Abstract
Schizophrenia is a mental disorder characterized by impairments in behavior, thought, and neurocognitive performance. We searched for susceptibility loci at a quantitative trait locus (QTL) previously reported for abstraction and mental flexibility (ABF), a cognitive function often compromised in schizophrenia patients and their unaffected relatives. Exome sequences were determined for 134 samples in 8 European American families from the original linkage study, including 25 individuals with schizophrenia or schizoaffective disorder. At chromosome 5q32–35.3, we analyzed 407 protein-altering variants for association with ABF and schizophrenia status. For replication, significant, Bonferroni-corrected findings were tested against cognitive traits in Mexican American families (n = 959), as well as interrogated for schizophrenia risk using GWAS results from the Psychiatric Genomics Consortium (PGC). From the gene SYNPO, rs6579797 (MAF = 0.032) shows significant associations with ABF (P = .015) and schizophrenia (P = .040), as well as jointly (P = .0027). In the Mexican American pedigrees, rs6579797 exhibits significant associations with IQ (P = .011), indicating more global effects on neurocognition. From the PGC results, other SYNPO variants were identified with near significant effects on schizophrenia risk, with a local linkage disequilibrium block displaying signatures of positive selection. A second missense variant within the QTL, rs17551608 (MAF = 0.19) in the gene WWC1, also displays a significant effect on schizophrenia in our exome sequences (P = .038). Remarkably, the protein products of SYNPO and WWC1 are interaction partners involved in AMPA receptor trafficking, a brain process implicated in synaptic plasticity. Our study reveals variants in these genes with significant effects on neurocognition and schizophrenia risk, identifying a potential pathogenic mechanism for schizophrenia spectrum disorders.
Key words: schizophrenia, cognition, SYNPO, WWC1, synaptic plasticity
Introduction
Schizophrenia is a complex, highly heritable brain disorder characterized by disturbances in behavior, thought, and emotion.1 Although a number of genes and neurobiological pathways have been implicated in linkage and genome-wide association studies, much of the genetic liability of schizophrenia remains to be explained,2,3 suggesting a polygenic architecture,4 likely confounded by the clinical heterogeneity of the disorder.5
Deficits in cognitive functions have been commonly observed in schizophrenia patients, and in smaller magnitude in unaffected family members, especially for executive function, learning, and memory,6–10 which may reflect innate, underlying differences that mediate the familial risk of schizophrenia. Analysis of such endophenotypes can delineate the psychiatric phenome and allow for identification of etiological mechanisms that are more proximate to gene action than disease endpoints.11 Thus, genes that moderately influence the risk of schizophrenia may exhibit substantially stronger effects on cognition, making them easier to detect at genome-wide significance levels.
This approach was employed by Almasy et al12 who conducted a genome-wide linkage screen of schizophrenia and cognitive performance in affected families, discovering a quantitative trait locus (QTL) for abstraction and mental flexibility (ABF) on chromosome 5q (log of odds [LOD] = 3.43; P = .011), with effects on schizophrenia risk revealed through bivariate analysis. Although several linkage studies of schizophrenia have implicated chromosome 5q,13–20 these findings provide insight into the potential neuropathology of the region, as the neural underpinnings for ABF have been localized primarily to the prefrontal brain circuitry,21 including the dorsolateral and superior prefrontal cortices,22 whose anatomical abnormalities and activity levels have been strongly associated with schizophrenia.23–27
The specific genetic variants contributing to this broad linkage region have yet to be determined. Previous endophenotype studies that have targeted schizophrenia candidate genes have been successful in identifying risk variants, including ones for cognitive traits.28–38 In this article, we investigate the QTL at chromosome 5q, focusing on nonsynonymous variation from 238 local genes. Exome sequencing was conducted on 134 samples from 8 European American families drawn from the original linkage study, including 25 diagnosed with schizophrenia or schizoaffective disorder. Significant SNP associations for ABF and schizophrenia were followed up in independent Mexican American families for select neurocognitive measurements, as well as examined among the GWAS results from the Psychiatric Genomics Consortium (PGC). We identify a number of potential risk loci, with implications for the neurobiological basis of cognitive impairment as observed in schizophrenia patients.
Methods
Family Samples
The Multiplex-Multigenerational Genetic Investigation of Schizophrenia (MGI) has been described previously.12,39 Families were recruited through a European American individual with schizophrenia, who had at least 1 first-degree relative with schizophrenia or schizoaffective disorder (SAD), depressed type. From the extended families, all the available first-, second- and third-degree relatives 15 years of age or older were invited to participate. MGI was approved by the Institutional Review Board of each of the 3 collaborating institutions, with all participants providing informed consent. In the case of minors under age 18 who provided assent, consent was obtained from a parent. Of the 43 MGI families (n = 676 participants), 8 of the largest, most densely affected ones were analyzed in this study. A total of 134 samples were exome sequenced, including 23 diagnosed with schizophrenia and 2 with SAD (see supplementary table S1 for pairwise familial relationships). Based on the original ABF linkage,12 5 of the families selected exhibit appreciable pedigree-specific LOD scores at the QTL, ranging from 0.16 to 0.33, representing 37.8% of the overall signal (LOD = 3.43; supplementary table S2).
Phenotyping
DSM-IV diagnoses were determined from: (1) the Diagnostic Interview for Genetics Studies, version 2.040; (2) the Family Interview for Genetics Studies41; and (3) reviews of medical records. Lifetime best-estimate diagnoses were arrived at by 2 investigators, each blind to the familial relationships among participants (kappa > 0.8). In total, 106 individuals were diagnosed with schizophrenia or SAD, with 75% undergoing treatment at the time of assessment. Effects of medication on neurocognitive measures have been shown to be negligible or subtle.42–44 In addition to schizophrenia, other psychiatric conditions identified included schizotypal personality disorder, psychotic disorder, and different forms of bipolar disorder.
Participants completed a computerized test battery45,46 designed to evaluate 9 neurocognitive domains.39 ABF was assessed using the Penn Conditional Exclusion Test (PCET),47 for which participants are required to select one of 4 shapes for exclusion based on a sorting principle. An efficiency score was calculated as the average z score for performance accuracy and speed.
Exome Sequencing
We used the Illumina TruSeq platform (Illumina) for sample preparation, exome enrichment, and sequencing on the Illumina HiSeq 2000 instrument. In total, 62Mb were sequenced, yielding uniform coverage of 201121 exons from 20794 genes. FASTQ files of demultiplexed paired sequencing reads of 100bp were produced by CASAVA 1.8 suite and mapped to the UCSC human genome reference assembly 19 (hg19) using BWA (v. 0.6.1).48 Mapped reads were analyzed with SAMtools (v. 0.1.12a)49 and Picard (v. 1.56) (http://picard.sourceforge.net) to mark likely PCR duplicates and ensure consistency of the mapped data, with the output processed with the GATK (v. 1.6) package50 (for more detail, see supplementary methods).
We called a total of 380895 high-quality SNPs, with an average of 35 reads per variant, each functionally annotated with ANNOVAR.51 At the QTL for ABF efficiency at 5q32–35.3, spanning approximately 35Mb, 6518 SNPs were called, encompassing 366 different genes. To focus on sites of potential functional relevance, sequence data were filtered for variant quality LOD scores of 4.0 or greater, have no missing genotype data, and represent nonsynonymous mutations, leaving 407 SNPs available for association analysis. Mendelian consistency of these loci was confirmed with Sequential Oligogenic Linkage Analysis Routines (SOLAR).52
Replication Samples
For any significant, Bonferroni-corrected associations for ABF and/or schizophrenia risk, replications were sought in independent Mexican American families from the Genetics of Brain Structure and Function (GOBSF) study for select neurocognitive measurements53,54: PCET accuracy (n = 519 subjects), Wechsler Adult Intelligence Scale II (WAIS-II; n = 430), Wechsler Test of Adult Reading (WTAR) (n = 264), California Verbal Learning Test (CVLT) total recall (n = 520), and CVLT delayed recall (n = 518). Unlike MGI, these families were not ascertained based on schizophrenia probands. Genotypes were obtained from whole genome sequences and imputed data (n = 959).55
For potential risk effects related to schizophrenia, we interrogated GWAS results from the PGC (available at http://www.med.unc.edu/pgc/downloads). Specifically, we examined 2 data sets: stage I, representing 17 population samples of European ancestry (n = 9394 cases and 12462 controls), with imputation based on HapMap3 reference panel56; and stage I plus additional Swedish cohorts (n = 5001 cases and 6243 controls), using 1000 Genomes phase 1 data for imputation.57
Statistical Analysis
All genetic analyses were performed in SOLAR, using a maximum likelihood (ML), variance decomposition approach. To evaluate ABF as an endophenotype to schizophrenia, both the genetic correlation and endophenotype ranking variable (ERV) were computed.58 SNP association testing was performed using measured genotype analyses.59 This single degree of freedom test assumes genetic additivity and compares a model saturated for both the random effects of kinship and the main effect of a SNP genotype to a null model with the SNP effect constrained to 0. Covariates include sex, age, age squared, and their interactions. P values were adjusted for multiple testing (n = 407 SNPs) via Bonferroni correction, corresponding to an alpha threshold of approximately 1.2×10−4. Bivariate models of ABF and schizophrenia were also examined for any SNPs of interest. Tail area-based false discovery rate (FDR) q-values were computed in the R package fdrtool.60 Multimarker, gene-based analyses were conducted using the sequence kernel association tests (SKAT) with the R script famSKAT.61 Diversity and neutrality test statistics were computed for genomic regions of interest using PopGenome,62 with coalescent simulations (1000 iterations) based on Hudson’s MS algorithm63 performed to evaluate significance of observed deviations from the neutral evolutionary model.
