Abstract
Cutaneous SCC (cSCC) is the most frequent skin cancer with metastatic potential and can manifest rapidly as a common side effect in patients receiving systemic kinase inhibitors. Here we use massively parallel exome and targeted level sequencing 132 sporadic cSCC, 39 squamoproliferative lesions and cSCC arising in patients receiving the BRAF inhibitor vemurafenib, as well as 10 normal skin samples to identify significant NOTCH1 mutation as an early event in squamous cell carcinogenesis. Bisected vemurafenib induced lesions revealed surprising heterogeneity with different activating HRAS and NOTCH1 mutations identified in two halves of the same cSCC suggesting polyclonal origin.
Immunohistochemical analysis using an antibody specific to nuclear NOTCH1 correlates with mutation status in sporadic cSCC and regions of NOTCH1 loss or down-regulation are frequently observed in normal looking skin. Our data indicate that NOTCH1 acts as a gatekeeper in human cSCC.
Introduction
Squamous cell carcinoma (SCC) causes >900,000 deaths worldwide and arises in stratifying epithelial tissues such as skin, lung, esophagus and cervix. Common mutation profiles and perturbed signaling events have been identified in SCC arising in different tissues, including loss of function NOTCH1 mutation in skin, lung and esophagus (Durinck et al, 2011; Stransky et al, 2011; Agrawal et al, 2012; Hammerman et al, 2012; Wang et al, 2012). Squamous cell carcinoma of the skin (cutaneous SCC, cSCC) is an increasing health problem with up to an estimated 400,000 new cases diagnosed in 2012 in the US and characterized by an upward trend with an increase of up to 200% over the past three decades (Karia et al, 2013). In Scotland cSCC is the fourth most frequent cancer registration behind basal cell carcinoma of the skin, lung and bronchus, and breast (ISD Scotland 2010 data: http://www.isdscotland.org/Health-Topics/Cancer/). Although it contributes only 0.37% overall cancer mortality, compared with 26% lung and bronchus and 7% breast (ISD Scotland), locoregional and metastatic cSCC has a dismal prognosis, with reported five year survival rates below 30% (Kwa et al, 1992; Cherpelis et al, 2002). Indeed, since tumors are visible from the outset, it is timely intervention, rather than tumor factors per se, which probably contributes to the relatively low mortality rate (Veness et al, 2007). In high risk populations, such as immune-suppressed organ transplant recipients, cSCC is both 100-fold more frequent and shows increased mortality (Shamanin et al, 1996; Harwood et al, 2013). Cutaneous side effects, particularly cSCC are commonly observed in patients receiving a range of targeted cancer therapies demonstrating that perturbation of signaling pathways which drive cancer progression frequently disrupt homeostasis in the skin (Belum et al, 2013). Of note, patients receiving BRAF or broad spectrum kinase inhibitors frequently manifest squamoproliferative lesions and cSCC (Arnault et al, 2009). Previous reports have identified key driver mutations (known as Mut-driver genes) in TP53 (Brash et al, 1991; Pierceall et al, 1991a), NOTCH receptors (Durinck et al, 2011; Wang et al, 2012) and RAS (Pierceall et al, 1991b; Daya-Grosjean et al, 1993) in sporadic cSCC and more recently a significant contribution of RAS mutations to kinase inhibitor-driven cSCC (Su et al, 2012). In order to define and assess the contribution of Mut-driver genes to a large number of sporadic cSCC and then compare this with kinase inhibitor induced cSCC and squamoproliferative lesions we have used exome capture and Illumina technology to sequence 20 sporadic cSCC followed by a targeted PCR amplification approach coupled with Roche 454 sequencing in a further 151 samples; 91 sporadic cSCC, 21 cSCC cell lines and 39 cSCC and squamoproliferative lesions isolated from patients receiving the BRAF inhibitor vemurafenib. In addition we sequenced 10 normal skin samples, 4 using exome capture and 6 using PCR amplification.
Results
Cutaneous SCC harbors a massive burden of mutation compared with common malignant tumor types
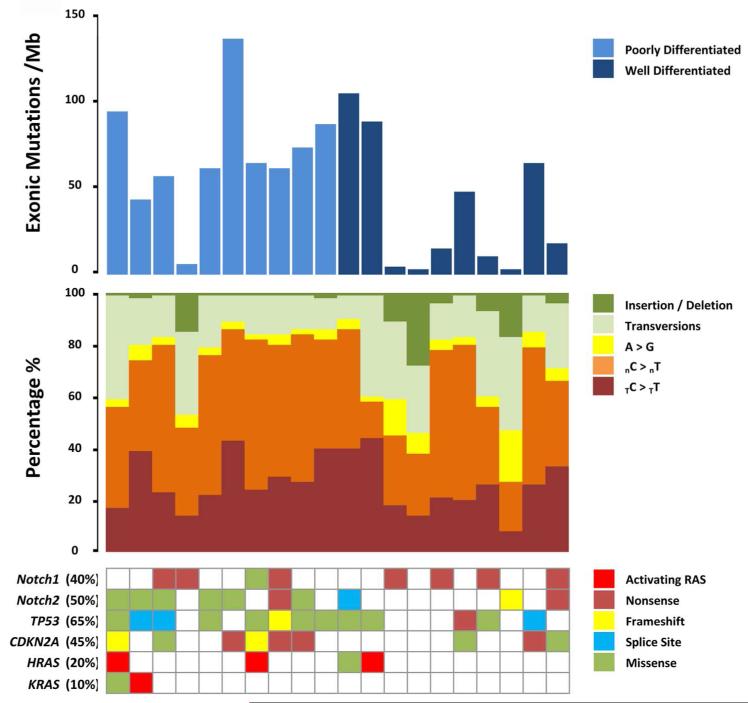
