DNA methylation markers, we identified a brain-related biological pathways that is affected in schizophrenia and can be identified using peripheral tissue. The pathway, related to precursor metabolites and signaling peptides, may provide new potential targets for schizophrenia research and treatment.
Supplementary Material
Supplementary material is available at http://schizophreniabulletin.oxfordjournals.org.
Funding
This work was supported by the National Institutes of Health (NIH/NCRR P41RR14075, K08 MH068540, NIH: 2P20GM103472-06 to J.L.); the Department of Energy (DE-FG02-99ER62764); the MIND Research Network, Morphometry BIRN (1U24, RR021382A); Function BIRN (U24RR021992-01, NIH.NCRR MO1 RR025758-01); the National Institute of General Medical Sciences and the National Institute of Biomedical Imaging and Bioengineering (NIGMS P20-GM103472 & NIBIB 2R01-EB000840 to V.C.); the National Association for Research in Schizophrenia and Affective Disorders (NARSAD award to S.E.), the German Ministry for Education, Research and Technology (BMBF, A.Z. 13N10777), and the Friedrich-Ebert foundation (scholarship to E.W.).
Supplementary Material
Acknowledgments
Veit Roessner has received lecture fees from Eli Lilly, Janssen-Cilag, Medice, Novartis and was a member of advisory boards of Eli Lilly, Novartis. Joshua Roffman has received research support from Pamlab. All other authors declare no biomedical financial interests or other potential conflict of interests.
References
- 1. Cross-Disorder Group of the Psychiatric Genomics Consortium. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. O'Donovan MC, Craddock NJ, Owen MJ. Genetics of psychosis; insights from views across the genome. Hum Genet. 2009;126:3–12. [DOI] [PubMed] [Google Scholar]
- 3. Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nishioka M, Bundo M, Kasai K, Iwamoto K. DNA methylation in schizophrenia: progress and challenges of epigenetic studies. Genome Med. 2012;4:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brenet F, Moh M, Funk P, et al. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One. 2011;6:e14524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ehrlich S, Walton E, Roffman JL, et al. Smoking, but not malnutrition, influences promoter-specific DNA methylation of the proopiomelanocortin gene in patients with and without anorexia nervosa. Can J Psychiatry. 2012;57:168–176. [DOI] [PubMed] [Google Scholar]
- 8. Ho SM, Tang WY, Belmonte de Frausto J, Prins GS. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006;66:5624–5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weaver IC, Cervoni N, Champagne FA, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. [DOI] [PubMed] [Google Scholar]
- 10. Weaver ICG, Champagne FA, Brown SE, et al. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J. Neurosci. 2005;25:11045–11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McGowan PO, Sasaki A, D'Alessio AC, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Melas PA, Rogdaki M, Ösby U, Schalling M, Lavebratt C, Ekström TJ. Epigenetic aberrations in leukocytes of patients with schizophrenia: association of global DNA methylation with antipsychotic drug treatment and disease onset. FASEB J. 2012;26:2712–2718. [DOI] [PubMed] [Google Scholar]
- 13. Ikegame T, Bundo M, Sunaga F, et al. DNA methylation analysis of BDNF gene promoters in peripheral blood cells of schizophrenia patients. Neurosci Res. 2013;77:208–214. [DOI] [PubMed] [Google Scholar]
- 14. Cheng J, Wang Y, Zhou K, et al. Male-specific association between dopamine receptor D4 gene methylation and schizophrenia. PLoS One. 2014;9:e89128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dempster EL, Pidsley R, Schalkwyk LC, et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20:4786–4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aberg KA, McClay JL, Nerella S, et al. Methylome-wide association study of schizophrenia: identifying blood biomarker signatures of environmental insults. JAMA Psychiatry. 2014;71:255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grayson DR, Jia X, Chen Y, et al. Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci U S A. 2005;102:9341–9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abdolmaleky HM, Cheng KH, Russo A, et al. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;134B:60–66. [DOI] [PubMed] [Google Scholar]
- 19. Abdolmaleky HM, Cheng KH, Faraone SV, et al. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum Mol Genet. 2006;15:3132–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mill J, Tang T, Kaminsky Z, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82:696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Horvath S, Zhang Y, Langfelder P, et al. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 2012;13:R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaminsky Z, Tochigi M, Jia P, et al. A multi-tissue analysis identifies HLA complex group 9 gene methylation differences in bipolar disorder. Mol Psychiatry. 2012;17:728–740. [DOI] [PubMed] [Google Scholar]
- 23. Davies MN, Volta M, Pidsley R, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13:R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pidsley R, Mill J. Epigenetic studies of psychosis: current findings, methodological approaches, and implications for postmortem research. Biol Psychiatry. 2011;69:146–156. [DOI] [PubMed] [Google Scholar]