Results
Descriptive Statistics and Heritability Estimates
Measures of ABF efficiency were available for 113 of the 134 sequenced individuals, with a mean of −0.45±0.11, ranging from −2.03 to 1.39, with no evidence of kurtosis (g 2 = −1.55). No significant differences are observed between the sexes. Age is negatively correlated with ABF (r = −.34; P = 2.0×10−4). For schizophrenia, affected individuals (n = 21) scored significantly worse for ABF efficiency (μ = −1.41±0.24) than unaffected individuals (μ = −0.24±0.12; P = 1.3×10−4). Both ABF and schizophrenia are significantly heritable, with respective estimates of 0.53±0.19 (P = 1.8×10−3) and 0.84±0.40 (P = 8.6×10−3). The genetic correlation between the traits is −0.19±0.11 (P = .34; ERV = 0.13), with a more robust genetic correlation of −0.47±0.15 (P = .021; ERV = 0.28) observed for the entire set of MGI families.
Genetic Associations at 5q32–35.3
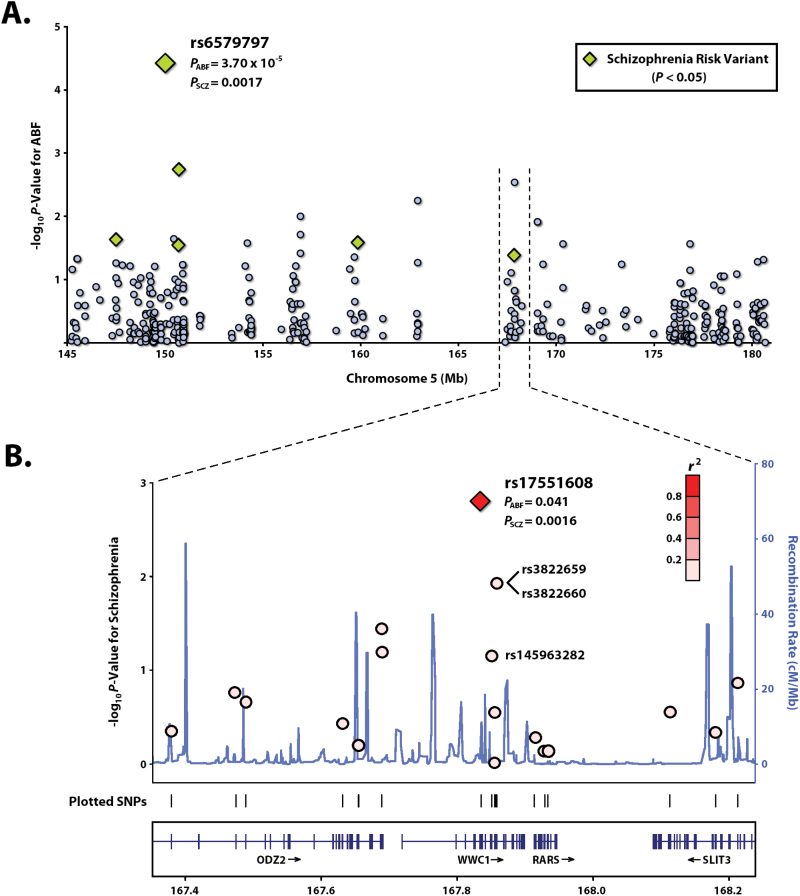
In total, 407 nonsynonymous variants from the QTL at 5q32–35.3 were tested for association with ABF (λ = 1.09; supplementary table S3). After Bonferroni adjustment for multiple testing, 1 SNP, rs6579797 (MAF = 0.032) from the gene SYNPO, was significantly associated with ABF, with the minor allele showing poorer performance (βABF = −2.01±0.48; P = 3.70×10−5; corrected P = .015; q = 0.014). This SNP, along with 23 others that exhibit nominal association with ABF (ie, P < .05), were also tested for association with schizophrenia, of which 7 show evidence of risk (table 1 and figure 1A). Of these, 2 SNPs remained significant after Bonferroni correction: the top hit for ABF, rs6579797 (βSCZ = 1.84±0.63; P = .0017; corrected P = .040) and the SNP rs17551608 (MAF = 0.19), located in WWC1, with its minor allele associated with improved ABF performance (βABF = 0.42±0.21; P = .041) and decreased liability for schizophrenia (β = −1.09±0.39; P = .0016; corrected P = .038). When considered jointly in a bivariate model, the 2 traits are significantly associated with both rs6579797 (βABF = −1.95±0.47; βSCZ = 1.75±0.55; P = 1.11×10−4) and rs17551608 (βABF = 0.45±0.19; βSCZ = −1.00±0.22; P = .0048). These 2 SNPs are in linkage equilibrium (r 2 = .019).
Table 1.
Top Association Results for ABF and Schizophrenia at 5q32–35.3
| Function Prediction | ABF | Schizophrenia | |||||||
|---|---|---|---|---|---|---|---|---|---|
| dbSNP 137 | Position (bp) | Gene | MAF | SIFT | PolyPhen2 | Beta (SE) | P Value | Beta (SE) | P Value |
| rs6579797a | 149998128 | SYNPO | 0.032 | Tolerated | Benign | −2.01 (0.48) | 3.7×10−5c | 1.84 (0.63) | .0017c |
| rs17660042 | 150666946 | SLC36A3 | 0.073 | Deleterious | Probably damaging | −1.02 (0.32) | .0018 | 0.94 (1.00) | .010 |
| rs2303063 | 147480027 | SPINK5 | 0.47 | Tolerated | Benign | 0.41 (0.18) | .023 | −0.52 (1.03) | .033 |
| rs2303067 | 147480955 | SPINK5 | 0.47 | Tolerated | Benign | 0.41 (0.18) | .023 | −0.52 (1.03) | .033 |
| rs2961944 | 159835658 | SLU7 | 0.20 | Tolerated | Benign | −0.40 (0.18) | .027 | 0.49 (0.24) | .037 |
| rs61740602 | 150646888 | GM2A | 0.12 | Tolerated | Benign | −0.56 (0.26) | .028 | 0.79 (0.29) | .0068 |
| rs17551608b | 167835539 | WWC1 | 0.19 | Deleterious | Possibly damaging | 0.42 (0.21) | .041 | −1.09 (0.39) | .0016c |
Note: The table lists nonsynonymous variants from 5q32–35.3 with at least nominal evidence (P < .05) for association with ABF and schizophrenia. SNP rs numbers are based on dbSNP build 137. MAFs are based on maximum likelihood estimates that account for familial relationships. Predicted effects of amino acid changes on protein function are based on the SIFT and PolyPhen2 algorithms, which were obtained with the Ensembl online tool Variant Effect Predictor (VEP). For the association results, positive beta estimates (SE in parentheses) for schizophrenia correspond to increased risk. All 7 of the SNP variants presented here show the expected directions of effect for the 2 traits (ie, decrease in ABF performance corresponds with an increase in schizophrenia risk, and vice versa). ABF, Abstraction and Mental Flexibility; MAFs, Minor Allele Frequencies;
aG199A; aspartic acid substituted for asparagine, D67N.
bC798T; arginine substituted for cysteine, R250C.
cSignificant after Bonferroni correction for multiple testing (α = .05): 407 tests for ABF; 24 tests for risk of schizophrenia.
Fig. 1.