Whole exome sequencing of 20 cSCC stratified by histological grade along with matched germline DNA (Supplementary Data Table 1) targeted 351,845 exons from 21,117 genes. The mean sequencing coverage across targeted bases was 64X, with 69% above 30X coverage. We identified a total of 20,671 non-silent (non-synonymous) mutations (mean of 1,034 and a median of 1,195 per tumor) with a mean total exonic mutations per tumor of 2,283 corresponding to a mean somatic mutation rate of 50 mutations per mega-base pair (Mbp) of DNA (Figure 1, Supplementary Data Table 2). The prevalent nucleotide change was C>T, 68% of all somatic mutations, consistent with UV damage. Frequency of mutation in known drivers of cSCC was consistent with previous studies (Figure 1). The burden of mutation we describe here is significantly greater than any other common malignant tumor type, with the exception of basal cell carcinomas (5 fold greater than lung cancer (Hammerman et al, 2012) for example; (Supplementary Data Figure 1A) and comparable to tumors with microsatellite instability (MSI) characterized by hypermutation and disruption of genes encoding DNA excision repair proteins and/or the DNA polymerase subunit POLE (TCGAN, 2012; Kandoth et al, 2013). Unlike tumors with MSI, cSCC mutation burden showed no discernible segregation with mutations in DNA excision repair pathway genes or the DNA polymerase subunit POLE (Supplementary Data Figure 1B). Basal cell carcinoma of the skin, with limited metastatic potential, harbors a 1.5 fold higher mutation rate (Jayaraman et al, 2013) than the cSCC we describe here suggesting that in vivo, epidermal keratinocytes require multiple mutations for transformation to occur.
Figure 1. High mutation rate and spectra indicative of ultraviolet radiation damage in 20 sporadic cSCC samples.
Upper bar graph shows exonic mutations per megabase for each individual tumor. Middle bar graph shows the spectrum of individual tumor somatic mutations as a percentage. Lower matrix depicts mutations (indicated by a filled square) in each individual tumor in genes previously reported to be frequently mutated in cSCC. Where more than one mutation is present in a single gene only one mutation type is given in the order delineated by color coded key; activating-Ras, nonsense, frameshift, splice site, missense.
We identified non-synonymous mutations in 68 genes in 40% or more of the discovery exome cohort (Supplementary Data Figure 2A) of which a significant proportion (72%) had larger open reading frames than NOTCH1 (Supplementary Data Table 3) making identification of new Mut-driver genes difficult. Indeed a large proportion of these genes were recently identified as likely passenger mutations from a meta-analysis of tumor exome and whole genome sequencing data (Lawrence et al, 2013). In this study later replication during S phase was identified as a factor increasing mutation by chance and of the 68 highly mutated genes described here, 62% have later replication timing than NOTCH1 (Supplementary Data Table 3). In keeping with a high background mutation rate unsupervised hierarchical clustering failed to separate cSCC based on histological grade or mutation profiles (Supplementary Data Figure 2B).
NOTCH and RAS mutation segregate in sporadic cSCC and BRAF-inhibitor induced squamoproliferative lesions and cSCC
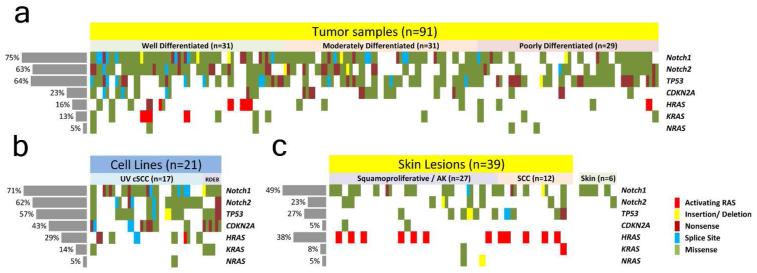
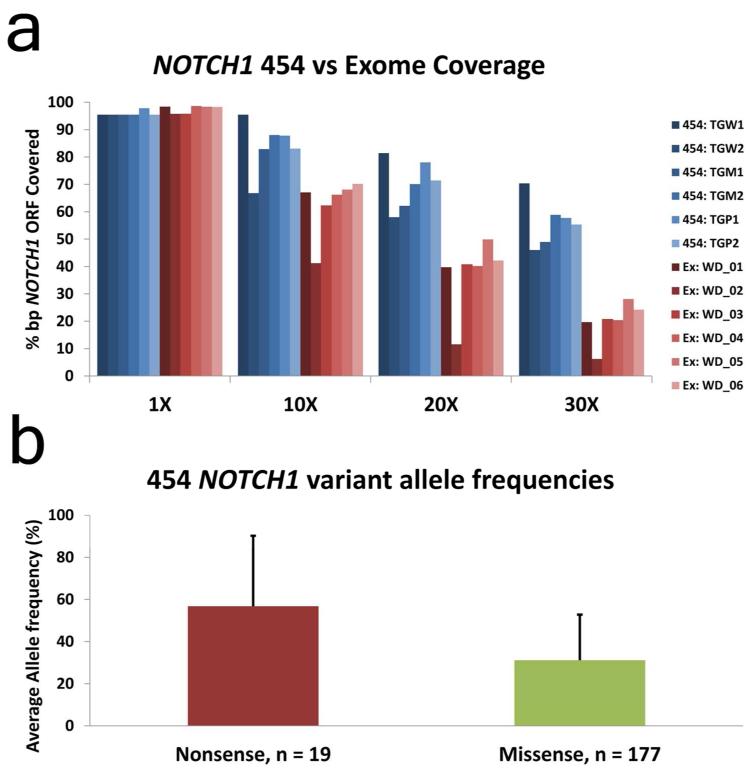
Next we pursued status of known, defined Mut-driver genes in a validation cohort of >100 sporadic cSCC samples (91 fresh frozen cSCC and 21 cell lines derived from cSCC) and 39 skin lesions arising in 7 patients receiving vemurafenib (Supplementary Data Table 4). We used 454 pyrosequencing and Fluidigm amplicon library preparation to re-sequence NOTCH1, NOTCH2, TP53, CDKN2A, HRAS, KRAS and NRAS (Figure 2A; Supplementary Data Tables 5-11). In total we sequenced 111 amplicons in 157 samples generating a mean sequencing coverage of 49X over 95 targeted exons. Using this approach we identified a similar mutation frequency in cSCC Mut-driver genes between the exome and 454 cohort of fresh frozen sporadic cSCC samples (n=91) with overall NOTCH1 and NOTCH2 combined mutation frequencies of 82%, TP53 63%, CDKN2A 28% and activating mutations in RAS at 11%. The increase in frequency of NOTCH1 mutation in the 454 data (75% compared with 45% in the exome study) is attributed to an overall low coverage for the exome capture sequencing of this gene (Figure 3A). This observation explains the higher proportion of non-sense mutations in NOTCH1 comparing exome capture to 454 sequencing as, interestingly, allelic frequency of non-sense mutations was nearly double that of missense mutation in the 454 cohort (Figure 3B). 