SUMMARY
Many functionally important interactions between genes and proteins involved in immunological diseases and processes are unknown. The exponential growth in public high-throughput data offers an opportunity to expand this knowledge. To unlock human-immunology-relevant insight contained in the global biomedical research effort, including all public high-throughput datasets, we performed immunological-pathway-focused Bayesian integration of a comprehensive, heterogeneous compendium comprising 38,088 genome-scale experiments. The distillation of this knowledge into immunological networks of functional relationships between molecular entities (ImmuNet), and tools to mine this resource, are accessible to the public at http://immunet.princeton.edu. The predictive capacity of ImmuNet, established by rigorous statistical validation, is easily accessed by experimentalists to generate data-driven hypotheses. We demonstrate the power of this approach through the identification of unique host-virus interaction responses, and we show how ImmuNet complements genetic studies by predicting disease-associated genes. ImmuNet should be widely beneficial for investigating the mechanisms of the human immune system and immunological diseases.
Graphical Abstract

INTRODUCTION
Many advances in immunology involve the identification of the functional roles of specific molecular entities (genes or proteins) in immunological diseases or immune pathways. These diseases and pathways emerge from a complex network of relationships among molecular entities. Numerous immunological disorders, for example inflammatory bowel disease (IBD), are now recognized to involve multiple genes and processes in the manifestation of the disease (Jostins et al., 2012). In addition, specific disease-associated genes can contribute to distinct phenotypes for different immunological diseases, suggesting the importance of context-specific functionalization of these genes (Hebbring, 2014). Consequently, improving systems-level knowledge of each immune process should further the understanding of the molecular basis for many immunological disorders. Although immunological research focusing on individual entities has been invaluable, the underlying genome-scale network of relationships remains largely unexplored. Mapping the functional association network of genes and proteins in the context of immunological processes is an important challenge for human immunology research.
In recent years, the public availability of high-throughput experimental data has grown exponentially (Bhattacharya et al., 2014; Brazma et al., 2003; Edgar et al., 2002; Heng et al., 2008; Stark et al., 2006). High-throughput experimental assays of gene expression, protein physical interaction, and localization have allowed researchers to measure cellular activity across multiple conditions, tissues, and diverse genetic backgrounds. In aggregate, these data hold critical insights that extend beyond the questions addressed in the experiments used to generate each individual dataset. For example, although cancer datasets contain rich information relevant to processes involved in immune surveillance (Dunn et al., 2004), there has been no practical way to mine them for immunological insight. Immunology research would benefit by more efficient extraction of and access to the information relevant to immunological diseases and pathways that is hidden within the global public data output and that can be resolved through simultaneous analysis of these thousands of diverse experiments.
One obstacle to harnessing the potential insight contained in the global research output for specific immunological questions is the difficulty of detecting relevant information from a large body of often conflicting data obtained from diverse experiments and assays. The complexity, heterogeneity, and variation in quality of high-throughput assays necessitate an approach that takes these factors into consideration. Integration and interpretation of this massive collection of datasets can be addressed by refined Bayesian approaches and rigorous, well-established statistical validation methods (Geisser, 1993; Hastie et al., 2011). These have proven uniquely suitable for extracting relevant information from heterogeneous sources, while being robust to noise (Alexeyenko and Sonnhammer, 2009; Huttenhower et al., 2009; Lee et al., 2011; Mostafavi et al., 2008; Snel et al., 2000; Troyanskaya et al., 2003).
The basic approach of Bayesian integration is to first select known information of the type one is trying to expand, such as a well-annotated specific immunological pathway. Then, each dataset in a large data compendium is evaluated for how well it can be used to reconstruct the targeted pathway. Based on this calculated accuracy and implied relevance, each dataset contributes to new predictions of the likelihood of functional relationships between molecular entities pertinent to the pathway or process of interest. There are two major advantages of this approach over other methods (e.g., rank aggregation [Kolde et al., 2012] or co-expression linkage [Lee et al., 2004; Stuart et al., 2003]) for summarizing data. First, individual datasets that contain no useful information due to their lack of relevance to the targeted pathway or their quality are statistically excluded. Second, the approach considers diverse types of measurements, ranging from global RNA expression to protein interaction studies, and rigorously selects the datasets that, in aggregate, best provide new insight about the pathway of interest. Although integrative approaches have begun to be applied to human biology (Hoffman et al., 2012; Huttenhower et al., 2009; Lee et al., 2011; Mostafavi et al., 2008; Park et al., 2015; Taşan et al., 2012; Troyanskaya et al., 2003), they have not been tailored to the context of the immune system and immune processes. Such context-specific integration is necessary to improve the relevance and accuracy of insight that can be obtained (Huttenhower et al., 2009). Toward this end, we transformed public datasets from 38,088 experiments, including genome-scale expression, physical interaction, and sequence studies, into an integrated map of immunological relationships among molecular entities. The resulting comprehensive web-accessible resource (ImmuNet) facilitates researchers’ use of the global data output to generate testable hypotheses for specific immunological research areas.