(A) Plot of association P values for missense SNPs from chromosome 5q32–35.3 (n = 407) for ABF. Variants that are also nominally associated with schizophrenia risk (P < .05) are represented as diamonds. (B) Regional plot of association results for dbSNP 137 variants from WWC1 and neighboring genes for schizophrenia. Recombination rate based on hg19 assembly for 1000 Genomes data (2012) for European populations. Plotted using LocusZoom.149
Based on SIFT64 and PolyPhen265 algorithms, which predict the effects of amino acid substitutions on protein function, 2 SNPs from table 1 are considered potentially deleterious: rs17551608, and rs17660042 from the gene SLC36A3 (P ABF = .0018; P SCZ = .010). Interestingly, in addition to rs17551608, 7 other missense variants were identified in WWC1 (figure 1B), representing a high concentration of protein-altering variation (top 10th percentile for genes in the region, accounting size). Of these 7 variants, 4 are observed among the 25 schizophrenia cases, 3 of which are predicted to impact protein function. The most noteworthy of these are: rs145963282 (MAF = 0.0083), which is nominally associated with ABF (βABF = −2.30±0.72; P = .0029), and nearly so with schizophrenia risk (βSCZ = 1.44±0.82; P = .070; bivariate association P = .026) and rs3822659 (MAF = 0.076), which shows increased risk of schizophrenia (βSCZ = 0.99±0.41; P = .012). Interestingly, genetic interactions between rs17551608 and these other WWC1 missense variants were detected, notably rs3822659 for both ABF (P = .054) and schizophrenia (P = .057), as well as rs61730019 (P ABF = .039; P SCZ = .0042; supplementary table S4), with no significant ablation of the main effects of rs17551608.
To assess the independence of these WWC1 variants, haplotypes were phased using MERLIN v. 1.1.2.66 With the exception of rs3822659, which is in perfect linkage disequilibrium (LD) with an adjacent SNP, rs3822660, each of the missense variants were phased to separate haplotypes. Collectively, the haplotypes account for 10.2% of the variation in ABF (P = 5.95×10−5) and 20.3% of the risk for schizophrenia (Kullback-Leibler R 2 value; P = 4.40×10−7) in our families. Based on multimarker SKAT analyses of ABF, WWC1 yielded the fifth strongest association among genes tested from the QTL region (P = .11; supplementary table S5), with the lone, nominally significant result belonging to the gene SLC36A3 (P = .029).
Replication of Neurocognitive Effects in GOBSF Families
The 2 SNPs showing significant effects on ABF and/or schizophrenia risk in our MGI families, rs6579797 (SYNPO) and rs17551608 (WWC1), were tested against neurocognitive measurements obtained in Mexican American pedigrees from GOBSF. Although PCET efficiency, representing a z score of performance accuracy and speed, was not assessed in these independent samples, accuracy scores were available (n = 519). However, we found no evidence of association with either rs6579797 (β = −0.041±0.18; P = .82) or rs17551608 (β = −0.14±0.12; P = .24).
Given the broad cognitive impairment typically observed in schizophrenia, which extends beyond executive functions, such as ABF, we examined a pair of IQ measures for general intelligence (table 2), revealing significant, detrimental effects of rs6579797 (WAIS-II: β = −6.28±2.79, P = .025; WTAR: β = −7.54±2.96, P = .011). Also, with previous research strongly implicating WWC1 in verbal memory performance (see “Discussion” section), we examined CVLT scores, with the WWC1 variant rs61730019 showing significant association with both delayed recall (P = .0031) and total recall (P = .0091).
Table 2.
Association Results for rs6579797 and WWC1 Variants for Select Cognitive Measures in Mexican American Families
| T2D-GENES Families (n = 959) | IQ | CVLT | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| WASI-II (n = 430) | WTAR VIQ (n = 264) | Total Recall (n = 520) | Delayed Recall (n = 518) | |||||||
| dbSNP 137 | Gene | MAF | Beta (SE) | P Value | Beta (SE) | P Value | Beta (SE) | P Value | Beta (SE) | P Value |
| rs6579797 | SYNPO | 0.022 | −6.28 (2.79) | .025 | −7.54 (2.96) | .011 | −1.55 (1.96) | .43 | −0.38 (0.56) | .50 |
| rs17551608 | WWC1 | 0.077 | −2.86 (1.78) | .11 | 0.74 (1.53) | .63 | −1.60 (1.26) | .20 | −0.52 (0.36) | .15 |
| rs61730019 | WWC1 | 0.015 | 1.58 (3.71) | .67 | 5.24 (3.71) | .16 | 7.63 (2.57) | .0031 | 1.93 (0.74) | .0091 |
Note: The table presents association results between SYNPO and WWC1 variants and measures of IQ and verbal memory in Mexican American families using whole genome sequences and imputed data from the T2D-GENES consortium. Two measures of IQ were tested for genetic association: WAIS II and WTAR verbal IQ. For verbal memory, total and delayed recalls for the CVLT were examined. MAFs are based on maximum likelihood estimates that account for familial relationships. Significant association P values (< .05) are highlighted in bold. CVLT, California Verbal Learning Test; IQ, Intelligent quoteint; MAFs, Minor allele frequencies; WAIS, Wechsler Adult Intelligence Scale; WTAR, Wechsler Test of Adult Reading.
PGC GWAS Results for Schizophrenia
To investigate the effects of SYNPO and WWC1 on schizophrenia, we interrogated GWAS results from the PGC. For stage I analyses (n = 21856), as reported by Ripke et al,56 rs6579797 (SYNPO) was neither directly genotyped nor imputed from HapMap 3 data, however a near significant risk effect was observed for a nearby tagging variant (r 2 = 1.0), intronic SNP rs9324647 (OR = 4.90±0.91; P = .080; MAF = 0.0046). More recently, the PGC sample collection has expanded to include large Swedish cohorts (total n = 32143), for which Ripke et al57 have reported updated association results (supplementary table S6). Based on 1000 Genomes phase 1 data, rs6579797 was successfully imputed but failed to show evidence of risk effects (OR = 1.04±0.070; P = .61; MAF = 0.02). For other nearby SYNPO variants (±10kb from rs6579797), a near significant association was identified for a rare intronic SNP, rs192542133 (OR = 0.79±0.08; P = .0025; corrected P = .059; MAF = 0.015). As for WWC1 variants, including rs17551608, no evidence was found.
Population Genetics and Signatures of Positive Selection
According to 1000 Genomes data, the putative risk allele of rs6579797 shows marked frequency differences between global populations (supplementary table S7). Among Europeans (EUR) and admixed Americans (AMR), the respective frequencies are 0.015 and 0.039, whereas higher frequencies are observed for African (AFR) and Asian (ASN) populations, around 0.25, yielding substantial pairwise F ST scores with the EUR groups (0.22 and 0.21, respectively). As for rs17551608, population differentiation is also evident, with its minor allele ranging in frequency from 0.16 in EUR populations to its complete absence in the ASN samples.
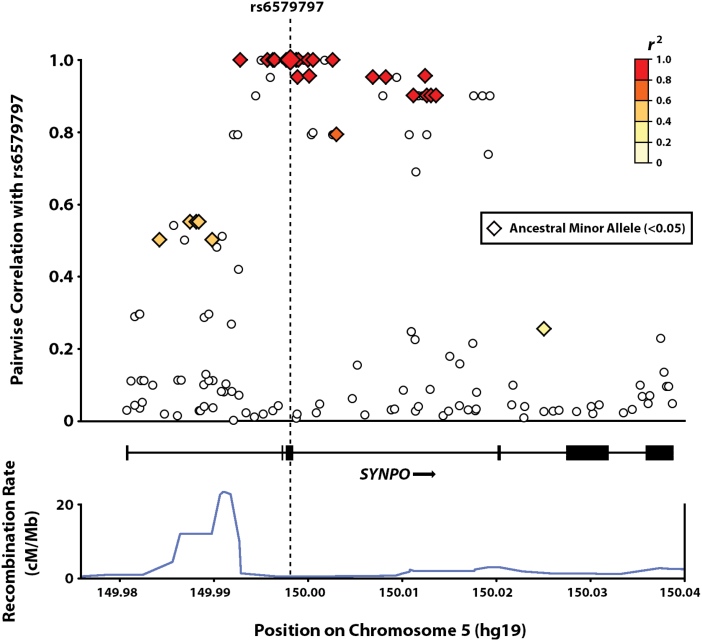
Interestingly, the minor risk allele (A) of rs6579797 appears to be the ancestral state, as determined from phylogenetic sequence alignments of nonhuman primate species, a possible signature of selection. To investigate this further, we computed the local LD structure of rs6579797 for the 5 EUR populations from 1000 Genomes (figure 2), observing a cluster of ancestral minor alleles in strong LD (r 2 > .9), indicative of an evolutionary sweep. Diversity and neutrality test statistics support this, as the EUR groups exhibit low nucleotide diversity within the LD block (π = 7.36), with highly significant deviations from the neutral evolutionary model, including Tajima’s D (P = .010) and Fu and Li’s F (P = .001) (table 3 and supplementary table S8). Similar negative deviations from neutrality are observed in admixed American populations, a likely reflection of their European origins.