21 primary cultures derived from cSCC tumors were also screened with the 454 platform and gave comparable mutation frequencies to tumor tissue with the exception of an increase in CDKN2A mutation, consistent with loss in culture (Loughran et al, 1997) (Figure 2B). Sequenced DNA of 39 squamoproliferative lesions or cSCC excised from 7 patients receiving the BRAF inhibitor vemurafenib showed a significant difference in overall mutation frequency (p<0.05) with the exception of KRAS and NRAS. Activating HRAS mutations, known to be frequent in such lesions (Su et al, 2012) were increased (38% versus 5%) while NOTCH1 mutations, although less frequent than sporadic cSCC (49% versus 75%), were greater still (49% NOTCH1 versus 38% HRAS) suggesting NOTCH1 mutation is required for vemurafenib driven keratinocyte clonal expansion (Figure 2C). Of the 16 samples harboring activating RAS mutations, 56% (9) also harbored a mutation in NOTCH1 or NOTCH2. In the sporadic cSCC cohort this figure was 100% (8/8). 97% of variants detected by exome capture and Illumina sequencing in the discovery cohort of 20 cSCC were confirmed using 454 pyrosequencing (data not shown).
Figure 2. 454 resequencing identifies prevalent mutation NOTCH1, NOTCH2 and TP53 in cSCC.
Upper Matrix depicts mutations in the given gene for each individual tumor sample. Where more than one mutation is present in a single gene two mutations are shown in the order delineated by key; activating RAS, frameshift, nonsense, splice site, missense. Lower left matrix details mutations in 21 keratinocyte cSCC cell lines and lower right matrix details mutations in 39 skin lesions excised from 7 patients receiving the BRAF inhibitor vemurafenib. Mutations identified in normal skin DNA isolated from 6 of these 7 patients are shown to the right of this matrix.
Figure 3. NOTCH1 coverage is lower in exome capture compared with PCR amplification and 454 sequencing.
A: Percentage of the NOTCH1 gene represented by 1X, 10X, 20X and 30X coverage in the first two samples sequenced using PCR amplification and 454 pyro-sequencing (454: TGM1 and 454: TGM2) compared with exome capture (Ex: WD_01 and Ex: WD_02).
B: The mean allelic frequency of non-sense and missense mutations identified in NOTCH1 from 91 cSCC fresh frozen samples. Error shows standard deviation. Student T-Test p-value = 7e-06.
NOTCH receptor and activating HRAS mutation identified in normal skin
Prevalent NOTCH and HRAS mutation identified in skin lesions rapidly promoted by BRAF inhibition suggests selection and expansion of pre-existing clonal populations harboring such mutation. Certainly experimental evidence suggesting that BRAF inhibition accelerates growth of cells harboring activating RAS mutation supports this notion (Heidorn et al, 2010; Poulikakos et al, 2010). We analysed NOTCH, TP53, CDKN2A and RAS mutation status in 10 normal skin samples. Four were used as control DNA for the exome sequencing as peripheral blood was not available for these patients (Materials and Methods; Supplementary Data Table 1) and 6 samples were obtained from patients receiving vemurafenib and were analysed by 454 sequencing (Supplementary Data Table 4).
In the exome sequenced samples we observed a HRAS G12D mutation and a single base pair deletion causing a frameshift in NOTCH1 in normal skin from patient PD_05, both of which met criteria used to call somatic mutations in the tumor samples (Materials and Methods). This normal skin sample was isolated from a site distant to the tumor (Supplementary Data Table 1) and neither of these mutations were present in the tumor sample or were found in public databases (Materials and Methods) or, in the case of the NOTCH1 deletion, in any other samples sequenced during this study. In the absence of peripheral blood or any other source of germ line DNA we cannot completely rule out that these are somatic mutations, however, we find the possibility of an activating HRAS or loss of function NOTCH1 present in the germline unlikely. Further interrogation of NOTCH, TP53, CDKN2A and RAS genes in the exome skin samples using IGV (Thorvaldsdottir et al, 2012) revealed two non-sense NOTCH1 mutations in PD_02 and PD_05 samples (E1270* and E124*) at frequencies or coverage below the detection criteria (2 from 10 reads and 5 from 12 reads respectively). A number of low frequency variants leading to activating mutations in all three RAS genes were observed across all four normal skin samples analysed with exome sequencing but only HRAS G12V in PD_08 was present in more than a single read albeit at a very low frequency (2 from 83 reads).