Through rigorous statistical assessments and real immunological applications, we show ImmuNet’s advantages over non-immune-specific networks or physical interaction databased networks alone. We further demonstrate ImmuNet’s utility in addressing immunological questions by providing insight into gene signatures associated with virus infection. Finally, we demonstrate the value and ease of use of ImmuNet to complement genetic studies for identifying disease-associated genes.
RESULTS
Development and Computational Validation of ImmuNet
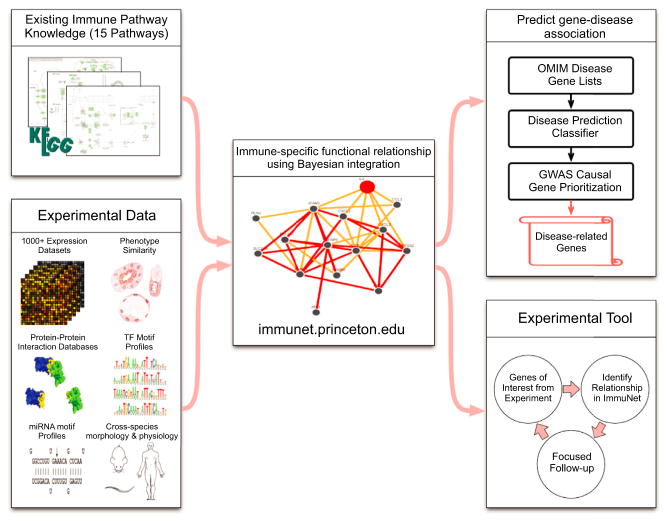
To develop the data-driven views of the entire immune landscape, we built genome-scale networks, each focused on an individual pathway of central importance to innate or adaptive immunity. For each of these pathway-based networks, a probabilistic Bayesian approach was used to determine a quantitative measure of the relevance of each of 1,013 datasets (comprising 38,088 genome-scale experiments). This approach entails selecting a public repository of well-annotated known relationships between molecular entities relevant for immunology that is then used as a training set for constructing the network of functional relationships. Bayesian integration assesses the degree to which each individual dataset in the compendium contains evidence for relationships within the training set and computes a corresponding weight for each dataset in constructing a network for making novel relevant inferences. The training sets selected for ImmuNet were the 15 expert-curated Kyoto Encyclopedia of Genes and Genomes (KEGG) immune-related pathways (Ogata et al., 1999). The selection of KEGG pathways for training Immu-Net was based on the superiority of the integrated networks obtained. As described in the Computational Methods, evaluation of the predictive capacity of networks based on two other publically available annotation and pathway repositories, the immune annotations in Gene Ontology (GO) (Harris et al., 2004) and the immune pathways in Reactome (Croft et al., 2011), indicated that networks using KEGG immunological pathways for training performed better than those using the alternative training sets. Full details of the method and its implementation can be found in the Computational Methods and in previous publications (Hibbs et al., 2007; Huttenhower et al., 2006, 2009).
This approach thus provides new information relevant to the relationships in each immunological pathway based on how well the input datasets recapitulate known relationships within the pathway. The method uses the quantification relevant for various types of datasets (e.g., high throughput binding evidence for physical protein associations, normalized correlation between each pair of genes in every microarray dataset, etc.; see the Supplemental Computational Methods) in assessing each dataset’s relevance. For example, correlated expression of beta chain of MHC class I molecules (B2M) and peptide transporter involved in antigen presentation (TAP1) in a microarray dataset will increase the weight of that dataset in generating the “antigen processing and presentation” network, because these two molecular entities are part of the corresponding KEGG pathway. Novel functional relationships between molecular entities in the context of that pathway are then predicted, taking into account the computed relevance of each dataset (Figure 1). Along with the 15 pathway-specific networks, we also constructed an “Immune Global” network that represents the aggregate of the information in the individual pathway networks. The resulting ImmuNet functional networks provide researchers with a data-driven summary view of the human immune system through the aggregate Immune Global functional network. In addition, when information most relevant to a specific immunological pathway is sought, ImmuNet provides this more granular context of specific immune pathways (e.g., chemokine signaling pathway functional network).