Fig. 2.
Plot of pairwise LD correlations for rs6579797 with local SYNPO variants, based on population samples of European ancestry from 1000 Genomes data. Ancestral minor alleles (frequency < 0.05) are highlighted.
Table 3.
Diversity and Neutrality Test Scores Based on 1000 Genomes Data for LD Block of SYNPO Variant rs6579797
| 1000 Genomes Populationsa | N | S | π | Tajima’s D | Fu and Li’s F | Fu and Li’s D | Fay and Wu’s H | Zeng’s E |
|---|---|---|---|---|---|---|---|---|
| African | 246 | 211 | 34.36 | −0.04 | 0.07 | 0.18 | 0.73 | −0.66 |
| Admixed American | 181 | 156 | 10.91 | −1.89** | −2.83** | −2.83** | 0.43c | −2.06** |
| East Asian | 286 | 107 | 26.59 | 1.66 | 1.53 | 0.90 | −4.44 | 5.14b |
| European | 379 | 116 | 7.36 | −1.75** | −3.02** | −3.37** | 0.44c | −1.90** |
| Total | 1092 | 298 | 23.04 | −1.19 | −2.15** | −2.81** | 0.69 | −1.59* |
Note: Diversity measurements (segregating sites [S], nucleotide diversity [π]), and neutrality test statistics, as determined by differences in unbiased estimators of θ = 4 Neμ, were computed for variants found within an observed LD block for rs6579797 in the gene SYNPO (pairwise r2 > .90, carrying ancestral minor alleles), corresponding to hg19 coordinates chr5: 149 992 784–150 013 606 (delineated by SNPs rs10074935 and rs61051686).
a1000 Genomes phase 1 version 3 data for 4 “super populations”: African (AFR), representing Yoruba in Nigeria (YRI), Luhya in Kenya (LWK), and Americans of African ancestry from southwestern United States (ASW); Admixed American (AMR), representing Colombians from Medellin, Colombia (CLM), Puerto Ricans (PUR), and Americans of Mexican ancestry from Los Angeles (MXL); East Asian (ASN), representing Han Chinese from Beijing (CHB), Southern Han Chinese (CHS), and Japanese from Tokyo (JPT); and European (EUR), representing Utah residents with Northern and Western European ancestry (CEU), Toscani in Italy (TSI), British in England and Scotland (GBR), Iberian population in Spain (IBS), and Finnish in Finland (FIN).
bFor the other tail of the distribution (ie, positive), this score is significant, with an empirical P value of .034.
cPositive deviation Fay and Wu’s H, a statistic that utilizes information from the intermediate- and high-frequency parts of the frequency spectrum, coupled with the negative score for Zeng’s E based on low- and high-frequency variant classes, suggests that the locus may be entering a recovery phase for these 2 populations.
*Empirical P value (1-tailed) <0.05, ** <0.01, based on an observed distribution for 1000 samples generated via coalescent simulation.
Discussion
Synaptopodin: Neurocognitive Implications for Schizophrenia Risk
From our analysis of exome sequence data for chromosome 5q32–35.3, a region encompassing a QTL for neurocognition,12 we identified nonsynonymous variants from 2 genes, SYNPO and WWC1, which are significantly associated with ABF and/or schizophrenia risk. Of these, rs6579797 (SYNPO) is particularly compelling. The minor allele is carried by 6 heterozygotes in 3 MGI families, representing a modest enrichment relative to EUR populations. Four of the carriers are affected, corresponding to significantly decreased ABF performance (corrected P = .015) and heightened risk for schizophrenia (corrected P = .040). Based on the original linkage results for ABF, 2 of the 3 families harboring the rs6579797 risk allele have pedigree-specific LODs of 0.33 and 0.28. In the third family, a near zero LOD score was observed, with 2 heterozygote carriers, both unaffected for schizophrenia, although one diagnosed with severe major depression with symptoms of psychosis.
With genome-wide microsatellite data available for many of the MGI samples,12 we estimated identity-by-descent sharing for chromosome 5, allowing us to impute ML genotypes (.95 probability threshold) for rs6579797 for an additional 82 individuals in our 8 study families (see supplementary methods). Combining the exome sequence data and imputed genotypes, the association signals at rs6579797 remained significant for both ABF (corrected P = .0031) and schizophrenia (corrected P = .024; supplementary table S9).
When examined in Mexican American pedigrees from GOBSF, the SYNPO variant showed significant association with IQ measures (smallest P = .011), indicating more generalized cognitive effects. This is consistent with the overlapping signals observed at the QTL, ranging in LODs from 1.05 to 1.70, for various neurocognitive traits: verbal memory, spatial processing, language and reasoning, and attention. Interestingly, when tested against these other measurements in the MGI families, rs6579797 showed significant detrimental effect on verbal memory accuracy (β = −1.44±0.45; P = .031). As for schizophrenia risk, we found suggestive evidence for SYNPO variants from GWAS results reported by the PGC, although rs6579797 displayed no association in the most current data.
Remarkably, SYNPO appears to be under selective pressure, further hinting at its potential relevance. The putative risk allele of rs6579797 represents the ancestral evolutionary state, yet is uncommon in EUR and AMR populations from 1000 Genomes. Pairwise correlations with rs6579797 reveal an LD block enriched with minor ancestral alleles, a potential footprint of an evolutionary sweep. Neutrality test statistics for EUR and AMR support this, revealing significant deviations from the neutral model. Notably, negative deviation was not observed for Fay and Wu’s H, which, when coupled with the negative score for Zeng’s E, suggests that the locus may be entering a recovery phase (ie, accumulation of neutral genetic variation).67 This finding adds to a growing list of genes implicated in neurocognition and brain development that appear to have undergone selection over the course of human evolution.68
Although SIFT and PolyPhen2 yield low probabilities that rs6579797 is damaging, both algorithms have false negative rates >10%69 and thus do not necessarily preclude it from having important functional consequences. The product of SYNPO, synaptopodin, is an actin-binding protein found in the dendritic spines of telencephalic neurons,70 with the D67N substitution encoded by rs6579797 situated within a PEST motif, which may serve as a molecular signal for proteasomal degradation.71 Interestingly, when we re-examined SYNPO variants excluded from our analyses (eg, those without SIFT and PolyPhen2 scores), we discovered a splice site variant, rs59962087, that is 469bp upstream of rs6579797 and in perfect LD, with an ancestral minor allele, thus representing another functional candidate for the observed association signal.
What makes this result compelling for the pathology of schizophrenia is that within the dendritic spine, synaptopodin is believed to be an essential component of the spine apparatus (SA), influencing local calcium storage72 and protein synthesis,73 with synaptopodin-deficient mice exhibiting deficits in synaptic plasticity and spatial learning.74,75More specifically, synaptopodin directly regulates the release of calcium76 and the accumulation of glutamate receptor 1 (GluR1) in the spine head, a subunit of the α-amino-3-hydroxyl-5-methyl-4-isoxazolepropionic acid (AMPA) receptor induced during long-term potentiation (LTP). This establishes a potential mechanistic link with synaptic plasticity,77 a key neural process that underlies ABF and mediates synaptic dynamics in the prefrontal cortex (PFC),78 a brain region strongly implicated in schizophrenia.79–87
Moreover, in a recent exome sequencing study by Timms et al,88 protein-altering variants in genes involved in N-methyl-d-aspartate (NMDA) receptor hypofunction were found to segregate in schizophrenia families, supporting the glutamatergic dysfunction hypothesis for schizophrenia,89,90 which ties in well with our findings. NMDA and AMPA are the 2 primary types of receptors activated by glutamate in the mammalian brain, each playing a critical, interrelated role in calcium-induced potentiation. Antagonists of NMDA receptors can replicate schizophrenia symptomatology in healthy people, including deficits in mental flexibility,91,92 whereas enhancers have been found to reduce negative features and improve cognition in patients.93,94 Studies of knockout mice have revealed impairments in behavioral flexibility,95–97 with effects on potentiation.98,99 In postmortem brain tissue of schizophrenia patients, altered mRNA and protein levels of glutamate receptors have been observed,100 including irregularities in AMPA receptor trafficking and localization, particularly in the PFC.101–104 Association studies of schizophrenia have identified a number of SNPs and copy number variants (CNVs) in genes involved in glutamatergic neurotransmission,105–109 including the synaptic adhesion molecule neurexin,110,111 which has been found to regulate AMPA receptor endocytosis and control excitatory synaptic strength.112
WWC1: Another Regulator of AMPA Receptor Trafficking
The other gene implicated in our analysis, WWC1, shows an enrichment of protein-altering variation in our MGI families, including 3 independent SNPs with significant or suggestive associations with ABF and schizophrenia, maintained by the imputed data (supplementary table S9): rs17551608, rs145963282, and rs3822659. Collectively, the WWC1 variants account for significant portions of the variability in ABF and schizophrenia risk, with evidence of genetic interaction effects. Interestingly, when affection status was broadened to include other schizophrenia spectrum diagnoses in these pedigrees, namely schizotypal personality disorder (n = 7) and psychosis disorder (n = 6), a stronger association was observed for rs17551608 (P = 3.80×10−4; β = −1.07±0.08). However, these findings were not independently replicated in the GOBSF families for PCET accuracy and IQ, as well as among the GWAS results for schizophrenia from the PGC, perhaps a reflection of the genetic loads carried by our multiplex families.