454 sequencing of 6 normal skin samples from patients receiving vemurafenib identified 5 NOTCH1 mutations and one NOTCH2 mutation which met the 454 detection criteria (Supplementary Data Tables 8 and 9; Figure 2C). Of these, a single NOTCH1 mutation was present at low allelic frequency in the skin adjacent to an SCC which harbored the identical mutation at considerably higher allelic frequency (14% vs 37%, Supplementary Data Table 8). Although an additional source of germ line DNA was not available to confirm whether these mutations were somatic or not, the low allelic frequency in normal skin and the fact that none of these mutations, other than the one found in the adjacent SCC were present in any of the other samples from the same patient strongly suggests that they are indeed somatic.
Vemurafenib induced squamoproliferative lesions and cSCC reveal surprising heterogeneity and independent activating HRAS mutations within the same lesion
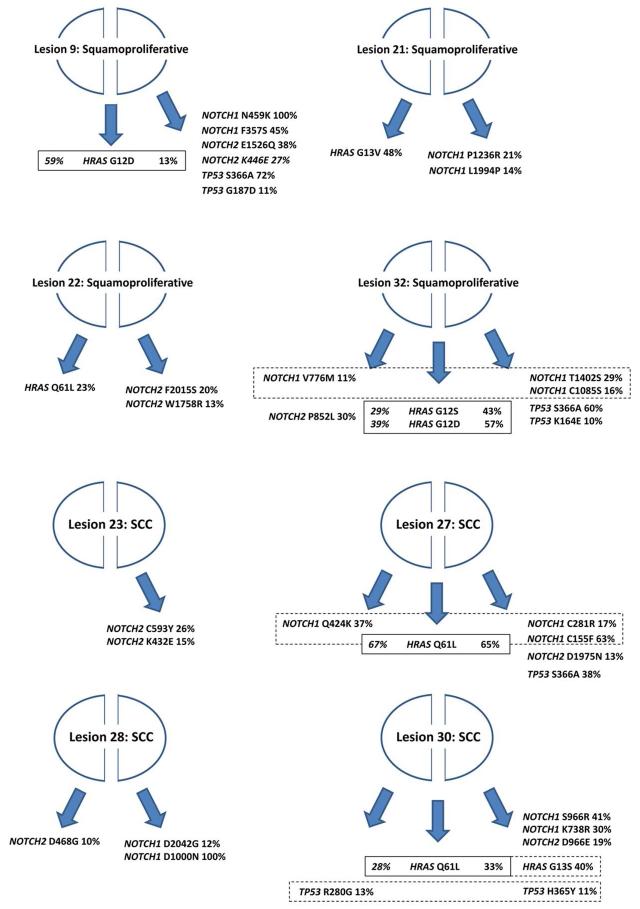
In order to assess heterogeneity of skin lesions arising in vemurafenib treated patients we isolated DNA from two separate halves of 8 samples and performed 454 sequencing as before. In some instances coverage at RAS loci was low and so we reduced the detection threshold for activating RAS mutations to 2 or more reads over 10% of total reads. Bisected vemurafenib induced lesions revealed surprising heterogeneity, with the same mutation being identified in only 4 of the 8 separate lesion halves. All shared mutations were in HRAS with no shared mutations observed in NOTCH1 or TP53. In one of the 8 samples different activating HRAS mutations were identified in the two separate halves of the same cSCC (Figure 4 and Supplementary Data Table 12).
Figure 4. Bisected vemurafenib induced squamoproliferative lesions and cSCC display significant heterogeneity.
Cartoons depict 8 bisected vemurafenib induced lesions and the resulting mutations identified in NOTCH1, NOTCH2, HRAS and TP53 from the isolated DNA. Solid lines enclose mutations in HRAS shared by both halves of the sample whilst dashed lines enclose different mutations in the same gene identified in the two halves of the sample. % indicates allelic frequency.
Activated NOTCH1 detected by immunohistochemistry correlates with mutation status in cSCC and readily identifies regions of normal skin negative for NOTCH1 signaling
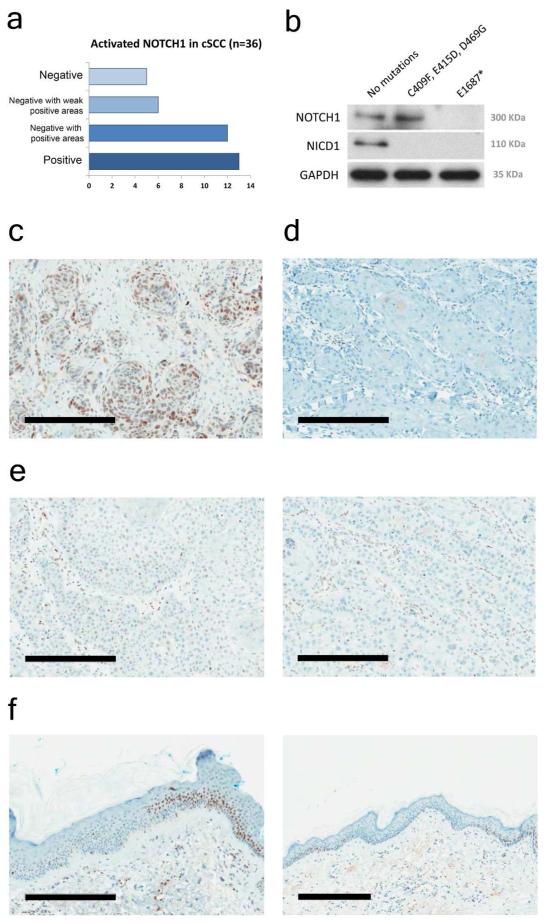
Next we utilized a recently developed immunohistochemical (IHC) stain using an antibody which detects a neoepitope created by the proteolytic cleavage event that activates NOTCH1 (Kluk et al, 2013). In total we analyzed 56 FFPE blocks from 36 cSCC samples for which we had mutation analysis (summarized in Figure 5A). Immunoblotting total cell lysates isolated from cSCC cell lines with defined mutations demonstrated the specificity of the antibody (Figure 5B). We observed positive nuclear NOTCH1 immuno-reactivity in all 6 cSCC samples where no mutations were identified (Figure 5C). In the only sample where non-sense mutation was identified at an allelic frequency of 100% we saw no IHC staining (Figure 5D). In cSCC samples where mutations in NOTCH1 were at heterozygous or subclonal allelic frequencies we frequently observed regions of weak staining (Figure 5E, left panel) as well regions of absent staining (Figure 5E, right panel). Finally we analyzed normal looking skin at the periphery of cSCC and readily detected patches with no or very weak IHC staining flanked by normal looking IHC staining (Figure 5F).