- 25. Yuferov V, Nielsen DA, Levran O, et al. Tissue-specific DNA methylation of the human prodynorphin gene in post-mortem brain tissues and PBMCs. Pharmacogenet Genomics. 2011;21:185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blümcke I, Thom M, Wiestler OD. Ammon’s horn sclerosis: a maldevelopmental disorder associated with temporal lobe epilepsy. Brain Pathol. 2002;12:199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pidsley R, Y Wong CC, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen YA, Lemire M, Choufani S, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. First M, Spitzer A, Gibbon M, Williams J. Structured Clinical Interview for DSM-IV-TR Axis I Disorders, Research Version, Nonpatient Edition. New York: New York State Psychiatric Institute; 2002. [Google Scholar]
- 31. Andreasen NC. Scale for the Assessment of Negative Symptoms. Iowa City: University of Iowa Press; 1983. [Google Scholar]
- 32. Andreasen NC. Scale for the Assessment of Positive Symptoms. Iowa City: University Iowa Press; 1984. [Google Scholar]
- 33. Addington D, Addington J, Maticka-Tyndale E. Assessing depression in schizophrenia: the Calgary Depression Scale. Br. J. Psychiatry. Suppl. 1993;22:39–44. [PubMed] [Google Scholar]
- 34. Ehrlich S, Morrow EM, Roffman JL, et al. The COMT Val108/158Met polymorphism and medial temporal lobe volumetry in patients with schizophrenia and healthy adults. Neuroimage. 2010;53:992–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fan S, Zhang X. CpG island methylation pattern in different human tissues and its correlation with gene expression. Biochem Biophys Res Commun. 2009;383:421–425. [DOI] [PubMed] [Google Scholar]
- 37. Hon GC, Rajagopal N, Shen Y, et al. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet. 2013;45:1198–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ladd-Acosta C, Pevsner J, Sabunciyan S, et al. DNA methylation signatures within the human brain. Am J Hum Genet. 2007;81:1304–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lokk K, Modhukur V, Rajashekar B, et al. DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome Biol. 2014;15:r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kundakovic M, Gudsnuk K, Herbstman JB, Tang D, Perera FP, Champagne FA. DNA methylation of BDNF as a biomarker of early-life adversity. [published online ahead of print November 10, 2014] Proc. Natl. Acad. Sci. U. S. A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ewald ER, Wand GS, Seifuddin F, et al. Alterations in DNA methylation of Fkbp5 as a determinant of blood-brain correlation of glucocorticoid exposure. Psychoneuroendocrinology. 2014;44:112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. [DOI] [PubMed] [Google Scholar]
- 44. Teltsh O, Kanyas K, Karni O, et al. Genome-wide linkage scan, fine mapping, and haplotype analysis in a large, inbred, Arab Israeli pedigree suggest a schizophrenia susceptibility locus on chromosome 20p13. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:209–215. [DOI] [PubMed] [Google Scholar]
- 45. Rubin LH, Carter CS, Bishop JR, et al. Peripheral vasopressin but not oxytocin relates to severity of acute psychosis in women with acutely-ill untreated first-episode psychosis. Schizophr Res. 2013;146:138–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Meyer-Lindenberg A, Domes G, Kirsch P, Heinrichs M. Oxytocin and vasopressin in the human brain: social neuropeptides for translational medicine. Nat Rev Neurosci. 2011;12:524–538. [DOI] [PubMed] [Google Scholar]
- 47. Matsuoka T, Sumiyoshi T, Tanaka K, et al. NC-1900, an arginine–vasopressin analogue, ameliorates social behavior deficits and hyperlocomotion in MK-801-treated rats: therapeutic implications for schizophrenia. Brain Res. 2005;1053:131–136. [DOI] [PubMed] [Google Scholar]
- 48. Bielsky IF, Hu SB, Ren X, Terwilliger EF, Young LJ. The V1a vasopressin receptor is necessary and sufficient for normal social recognition: a gene replacement study. Neuron. 2005;47:503–513. [DOI] [PubMed] [Google Scholar]
- 49. Biedermann F, Frajo-Apor B, Hofer A. Theory of mind and its relevance in schizophrenia. Curr. Opin. Psychiatry. 2012;25:71–75. [DOI] [PubMed] [Google Scholar]
- 50. Dantzer R, Bluthe RM, Koob GF, Le Moal M. Modulation of social memory in male rats by neurohypophyseal peptides. Psychopharmacology (Berl). 1987;91:363–368. [DOI] [PubMed] [Google Scholar]
- 51. Le Moal M, Dantzer R, Michaud B, Koob GF. Centrally injected arginine vasopressin (AVP) facilitates social memory in rats. Neurosci Lett. 1987;77:353–359. [DOI] [PubMed] [Google Scholar]
- 52. Demontis D, Nyegaard M, Christensen JH, et al. The gene encoding the melanin-concentrating hormone receptor 1 is associated with schizophrenia in a Danish case-control sample. Psychiatr Genet. 2012;22:62–69. [DOI] [PubMed] [Google Scholar]
- 53. Sütt S, Altpere A, Reimets R, et al. Wfs1-deficient animals have brain-region-specific changes of Na+, K+-ATPase activity and mRNA expression of α1 and β1 subunits. J Neurosci Res. 2015;93:530–537. [DOI] [PubMed] [Google Scholar]
- 54. Andreasen NC, Pressler M, Nopoulos P, Miller D, Ho BC. Antipsychotic dose equivalents and dose-years: a standardized method for comparing exposure to different drugs. Biol Psychiatry. 2010;67:255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.