Nonetheless, WWC1 remains intriguing. Its protein product, the WW domain containing protein 1 (WWC1), a postsynaptic scaffolding molecule expressed in the human brain, exhibits protein-protein interactions (PPIs) with other postsynaptic proteins, most notably dendrin and synaptopodin,70 via its WW domains.113,114 Other binding features include a C2-like motif that interacts with phospholipids115 and a region that binds protein kinase C (PKC)ζ,116 a molecule integral for neuronal plasticity117 and known to affect long-term memory.118 Based on the online database BioGRID (v. 3.2.102),119 other PPIs have been detected for synaptopodin and WWC1, including proteins of genes implicated in schizophrenia risk (supplementary table S10).120–127 However, no genetic interaction effects were detected between rs6579797 and WWC1 missense variants for ABF or schizophrenia risk, as well as with other variants from these PPI networks (supplementary table S11).
The influence of WWC1 on neurocognition, however, is well supported. In a seminal paper by Papassotiropoulos et al,128 an intronic SNP within WWC1, rs17070145, was reported to be associated with human memory, with allelic differences in hippocampal activations during memory tasks. This finding has been replicated in multiple studies involving both healthy subjects and patients with mild cognitive impairment,129–134 with differential effects on memory in psychotic individuals,135 as well as a predisposition for late-onset Alzheimer’s disease.136 Remarkably, rs17070145 has also been linked to cognitive flexibility, with tobacco use possibly modulating this effect,137 a notable interaction given its prevalence among schizophrenia patients,138 although we found no such effects. However, the chromosome 5 QTL does show suggestive linkage for verbal memory accuracy in MGI, as measured by the Penn Word Memory Test (LOD = 1.50, P = .0043), with a near significant association (β = −1.05±0.55, P = .059) detected for the WWC1 missense variant rs61730019 (MAF = 0.061), that also displays significant effects on CVLT total recall (P = .0031) and delayed recall (.0091) scores in GOBSF.
Despite the convincing case for its neurocognitive implications, how WWC1 controls higher brain function at the molecular level remains to be elucidated. Research has shown that it binds to PICK1,139 a synaptic protein involved in AMPA receptor trafficking140 and considered crucial for hippocampal synaptic plasticity,141 as WWC1 knockdown accelerates the rate of AMPA receptor recycling, with knockout mice exhibiting profound learning and memory impairment. It has been hypothesized that WWC1 may serve as a docking station for AMPA receptors,142 mediating linkage between endosomes containing phosphatidylinositol-3-phosphate, a key regulator of vesicular traffic in excitatory neurons,143 and components of the postsynaptic cytoskeleton that include dendrin and synaptopodin.
Conclusion
From our analysis of exome sequence data from chromosome 5q32–35.3, a region linked to neurocognition in families impacted by schizophrenia, we identify missense variants in 2 genes involved in AMPA receptor trafficking and neuronal plasticity, SYNPO and WWC1, that are associated with ABF performance and schizophrenia susceptibility. When examined in Mexican American pedigrees, the SYNPO variant, rs6579797, shows deleterious effects on general intelligence, with evidence of selection operating at this locus. Thus, these findings suggest that disruptions in AMPA receptor turnover in the postsynaptic cell have important pathological consequences on neurocognition, lending support to the glutamatergic dysfunction hypothesis. Recognizing that functional improvement in schizophrenia patients is likely to require treatment of cognitive capabilities,144,145 as global impairment represents a core feature,146 augmenting synaptic transmission and plasticity may have therapeutic potential, with a diverse class of allosteric agents available for modulating AMPA receptor activity.147 Of course, schizophrenia is a highly complex disorder involving perhaps thousands of risk alleles4 from genes involved in other neurotransmitter systems, with increasing evidence for the central importance of calcium channel signaling,148 thus necessitating an integrated, neurobiological approach to developing effective treatment strategies for this devastating mental illness.
Supplementary Material
Supplementary material (references 150–152 are cited in the supplementary material) is available at http://schizophreniabulletin.oxfordjournals.org.
Funding
This work was supported by the National Institutes of Health (MH061622, MH042191, and MH063480 for the MGI study; MH0708143, MH078111, and MH083824 for the GOBSF study; MH059490 for the development of SOLAR; and C06 RR017515) and Texas Biomedical Research Institute Forum Grant (11–4311 to M.Z.K).
Supplementary Material
Acknowledgments
We thank the participants of the MGI and GOBSF studies, as well as our research staffs. We would also like to recognize the invaluable comments and suggestions we received by the anonymous reviewers of this article. The authors have declared that there are no conflicts of interest in relation to the subject of this study.
References
- 1. American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4th Ed. Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- 2. Visscher PM, Goddard ME, Derks EM, Wray NR. Evidence-based psychiatric genetics, AKA the false dichotomy between common and rare variant hypotheses. Mol Psychiatry. 2012;17:474–485. [DOI] [PubMed] [Google Scholar]
- 3. Lee SH, DeCandia TR, Ripke S, et al. ; Schizophrenia Psychiatric Genome-Wide Association Study Consortium (PGC-SCZ); International Schizophrenia Consortium (ISC); Molecular Genetics of Schizophrenia Collaboration (MGS). Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44:247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Purcell SM, Wray NR, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lett TA, Chakravarty MM, Felsky D, et al. The genome-wide supported microRNA-137 variant predicts phenotypic heterogeneity within schizophrenia. Mol Psychiatry. 2013;18:443–450. [DOI] [PubMed] [Google Scholar]
- 6. Saykin AJ, Gur RC, Gur RE, et al. Neuropsychological function in schizophrenia. Selective impairment in memory and learning. Arch Gen Psychiatry. 1991;48:618–624. [DOI] [PubMed] [Google Scholar]
- 7. Elvevåg B, Goldberg TE. Cognitive impairment in schizophrenia is the core of the disorder. Crit Rev Neurobiol. 2000;14:1–21. [PubMed] [Google Scholar]
- 8. Egan MF, Goldberg TE, Gscheidle T, et al. Relative risk for cognitive impairments in siblings of patients with schizophrenia. Biol Psychiatry. 2001;50:98–107. [DOI] [PubMed] [Google Scholar]
- 9. Conklin HM, Curtis CE, Calkins ME, Iacono WG. Working memory functioning in schizophrenia patients and their first-degree relatives: cognitive functioning shedding light on etiology. Neuropsychologia. 2005;43:930–942. [DOI] [PubMed] [Google Scholar]
- 10. Thompson JL, Watson JR, Steinhauer SR, Goldstein G, Pogue-Geile MF. Indicators of genetic liability to schizophrenia: a sibling study of neuropsychological performance. Schizophr Bull. 2005;31:85–96. [DOI] [PubMed] [Google Scholar]
- 11. Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. [DOI] [PubMed] [Google Scholar]
- 12. Almasy L, Gur RC, Haack K, et al. A genome screen for quantitative trait loci influencing schizophrenia and neurocognitive phenotypes. Am J Psychiatry. 2008;165:1185–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Silverman JM, Greenberg DA, Altstiel LD, et al. Evidence of a locus for schizophrenia and related disorders on the short arm of chromosome 5 in a large pedigree. Am J Med Genet. 1996;67:162–171. [DOI] [PubMed] [Google Scholar]
- 14. Straub RE, MacLean CJ, O’Neill FA, Walsh D, Kendler KS. Support for a possible schizophrenia vulnerability locus in region 5q22-31 in Irish families. Mol Psychiatry. 1997;2:148–155. [DOI] [PubMed] [Google Scholar]
- 15. Shaw SH, Kelly M, Smith AB, et al. A genome-wide search for schizophrenia susceptibility genes. Am J Med Genet. 1998;81:364–376. [DOI] [PubMed] [Google Scholar]
- 16. Gurling HM, Kalsi G, Brynjolfson J, et al. Genomewide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q32.2, 5q33.2, and 8p21-22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3-24 and 20q12.1-11.23. Am J Hum Genet. 2001;68:661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DeLisi LE, Mesen A, Rodriguez C, et al. Genome-wide scan for linkage to schizophrenia in a Spanish-origin cohort from Costa Rica. Am J Med Genet. 2002;114:497–508. [DOI] [PubMed] [Google Scholar]