Figure 5. NOTCH1 Western blotting and immunohistochemistry correlates with mutation status and identifies regions of normal looking skin negative for NOTCH1 signaling.
Standard 4-micron paraffin embedded tissue sections were stained using the Ventana Benchmark XT platform (Ventana Medical Systems, Inc., Tucson, AZ.) with extended heat induced epitope retrieval (CC1 Buffer). Slides were incubated for 1hr at room temperature with anti-NICD1 rabbit monoclonal antibody (clone D3B8, catalog #4147, Cell Signaling Technology, Beverly, MA; final concentration, 8.5microgram/mL). Signals were then amplified (Ventana Amplification Kit, #760-080) and visualized (Ventana Ultraview Universal DAB detection kit, #760-500) per the manufacturer’s instructions. A: Summary of immune-reactivity of 36 cSCC samples with anti-NICD1 antibody. B: Western blotting with 50μg of total cell lysate from SCCIC12 (no mutations), SCCT2 (missense, loss-of-function mutations) and SCCIC8 (homozygous PTC mutation) were resolved by 4-12% SDS-PAGE using standard techniques. The proteins were then transferred to a nitrocellulose membrane and visualized using antibodies recognizing full length NOTCH1, NICD1 and GAPDH. C: Immunohistochemistry data from sample TGP8, no NOTCH1 mutations. D: Sample TGW29, 100% STOP mutation. E: Sample TGM4, P391S 55%, G4247D 22%, P1860L 37% mutations. Left panel shows weakly positive region of the tumor while right panel shows negative region of the same tumor. F: Normal looking peri-tumor skin showing regions negative for NOTCH1 signaling adjacent to regions displaying the expected normal distribution of activated NOTCH1. Right panel shows extent of NOTCH1 negative patch flanked by positive NOTCH1 skin. All bars C-F = 300μm.
Discussion
Here we present exome level sequencing of a discovery set of 20 cSCC and validation by targeted deep sequencing of NOTCH1, NOTCH2, TP53, CDKN2A and RAS in over 150 cSCC and squamoproliferative lesions as well as 10 matched normal skin samples. Our initial discovery series identifies a massive burden of mutation far exceeding that of any other commonly registered cancer with metastatic potential sequenced to date; 5 fold greater than lung cancer, and an order of magnitude greater than breast (Hammerman et al, 2012; Stephens et al, 2012). The mutation rate of 50 per Mbp is in line with an earlier study of 8 cSCC which identified a mean mutation rate of 33 per Mbp (Durinck et al, 2011). Even compared to a subset of colorectal cancers with MSI (TCGAN, 2012), cSCC tumors harbor a greater median number of non-synonymous mutations (Supplementary Data Figure 1A). MSI tumors are associated with a high base pair substitution rate, low copy number variation and low TP53 mutation (Boland et al, 1998; Soreide et al, 2006). Mutation and epigenetic silencing of DNA excision repair pathway genes and hotspot mutations in the DNA polymerase subunit POLE have recently been shown to be prevalent in highly mutated colorectal and endometrial carcinomas (TCGAN, 2012; Kandoth et al, 2013). In contrast, cSCC exhibit high copy number variation (Ashton et al, 2005; Purdie et al, 2009), TP53 mutation (Dumaz et al, 1993) and low frequency of DNA repair/ polymerase mutation (Supplementary Data Figure 2B and (Perret et al, 2010)). Although we have not explored the possibility that methylation acts as a mechanism of silencing the expression of the mismatch repair (MMR) genes MLH1 or MSH2, (commonly identified in MSI (Kandoth et al, 2013)), a recent study of 86 cSCC identified protein expression in all tumors indicating competent MMR (Perret et al, 2010). The high rate of C>T transitions (68%) confirms UV radiation as the primary cause of cSCC mutation (Brash et al, 1991). Our data identifying TP53 mutation in all but one of the heavily mutated tumors (Figure 1A) would agree with a loss of wild type TP53 preceding massive expansion of mutation as reported previously (Durinck et al, 2011). The high mutation rate coupled with a similar burden identified in the most common tumor of the skin, basal cell carcinoma (Jayaraman et al, 2013), suggests that either epidermal keratinocytes are inherently resistant to transformation or that skin acts as a potent tumor suppressor keeping clonal expansion of tumor initiating cells at bay. Previous studies identifying only one or two oncogenes required for efficient transformation of primary human keratinocytes (Halbert et al, 1991; Lazarov et al, 2002) would argue the latter and recent data implicating an altered dermal microenvironment driving cSCC arising in patients with the genetic blistering disease Recessive Dystrophic Epidermolysis Bullosa would also agree with this line of reasoning (Ng et al, 2012). In addition to these two independent observations the fact that perturbation of major signaling pathways, such as MAPK, leads to rapid clonal expansion of mutated keratinocytes in the skin would also point to the tumor suppressive properties of normal homeostatic human skin.
