Abstract
Connexins and their channels are involved in the control of all aspects of the cellular life cycle, ranging from cell growth to cell death, by mediating extracellular, intercellular and intracellular communication. These multifaceted aspects of connexin-related cellular signaling obviously require strict regulation. While connexin channel activity is mainly directed by posttranslational modifications, connexin expression as such is managed by classical cis/trans mechanisms. Over the past few years, it has become clear that connexin production is equally dictated by epigenetic actions. This paper provides an overview of the role of major determinants of the epigenome, including DNA methylation, histone acetylation and microRNA species, in connexin expression.
Keywords: connexin, hemichannel, gap junction, DNA methylation, histone acetylation, microRNA
1. Introduction
Connexins have been first described about 50 years ago and currently this protein family fosters 21 members in human [1,2]. They are all named based on their molecular weight as predicted by cDNA sequencing and share a common architecture consisting of 4 transmembrane regions, 2 extracellular loops, 1 cytosolic loop, 1 cytosolic aminoterminal tail and 1 cytosolic carboxyterminal tail [3,4]. Throughout the years, connexins have been identified as major goalkeepers of tissue homeostasis by acting at 3 communication levels. First, at the intracellular level, connexins can physically interact with regulators of the cellular life cycle, such as cyclins or B-cell lymphoma 2 proteins, or can affect their expression. Second, connexins form hexameric structures called hemichannels that create pores at the membrane plasma, providing a pathway for extracellular communication. Hemichannels link the cytosol of individual cells and their extracellular environment, and convey small (i.e. less than 1 kilodalton) and hydrophilic substances, such as second messengers and ions. Third, 2 hemichannels of adjacent cells can dock and generate a gap junction that mediates direct intercellular trafficking of permeants similar to those involved in hemichannel signaling [5]. Inherent to their role as critical determinants of all aspects of tissue homeostasis, connexins and their channels are also frequently involved in disease. In fact, although controversial, it seems that hemichannels, unlike their full channel gap junction counterparts, preferentially open up in pathological circumstances, including cell death and inflammation [3,6].
The activity and thus the opening of connexin-based channels are regulated by a plethora of mechanisms. Short-term control, so-called gating, mainly relies on posttranslational modifications of connexin proteins. Connexins can undergo several of such modifications, including glycosylation, N-acetylation, ubiquitination, lipidation, hydroxylation, methylation, deamidation, S-nitrosylation, sumoylation and phosphorylation [2,7]. The latter, typically occurring at their carboxyterminal tail, has been best studied thus far and may have various effects depending on the identity of the connexin and kinase as well as the cellular context [2]. Long-term control of hemichannel and gap junction activity involves regulation of connexin expression. The architecture of most connexin genes is simple, consisting of a first exon that harbors the 5′-untranslated region (UTR), which is separated by an intron of varying length from a second exon, bearing the complete coding sequence and the 3′-UTR. Connexin gene promoters display binding affinity for several general transcription factors, including activator protein 1, yin yang 1 and specificity protein 1. Furthermore, a number of tissue-specific transcription factors control connexin gene transcription, like hepatocyte nuclear factor 1α in liver [8,9].
In the last decade, epigenetic mechanisms, including DNA methylation, histone acetylation and microRNA-related control, have also joined in as master regulators of connexin expression. These mechanisms will be discussed in detail in the following sections. Specific attention will be paid to microRNA-related control, which has become particularly studied in the connexin field in recent years.
2. DNA methylation
Hypermethylation of gene promoters, catalyzed by DNA methyltransferase (DNMT) enzymes, is typically linked to transcriptional silencing [10]. The negative correlation between DNA methylation and gene expression is mediated by methylated DNA-binding proteins that concentrate at hypermethylated CpG dinucleotides and that recruit transcriptional suppressors [11].
The role of DNA methylation in the control of connexin expression has been mainly studied in a pathological context. In a rat model of D-galactose-induced presbycusis, gradual downregulation of Cx26 expression was seen in cochlear tissue, which was associated with increased methylation of its gene promoter [12]. Along the same line, Cx32 and Cx43 mRNA levels progressively decrease in human gastric mucosa during Helicobacter pylori infection, an event that goes hand in hand with hypermethylation of their gene promoters [13]. It should be mentioned that the latter is probably not due to infection per se, but rather to tumor progression. Indeed, methylation-driven suppression of connexin expression has been most extensively demonstrated for carcinogenic events. Abrogation of expression of Cx26, Cx32, Cx36, Cx43 and Cx45 has been associated with the accumulation of methylated CpG dinucleotides in the corresponding connexin gene promoters in a variety of human malignant cells, including lung cancer cells [14,15], renal carcinoma cells [16-18], esophageal cancer cells [19], breast cancer cells [20], nasopharyngeal cancer cells [21], colon cancer cells [22,23] and glioma cells [24] as well as in other species [25-29]. Accordingly, hypomethylating agents that act by inhibiting DNMTs, such as the drugs 5-azacytidine and 5-aza-2-deoxycytidine (i.e. decitabine), have been found to upregulate connexin expression in many of those cancer cells (Table 1), which often results in enhanced gap junction activity [16,21], although this occurs in a cell type-dependent and connexin-specific fashion [16,19,20,25,30]. However, methylation of connexin gene promoters, including Cx30, Cx36 and Cx37, during cancer is not always accompanied by their downregulated expression [23]. In physiological conditions, such as during murine embryogenesis, decitabine was even found to suppress the production of Cx31, Cx43 and Cx45 in mouse embryos [31].
Table 1.
Effects of DNA methyltransferase inhibitors on connexin expression.
| Model | DNMTi | Upregulation | Downregulation | No effect | Reference |
|---|---|---|---|---|---|
| Human colon cancer cells | DAC | Cx43** | [23] | ||
| Human esophageal cancer cells | DAC | Cx26**/Cx43** | [19] | ||
| Human lung carcinoma cells | DAC | Cx26** | [14] | ||
| Human renal carcinoma cells | DAC | Cx32*,** | [16,109] | ||
| EG | Cx32* | [18] | |||
| Human proximal tubular cells | DAC | Cx32*,** | [16] | ||
| Human breast cancer cells | DAC | Cx26** | [20] | ||
| DAC | Cx26** | [33] | |||
| Human cervical carcinoma cells | DAC | Cx43* | [110] | ||
| Human nasopharyngeal cancer cells | DAC | Cx43*,** | [21] | ||
| Rat liver epithelial cells | DAC | Cx43** | Cx32** | [25] | |
| Mouse pancreatic cancer cells | AZA | Cx36** | [30] | ||
| Mouse pituitary corticotrophic cells | AZA | Cx36** | [30] | ||
| Mouse neuronal cells | AZA | Cx36** | [30] | ||
| Mouse fibroblasts | AZA | Cx36** | [30] | ||
| Mouse embryonic cells | DAC | Cx31**/Cx43**/Cx45** | [31] |
protein level;
mRNA level; AZA, 5-azacytidine; Cx, connexin; DAC, decitabine; DNMTi, DNA methyltransferase inhibitor; EG, epigallocatechin-3-gallate.
Although the mechanism of DNA methylation causing connexin gene silencing is clear, the true triggers of this epigenetic process remain more elusive. Decreased expression of connexins, in casu Cx26, in liver cancer has been casually linked to elevated DNMT1 mRNA levels [26]. Furthermore, aberrant binding of transcription factors to methylated connexin gene promoters could underlie poor connexin expression in cancer cells. In this respect, decreased Cx43 gene transcription in human non-small cell lung cancer cells is accompanied by DNA methylation and correlates with reduced binding of activator protein 1 to the its gene promoter [32]. Furthermore, methylated CpG dinucleotides are preferentially located in the specificity protein 1 cis-acting elements of the Cx26 gene promoter and the Cx32 gene promoter in human breast cancer cells [33] and rat liver cancer cells [25], respectively.
3. Histone modifications
The vast majority of data that show the involvement of histone modifications in connexin expression comes from studies using modifiers of these particular epigenetic mechanisms. This is specifically the case for reversible histone acetylation, which is catalyzed by histone acetyltransferases and that usually is paralleled by transcriptional activation through chromatin decondensation, while the opposite reaction, driven by histone deacetylase (HDAC) enzymes, frequently underlies suppression of gene expression [34].