- 18. Devlin B, Bacanu SA, Roeder K, et al. Genome-wide multipoint linkage analyses of multiplex schizophrenia pedigrees from the oceanic nation of Palau. Mol Psychiatry. 2002;7:689–694. [DOI] [PubMed] [Google Scholar]
- 19. Sklar P, Pato MT, Kirby A, et al. Genome-wide scan in Portuguese Island families identifies 5q31-5q35 as a susceptibility locus for schizophrenia and psychosis. Mol Psychiatry. 2004;9:213–218. [DOI] [PubMed] [Google Scholar]
- 20. Ng MY, Levinson DF, Faraone SV, et al. Meta-analysis of 32 genome-wide linkage studies of schizophrenia. Mol Psychiatry. 2009;14:774–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krawczyk DC. Contributions of the prefrontal cortex to the neural basis of human decision making. Neurosci Biobehav Rev. 2002;26:631–664. [DOI] [PubMed] [Google Scholar]
- 22. Liang P, Wang Z, Yang Y, Jia X, Li K. Functional disconnection and compensation in mild cognitive impairment: evidence from DLPFC connectivity using resting-state fMRI. PLoS One. 2011;6:e22153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weinberger DR, Berman KF, Zec RF. Physiologic dysfunction of dorsolateral prefrontal cortex in schizophrenia. I. Regional cerebral blood flow evidence. Arch Gen Psychiatry. 1986;43:114–124. [DOI] [PubMed] [Google Scholar]
- 24. Minzenberg MJ, Laird AR, Thelen S, Carter CS, Glahn DC. Meta-analysis of 41 functional neuroimaging studies of executive function in schizophrenia. Arch Gen Psychiatry. 2009;66:811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shashi V, Kwapil TR, Kaczorowski J, et al. Evidence of gray matter reduction and dysfunction in chromosome 22q11.2 deletion syndrome. Psychiatry Res. 2010;181:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Veelen NM, Vink M, Ramsey NF, Kahn RS. Left dorsolateral prefrontal cortex dysfunction in medication-naive schizophrenia. Schizophr Res. 2010;123:22–29. [DOI] [PubMed] [Google Scholar]
- 27. Ursu S, Kring AM, Gard MG, et al. Prefrontal cortical deficits and impaired cognition-emotion interactions in schizophrenia. Am J Psychiatry. 2011;168:276–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weinshilboum RM, Otterness DM, Szumlanski CL. Methylation pharmacogenetics: catechol O-methyltransferase, thiopurine methyltransferase, and histamine N-methyltransferase. Annu Rev Pharmacol Toxicol. 1999;39:19–52. [DOI] [PubMed] [Google Scholar]
- 29. Chen J, Lipska BK, Halim N, et al. Functional analysis of genetic variation in catechol-O-methyltransferase (COMT): effects on mRNA, protein, and enzyme activity in postmortem human brain. Am J Hum Genet. 2004;75:807–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goldberg TE, Egan MF, Gscheidle T, et al. Executive subprocesses in working memory: relationship to catechol-O-methyltransferase Val158Met genotype and schizophrenia. Arch Gen Psychiatry. 2003;60:889–896. [DOI] [PubMed] [Google Scholar]
- 31. Blasi G, Mattay VS, Bertolino A, et al. Effect of catechol-O-methyltransferase val158met genotype on attentional control. J Neurosci. 2005;25:5038–5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bruder GE, Keilp JG, Xu H, et al. Catechol-O-methyltransferase (COMT) genotypes and working memory: associations with differing cognitive operations. Biol Psychiatry. 2005;58:901–907. [DOI] [PubMed] [Google Scholar]
- 33. Aguilera M, Barrantes-Vidal N, Arias B, et al. Putative role of the COMT gene polymorphism (Val158Met) on verbal working memory functioning in a healthy population. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:898–902. [DOI] [PubMed] [Google Scholar]
- 34. Burdick KE, Goldberg TE, Funke B, et al. DTNBP1 genotype influences cognitive decline in schizophrenia. Schizophr Res. 2007;89:169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baek JH, Kim JS, Ryu S, et al. Association of genetic variations in DTNBP1 with cognitive function in schizophrenia patients and healthy subjects. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:841–849. [DOI] [PubMed] [Google Scholar]
- 36. Burdick KE, Hodgkinson CA, Szeszko PR, et al. DISC1 and neurocognitive function in schizophrenia. Neuroreport. 2005;16:1399–1402. [DOI] [PubMed] [Google Scholar]
- 37. Carless MA, Glahn DC, Johnson MP, et al. Impact of DISC1 variation on neuroanatomical and neurocognitive phenotypes. Mol Psychiatry. 2011;16:1096–1104, 1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Prasad KM, Almasy L, Gur RC, et al. RGS4 polymorphisms associated with variability of cognitive performance in a family-based schizophrenia sample. Schizophr Bull. 2010;36:983–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gur RE, Nimgaonkar VL, Almasy L, et al. Neurocognitive endophenotypes in a multiplex multigenerational family study of schizophrenia. Am J Psychiatry. 2007;164:813–819. [DOI] [PubMed] [Google Scholar]
- 40. Nurnberger JI, Blehar MC, Kaufmann CA, et al. Diagnostic interview for genetic studies: rationale, unique features, and training. NIMH Genetic Initiative. Arch Gen Psychiatry. 1994;51:849–859. [DOI] [PubMed] [Google Scholar]
- 41. Maxwell ME. Manual for the Family Interview for Genetic Studies (FIGS). Bethesda, MD: Clinical Neurogenetics Branch, Intramural Research Program, National Institute of Mental Health; 1992. [Google Scholar]
- 42. Saykin AJ, Shtasel DL, Gur RE, et al. Neuropsychological deficits in neuroleptic naive patients with first-episode schizophrenia. Arch Gen Psychiatry. 1994;51:124–131. [DOI] [PubMed] [Google Scholar]
- 43. Censits DM, Ragland JD, Gur RC, Gur RE. Neuropsychological evidence supporting a neurodevelopmental model of schizophrenia: a longitudinal study. Schizophr Res. 1997;24:289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Goldberg TE, Goldman RS, Burdick KE, et al. Cognitive improvement after treatment with second-generation antipsychotic medications in first-episode schizophrenia: is it a practice effect? Arch Gen Psychiatry. 2007;64:1115–1122. [DOI] [PubMed] [Google Scholar]
- 45. Gur RC, Ragland JD, Moberg PJ, et al. Computerized neurocognitive scanning: II. The profile of schizophrenia. Neuropsychopharmacology. 2001;25:777–788. [DOI] [PubMed] [Google Scholar]
- 46. Gur RC, Ragland JD, Moberg PJ, et al. Computerized neurocognitive scanning: I. Methodology and validation in healthy people. Neuropsychopharmacology. 2001;25:766–776. [DOI] [PubMed] [Google Scholar]
- 47. Kurtz MM, Ragland JD, Moberg PJ, Gur RC. The Penn Conditional Exclusion Test: a new measure of executive-function with alternate forms of repeat administration. Arch Clin Neuropsychol. 2004;19:191–201. [DOI] [PubMed] [Google Scholar]
- 48. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li H, Handsaker B, Wysoker A, et al. ; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Olvera RL, Bearden CE, Velligan DI, et al. Common genetic influences on depression, alcohol, and substance use disorders in Mexican-American families. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Glahn DC, Kent JW, Jr, Sprooten E, et al. Genetic basis of neurocognitive decline and reduced white-matter integrity in normal human brain aging. Proc Natl Acad Sci USA. 2013;110:19006–19011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Flannick J, Thorleifsson G, Beer NL, et al. ; Go-T2D Consortium; T2D-GENES Consortium. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat Genet. 2014;46:357–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ripke S, Sanders AR, Kendler KS, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ripke S, O’Dushlaine C, Chambert K, et al. ; Multicenter Genetic Studies of Schizophrenia Consortium; Psychosis Endophenotypes International Consortium; Wellcome Trust Case Control Consortium 2. genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Glahn DC, Curran JE, Winkler AM, et al. High dimensional endophenotype ranking in the search for major depression risk genes. Biol Psychiatry. 2012;71:6–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Boerwinkle E, Chakraborty R, Sing CF. The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann Hum Genet. 1986;50:181–194. [DOI] [PubMed] [Google Scholar]
- 60. Strimmer K. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008;24:1461–1462. [DOI] [PubMed] [Google Scholar]
- 61. Chen H, Meigs JB, Dupuis J. Sequence kernel association test for quantitative traits in family samples. Genet Epidemiol. 2013;37:196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pfeifer B, Wittelsbürger U, Ramos-Onsins SE, Lercher MJ. PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol Biol Evol. 2014;31:1929–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. [DOI] [PubMed] [Google Scholar]