Figure 1. ImmuNet Development and Selected Applications.
Data from more than 38,000 experiments (including mRNA expression, protein interaction assays, and phenotypic assays) were collected from public repositories and systematically processed (see Supplemental Computational Methods). These data and curated immune pathway prior knowledge from KEGG were used as input to infer 15 immune-specific functional relationship networks and an overall Immune Global context averaged network. Each immune-specific functional network predicts functional association between molecular entities (genes or proteins) specific to a particular immune biological process (e.g., antigen processing and presentation). ImmuNet leverages this massive data compendium to predict novel immune process or immune disease associations. See Supplemental Computational Methods for full information on the data compendium and integration.
To determine the ability of ImmuNet to capture immune process-specific interactions, we conducted a cross-validation-based evaluation by using statistically rigorous, well-established approaches (Geisser, 1993; Hastie et al., 2011). We withheld a randomly selected third of the known pathway-determined immune relationships and integrated the data compendium as if this held-out information were unavailable. We then tested the ability of the resulting network to predict the held-out relationships. The procedure was repeated with each non-overlapping third of the data. Thus, ImmuNet’s accuracy in predicting this held-out information provided an estimate of its ability to make novel predictions.
ImmuNet was able to accurately recapitulate relationships among molecular entities that were held out from each of the 15 KEGG immune pathways (Figure 2A). The predictions obtained with this data-driven approach represent a dramatic improvement over chance (range p = 10−266 to p = 10−10, see Table S1 for all p values and associated methods for the computation of the statistical significance). This analysis indicates that the ImmuNet integration can correctly make predictions that were not used in its construction and therefore can make novel predictions.
Figure 2. ImmuNet Accurately Recapitulates Known Functional Relationships in Immune Pathways.

(A) ImmuNet networks were evaluated via 3-fold cross-validation. For each pathway, one-third of the pathway data was iteratively omitted when constructing the network and the accuracy of this network in predicting the held-out information was tested. The panel shows the successful recovery of held-out immune data when we used the standard area-under receiver operator curve (AUC) metric that reflects both specificity and sensitivity (Hastie et al., 2011). Bar plots represent the mean ± SEM of the three cross-validations. (B) Using 3-fold cross-validation, the performance of the ImmuNet global network was compared to two standard non-immune-specific networks, BioGRID PPI network and a functional integration Human Global network. Boxplots represent the AUC performance distribution of each network at recovering known immune relationships (from the 15 KEGG contexts) that were held out during the training of each network. ImmuNet significantly outperformed the other two networks. p values are based on Wilcoxon signed-rank test.
We also compared the results obtained from ImmuNet to those obtained from two non-immune networks built using well-established approaches: the network comprised of experimentally determined protein-protein physical interactions (PPI) curated in the BioGRID database (Stark et al., 2006) and a non-immune-specific Bayesian functional integration network (Human Global, see Supplemental Computational Methods), which is an updated version of a previously reported network (Huttenhower et al., 2009), that is now based on the exact data compendium used for generating ImmuNet. The Immune Global Network significantly outperformed the experimental physical interaction PPI network (~20% improvement, p < 3 × 10−9; Wilcoxon signed-rank test) as well as the Human Global network (~17% improvement, p < 2 × 10−8, Figure 2B). The Immune Global Network is based on data from the PPI network as well as other data sources, so this comparison supports the power of this approach in integrating insight contained in extremely diverse types of measurements. The improved ability of the ImmuNet networks to predict the KEGG immunological pathways underscores the robustness of the Bayesian integration to extract relevant pertinent information and increase predictive accuracy despite the heterogeneous and noisy nature of the underlying data compendium. Overall, these results demonstrate that developing an immunologically based network improves the ability to predict new immune-specific functional relationships among molecular entities.