Inhibitors of HDAC enzymes have been demonstrated to increase connexin production [35-47], often associated with induction of gap junction opening [35-41,44,48-50] in various experimental settings (Table 2). As seems to hold for DNMT inhibitors, the effects of HDAC inhibitors are dictated by the nature of the connexin species and epigenetic modifier as well as the cellular environment. In this respect, the prototypical class I and II HDAC inhibitor trichostatin A positively affects Cx36 expression in mouse pancreatic cell lines, but not in mouse fibroblasts, neuronal cells and pituitary cells [30]. On the other hand, sodium butyrate and 4-phenylbutyrate, but not trichostatin A, increase Cx43 protein levels in human nasopharyngeal tumor cells [51]. Also, suberoylanilide hydroxamic acid leaves Cx43 and Cx45 unaffected, but upregulates Cx32 and Cx37 expression and simultaneously reduces Cx40 protein amounts in cardiodystrophic mice [52]. HDAC inhibitors hereby directly affect connexin gene transcription. Indeed, trichostatin A-mediated induction of Cx43 in human prostate cancer cells relies on the recruitment of p300/cyclic adenosine monophosphate response element-binding protein, a transcriptional coactivator displaying histone acetyltransferase activity, and the transcription factors activator protein 1 and specificity protein 1 to the Cx43 gene promoter. This is associated with hyperacetylation of histone H4 surrounding binding sequences of both transcription factors [37]. Likewise, suberoylanilide hydroxamic acid triggers accumulation of acetylated histones H3 and H4 in the Cx43 gene locus, leading to its enhanced expression in human peritoneal mesothelial cells [41]. Cx36 expression in pancreatic cells is controlled, at least in part, by the RE-1 silencing transcription factor, a transcriptional repressor consisting of 2 independently acting HDAC-recruiting repression domains [53,54]. Active Cx36 production in these cells is featured by the presence of trimethylated lysine 4 residues in histone H4 near its gene promoter, an epigenetic marker of actively transcribed genes, and is inducible by trichostatin A [30].
Table 2.
Effects of histone deacetylase inhibitors on connexin expression.
| Model | HDACi | Upregulation | Downregulation | No effect | Reference |
|---|---|---|---|---|---|
| Human glioblastoma cells | NaB | Cx43* | [111] | ||
| 4-PB | Cx43* | [36] | |||
| Human pancreatic cancer cells | 4-PB | Cx43* | [42] | ||
| SR | Cx43* | [43] | |||
| Human prostate carcinoma cells | TSA | Cx43*,** | [37] | ||
| Human prostate epithelial cells | TSA | Cx43*,** | [37] | ||
| Human cervical carcinoma cells | 4-B | Cx43* | [44] | ||
| Human nasopharyngeal tumor cells | 4-PB/NaB | Cx43* | [51] | ||
| TSA | Cx43* | [51] | |||
| Human peritoneal mesothelial cells | HMBA | Cx43*,** | [39,40] | ||
| SAHA | Cx43*,** | [41] | |||
| Human lung artery endothelial cells | MC1568 | Cx37**/Cx40** | [45] | ||
| Human embryonic kidney cells | 4-PB | Cx43* | [44] | ||
| Human neural progenitor cells | 4-PB/TSA | Cx43* | [38] | ||
| Human liver cancer cells | TSA | Cx43** | Cx26**/Cx32** | [112] | |
| Rat glioma cells | NaB | Cx43* | [111] | ||
| 4-PB | Cx43* | [35] | |||
| Rat colon cancer cells | NaB | Cx43** | [113] | ||
| Rat transformed epithelial cells | SAHA | Cx43*,** | [41] | ||
| Rat hepatocytes | TSA | Cx32*/Cx43* | Cx26* | [49] | |
| J-1 | Cx32* | Cx26*/Cx43* | [50] | ||
| Rat corpora smooth muscle cells | 4-PB | Cx43** | Cx43* | [64] | |
| Mouse pancreatic cancer cells | TSA | Cx36** | [30] | ||
| Mouse pituitary corticotrophic cells | TSA | Cx36** | [30] | ||
| Mouse neuronal cells | TSA | Cx36** | [30] | ||
| Mouse fibroblasts | TSA | Cx36** | [30] | ||
| Mouse embryonic cells | TSA | Cx43** | [55] | ||
| Cardiodystrophic mdx mice | SAHA | Cx32*/Cx37* | Cx40* | Cx43*/Cx45* | [52] |
| HopX cardial hypetrophic mice | TSA | Cx40* | Cx43** | [47] |
protein level;
mRNA level; 4-PB, 4-phenylbutyrate; Cx, connexin; HDACi, histone deacetylase inhibitor; HMBA, hexamethylene bisacetamide; J-1, 5-(4-dimethylaminobenzoyl)aminovaleric acid hydroxamide; NaB, sodium butyrate; SAHA, suberoylanilide hydroxamic acid; SR, sulforaphane; TSA, trichostatin A.
A number of reports have documented the identity of the HDAC enzymes that are involved in the regulation of connexin expression. Specific deletion of HDAC1 and exposure to trichostatin A diminish Cx43 mRNA levels in mouse embryonic stem cells. In fact, loss of HDAC1 increases trimethylation of lysine 9 residues in histone H3 surrounding the Cx43 gene promoter region, an epigenetic signature of silenced genes, and only slightly reduces histone H3 and H4 acetylation. This indicates that the Cx43 gene requires both HDAC1 presence and activity for its transcription, but histones H3 and H4 are merely minor targets in this regulatory process [55]. In line with this finding, silencing of HDAC1 production substantially decreases expression of Cx43 in murine induced pluripotent stem cells [56]. By contrast, small interfering RNA-mediated knockdown of HDAC1 during differentiation of rat bone mesenchymal stem cells into cardiomyocytes increases Cx43 expression [57]. Upon transfection, the breast cancer metastasis suppressor 1 protein localizes in the cell nucleus, and restores gap junction activity in human breast cancer cells [58-60] and in melanoma cells [61]. In the former case, this coincides with elevated Cx43 mRNA levels and concomitant Cx32 gene transcription [58,60]. It has been further found that breast cancer metastasis suppressor 1 interacts with the large mammalian Sin3 HDAC complex, which contains both HDAC1 and HDAC2, but also forms smaller complexes with HDAC1 [62]. The mammalian Sin3 HDAC complex is also involved in the repression of Cx43 expression in human telomerase-immortalized myometrial cells by progesterone. The latter hereby binds to the Cx43 gene promoter through a protein-protein interaction with activator protein 1 [63]. MC1568, a selective class II HDAC inhibitor, promotes expression of Cx37 and Cx43 in lung artery endothelial cells of pulmonary arterial hypertension patients. This results from inhibition of HDAC4 and HDAC5, both which regulate the activity of the transcription factor myocyte enhancer factor 2, known to control expression of Cx37 and Cx40. This is further substantiated by the observation that experimental suppression of HDAC4 and HDAC5 upregulates production of both connexins in these cells [45].
It should be mentioned that HDAC inhibitors can act at levels of connexin expression other than the transcriptional one. Thus, trichostatin A enhances gap junction opening in cultured rat hepatocytes, a finding associated with differential effects on Cx26, Cx32 and Cx43 protein contents, but not with alterations in the corresponding mRNA amounts [49]. 4-phenylbutryate increases gap junction activity in rat corpora smooth muscle cell cultures, with no effect on Cx43 protein levels and even a decline in Cx43 mRNA transcript number. This could point to stabilization of the existing Cx43 pool or alterations in functional channel amounts [64]. In addition, HDAC inhibitors can interfere with posttranslational connexin control, as they both increase [36,39-41] and decrease [38] the abundance of phosphorylated Cx43 isoforms in different cell types. Sodium butyrate prevents tumor promoter-mediated inhibition of gap junction activity via extracellular signal-regulated kinase 1/2 inactivation, while trichostatin A restores gap junctional communication and induces Cx43 hyperphosphorylation by preventing p38 mitogen-activated protein kinase in cultured rat liver epithelial cells [65]. HDAC inhibitors may also affect subcellular localization of connexin proteins both in vitro [49,50] and in vivo [52]. Curiously, the interaction between histone acetylation and connexins can also occur in the opposite direction. In this regard, transfection of metastatic human pulmonary giant cells carcinoma cells with the gene encoding Cx43 increases acetylation of histones H3 and H4 in the promoter of the follistatin-like 1 gene, which in turns affects invasive and metastatic potential [66].
4. MiRNA-related control
In the last few years, microRNA (miRNA) species have emerged as critical posttranscriptional regulators of connexin expression. Following their synthesis in the cell nucleus and processing in the cytoplasm, miRNAs bind to complementary sequences in target mRNA molecules and either suppress their translation or cleave mRNAs as such [67].