- 64. Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. [DOI] [PubMed] [Google Scholar]
- 65. Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. [DOI] [PubMed] [Google Scholar]
- 67. Zeng K, Fu YX, Shi S, Wu CI. Statistical tests for detecting positive selection by utilizing high-frequency variants. Genetics. 2006;174:1431–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gittelman RM, Hun E, Ay F, et al. Comprehensive identification and analysis of human accelerated regulatory DNA. Genome Res. 2015;25:1245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Frousios K, Iliopoulos CS, Schlitt T, Simpson MA. Predicting the functional consequences of non-synonymous DNA sequence variants–evaluation of bioinformatics tools and development of a consensus strategy. Genomics. 2013;102:223–228. [DOI] [PubMed] [Google Scholar]
- 70. Mundel P, Heid HW, Mundel TM, Krüger M, Reiser J, Kriz W. Synaptopodin: an actin-associated protein in telencephalic dendrites and renal podocytes. J Cell Biol. 1997;139:193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Spencer ML, Theodosiou M, Noonan DJ. NPDC-1, a novel regulator of neuronal proliferation, is degraded by the ubiquitin/proteasome system through a PEST degradation motif. J Biol Chem. 2004;279:37069–37078. [DOI] [PubMed] [Google Scholar]
- 72. Svoboda K, Mainen ZF. Synaptic [Ca2+]: intracellular stores spill their guts. Neuron. 1999;22:427–430. [DOI] [PubMed] [Google Scholar]
- 73. Pierce JP, van Leyen K, McCarthy JB. Translocation machinery for synthesis of integral membrane and secretory proteins in dendritic spines. Nat Neurosci. 2000;3:311–313. [DOI] [PubMed] [Google Scholar]
- 74. Deller T, Korte M, Chabanis S, et al. Synaptopodin-deficient mice lack a spine apparatus and show deficits in synaptic plasticity. Proc Natl Acad Sci USA. 2003;100:10494–10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jedlicka P, Schwarzacher SW, Winkels R, et al. Impairment of in vivo theta-burst long-term potentiation and network excitability in the dentate gyrus of synaptopodin-deficient mice lacking the spine apparatus and the cisternal organelle. Hippocampus. 2009;19:130–140. [DOI] [PubMed] [Google Scholar]
- 76. Korkotian E, Segal M. Synaptopodin regulates release of calcium from stores in dendritic spines of cultured hippocampal neurons. J Physiol. 2011;589:5987–5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Vlachos A, Korkotian E, Schonfeld E, Copanaki E, Deller T, Segal M. Synaptopodin regulates plasticity of dendritic spines in hippocampal neurons. J Neurosci. 2009;29:1017–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Stokes MG, Kusunoki M, Sigala N, Nili H, Gaffan D, Duncan J. Dynamic coding for cognitive control in prefrontal cortex. Neuron. 2013;78:364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Goldman-Rakic PS. Working memory dysfunction in schizophrenia. J Neuropsychiatry Clin Neurosci. 1994;6:348–357. [DOI] [PubMed] [Google Scholar]
- 80. Eisenberg DP, Berman KF. Executive function, neural circuitry, and genetic mechanisms in schizophrenia. Neuropsychopharmacology. 2010;35:258–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry. 2000;57:65–73. [DOI] [PubMed] [Google Scholar]
- 82. Salisbury DF, Kuroki N, Kasai K, Shenton ME, McCarley RW. Progressive and interrelated functional and structural evidence of post-onset brain reduction in schizophrenia. Arch Gen Psychiatry. 2007;64:521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fornito A, Yücel M, Patti J, Wood SJ, Pantelis C. Mapping grey matter reductions in schizophrenia: an anatomical likelihood estimation analysis of voxel-based morphometry studies. Schizophr Res. 2009;108:104–113. [DOI] [PubMed] [Google Scholar]
- 84. Zhou Y, Liang M, Jiang T, et al. Functional dysconnectivity of the dorsolateral prefrontal cortex in first-episode schizophrenia using resting-state fMRI. Neurosci Lett. 2007;417:297–302. [DOI] [PubMed] [Google Scholar]
- 85. Camchong J, MacDonald AW, 3rd, Bell C, Mueller BA, Lim KO. Altered functional and anatomical connectivity in schizophrenia. Schizophr Bull. 2011;37:640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Cole MW, Anticevic A, Repovs G, Barch D. Variable global dysconnectivity and individual differences in schizophrenia. Biol Psychiatry. 2011;70:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sehatpour P, Dias EC, Butler PD, et al. Impaired visual object processing across an occipital-frontal-hippocampal brain network in schizophrenia: an integrated neuroimaging study. Arch Gen Psychiatry. 2010;67:772–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Timms AE, Dorschner MO, Wechsler J, et al. Support for the N-methyl-D-aspartate receptor hypofunction hypothesis of schizophrenia from exome sequencing in multiplex families. JAMA Psychiatry. 2013;70:582–590. [DOI] [PubMed] [Google Scholar]
- 89. Konradi C, Heckers S. Molecular aspects of glutamate dysregulation: implications for schizophrenia and its treatment. Pharmacol Ther. 2003;97:153–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Coyle JT. NMDA receptor and schizophrenia: a brief history. Schizophr Bull. 2012;38:920–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Javitt DC, Zukin SR. Recent advances in the phencyclidine model of schizophrenia. Am J Psychiatry. 1991;148:1301–1308. [DOI] [PubMed] [Google Scholar]
- 92. Krystal JH, Karper LP, Seibyl JP, et al. Subanesthetic effects of the noncompetitive NMDA antagonist, ketamine, in humans. Psychotomimetic, perceptual, cognitive, and neuroendocrine responses. Arch Gen Psychiatry. 1994;51:199–214. [DOI] [PubMed] [Google Scholar]
- 93. Coyle JT, Balu D, Benneyworth M, Basu A, Roseman A. Beyond the dopamine receptor: novel therapeutic targets for treating schizophrenia. Dialogues Clin Neurosci. 2010;12:359–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tsai GE, Lin PY. Strategies to enhance N-methyl-D-aspartate receptor-mediated neurotransmission in schizophrenia, a critical review and meta-analysis. Curr Pharm Des. 2010;16:522–537. [DOI] [PubMed] [Google Scholar]
- 95. Kirchner L, Weitzdoerfer R, Hoeger H, et al. Impaired cognitive performance in neuronal nitric oxide synthase knockout mice is associated with hippocampal protein derangements. Nitric Oxide. 2004;11:316–330. [DOI] [PubMed] [Google Scholar]
- 96. Brigman JL, Feyder M, Saksida LM, Bussey TJ, Mishina M, Holmes A. Impaired discrimination learning in mice lacking the NMDA receptor NR2A subunit. Learn Mem. 2008;15:50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kim JI, Lee HR, Sim SE, et al. PI3Kγ is required for NMDA receptor-dependent long-term depression and behavioral flexibility. Nat Neurosci. 2011;14:1447–1454. [DOI] [PubMed] [Google Scholar]
- 98. Brigman JL, Wright T, Talani G, et al. Loss of GluN2B-containing NMDA receptors in CA1 hippocampus and cortex impairs long-term depression, reduces dendritic spine density, and disrupts learning. J Neurosci. 2010;30:4590–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Erickson MA, Maramara LA, Lisman J. A single brief burst induces GluR1-dependent associative short-term potentiation: a potential mechanism for short-term memory. J Cogn Neurosci. 2010;22:2530–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Weickert CS, Fung SJ, Catts VS, et al. Molecular evidence of N-methyl-D-aspartate receptor hypofunction in schizophrenia. Mol Psychiatry. 2013;18:1185–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Toyooka K, Iritani S, Makifuchi T, et al. Selective reduction of a PDZ protein, SAP-97, in the prefrontal cortex of patients with chronic schizophrenia. J Neurochem. 2002;83:797–806. [DOI] [PubMed] [Google Scholar]
- 102. Dracheva S, McGurk SR, Haroutunian V. mRNA expression of AMPA receptors and AMPA receptor binding proteins in the cerebral cortex of elderly schizophrenics. J Neurosci Res. 2005;79:868–878. [DOI] [PubMed] [Google Scholar]
- 103. Beneyto M, Meador-Woodruff JH. Lamina-specific abnormalities of AMPA receptor trafficking and signaling molecule transcripts in the prefrontal cortex in schizophrenia. Synapse. 2006;60:585–598. [DOI] [PubMed] [Google Scholar]
- 104. Hammond JC, McCullumsmith RE, Funk AJ, Haroutunian V, Meador-Woodruff JH. Evidence for abnormal forward trafficking of AMPA receptors in frontal cortex of elderly patients with schizophrenia. Neuropsychopharmacology. 2010;35:2110–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Allen NC, Bagade S, McQueen MB, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40:827–834. [DOI] [PubMed] [Google Scholar]