To investigate whether biological datasets stemming from non-immunological studies contain immune-related signals, we assessed the difference in performance for the ImmuNet networks generated using the entire compendium of data with those generated using only data from immune-related experiments (see Supplemental Computational Methods). Using the hold-out of known examples procedure as described above, our results (Figure S1) showed a 12% improvement in performance (p < 3 × 10−9; Wilcoxon signed-rank test) over networks generated using only data from immune-related experiments, demonstrating the benefit of using the complete compendium of experimental data. Because Bayesian integration is able to automatically infer each dataset’s relevance to immune biology or a specific immune pathway, using the larger, more complete data compendium becomes a prime opportunity for generating more informative networks.
Interrogating the ImmuNet Resource
We have shown computationally that ImmuNet can provide new insight into immunological relationships among molecular entities. In order to make this a flexible and convenient resource for hypothesis generation by immunologists, we have developed an intuitive interface and tools to allow researchers to address a wide variety of questions that pivot on predicting functional relationships. We illustrate how ImmuNet can be applied to generate hypotheses that might help guide further investigation.
The production of type I interferons and the activation of cellular apoptosis have both been associated with the immune responses to influenza A virus (IAV) infection. Interferons, which are released by cells infected by pathogen or cells stimulated by activation of pathogen-associated molecular pattern (PAMP) receptors, act on other cells to inhibit virus infection and replication (Koerner et al., 2007). Cell apoptosis initiated during antigen presentation has been proposed to impair IAV dissemination (Mok et al., 2007) and to enhance T-cell-mediated immunity via antigen cross-presentation (Albert et al., 1998). We have found that when human dendritic cells were infected in vitro with various H1N1 IAV strains, the seasonal viruses, but not the pandemic viruses, induced cell death (Hartmann et al., 2014). To further our understanding of the relationship of cell death and interferon-mediated antiviral responses, we queried Immu-Net to identify new targets that connect these processes.
To interrogate the ImmuNet resource, we selected representative genes or proteins for each process that were then used to assemble a subnetwork showing their overall shared relationships to molecular entities not included in the query. The use of ImmuNet thus provides a bridge from the researcher’s own knowledge to the inference of novel, relevant global-data-derived hypotheses. Although multiple molecular entities can be used for any query, for simplicity we restrict this example to two entities. To address interferon and cell death pathway crosstalk in antiviral responses, we queried ImmuNet using IFNAR1, a type I interferon receptor component, to represent interferon signaling, and FAS, the cell surface death receptor, to represent cell death processes. The predicted subnetwork resulting from this query is shown in Figure 3A. One molecular entity that was retrieved, the prostaglandin receptor PTGER2, suggested the possible involvement of prostaglandins in the modulation of cell death and interferon-mediated antiviral processes. Supporting this ImmuNet-derived inference, a recent report in this journal has demonstrated that IAV upregulates production of the PTGER2 ligand, prostaglandin E2 (PGE2), to evade host type I interferon-mediated immunity and to decrease apoptosis in alveolar macrophages (Coulombe et al., 2014).
Figure 3. Illustration of the Use of Immune-Specific Functional Networks.

(A) High-confidence subnetwork obtained by querying the ImmuNet hematopoietic cell lineage network with IFNAR1 (Interferon receptor 1) and FAS (Cell surface death receptor). The subnetwork obtained predicted that the processes reflected by the query genes are functionally related to PTGER2 and MNDA. The visualization parameters used to generate the graph shown are minimum relationship confidence = 0.61 and maximum number of genes = 21. (B) The relationship of MNDA to cell death in the context of antiviral responses was evaluated by comparing its induction by infection of DCs by IAV that induce cell death (seasonal viruses NC/99; TX/91) or do not induce cell death (pandemic viruses Brevig, Cal/09) in these cells. Data shown is 8 hr after infection at MOI = 1, normalized to the levels obtained with vehicle-treated cells. Notably, MNDA is differentially induced by the two virus groups. Bar plots represent the mean ± SEM gene expression fold-change of three replicate infections.
See also Table S2.