A plethora of miRNAs have been reported to directly bind to the 3′-UTR region of Cx43 mRNA and thereby to suppress its translation (Table 3). This type of regulation has been studied both in a physiological and a pathological context. Regarding the former, miR-206 production is upregulated upon perinatal skeletal muscle development in mice in vivo and both miR-1 and miR-206 downregulate Cx43 expression during myoblast fusion in vitro [68,69]. Mice that overexpress miR-206 show decreased Cx43 expression and impaired bone formation [70]. Similarly, Cx43 levels increase during differentiation of bone cells, a process counteracted by miR-23a [71]. Of note, miRNAs may be involved in establishing gender-specific differences in connexin production. This has been shown for miR-1, which regulates Cx43, being expressed to a higher extent in female rat cardiomyocytes compared to male counterparts [72].
Table 3.
MicroRNA species experimentally shown to directly bind the 3′-UTR of Cx43 mRNA in different cell types.
| MiRNA species | Cell type | Reference |
|---|---|---|
| MiR-1 | Human breast cancer cells | [90] |
| Rat myocardial cells | [80,82] | |
| MiR-19a/b | Human embryonic kidney cells | [86] |
| MiR-20a | Human prostate cancer cells | [89] |
| MiR-23a | Human osteosarcoma cells | [71] |
| Human breast cancer cells | [90] | |
| MiR-125b | Human embryonic kidney cells | [87] |
| MiR-130a | Mouse embryonic fibroblasts | [85] |
| Mouse cardiomyocyte tumor cells | [85] | |
| MiR-186 | Human breast cancer cells | [90] |
| MiR-200a | Human breast cancer cells | [90] |
| MiR-206 | Human breast cancer cells | [90,91] |
| Rat vascular smooth muscle cells | [92] | |
| Mouse corneal cells | [92] | |
| Mouse osteoblasts | [70] | |
| Mouse myoblasts | [68,69] | |
| MiR-218 | Human nasopharyngeal carcinoma | [114] |
| Human breast cancer cells | [114] | |
| Human cervical cancer cells | [114] | |
| MiR-222 | Human glioblastoma cells | [88] |
| Rat vascular smooth muscle cells | [92] | |
| MiR-381 | Human breast cancer cells | [90] |
| MiR-1298 | Rat vascular smooth muscle cells | [98] |
miRNA, microRNA.
MicroRNAs can also act as positive regulators of connexin expression. In this light, miR-145 upregulates Cx43 production upon differentiation of human corneal epithelial progenitor cells [73]. Likewise, miR-208a seems to promote cardiac Cx40 expression [74]. Furthermore, microRNAs can indirectly affect connexin production. Thus, miR-103/107 directly targets the expression of receptor M type protein tyrosine phosphatase in limbal derived corneal epithelial cells, which in turns affects Cx43-based gap junctions [75]. Similarly, myocardin downregulates Cx43 expression via miR-1 upregulation in bladder capacity during development [76]. Also, miR-200 regulates production zinc finger E-box binding homeobox proteins 1 and 2, which transcriptionally repress Cx43 expression in human myometrial cells [77]. From the pathological perspective, miRNAs underlie modifications in connexin production during the onset and progression of several diseases, in particular cardiac pathologies. MiR-1 gained quite some attention in this respect. Its overexpression slows down conduction and depolarizes the cytoplasmic membrane [78], resulting in atrioventricular block in rodents [79]. This is due, at least in part, to the direct negative impact of miR-1 on cardiac Cx43 production [79,80]. Furthermore, hypertrophic stimulation of cardiomyocytes induces miR-1 downregulation both in vitro and in vivo, subsequently modifying the expression of Cx43, which in turn is phosphorylated by the hypertrophic stress-induced mitogen-activated protein kinase and as such displaced from the gap junction configuration [80,81]. Aberrant production or processing of miR-1 accompanied by altered Cx43 levels has also been observed in viral myocarditis in mouse [82] and in myotonic dystrophy in human [83]. In addition, miR-1 as well as miR-206 are downregulated in patients suffering from tetralogy of Fallot [84]. Other miRNAs have also been related to cardiac tachycardia and/or arrhythmias, such as miR-130a [85] and miR-19a/b [86], respectively. Several studies have documented roles for miRNAs as tumor suppressors or promoters. MiR-125b [87] and miR221/222 [88] promote cell cycling and/or invasion in glioma cell cultures, thereby suppressing Cx43 expression. Likewise, miR-20a is highly expressed in human cancer cells and negatively affects Cx43 production [89]. Both miR-200a [90] and miR-206 [91] also modify Cx43 expression during carcinogenesis. More recently, it was found that miR-206 and miR-1 diminish Cx43 levels during experimentally induced alkali burn injury in mouse cornea [92] and in chronic neuropathy in rat sciatic nerves [93], respectively.
It should be mentioned that the interaction between miRNAs and connexins can occur in both directions. Indeed, forced expression of Cx43 in glioma cell cultures antagonizes miR-125b-mediated cell growth [87]. On the other hand, silencing of Cx43 production reverses the protective effect of miR-206 downregulation in alkali-burned cornea [92]. Interestingly, besides acting as regulators of the production of their building stones, miRNAs can also permeate gap junctions. In this regard, miR-5096 is conveyed via gap junctions between glioma cells and as such exerts proinvasive effects [94]. Antiproliferative miR-124-3p travels through Cx43-based gap junctions in glioblastoma cells [95]. Gap junctions also transfer miR-210 in cocultures of mesenchymal stem cells and cardiomyocytes [96] as well as miR-142 and miR-223 between macrophages and hepatocellular carcinoma cells [97].
5. Conclusions and perspectives
Connexins and their channels control all facets of the cellular life cycle by acting at multiple communication platforms [5]. A strict and well-coordinated regulation is compulsory their appropriate expression and functioning. Considerable efforts have yet been focused throughout the years on the elucidation of the cis/trans machinery that drives connexin gene transcription [8,9]. A large body of evidence also points to the involvement of epigenetic phenomena in this process, including DNA methylation and histone acetylation at the pretranscriptional level and miRNAs at the posttranscriptional level. These mechanisms may also act in concert while controling connexin expression. This has been recently exemplified for miR-1298, which is regulated by DNA methylation and that directly binds to Cx43 mRNA in vascular smooth muscle cells, resulting in its reduced expression and gap junction channel activity [98]. As a matter of fact, a major challenge lies ahead in deciphering the global epigenetic codes that determine connexin expression, including other histone modifications, such as histone methylation, which also emerge as regulators of connexin production [99]. Such research is often complicated by the observation the methylation and acetylation not only affect connexin genes, but also their proteins. In this respect, HDAC3, HDAC4, HDAC5 and p300/cyclic adenosine monophosphate response element-binding protein colocalizes with Cx43 in cardiac tissue of dystrophic mice. They control Cx43 protein acetylation, which in turn determines its interaction with other junctional proteins, such as N-cadherin, and association with gap junctions [46]. Furthermore, bioinformatic analysis showed that specific Cx26 gene mutations known to be associated with human disease can directly trigger loss or gain of posttranslational Cx26 methylation [100].
Epigenetic modifiers are indispensable tools during further research of the role of the epigenome in connexin expression. In addition to the conventional and widely used inhibitors of DNMT and HDAC enzymes, such as decitabine and trichostatin A, respectively, several dietary compounds have been characterized as epigenetic modifiers that affect connexin expression. Epigallocatechin-3-gallate, a major constituent of green tea, decreases DNA methylation in the Cx32 gene promoter and increases its protein levels in human renal carcinoma cells [18]. Sulforaphane, an organosulfur HDAC inhibitor present in cruciferous vegetables, upregulates Cx43 protein amounts and induces gap junction opening in human cancer cells by affecting the phosphorylation status [43]. Resveratrol, which acts on histone acetylation and that is found in grapes and red wine, opens gap junctions in human glioblastoma cells [101] rat liver epithelial cells [102-104]. Like sulforaphane, this occurs independently of changes in Cx43 mRNA levels [103] and is allied with altered Cx43 phosphorylation [101-103].
Overall, the interplay between epigenetic mechanisms and their modifiers on the one hand, and connexin expression and signaling on the other hand has been predominantly studied in pathological scenarios. Typically, the epigenetic machinery during cancer triggers the silencing of tumor suppressor genes, including those coding for connexins [105,106]. Connexins have indeed repeatedly been demonstrated to possess potent antitumor properties by inducing cell cycle arrests, differentiation and apoptosis in neoplastic cells [5,106,107]. Upregulation of connexin expression using epigenetic modifiers may therefore represent an attractive anticancer therapy [106,108]. In addition, HDAC inhibitors have lately gained attention for the potential treatment of cardiac diseases, thereby also affecting connexins. In cardiodystrophic mice, Cx40 protein production is increased, while Cx43 shows lateralization. Administration of suberoylanilide hydroxamic acid to these animals restores normal Cx40 protein amounts and reestablishes the physiological Cx43 distribution pattern [52]. Likewise, trichostatin A reverses atrial arrhythmia inducibility and fibrosis in cardiac hypertrophy by normalizing Cx40 production [47]. It can be anticipated that further exploration of the effects of epigenetic modifiers on connexin production and channel activity in the upcoming years will open promising perspectives for the therapy of many other diseases.