Our study confirms NOTCH receptor mutation as a major tumor suppressor mechanism in cSCC (Wang et al, 2012), giving an overall frequency in 112 tumor samples of 82% (in 91 fresh frozen cSCC and 21 cell lines analyzed by 454 sequencing), and identifies NOTCH1 as a tumor suppressor important for HRAS driven skin carcinogenesis, being the only other Mut-driver gene identified in this study as being highly mutated in lesions arising in patients taking the BRAF inhibitor vemurafenib (Figure 2). It is important to note that we cannot rule out other mutation events in genes not included in our targeted sequencing and that a comprehensive analysis through exome or whole genome sequencing would be of great value in understanding the mutation landscape of these lesions. Interestingly, activating RAS mutations co-segregated with NOTCH1 mutation in the 454 cohort of 112 cSCC (Figure 2). Overall activated RAS mutation frequency in this cohort was 11% in agreement with recent studies in similarly sized sample sets (Oberholzer et al, 2012). Loss of NOTCH signaling in concert with recombinant oncogenic RAS has previously been shown to promote tumor development of normal human primary keratinocytes inoculated into immune compromised mice (Lefort et al, 2007). Here we provide convincing evidence that this indeed is evident in vivo.
Field cancerization is a concept introduced by Slaughter and colleagues in 1953 to describe the observation that multiple primary SCC arise within close proximity suggestive of a predisposing area of tissue, or field (Slaughter et al, 1953). Molecular evidence to support this notion followed in the late 1990s primarily concentrating on TP53 mutation (Franklin et al, 1997) but also describing unique epigenetic phenomena (Suzuki et al, 2004). Certainly in the skin field cancerization characterized by TP53 mutation has been well described (Jonason et al, 1996; Stahl et al, 2010). In our study we were unable to find any evidence of TP53 mutation in the 10 normal skin samples but readily identified NOTCH receptor mutation (70%) of which a proportion (2/7) were frameshift or PTC mutations. In support of this molecular data, IHC analysis revealed regions of normal looking skin with loss or severely reduced NOTCH1 signaling (Figure 5F). Recent data show that in murine models inhibition of NOTCH signaling in the dermal compartment can induce spontaneous carcinogenesis (Hu et al, 2012) and that mosaic loss of NOTCH1 in the epidermal compartment can induce spontaneous tumors competent in NOTCH1 signaling (Demehri et al, 2009). Collectively and considering the high frequency of NOTCH receptor mutation identified here we would consider NOTCH mutation as an early event in skin carcinogenesis similar to APC loss in the colon (Powell et al, 1992).
Lastly, although our exome sequencing failed to identify further definitive Mut-driver genes in cSCC (largely due to the heavily mutated nature of the tumor and the small sample size in the initial exome capture cohort) we note that within the 68 genes mutated in 40% or more samples FAT1 and CACNA1C were second only to NOTCH1 in the number of tumors harboring non-sense mutations (5 and 6 compared with 7). FAT1 was recently identified in a transposon based tumor suppressor screen in cSCC (Quintana et al, 2013) and as a novel Mut-driver gene in HNSCC (Morris et al, 2013; Pickering et al, 2013).
In conclusion, we present a comprehensive mutation analysis of NOTCH1, NOTCH2, TP53, CDKN2A and RAS in over 170 cSCC, squamoproliferative lesions and normal skin samples demonstrating that NOTCH receptors are significantly mutated; 82% of sporadic cSCC (n=112); 49% of squamoproliferative lesions arising in patients receiving vemurafenib (n=39) and 70% of normal skin samples (n=10, 4 peri-lesional and 6 separate from lesion). Although clearly a larger study is required, we do show in one instance that normal skin adjacent to cSCC can harbor the same NOTCH1 mutation at significantly lower allelic frequency suggesting that field effect can be driven by NOTCH mutation in a clinical setting. We also demonstrate oncogenic HRAS mutations are present in normal human skin, again albeit in a single sample. Overall these data attest to the tumor suppressive qualities of normal human skin and that perturbation of major signaling pathways in this tissue can lead to rapid expansion of pre-existing tumor initiating clonal populations, highlighting NOTCH1 mutation as a gatekeeper event in squamous carcinogenesis of the skin.
Materials & Methods
Samples
Ethical approval for this investigation was obtained from the East London and City Health Authority local ethics committee and the study was conducted according to the Declaration of Helsinki Principles. All patients participating in this study provided written, informed consent. Punch biopsies of cSCC tissue were taken after surgical excision of tumors and immediately snap-frozen in liquid nitrogen with the remainder of the tumor sent for formalin fixation and histopathologic diagnosis. In order to enrich for tumor cell populations, fresh frozen cSCC biopsies were laser capture microdissected using the Zeiss Palm Microbeam microscope (Zeiss, UK). Depending on sample size and tumor purity as estimated from a reference H&E slide, between 30-60 sections of 8 μm thickness were cut onto 1.0 mm PEN membrane slides (Zeiss, UK), stained in 0.05% acid fuchsin (Acros Organics, NJ) in distilled water and 0.05% toluidine blue O (Acros Organics, NJ) in 70% ethanol and microdissected, with tumor cells collected into 180 μl ATL buffer (Qiagen, Crawley, UK). DNA extraction was performed using the QIAamp DNA micro kit (Qiagen, Crawley, UK) according to manufacturer’s instructions. To provide a source of germline DNA, paired venous blood samples or normal skin were obtained concomitantly with cSCC tissue and stored at −80 prior to DNA extraction using the protocol above in the case of normal skin or the QIAamp DNA blood mini kit (Qiagen, Crawley, UK). Squamoproliferative lesions included in this series were all benign keratinocyte hyperproliferative lesions with or without features suggestive of viral infection but with no evidence of dysplasia or invasive malignancy.