This example also shows how one can use ImmuNet to generate global-data-driven hypotheses for future study. For example, the subnetwork showed that myeloid cell nuclear differentiation antigen (MNDA) was linked to both FAS and IFNAR1. MNDA was originally identified by its association with myeloid leukemia (Briggs et al., 2006; Hofmann et al., 2002; Pradhan et al., 2004). MNDA is a member of the Pyrin and HIN domain (PYHIN) family of proteins, other members of which have recently been identified as viral pathogen recognition receptors (PRR) (Connolly and Bowie, 2014; Schattgen and Fitzgerald, 2011). Based on the ImmuNet-predicted subnetwork and subsequent review of the literature, we speculated that MNDA might represent a PRR that functions at the interface of interferon and cell death antiviral processes. Because specific IAV strains differ in their capacity to induce cell death in dendritic cells (Hartmann et al., 2014), this hypothesis motivated us to compare the induction of MNDA expression in IAV strains that induce or do not induce cell death. We found experimentally that MNDA expression was induced only by the two pandemic IAV strains, which do not cause cell death in these cells, and not by the cell-death-inducing seasonal IAV strains (Figure 3B, see Supplemental Computational Methods). These results are consistent with the hypothesis generated from ImmuNet that MNDA is functionally related to cell death and antiviral processes, and suggest that further study of this molecular target and its role in the differential response to IAV infection in these cells is warranted.
ImmuNet also allows the relative importance of the specific datasets that underlie inferred relationships to be reviewed by clicking on the corresponding edge in the network. Notably, for both PTGER2 and MNDA, the vast majority of datasets underlying their predicted relationships to FAS, as well as to other nodes in this subnetwork, have little apparent relevance to immunology (see Table S2 for an example). Thus the use of ImmuNet allows the researcher to rapidly develop global-data-driven hypotheses based on a wealth of experiments that would otherwise never be considered in order to help prioritize directions for further study.
Using ImmuNet for Gene Signature Prediction
We next show an application of ImmuNet to investigate gene program responses to virus infection. After cell infection, different viruses elicit specific host gene expression responses (Huang et al., 2001). The specificity of the responses results in part because each virus can differ in its ability to suppress particular host defense mechanisms via viral antagonists. Virus immune antagonists, which help pathogenic viruses to evade the host immune response, differ in their molecular targets, even among closely related viruses (García-Sastre, 2011; Nemeroff et al., 1998; Noah et al., 2003). In order to facilitate the identification of viral immune antagonist mechanisms, it would be useful to identify “expected” response genes that are missing in the response to a specific virus. These differential-absence signature genes represent candidate targets for novel immune antagonism mechanisms. Finding a differential absence signature for a particular virus experimentally would require, in principle, comparison of the gene response to the virus of interest with the expression patterns elicited by a myriad of possible immune-response-eliciting stimuli. Such an undertaking would be arduous, and prioritizing and interpreting any results obtained would be a daunting bioinformatics project. We show that using ImmuNet to draw evidence from the massive data already available in the public domain greatly facilitates this type of inquiry.
To illustrate this approach, we focused on the H1N1 influenza A/New Caledonia/99 virus strain (NC/99). Beginning with a seed set of 183 genes that characterizes the early immune response to NC/99 infection in monocyte-derived dendritic cells (Zaslavsky et al., 2013), we used ImmuNet and the support vector machine (SVM) algorithm (Noble, 2006) to generate a prioritized list of putative differential-absence genes (Figure 4A). The SVM classifier identifies which ImmuNet functional network patterns are predictive of genes that are functionally similar to the seed genes. That is, the classifier learns the functional network connectivity properties that characterize the NC/99 early response genes and applies them to predict other genes that would be expected to be regulated (see the Computational Methods).
Figure 4. Functional Networks Predict Specificity of Influenza Viral Infection.
(A) Pathogenic viruses, such as the NC/99 IAV strain, have developed immune antagonist mechanisms to suppress components of the antiviral response gene program. Genes induced in human DCs infected with NC/99 (Zaslavsky et al., 2013) were used as input for an ImmuNet-based method to predict differential absence genes. With these 183 genes as positive examples, an SVM classifier was trained to identify genes in the Immune Global network that were closely related to the seed set but were not induced by NC/99. The absence of these “expected” genes identified them as candidates for NC/99 immune antagonist mechanisms.
(B) DCs were infected at MOI = 1 with NC/99 or NDV. Infectivity was assayed by immunostaining of viral proteins (NP for NC/99 and HN for NDV). Antiviral gene MX1 was induced by each virus, assayed by RT-PCR, indicating virus detection and initiation of cellular responses.
(C) Expression levels of the 16 SVM-classifier top-ranked “expected” absence genes 8 hr after NC/99 or NDV infection were assayed. Seven of the predicted genes were significantly higher after NDV infection at 8 hr (p < 0.05). Data represent mean ± SEM from three independent experiments, each performed in triplicate.