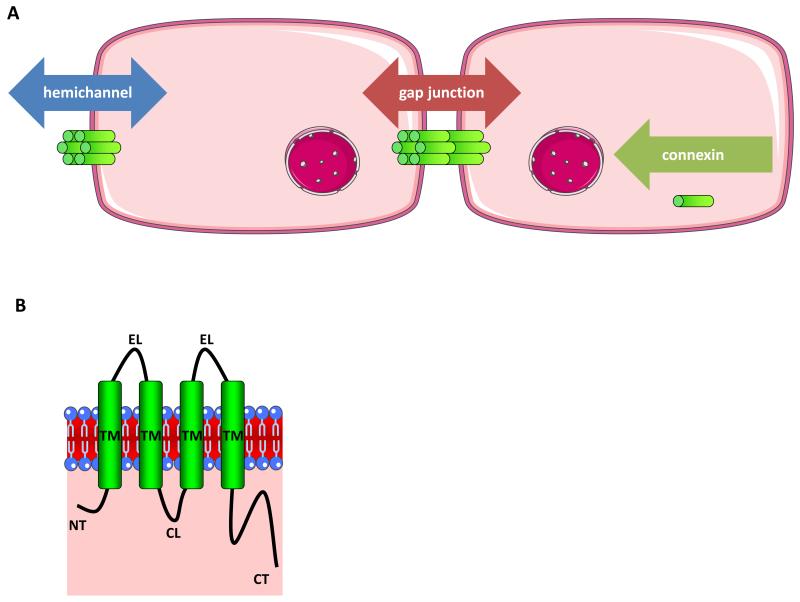
Figure 1.
A. Architecture of connexin channels and aspects of connexin signaling. Gap junctions are formed by the interaction between 2 hemichannels of adjacent cells and mediate intercellular communication (red arrow). Hemichannels are built up by 6 connexin proteins and support extracellular communication (blue arrow). Connexins as such may be involved in intacellular communication (green arrow).
B. Topology of connexin proteins. Connexins all consist of 4 transmembrane domains (TM), 2 extracellular loops (EL), 1 cytosolic loop (CL), 1 carboxyterminal cytosolic (CT) and aminoterminal (NT) tail.
Highlights.
- Connexin expression is controlled by DNA methylation, histone acetylation and microRNAs.
- The role of epigenetics in the regulation of connexin expression has been mainly studied in vitro.
- Epigenetically modifying connexin expression might be a potential clinical therapy for several pathologies.
Acknowledgements
This work was funded by grants of the European Research Council (Starting Grant 335476), the Fund for Scientific Research-Flanders (FWO grants G009514N and G010214N), the University Hospital of the Vrije Universiteit Brussel-Belgium (“Willy Gepts Fonds” UZ-VUB), the University of São Paulo-Brazil and the Foundation for Research Support of the State of São Paulo-Brazil (FAPESP SPEC grant 2013/50420-6).
List of abbreviations
- 4-PB
4-phenylbutyrate
- AZA
5-azacytidine
- Cx
connexin
- DAC
decitabine
- DNMT(i)
DNA methyltransferase (inhibitor)
- EG
epigallocatechin-3-gallate
- HDAC(i)
histone deacetylase (inhibitor)
- HMBA
hexamethylene bisacetamide
- J-1
5-(4-dimethylaminobenzoyl)aminovaleric acid hydroxamide
- miRNA
microRNA
- NaB
sodium butyrate
- SAHA
suberoylanilide hydroxamic acid
- SR
sulforaphane
- TSA
trichostatin A
- UTR
untranslated region
References
- [1].Kar R, Batra N, Riquelme MA, Jiang JX. Biological role of connexin intercellular channels and hemichannels. Arch. Biochem. Biophys. 2012;524:2–15. doi: 10.1016/j.abb.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nielsen MS, Axelsen LN, Sorgen PL, Verma V, Delmar M, Holstein-Rathlou NH. Gap junctions. Compr. Physiol. 2012;2:1981–2035. doi: 10.1002/cphy.c110051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Decrock E, Vinken M, De Vuyst E, Krysko DV, D'Herde K, Vanhaecke T, Vandenabeele P, Rogiers V, Leybaert L. Connexin-related signaling in cell death: to live or let die? Cell Death Differ. 2009;16:524–536. doi: 10.1038/cdd.2008.196. [DOI] [PubMed] [Google Scholar]
- [4].Vinken M, Doktorova T, Decrock E, Leybaert L, Vanhaecke T, Rogiers V. Gap junctional intercellular communication as a target for liver toxicity and carcinogenicity. Crit. Rev. Biochem. Mol. Biol. 2009;44:201–222. doi: 10.1080/10409230903061215. [DOI] [PubMed] [Google Scholar]
- [5].Vinken M, Vanhaecke T, Papeleu P, Snykers S, Henkens T, Rogiers V. Connexins and their channels in cell growth and cell death. Cell. Signal. 2006;18:592–600. doi: 10.1016/j.cellsig.2005.08.012. [DOI] [PubMed] [Google Scholar]
- [6].Chandrasekhar A, Bera AK. Hemichannels: permeants and their effect on development, physiology and death. Cell Biochem. Funct. 2012;30:89–100. doi: 10.1002/cbf.2794. [DOI] [PubMed] [Google Scholar]
- [7].Johnstone SR, Billaud M, Lohman AW, Taddeo EP, Isakson BE. Posttranslational modifications in connexins and pannexins. J. Membr. Biol. 2012;245:319–332. doi: 10.1007/s00232-012-9453-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Oyamada M, Oyamada Y, Takamatsu T. Regulation of connexin expression. Biochim. Biophys. Acta. 2005;1719:6–23. doi: 10.1016/j.bbamem.2005.11.002. [DOI] [PubMed] [Google Scholar]
- [9].Oyamada M, Takebe K, Oyamada Y. Regulation of connexin expression by transcription factors and epigenetic mechanisms. Biochim. Biophys. Acta. 2013;1828:118–133. doi: 10.1016/j.bbamem.2011.12.031. [DOI] [PubMed] [Google Scholar]
- [10].Szyf M. The dynamic epigenome and its implications in toxicology. Toxicol. Sci. 2007;100:7–23. doi: 10.1093/toxsci/kfm177. [DOI] [PubMed] [Google Scholar]
- [11].Géranton SM, Tochiki KK. Regulation of gene expression and pain states by epigenetic mechanisms. Prog. Mol. Biol. Transl. Sci. 2015;131:147–183. doi: 10.1016/bs.pmbts.2014.11.012. [DOI] [PubMed] [Google Scholar]
- [12].Wu X, Wang Y, Sun Y, Chen S, Zhang S, Shen L, Huang X, Lin X, Kong W. Reduced expression of connexin26 and its DNA promoter hypermethylation in the inner ear of mimetic aging rats induced by d-galactose. Biochem. Biophys. Res. Commun. 2014;452:340–346. doi: 10.1016/j.bbrc.2014.08.063. [DOI] [PubMed] [Google Scholar]
- [13].Wang Y, Huang LH, Xu CX, Xiao J, Zhou L, Cao D, Liu XM, Qi Y. Connexin32 and 43 promoter methylation in Helicobacter pylori-associated gastric tumorigenesis. World J. Gastroenterol. 2014;20:11770–11779. doi: 10.3748/wjg.v20.i33.11770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chen Y, Hühn D, Knösel T, Pacyna-Gengelbach M, Deutschmann N, Petersen I. Downregulation of connexin26 in human lung cancer is related to promoter methylation. Int. J. Cancer. 2005;113:14–21. doi: 10.1002/ijc.20498. [DOI] [PubMed] [Google Scholar]
- [15].Jinn Y, Inase N. Connexin43, E-cadherin, beta-catenin and ZO-1 expression, and aberrant methylation of the connexin43 gene in NSCLC. Anticancer Res. 2010;30:2271–2278. [PubMed] [Google Scholar]
- [16].Hirai A, Yano T, Nishikawa K, Suzuki K, Asano R, Satoh H, Hagiwara K, Yamasaki H. Downregulation of connexin32 gene expression through DNA methylation in a human renal cell carcinoma cell. Am. J. Nephrol. 2003;23:172–177. doi: 10.1159/000070653. [DOI] [PubMed] [Google Scholar]
- [17].Yano T, Ito F, Kobayashi K, Yonezawa Y, Suzuki K, Asano R, Hagiwara K, Nakazawa H, Toma H, Yamasaki H. Hypermethylation of the CpG island of connexin32, a candiate tumor suppressor gene in renal cell carcinomas from hemodialysis patients. Cancer Lett. 2004;208:137–142. doi: 10.1016/j.canlet.2003.11.029. [DOI] [PubMed] [Google Scholar]
- [18].Sato A, Sekine M, Kobayashi M, Virgona N, Ota M, Yano T. Induction of the connexin32 gene by epigallocatechin-3-gallate potentiates vinblastine-induced cytotoxicity in human renal carcinoma cells. Chemotherapy. 2013;59:192–199. doi: 10.1159/000354715. [DOI] [PubMed] [Google Scholar]
- [19].Loncarek J, Yamasaki H, Levillain P, Milinkevitch S, Mesnil M. The expression of the tumor suppressor gene connexin26 is not mediated by methylation in human esophageal cancer cells. Mol. Carcinog. 2003;36:74–81. doi: 10.1002/mc.10102. [DOI] [PubMed] [Google Scholar]
- [20].Singal R, Tu ZJ, Vanwert JM, Ginder GD, Kiang DT. Modulation of the connexin26 tumor suppressor gene expression through methylation in human mammary epithelial cell lines. Anticancer Res. 2000;20:59–64. [PubMed] [Google Scholar]