Sequencing, mapping and variant analysis
Detailed methods are provided in the supplementary data. Briefly, exome capture and sequencing was provided by the EASIH - University of Cambridge supported by NIHR - Cambridge BRC using Agilent SureSelect Human All Exon 50 Mb according to the manufacturers’ instructions, coupled with Illumina HiSeq technology. Exome capture variant detection required a minimum read depth at a position to make a call of 10, a minimum of 4 supporting reads and a minimum variant allele frequency threshold of 0.2. 454 pyrosequencing was performed using the GS Junior system (Roche/454 Life Sciences) and Fluidigm (Fluidigm Corporation, San Francisco, CA) PCR amplicon libraries as template according to recommended guidelines (Roche, Mannheim, Germany). 454 variant detection required a minimum of 4 supporting reads and a minimum variant allele frequency threshold of 0.1.
Statistics
Chi-Square test was used where permitted (NOTCH1, NOTCH2, TP53, and HRAS) and Fischer’s exact test was used where numbers were too low (CDKN2A, KRAS, and NRAS) to determine whether mutation frequencies between sporadic cSCC (n=91) and vemurafenib induced lesions (n=39) significantly differed.
Supplementary Material
Acknowledgements
This work was funded by Cancer Research UK and The British Skin Foundation. I.M.L is funded by the European Research Council and a strategic grant from the Wellcome Trust. We would like to thank Susan Bray and Tayside Tissue Bank for assistance with IHC.
Abbreviations used
- (SCC)
squamous cell carcinoma
- (cSCC)
cutaneous squamous cell carcinoma
- (MSI)
microsatellite instability
- (IHC)
immunohistochemical
Footnotes
Conflict of interest The authors state no conflict of interest.
References
- Agrawal N, et al. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov. 2012;2:899–905. doi: 10.1158/2159-8290.CD-12-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnault JP, Wechsler J, Escudier B, et al. Keratoacanthomas and squamous cell carcinomas in patients receiving sorafenib. J. Clin. Oncol. 2009;27:e59–61. doi: 10.1200/JCO.2009.23.4823. [DOI] [PubMed] [Google Scholar]
- Ashton KJ, Carless MA, Griffiths LR. Cytogenetic alterations in nonmelanoma skin cancer: a review. Genes Chromosomes Cancer. 2005;43:239–48. doi: 10.1002/gcc.20183. [DOI] [PubMed] [Google Scholar]
- Belum VR, Fontanilla Patel H, Lacouture ME, Rodeck U. Skin toxicity of targeted cancer agents: mechanisms and intervention. Future Oncol. 2013;9:1161–70. doi: 10.2217/fon.13.62. [DOI] [PubMed] [Google Scholar]
- Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–57. [PubMed] [Google Scholar]
- Brash DE, Rudolph JA, Simon JA, et al. A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc Natl Acad Sci U S A. 1991;88:10124–8. doi: 10.1073/pnas.88.22.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherpelis BS, Marcusen C, Lang PG. Prognostic factors for metastasis in squamous cell carcinoma of the skin. Dermatol Surg. 2002;28:268–73. doi: 10.1046/j.1524-4725.2002.01169.x. [DOI] [PubMed] [Google Scholar]
- Daya-Grosjean L, Robert C, Drougard C, Suarez H, Sarasin A. High mutation frequency in ras genes of skin tumors isolated from DNA repair deficient xeroderma pigmentosum patients. Cancer Res. 1993;53:1625–9. [PubMed] [Google Scholar]
- Demehri S, Turkoz A, Kopan R. Epidermal Notch1 loss promotes skin tumorigenesis by impacting the stromal microenvironment. Cancer Cell. 2009;16:55–66. doi: 10.1016/j.ccr.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumaz N, Drougard C, Sarasin A, Daya-Grosjean L. Specific UV-induced mutation spectrum in the p53 gene of skin tumors from DNA-repair-deficient xeroderma pigmentosum patients. Proc Natl Acad Sci U S A. 1993;90:10529–33. doi: 10.1073/pnas.90.22.10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, et al. Temporal dissection of tumorigenesis in primary cancers. Cancer Discov. 2011;1:137–43. doi: 10.1158/2159-8290.CD-11-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin WA, et al. Widely dispersed p53 mutation in respiratory epithelium. A novel mechanism for field carcinogenesis. J Clin Invest. 1997;100:2133–2137. doi: 10.1172/JCI119748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbert CL, Demers GW, Galloway DA. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J Virol. 1991;65:473–8. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerman P, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood CA, et al. A surveillance model for skin cancer in organ transplant recipients: a 22-year prospective study in an ethnically diverse population. Am J Transplant. 2013;13:119–29. doi: 10.1111/j.1600-6143.2012.04292.x. [DOI] [PubMed] [Google Scholar]
- Heidorn SJ, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–21. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, et al. Multifocal epithelial tumors and field cancerization from loss of mesenchymal CSL signaling. Cell. 2012;149:1207–20. doi: 10.1016/j.cell.2012.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman SS, Rayhan DJ, Hazany S, Kolodney MS. Mutational Landscape of Basal Cell Carcinomas by Whole-Exome Sequencing. J Invest Dermatol. 2013 doi: 10.1038/jid.2013.276. doi: 10.1038/jid.2013.276. [DOI] [PubMed] [Google Scholar]