- 106. Bass NJ, Datta SR, McQuillin A, et al. Evidence for the association of the DAOA (G72) gene with schizophrenia and bipolar disorder but not for the association of the DAO gene with schizophrenia. Behav Brain Funct. 2009;5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Nicodemus KK, Law AJ, Luna A, et al. A 5’ promoter region SNP in NRG1 is associated with schizophrenia risk and type III isoform expression. Mol Psychiatry. 2009;14:741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kirov G, Pocklington AJ, Holmans P, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Yokley JL, Prasad KM, Chowdari KV, et al. Genetic associations between neuregulin-1 SNPs and neurocognitive function in multigenerational, multiplex schizophrenia families. Psychiatr Genet. 2012;22:70–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Reichelt AC, Rodgers RJ, Clapcote SJ. The role of neurexins in schizophrenia and autistic spectrum disorder. Neuropharmacology. 2012;62:1519–1526. [DOI] [PubMed] [Google Scholar]
- 112. Aoto J, Martinelli DC, Malenka RC, Tabuchi K, Südhof TC. Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell. 2013;154:75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Duning K, Schurek EM, Schlüter M, et al. KIBRA modulates directional migration of podocytes. J Am Soc Nephrol. 2008;19:1891–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kremerskothen J, Kindler S, Finger I, Veltel S, Barnekow A. Postsynaptic recruitment of Dendrin depends on both dendritic mRNA transport and synaptic anchoring. J Neurochem. 2006;96:1659–1666. [DOI] [PubMed] [Google Scholar]
- 115. Johannsen S, Duning K, Pavenstädt H, Kremerskothen J, Boeckers TM. Temporal-spatial expression and novel biochemical properties of the memory-related protein KIBRA. Neuroscience. 2008;155:1165–1173. [DOI] [PubMed] [Google Scholar]
- 116. Kremerskothen J, Plaas C, Büther K, et al. Characterization of KIBRA, a novel WW domain-containing protein. Biochem Biophys Res Commun. 2003;300:862–867. [DOI] [PubMed] [Google Scholar]
- 117. Sacktor TC. PKMzeta, LTP maintenance, and the dynamic molecular biology of memory storage. Prog Brain Res. 2008;169:27–40. [DOI] [PubMed] [Google Scholar]
- 118. Serrano P, Yao Y, Sacktor TC. Persistent phosphorylation by protein kinase Mzeta maintains late-phase long-term potentiation. J Neurosci. 2005;25:1979–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Chatr-Aryamontri A, Breitkreutz BJ, Heinicke S, et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2013;41:D816–D823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Karlsson R, Graae L, Lekman M, et al. MAGI1 copy number variation in bipolar affective disorder and schizophrenia. Biol Psychiatry. 2012;71:922–930. [DOI] [PubMed] [Google Scholar]
- 121. Martins-de-Souza D, Gattaz WF, Schmitt A, et al. Prefrontal cortex shotgun proteome analysis reveals altered calcium homeostasis and immune system imbalance in schizophrenia. Eur Arch Psychiatry Clin Neurosci. 2009;259:151–163. [DOI] [PubMed] [Google Scholar]
- 122. Lee SA, Tsao TT, Yang KC, et al. Construction and analysis of the protein-protein interaction networks for schizophrenia, bipolar disorder, and major depression. BMC Bioinformatics. 2011;12 Suppl 13:S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Jia Y, Yu X, Zhang B, et al. An association study between polymorphisms in three genes of 14-3-3 (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein) family and paranoid schizophrenia in northern Chinese population. Eur Psychiatry. 2004;19:377–379. [DOI] [PubMed] [Google Scholar]
- 124. Lipska BK, Peters T, Hyde TM, et al. Expression of DISC1 binding partners is reduced in schizophrenia and associated with DISC1 SNPs. Hum Mol Genet. 2006;15:1245–1258. [DOI] [PubMed] [Google Scholar]
- 125. Kang E, Burdick KE, Kim JY, et al. Interaction between FEZ1 and DISC1 in regulation of neuronal development and risk for schizophrenia. Neuron. 2011;72:559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Potkin SG, Turner JA, Guffanti G, et al. ; FBIRN. A genome-wide association study of schizophrenia using brain activation as a quantitative phenotype. Schizophr Bull. 2009;35:96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Wang Q, Charych EI, Pulito VL, et al. The psychiatric disease risk factors DISC1 and TNIK interact to regulate synapse composition and function. Mol Psychiatry. 2011;16:1006–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Papassotiropoulos A, Stephan DA, Huentelman MJ, et al. Common Kibra alleles are associated with human memory performance. Science. 2006;314:475–478. [DOI] [PubMed] [Google Scholar]
- 129. Almeida OP, Schwab SG, Lautenschlager NT, et al. KIBRA genetic polymorphism influences episodic memory in later life, but does not increase the risk of mild cognitive impairment. J Cell Mol Med. 2008;12:1672–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Nacmias B, Bessi V, Bagnoli S, et al. KIBRA gene variants are associated with episodic memory performance in subjective memory complaints. Neurosci Lett. 2008;436:145–147. [DOI] [PubMed] [Google Scholar]
- 131. Schaper K, Kolsch H, Popp J, Wagner M, Jessen F. KIBRA gene variants are associated with episodic memory in healthy elderly. Neurobiol Aging. 2008;29:1123–1125. [DOI] [PubMed] [Google Scholar]
- 132. Bates TC, Price JF, Harris SE, et al. Association of KIBRA and memory. Neurosci Lett. 2009;458:140–143. [DOI] [PubMed] [Google Scholar]
- 133. Preuschhof C, Heekeren HR, Li SC, Sander T, Lindenberger U, Bäckman L. KIBRA and CLSTN2 polymorphisms exert interactive effects on human episodic memory. Neuropsychologia. 2010;48:402–408. [DOI] [PubMed] [Google Scholar]
- 134. Milnik A, Heck A, Vogler C, Heinze HJ, de Quervain DJ, Papassotiropoulos A. Association of KIBRA with episodic and working memory: a meta-analysis. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:958–969. [DOI] [PubMed] [Google Scholar]
- 135. Vassos E, Bramon E, Picchioni M, et al. Evidence of association of KIBRA genotype with episodic memory in families of psychotic patients and controls. J Psychiatr Res. 2010;44:795–798. [DOI] [PubMed] [Google Scholar]
- 136. Corneveaux JJ, Liang WS, Reiman EM, et al. Evidence for an association between KIBRA and late-onset Alzheimer’s disease. Neurobiol Aging. 2010;31:901–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Zhang H, Kranzler HR, Poling J, Gruen JR, Gelernter J. Cognitive flexibility is associated with KIBRA variant and modulated by recent tobacco use. Neuropsychopharmacology. 2009;34:2508–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Winterer G. Why do patients with schizophrenia smoke? Curr Opin Psychiatry. 2010;23:112–119. [DOI] [PubMed] [Google Scholar]
- 139. Makuch L, Volk L, Anggono V, et al. Regulation of AMPA receptor function by the human memory-associated gene KIBRA. Neuron. 2011;71:1022–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Citri A, Bhattacharyya S, Ma C, et al. Calcium binding to PICK1 is essential for the intracellular retention of AMPA receptors underlying long-term depression. J Neurosci. 2010;30:16437–16452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Terashima A, Pelkey KA, Rah JC, et al. An essential role for PICK1 in NMDA receptor-dependent bidirectional synaptic plasticity. Neuron. 2008;57:872–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Duning K, Wennmann DO, Bokemeyer A, et al. Common exonic missense variants in the C2 domain of the human KIBRA protein modify lipid binding and cognitive performance. Transl Psychiatry. 2013;3:e272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Shepherd JD, Huganir RL. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu Rev Cell Dev Biol. 2007;23:613–643. [DOI] [PubMed] [Google Scholar]
- 144. Buchanan RW, Freedman R, Javitt DC, Abi-Dargham A, Lieberman JA. Recent advances in the development of novel pharmacological agents for the treatment of cognitive impairments in schizophrenia. Schizophr Bull. 2007;33:1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Barch DM, Carter CS, Arnsten A, et al. Selecting paradigms from cognitive neuroscience for translation into use in clinical trials: proceedings of the third CNTRICS meeting. Schizophr Bull. 2009;35:109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Schaefer J, Giangrande E, Weinberger DR, Dickinson D. The global cognitive impairment in schizophrenia: consistent over decades and around the world. Schizophr Res. 2013;150:42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Traynelis SF, Wollmuth LP, McBain CJ, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Purcell SM, Moran JL, Fromer M, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.