- [21].Yi ZC, Wang H, Zhang GY, Xia B. Downregulation of connexin43 in nasopharyngeal carcinoma cells is related to promoter methylation. Oral Oncol. 2007;43:898–904. doi: 10.1016/j.oraloncology.2006.11.004. [DOI] [PubMed] [Google Scholar]
- [22].Ahmed D, Lothe RA, Rivedal E, Lind GE. Quantitative validation of GJC1 promoter hypermethylation in benign and malignant colorectal tumors. Endocr. Relat. Cancer. 2011;18:C31–C34. doi: 10.1530/ERC-11-0204. [DOI] [PubMed] [Google Scholar]
- [23].Sirnes S, Honne H, Ahmed D, Danielsen SA, Rognum TO, Meling GI, Leithe E, Rivedal E, Lothe RA, Lind GE. DNA methylation analyses of the connexin gene family reveal silencing of GJC1 (Connexin45) by promoter hypermethylation in colorectal cancer. Epigenetics. 2011;6:602–609. doi: 10.4161/epi.6.5.15237. [DOI] [PubMed] [Google Scholar]
- [24].Yu SC, Xiao HL, Jiang XF, Wang QL, Li Y, Yang XJ, Ping YF, Duan JJ, Jiang JY, Ye XZ, Xu SL, Xin YH, Yao XH, Chen JH, Chu WH, Sun W, Wang B, Wang JM, Zhang X, Bian XW. Connexin43 reverses malignant phenotypes of glioma stem cells by modulating E-cadherin. Stem Cells. 2012;30:108–120. doi: 10.1002/stem.1685. [DOI] [PubMed] [Google Scholar]
- [25].Piechocki MP, Burk RD, Ruch RJ. Regulation of connexin32 and connexin43 gene expression by DNA methylation in rat liver cells. Carcinogenesis. 1999;20:401–406. doi: 10.1093/carcin/20.3.401. [DOI] [PubMed] [Google Scholar]
- [26].Shimizu K, Onishi M, Sugata E, Sokuza Y, Mori C, Nishikawa T, Honoki K, Tsujiuchi T. Disturbance of DNA methylation patterns in the early phase of hepatocarcinogenesis induced by a choline-deficient L-amino acid-defined diet in rats. Cancer Sci. 2007;98:1318–1322. doi: 10.1111/j.1349-7006.2007.00564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tsujiuchi T, Shimizu K, Itsuzaki Y, Onishi M, Sugata E, Fujii H, Honoki K. CpG site hypermethylation of E-cadherin and connexin26 genes in hepatocellular carcinomas induced by a choline-deficient L-amino acid-defined diet in rats. Mol. Carcinog. 2007;46:269–274. doi: 10.1002/mc.20268. [DOI] [PubMed] [Google Scholar]
- [28].Shimizu K, Shimoichi Y, Hinotsume D, Itsuzaki Y, Fujii H, Honoki K, Tsujiuchi T. Reduced expression of the connexin26 gene and its aberrant DNA methylation in rat lung adenocarcinomas induced by N-nitrosobis(2-hydroxypropyl)amine. Mol. Carcinog. 2006;45:710–714. doi: 10.1002/mc.20207. [DOI] [PubMed] [Google Scholar]
- [29].Borinstein SC, Conerly M, Dzieciatkowski S, Biswas S, Washingto MK, Trobridge P, Henikoff S, Grady WM. Aberrant DNA methylation occurs in colon neoplasms arising in the azoxymethane colon cancer model. Mol. Carcinog. 2010;49:94–103. doi: 10.1002/mc.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hohl M, Thiel G. Cell type-specific regulation of RE-1 silencing transcription factor (REST) target genes. Eur. J. Neurosci. 2005;22:2216–2230. doi: 10.1111/j.1460-9568.2005.04404.x. [DOI] [PubMed] [Google Scholar]
- [31].Yu JN, Xue CY, Wang XG, Lin F, Liu CY, Lu FZ, Liu HL. 5-AZA-2'-deoxycytidine (5-AZA-CdR) leads to downregulation of Dnmt1o and gene expression in preimplantation mouse embryos. Zygote. 2009;17:137–145. doi: 10.1017/S0967199408005169. [DOI] [PubMed] [Google Scholar]
- [32].Chen JT, Cheng YW, Chou MC, Sen-Lin T, Lai WW, Ho WL, Lee H. The correlation between aberrant connexin43 mRNA expression induced by promoter methylation and nodal micrometastasis in non-small cell lung cancer. Clin. Cancer Res. 2003;9:4200–4204. [PubMed] [Google Scholar]
- [33].Tan LW, Bianco T, Dobrovic A. Variable promoter region CpG island methylation of the putative tumor suppressor gene connexin26 in breast cancer. Carcinogenesis. 2002;23:231–236. doi: 10.1093/carcin/23.2.231. [DOI] [PubMed] [Google Scholar]
- [34].Olzscha H, Sheikh S, La Thangue NB. Deacetylation of chromatin and gene expression regulation: a new target for epigenetic therapy. Crit. Rev. Oncog. 2015;20:1–17. doi: 10.1615/critrevoncog.2014012463. [DOI] [PubMed] [Google Scholar]
- [35].Ammerpohl O, Thormeyer D, Khan Z, Appelskog IB, Gojkovic Z, Almqvist PM, Ekström TJ. HDACi phenylbutyrate increases bystander killing of HSV-tk transfected glioma cells. Biochem. Biophys. Res. Commun. 2004;324:8–14. doi: 10.1016/j.bbrc.2004.09.016. [DOI] [PubMed] [Google Scholar]
- [36].Asklund T, Appelskog IB, Ammerpohl O, Ekström TJ, Almqvist PM. Histone deacetylase inhibitor 4-phenylbutyrate modulates glial fibrillary acidic protein and connexin43 expression, and enhances gap-junction communication, in human glioblastoma cells. Eur. J. Cancer. 2004;40:1073–1081. doi: 10.1016/j.ejca.2003.11.034. [DOI] [PubMed] [Google Scholar]
- [37].Hernandez M, Shao Q, Yang XJ, Luh SP, Kandouz M, Batist G, Laird DW, Alaoui-Jamali MA. A histone deacetylation-dependent mechanism for transcriptional repression of the gap junction gene Cx43 in prostate cancer cells. Prostate. 2006;66:1151–1161. doi: 10.1002/pros.20451. [DOI] [PubMed] [Google Scholar]
- [38].Khan Z, Akhtar M, Asklund T, Juliusson B, Almqvist PM, Ekström TJ. HDAC inhibition amplifies gap junction communication in neural progenitors: potential for cell-mediated enzyme prodrug therapy. Exp. Cell Res. 2007;31:2958–2967. doi: 10.1016/j.yexcr.2007.05.004. [DOI] [PubMed] [Google Scholar]
- [39].Ogawa T, Hayashi T, Kyoizumi S, Ito T, Trosko JE, Yorioka N. Upregulation of gap junctional intercellular communication by hexamethylene bisacetamide in cultured human peritoneal mesothelial cells. Lab. Invest. 1999;79:1511–1520. [PubMed] [Google Scholar]
- [40].Ogawa T, Hayashi T, Yorioka N, Kyoizumi S, Trosko JE. Hexamethylene bisacetamide protects peritoneal mesothelial cells from glucose. Kidney Int. 2001;60:996–1008. doi: 10.1046/j.1523-1755.2001.060003996.x. [DOI] [PubMed] [Google Scholar]
- [41].Ogawa T, Hayashi T, Tokunou M, Nakachi K, Trosko JE, Chang CC, Yorioka N. Suberoylanilide hydroxamic acid enhances gap junctional intercellular communication via acetylation of histone containing connexin43 gene locus. Cancer Res. 2005;65:9771–9778. doi: 10.1158/0008-5472.CAN-05-0227. [DOI] [PubMed] [Google Scholar]
- [42].Dovzhanskiy DI, Hartwig W, Lázár NG, Schmidt A, Felix K, Straub BK, Hackert T, Krysko DV, Werner J. Growth inhibition of pancreatic cancer by experimental treatment with 4-phenylbutyrate is associated with increased expression of connexin43. Oncol. Res. 2012;20:103–111. doi: 10.3727/096504012x13477145152959. [DOI] [PubMed] [Google Scholar]