- Jonason AS, et al. Frequent clones of p53-mutated keratinocytes in normal human skin. Proc Natl Acad Sci U S A. 1996;93:14025–9. doi: 10.1073/pnas.93.24.14025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karia PS, Han J, Schmults CD. Cutaneous squamous cell carcinoma: estimated incidence of disease, nodal metastasis, and deaths from disease in the United States, 2012. J Am Acad Dermatol. 2013;68:957–66. doi: 10.1016/j.jaad.2012.11.037. [DOI] [PubMed] [Google Scholar]
- Kluk MJ, Ashworth T, Wang H, et al. Gauging NOTCH1 Activation in Cancer Using Immunohistochemistry. PLOS One. 2013;8:e67306. doi: 10.1371/journal.pone.0067306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwa RE, Campana K, Moy RL. Biology of cutaneous squamous cell carcinoma. J Am Acad Dermatol. 1992;26:1–26. doi: 10.1016/0190-9622(92)70001-v. [DOI] [PubMed] [Google Scholar]
- Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarov M, et al. CDK4 coexpression with Ras generates malignant human epidermal tumorigenesis. Nat Med. 2002;8:1105–14. doi: 10.1038/nm779. [DOI] [PubMed] [Google Scholar]
- Lefort K, et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev. 2007;21:562–77. doi: 10.1101/gad.1484707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran O, et al. Evidence for the inactivation of multiple replicative lifespan genes in immortal human squamous cell carcinoma keratinocytes. Oncogene. 1997;14:1955–64. doi: 10.1038/sj.onc.1201028. [DOI] [PubMed] [Google Scholar]
- Morris LG, et al. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat Genet. 2013;45:253–61. doi: 10.1038/ng.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng YZ, et al. Fibroblast-derived dermal matrix drives development of aggressive cutaneous squamous cell carcinoma in patients with recessive dystrophic epidermolysis bullosa. Cancer Res. 2012;72:3522–34. doi: 10.1158/0008-5472.CAN-11-2996. [DOI] [PubMed] [Google Scholar]
- Oberholzer PA, et al. RAS mutations are associated with the development of cutaneous squamous cell tumors in patients treated with RAF inhibitors. J Clin Oncol. 2012;30:316–21. doi: 10.1200/JCO.2011.36.7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrett CM, et al. Expression of DNA mismatch repair proteins and MSH2 polymorphisms in nonmelanoma skin cancers of organ transplant recipients. Br J Dermatol. 2010;162:732–42. doi: 10.1111/j.1365-2133.2009.09550.x. [DOI] [PubMed] [Google Scholar]
- Pickering CR, et al. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov. 2013;3:770–81. doi: 10.1158/2159-8290.CD-12-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierceall WE, Mukhopadhyay T, Goldberg LH, Ananthaswamy HN. Mutations in the p53 tumor suppressor gene in human cutaneous squamous cell carcinomas. Mol Carcinog. 1991a;4:445–9. doi: 10.1002/mc.2940040606. [DOI] [PubMed] [Google Scholar]
- Pierceall WE, Goldberg LH, Tainsky MA, Mukhopadhyay T, Ananthaswamy HN. Ras gene mutation and amplification in human nonmelanoma skin cancers. Mol Carcinog. 1991b;4:196–202. doi: 10.1002/mc.2940040306. [DOI] [PubMed] [Google Scholar]
- Poulikakos PI, et al. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–30. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell SM, et al. APC mutations occur early during colorectal tumorigenesis. Nature. 1992;359:235–7. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- Purdie KJ, et al. Single nucleotide polymorphism array analysis defines a specific genetic fingerprint for well-differentiated cutaneous SCCs. J Invest Dermatol. 2009;129:1562–8. doi: 10.1038/jid.2008.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana RM, et al. A transposon-based analysis of gene mutations related to skin cancer development. J Invest Dermatol. 2013;133:239–48. doi: 10.1038/jid.2012.245. [DOI] [PubMed] [Google Scholar]
- Shamanin V, et al. Human papillomavirus infections in nonmelanoma skin cancers from renal transplant recipients and nonimmunosuppressed patients. J Natl Cancer Inst. 1996;88:802–11. doi: 10.1093/jnci/88.12.802. [DOI] [PubMed] [Google Scholar]
- Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963–8. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Soreide K, Janssen EA, Soiland H, Korner H, Baak JP. Microsatellite instability in colorectal cancer. Br J Surg. 2006;93:395–406. doi: 10.1002/bjs.5328. [DOI] [PubMed] [Google Scholar]
- Ståhl PL, et al. Sun-induced nonsynonymous p53 mutations are extensively accumulated and tolerated in normal appearing human skin. J Invest Dermatol. 2010;131:504–8. doi: 10.1038/jid.2010.302. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–4. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su F, et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N Engl J Med. 2012;366:207–15. doi: 10.1056/NEJMoa1105358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- TCGAN Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2012;14:178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veness MJ, Porceddu S, Palme CE, Morgan GJ. Cutaneous head and neck squamous cell carcinoma metastatic to parotid and cervical lymph nodes. Head Neck. 2007;29:621–31. doi: 10.1002/hed.20576. [DOI] [PubMed] [Google Scholar]
- Wang NJ, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci U S A. 2012;108:17761–6. doi: 10.1073/pnas.1114669108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.