- [43].Forster T, Rausch V, Zhang Y, Isayev O, Heilmann K, Schoensiegel F, Liu L, Nessling M, Richter K, Labsch S, Nwaeburu CC, Mattern J, Gladkich J, Giese N, Werner J, Schemmer P, Gross W, Gebhard MM, Gerhauser C, Schaefer M, Herr I. Sulforaphane counteracts aggressiveness of pancreatic cancer driven by dysregulated Cx43-mediated gap junctional intercellular communication. Oncotarget. 2014;5:1621–1634. doi: 10.18632/oncotarget.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kaufman J, Gordon C, Bergamaschi R, Wang HZ, Cohen IS, Valiunas V, Brink PR. The effects of the histone deacetylase inhibitor 4-phenylbutyrate on gap junction conductance and permeability. Front. Pharmacol. 2013;4:111. doi: 10.3389/fphar.2013.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kim J, Hwangbo C, Hu X, Kang Y, Papangeli I, Mehrotra D, Park H, Ju H, McLean DL, Comhair SA, Erzurum SC, Chun HJ. Restoration of impaired endothelial myocyte enhancer factor 2 function rescues pulmonary arterial hypertension. Circulation. 2015;131:190–199. doi: 10.1161/CIRCULATIONAHA.114.013339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Colussi C, Rosati J, Straino S, Spallotta F, Berni R, Stilli D, Rossi S, Musso E, Macchi E, Mai A, Sbardella G, Castellano S, Chimenti C, Frustaci A, Nebbioso A, Altucci L, Capogrossi MC, Gaetano C. Nε-lysine acetylation determines dissociation from gap junctions and lateralization of connexin43 in normal and dystrophic heart. Proc. Natl. Acad. Sci. USA. 2011;108:2795–2800. doi: 10.1073/pnas.1013124108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Liu F, Levin MD, Petrenko NB, Lu MM, Wang T, Yuan LJ, Stout AL, Epstein JA, Patel VV. Histone deacetylase inhibition reverses atrial arrhythmia inducibility and fibrosis in cardiac hypertrophy independent of angiotensin. J. Mol. Cell. Cardiol. 2008;45:715–723. doi: 10.1016/j.yjmcc.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ammerpohl O, Trauzold A, Schniewind B, Griep U, Pilarsky C, Grutzmann R, Saeger HD, Janssen O, Sipos B, Kloppel G, Kalthoff H. Complementary effects of HDAC inhibitor 4-PB on gap junction communication and cellular export mechanisms support restoration of chemosensitivity of PDAC cells. Br. J. Cancer. 2007;96:73–81. doi: 10.1038/sj.bjc.6603511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Vinken M, Henkens T, Vanhaecke T, Papeleu P, Geerts A, Van Rossen E, Chipman JK, Meda P, Rogiers V. Trichostatin A enhances gap junctional intercellular communication in primary cultures of adult rat hepatocytes. Toxicol. Sci. 2006;91:484–492. doi: 10.1093/toxsci/kfj152. [DOI] [PubMed] [Google Scholar]
- [50].Vinken M, Henkens T, Snykers S, Lukaszuk A, Tourwé D, Rogiers V, Vanhaecke T. The novel histone deacetylase inhibitor 4-Me2N-BAVAH differentially affects cell junctions between primary hepatocytes. Toxicology. 2007;236:92–102. doi: 10.1016/j.tox.2007.04.003. [DOI] [PubMed] [Google Scholar]
- [51].Hattori Y, Fukushima M, Maitani Y. Non-viral delivery of the connexin43 gene with histone deacetylase inhibitor to human nasopharyngeal tumor cells enhances gene expression and inhibits in vivo tumor growth. Int. J. Oncol. 2007;30:1427–1439. [PubMed] [Google Scholar]
- [52].Colussi C, Berni R, Rosati J, Straino S, Vitale S, Spallotta F, Baruffi S, Bocchi L, Delucchi F, Rossi S, Savi M, Rotili D, Quaini F, Macchi E, Stilli D, Musso E, Mai A, Gaetano C, Capogrossi MC. The histone deacetylase inhibitor suberoylanilide hydroxamic acid reduces cardiac arrhythmias in dystrophic mice. Cardiovasc. Res. 2010;87:73–82. doi: 10.1093/cvr/cvq035. [DOI] [PubMed] [Google Scholar]
- [53].Martin D, Tawadros T, Meylan L, Abderrahmani A, Condorelli DF, Waeber G, Haefliger JA. Critical role of the transcriptional repressor neuron-restrictive silencer factor in the specific control of connexin36 in insulin-producing cell lines. J. Biol. Chem. 2003;278:53082–53089. doi: 10.1074/jbc.M306861200. [DOI] [PubMed] [Google Scholar]
- [54].Martin D, Allagnat F, Chaffard G, Caille D, Fukuda M, Regazzi R, Abderrahmani A, Waeber G, Meda P, Maechler P, Haefliger JA. Functional significance of repressor element 1 silencing transcription factor (REST) target genes in pancreatic beta cells. Diabetologia. 2008;51:1429–1439. doi: 10.1007/s00125-008-0984-1. [DOI] [PubMed] [Google Scholar]
- [55].Zupkovitz G, Tischler J, Posch M, Sadzak I, Ramsauer K, Egger G, Grausenburger R, Schweifer N, Chiocca S, Decker T, Seiser C. Negative and positive regulation of gene expression by mouse histone deacetylase 1. Mol. Cell. Biol. 2006;26:7913–7928. doi: 10.1128/MCB.01220-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hoxha E, Lambers E, Xie H, De Andrade A, Krishnamurthy P, Wasserstrom JA, Ramirez V, Thal M, Verma SK, Soares MB, Kishore R. Histone deacetylase 1 deficiency impairs differentiation and electrophysiological properties of cardiomyocytes derived from induced pluripotent cells. Stem Cells. 2012;30:2412–2422. doi: 10.1002/stem.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lu DF, Wang Y, Su ZZ, Zeng ZH, Xing XW, He ZY, Zhang C. Knockdown of the HDAC1 promotes the directed differentiation of bone mesenchymal stem cells into cardiomyocytes. PLoS One. 2014;9:e92179. doi: 10.1371/journal.pone.0092179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kapoor P, Saunders MM, Li Z, Zhou Z, Sheaffer N, Kunze EL, Samant RS, Welch DR, Donahue HJ. Breast cancer metastatic potential: correlation with increased heterotypic gap junctional intercellular communication between breast cancer cells and osteoblastic cells. Int. J. Cancer. 2004;111:693–697. doi: 10.1002/ijc.20318. [DOI] [PubMed] [Google Scholar]
- [59].Samant RS, Seraj MJ, Saunders MM, Sakamaki TS, Shevde LA, Harms JF, Leonard TO, Goldberg SF, Budgeon L, Meehan WJ, Winter CR, Christensen ND, Verderame MF, Donahue HJ, Welch DR. Analysis of mechanisms underlying BRMS1 suppression of metastasis. Clin. Exp. Metastasis. 2001;18:683–693. doi: 10.1023/a:1013124725690. [DOI] [PubMed] [Google Scholar]
- [60].Saunders MM, Seraj MJ, Li Z, Zhou Z, Winter CR, Welch DR, Donahue HJ. Breast cancer metastatic potential correlates with a breakdown in homospecific and heterospecific gap junctional intercellular communication. Cancer Res. 2001;61:1765–1767. [PubMed] [Google Scholar]
- [61].Shevde LA, Samant RS, Goldberg SF, Sikaneta T, Alessandrini A, Donahue HJ, Mauger DT, Welch DR. Suppression of human melanoma metastasis by the metastasis suppressor gene, BRMS1. Exp. Cell Res. 2002;273:229–239. doi: 10.1006/excr.2001.5452. [DOI] [PubMed] [Google Scholar]
- [62].Meehan WJ, Samant RS, Hopper JE, Carrozza MJ, Shevde LA, Workman JL, Eckert KA, Verderame MF, Welch DR. Breast cancer metastasis suppressor 1 (BRMS1) forms complexes with retinoblastoma-binding protein 1 (RBP1) and the mSin3 histone deacetylase complex and represses transcription. J. Biol. Chem. 2004;279:1562–1569. doi: 10.1074/jbc.M307969200. [DOI] [PubMed] [Google Scholar]
- [63].Xie N, Liu L, Li Y, Yu C, Lam S, Shynlova O, Gleave M, Challis JR, Lye S, Dong X. Expression and function of myometrial PSF suggest a role in progesterone withdrawal and the initiation of labor. Mol. Endocrinol. 2012;26:1370–1379. doi: 10.1210/me.2012-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Wang HZ, Rosati B, Gordon C, Valiunas V, McKinnon D, Cohen IS, Brink PR. Inhibition of histone deacetylase (HDAC) by 4-phenylbutyrate results in increased junctional conductance between rat corpora smooth muscle cells. Front. Pharmacol. 2015;6:9. doi: 10.3389/fphar.2015.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Jung JW, Cho SD, Ahn NS, Yang SR, Park JS, Jo EH, Hwang JW, Aruoma OI, Lee YS, Kang KS. Effects of the histone deacetylases inhibitors sodium butyrate and trichostatin A on the inhibition of gap junctional intercellular communication by H2O2- and 12-O-tetradecanoylphorbol-13-acetate in rat liver epithelial cells. Cancer Lett. 2006;241:301–308. doi: 10.1016/j.canlet.2005.10.029. [DOI] [PubMed] [Google Scholar]
- [66].Zhao W, Han HB, Zhang ZQ. Suppression of lung cancer cell invasion and metastasis by connexin43 involves the secretion of follistatin-like 1 mediated via histone acetylation. Int. J. Biochem. Cell Biol. 2011;43:1459–1468. doi: 10.1016/j.biocel.2011.06.009. [DOI] [PubMed] [Google Scholar]
- [67].Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Anderson C, Catoe H, Werner R. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res. 2006;34:5863–5871. doi: 10.1093/nar/gkl743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J. Cell Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Inose H, Ochi H, Kimura A, Fujita K, Xu R, Sato S, Iwasaki M, Sunamura S, Takeuchi Y, Fukumoto S, Saito K, Nakamura T, Siomi H, Ito H, Arai Y, Shinomiya K, Takeda S. A microRNA regulatory mechanism of osteoblast differentiation. Proc. Natl. Acad. Sci. USA. 2009;106:20794–20799. doi: 10.1073/pnas.0909311106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Gindin Y, Jiang Y, Francis P, Walker RL, Abaan OD, Zhu YJ, Meltzer PS. MiR-23a impairs bone differentiation in osteosarcoma via downregulation of GJA1. Front. Genet. 2015;6:233. doi: 10.3389/fgene.2015.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Stauffer BL, Sobus RD, Sucharov CC. Sex differences in cardiomyocyte connexin43 expression. J. Cardiovasc. Pharmacol. 2011;58:32–39. doi: 10.1097/FJC.0b013e31821b70b4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Lee SK, Teng Y, Wong HK, Ng TK, Huang L, Lei P, Choy KW, Liu Y, Zhang M, Lam DS, Yam GH, Pang CP. MicroRNA-145 regulates human corneal epithelial differentiation. PLoS One. 2011;6:e21249. doi: 10.1371/journal.pone.0021249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, Willis MS, Selzman CH, Wang DZ. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J. Clin. Invest. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Peng H, Park JK, Katsnelson J, Kaplan N, Yang W, Getsios S, Lavker RM. MicroRNA-103/107 family regulates multiple epithelial stem cell characteristics. Stem Cells. 2015;33:1642–1656. doi: 10.1002/stem.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Imamura M, Sugino Y, Long X, Slivano OJ, Nishikawa N, Yoshimura N, Miano JM. Myocardin and microRNA-1 modulate bladder activity through connexin43 expression during postnatal development. J. Cell. Physiol. 2013;228:1819–1826. doi: 10.1002/jcp.24333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Renthal NE, Chen CC, Williams KC, Gerard RD, Prange-Kiel J, Mendelson CR. miR-200 family and targets, ZEB1 and ZEB2, modulate uterine quiescence and contractility during pregnancy and labor. Proc. Natl. Acad. Sci. USA. 2010;107:20828–20833. doi: 10.1073/pnas.1008301107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H, Chen G, Wang Z. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat. Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- [79].Zhang Y, Sun L, Zhang Y, Liang H, Li X, Cai R, Wang L, Du W, Zhang R, Li J, Wang Z, Ma N, Wang X, Du Z, Yang B, Gao X, Shan H. Overexpression of microRNA-1 causes atrioventricular block in rodents. Int. J. Biol. Sci. 2013;9:455–462. doi: 10.7150/ijbs.4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Curcio A, Torella D, Iaconetti C, Pasceri E, Sabatino J, Sorrentino S, Giampà S, Micieli M, Polimeni A, Henning BJ, Leone A, Catalucci D, Ellison GM, Condorelli G, Indolfi C. MicroRNA-1 downregulation increases connexin43 displacement and induces ventricular tachyarrhythmias in rodent hypertrophic hearts. PLoS One. 2013;8:e70158. doi: 10.1371/journal.pone.0070158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Yang H, Wu C, Xiao Y, Zhou S. Connexin and fibrosis related microRNAs in complex fractionated atrial electrograms. Arch. Med. Sci. 2015;11:679–682. doi: 10.5114/aoms.2015.52375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Xu HF, Ding YJ, Shen YW, Xue AM, Xu HM, Luo CL, Li BX, Liu YL, Zhao ZQ. MicroRNA- 1 represses Cx43 expression in viral myocarditis. Mol. Cell. Biochem. 2012;362:141–148. doi: 10.1007/s11010-011-1136-3. [DOI] [PubMed] [Google Scholar]
- [83].Rau F, Freyermuth F, Fugier C, Villemin JP, Fischer MC, Jost B, Dembele D, Gourdon G, Nicole A, Duboc D, Wahbi K, Day JW, Fujimura H, Takahashi MP, Auboeuf D, Dreumont N, Furling D, Charlet-Berguerand N. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy, Nat. Struct. Mol. Biol. 2011;18:840–845. doi: 10.1038/nsmb.2067. [DOI] [PubMed] [Google Scholar]
- [84].Wu Y, Ma XJ, Wang HJ, Li WC, Chen L, Ma D, Huang GY. Expression of Cx43-related microRNAs in patients with tetralogy of Fallot. World J. Pediatr. 2014;10:138–144. doi: 10.1007/s12519-013-0434-0. [DOI] [PubMed] [Google Scholar]
- [85].Osbourne A, Calway T, Broman M, McSharry S, Earley J, Kim GH. Downregulation of connexin43 by microRNA-130a in cardiomyocytes results in cardiac arrhythmias. J. Mol. Cell. Cardiol. 2014;74:53–63. doi: 10.1016/j.yjmcc.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Danielson LS, Park DS, Rotllan N, Chamorro-Jorganes A, Guijarro MV, Fernandez-Hernando C, Fishman GI, Phoon CK, Hernando E. Cardiovascular dysregulation of miR-17-92 causes a lethal hypertrophic cardiomyopathy and arrhythmogenesis. FASEB J. 2013:1460–1467. doi: 10.1096/fj.12-221994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Jin Z, Xu S, Yu H, Yang B, Zhao H, Zhao G. miR-125b inhibits connexin43 and promotes glioma growth. Cell. Mol. Neurobiol. 2013;33:1143–1148. doi: 10.1007/s10571-013-9980-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Hao J, Zhang C, Zhang A, Wang K, Jia Z, Wang G, Han L, Kang C, Pu P. MiR-221/222 is the regulator of Cx43 expression in human glioblastoma cells. Oncol. Rep. 2012;27:1504–1510. doi: 10.3892/or.2012.1652. [DOI] [PubMed] [Google Scholar]
- [89].Li X, Pan JH, Song B, Xiong EQ, Chen ZW, Zhou ZS, Su YP. Suppression of Cx43 expression by miR-20a in the progression of human prostate cancer. Cancer Biol. Ther. 2012;13:890–898. doi: 10.4161/cbt.20841. [DOI] [PubMed] [Google Scholar]
- [90].Ming J, Zhou Y, Du J, Fan S, Pan B, Wang Y, Fan L, Jiang J. Identification of miR-200a as a novel suppressor of connexin 43 in breast cancer cells. Biosci. Rep. 2015;35:e00251. doi: 10.1042/BSR20150153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Lin CC, Sharma SB, Farrugia MK, McLaughlin SL, Ice RJ, Loskutov YV, Pugacheva EN, Brundage KM, Chen D, Ruppert JM. Kruppel-like factor 4 signals through microRNA-206 to promote tumor initiation and cell survival. Oncogenesis. 2015;4:e155. doi: 10.1038/oncsis.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Li X, Zhou H, Tang W, Guo Q, Zhang Y. Transient downregulation of microRNA-206 protects alkali burn injury in mouse cornea by regulating connexin43. Int. J. Clin. Exp. Pathol. 2015;8:2719–2727. [PMC free article] [PubMed] [Google Scholar]
- [93].Neumann E, Hermanns H, Barthel F, Werdehausen R, Brandenburger T. Expression changes of microRNA-1 and its targets connexin43 and brain-derived neurotrophic factor in the peripheral nervous system of chronic neuropathic rats. Mol. Pain. 2015;11:39. doi: 10.1186/s12990-015-0045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Hong X, Sin WC, Harris AL, Naus CC. Gap junctions modulate glioma invasion by direct transfer of microRNA. Oncotarget. 2015;6:15566–15577. doi: 10.18632/oncotarget.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Suzhi Z, Liang T, Yuexia P, Lucy L, Xiaoting H, Yuan Z, Qin W. Gap junctions enhance the antiproliferative effect of microRNA-124-3p in glioblastoma cells. J. Cell. Physiol. 2015;230:2476–2488. doi: 10.1002/jcp.24982. [DOI] [PubMed] [Google Scholar]
- [96].Kim HW, Jiang S, Ashraf M, Haider KH. Stem cell-based delivery of hypoxamir-210 to the infarcted heart: implications on stem cell survival and preservation of infarcted heart function. J. Mol. Med. 2012;90:997–1010. doi: 10.1007/s00109-012-0920-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Aucher A, Rudnicka D, Davis DM. MicroRNAs transfer from human macrophages to hepato-carcinoma cells and inhibit proliferation. J. Immunol. 2013;191:6250–6260. doi: 10.4049/jimmunol.1301728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Hu W, Wang M, Yin H, Yao C, He Q, Yin L, Zhang C, Li W, Chang G, Wang S. MicroRNA-1298 is regulated by DNA methylation and affects vascular smooth muscle cell function by targeting connexin43. Cardiovasc. Res. 2015;107:534–545. doi: 10.1093/cvr/cvv160. [DOI] [PubMed] [Google Scholar]
- [99].Li X, Su Y, Pan J, Zhou Z, Song B, Xiong E, Chen Z. Connexin 26 is downregulated by KDM5B in the progression of bladder cancer. Int. J. Mol. Sci. 2013;14:7866–7879. doi: 10.3390/ijms14047866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Yilmaz A. A, Bioinformatic analysis of GJB2 gene missense mutations. Cell. Biochem. Biophys. 2015 doi: 10.1007/s12013-014-0385-7. in press. [DOI] [PubMed] [Google Scholar]
- [101].Leone S, Fiore M, Lauro MG, Pino S, Cornetta T, Cozzi R. Resveratrol and X rays affect gap junction intercellular communications in human glioblastoma cells. Mol. Carcinog. 2008;47:587–598. doi: 10.1002/mc.20416. [DOI] [PubMed] [Google Scholar]
- [102].Kim JH, Lee BK, Lee KW, Lee HJ. Resveratrol counteracts gallic acid-induced downregulation of gap junction intercellular communication. J. Nutr. Biochem. 2009;20:149–154. doi: 10.1016/j.jnutbio.2008.01.008. [DOI] [PubMed] [Google Scholar]
- [103].Nielsen M, Ruch RJ, Vang O. Resveratrol reverses tumor-promoter-induced inhibition of gap-junctional intercellular communication. Biochem. Biophys. Res. Commun. 2000;275:804–809. doi: 10.1006/bbrc.2000.3378. [DOI] [PubMed] [Google Scholar]
- [104].Upham BL, Guzvić M, Scott J, Carbone JM, Blaha L, Coe C, Li LL, Rummel AM, Trosko JE. Inhibition of gap junctional intercellular communication and activation of mitogen-activated protein kinase by tumor-promoting organic peroxides and protection by resveratrol. Nutr. Cancer. 2007;57:38–47. doi: 10.1080/01635580701268188. [DOI] [PubMed] [Google Scholar]
- [105].Leithe E, Sirnes S, Omori Y, Rivedal E. Downregulation of gap junctions in cancer cells. Crit. Rev. Oncog. 2006;12:225–256. doi: 10.1615/critrevoncog.v12.i3-4.30. [DOI] [PubMed] [Google Scholar]
- [106].Pointis G, Fiorini C, Gilleron J, Carette D, Segretain D. Connexins as precocious markers and molecular targets for chemical and pharmacological agents in carcinogenesis. Curr. Med. Chem. 2007;14:2288–2303. doi: 10.2174/092986707781696564. [DOI] [PubMed] [Google Scholar]
- [107].Czyz J. The stage-specific function of gap junctions during tumourigenesis. Cell. Mol. Biol. Lett. 2008;13:92–102. doi: 10.2478/s11658-007-0039-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Vinken M, Henkens T, De Rop E, Fraczek J, Vanhaecke T, Rogiers V. Biology and pathobiology of gap junctional channels in hepatocytes. Hepatology. 2008;47:1077–1088. doi: 10.1002/hep.22049. [DOI] [PubMed] [Google Scholar]
- [109].Hagiwara H, Sato H, Ohde Y, Takano Y, Seki T, Ariga T, Hokaiwado N, Asamoto M, Shirai T, Nagashima Y, Yano T. 5-Aza-2'-deoxycytidine suppresses human renal carcinoma cell growth in a xenograft model via upregulation of the connexin32 gene. Br. J. Pharmacol. 2008;153:1373–1381. doi: 10.1038/bjp.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].King TJ, Fukushima LH, Donlon TA, Hieber AD, Shimabukuro KA, Bertram JS. Correlation between growth control, neoplastic potential and endogenous connexin43 expression in HeLa cell lines: implications for tumor progression. Carcinogenesis. 2000;21:311–315. doi: 10.1093/carcin/21.2.311. [DOI] [PubMed] [Google Scholar]
- [111].Robe PA, Jolois O, N'Guyen M, Princen F, Malgrange B, Merville MP, Bours V. Modulation of the HSV-TK/ganciclovir bystander effect by n-butyrate in glioblastoma: correlation with gap junction intercellular communication. Int. J. Oncol. 2004;25:187–192. [PubMed] [Google Scholar]
- [112].Yamashita Y, Shimada M, Harimoto N, Tanaka S, Shirabe K, Ijima H, Nakazawa K, Fukuda J, Funatsu K, Maehara Y. cDNA microarray analysis in hepatocyte differentiation in Huh7 cells. Cell Transplant. 13:793–799. doi: 10.3727/000000004783983396. (204) [DOI] [PubMed] [Google Scholar]
- [113].Germann A, Dihlmann S, Hergenhahn M, Doeberitz M, Koesters R. Expression profiling of CC531 colon carcinoma cells reveals similar regulation of beta-catenin target genes by both butyrate and aspirin. Int. J. Cancer. 2003;106:187–197. doi: 10.1002/ijc.11215. [DOI] [PubMed] [Google Scholar]
- [114].Alajez NM, Lenarduzzi M, Ito E, Hui AB, Shi W, Bruce J, Yue S, Huang SH, Xu W, Waldron J, O'Sullivan B, Liu FF. MiR-218 suppresses nasopharyngeal cancer progression through downregulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res. 2011;71:2381–2391. doi: 10.1158/0008-5472.CAN-10-2754. [DOI] [PubMed] [Google Scholar]