We selected the top 16 genes identified by the classifier that were highly connected to the NC/99 response but were not regulated by the NC/99 infection at any time point. To support the hypothesis that this NC/99 differential absence signature is enriched in genes that are subject to NC/99 viral antagonism mechanisms, we quantified the response of these top 16 Immu-Net-predicted differential absence genes to infection by Newcastle Disease Virus (NDV), an avian pathogen that lacks human immune antagonist activity (Park et al., 2003a, 2003b; Zaslavsky et al., 2010). We first established that infection of monocyte-derived dendritic cells with either NC/99 or NDV yielded comparable infectivity (Figure 4B, top panel, see the Supplemental Computational Methods) and elicited an antiviral response, as indicated by the induction of the antiviral gene MX1 (Figure 4B, bottom panel). Of the 16 genes included in the ImmuNet-predicted NC/99 differential absence signature, seven genes were induced in NDV-infected cells at 8 hr, but were not induced by NC/99 at either 4 hr (data not shown) or 8 hr (Figure 4C, left). We also evaluated the chances of finding this high a proportion of differentially regulated genes without benefiting from the predictive ability of ImmuNet. To assess this, we selected genes that were not induced by NC/99 infection, but were annotated in Gene Ontology (GO) as immunological genes. Only 2% of this set were reported as regulated by NDV (Zaslavsky et al., 2010), in comparison with the 44% of ImmuNet-predicted genes that were regulated (p < 3 × 10−16, proportion difference 99% CI [0.07, 0.77], Pearson’s chi-square test).
Among the immune genes that were hypothesized to be specifically targeted by NC/99 antagonism, we found IFI6, which has been previously implicated in more targeted antiviral specificity (Schoggins et al., 2011), and BST2 (also known as tetherin), which is an interferon inducible host protein that, when not suppressed, interferes with budding of IAV (García-Sastre, 2011). Furthermore, ImmuNet identified interesting targets for study that are not known to have immunological roles. Notably, three of the ImmuNet-predicted differential absence genes that were regulated by NDV were not annotated to immunological processes in GO. This indicates the value of ImmuNet in expanding the known universe of genes and proteins involved in immunological processes.
Overall, this analysis indicates that ImmuNet provides an efficient and specific approach to use the global research data compendium to guide studies toward identifying virus-strain-specific immune antagonist mechanisms. In order to allow the non-computational immunology researcher to perform this type of classifier analysis, we have developed a user-friendly SVM tool available through the ImmuNet website.
Using ImmuNet to Predict Disease-Associated Genes
We next examined whether ImmuNet networks could be used as a functional summary of the public data compendium to facilitate the identification of disease-associated genes. We implemented a set of SVM classifiers to predict disease-associated genes for immune-related disease groups, including inflammatory bowel disease (IBD), rheumatoid arthritis (RA), and common variable immune deficiency (CVID). In order to build each disease classifier, the genes known to be associated with the disease were selected from the Online Mendelian Inheritance in Man catalog (OMIM) (Table S3; Hamosh et al., 2005). Genes used as negative examples for training were selected at random from all non-immune-disease-associated genes in OMIM. The classifier then provided the ability to rank all genes in the ImmuNet resource by their probability of being associated with the disease of interest.
The validity of the predictions and the comparative performance of the immune-specific ImmuNet network with the PPI network and the human global non-immune-specific Bayesian data integration were tested using the cross-validation approach described previously. In this systematic cross-validation evaluation, we iteratively withheld a subset of known disease genes from the OMIM training sets and assessed how well the predictions recapitulated these held-out known disease-associated genes not used in building the evaluation classifier. The ImmuNet predictions showed high accuracy (AUCs = 0.75 to 0.88; see Figure 5A) and significantly outperformed those using the PPI network (p < 0.007; Wilcoxon signed-rank test) and the non-immune-specific functional network (p < 0.005, Figure S2A). This evaluation suggests that the ImmuNet-based disease classifiers should be accurate in predicting new disease-associated genes.
Figure 5. Immune-Specific Functional Networks Accurately Predict Gene-Disease Associations.
To predict disease-associated genes, SVM classifiers were trained using ImmuNet with OMIM genes annotated to CVID, IBD, and RA.
(A) Results of 3-fold cross-validation, in which each classifier was trained without one-third of the known positives, and the accuracy of predicting this held-out information was evaluated by receiver-operator AUC.
(B) Prediction of GWAS-associated genes by ImmuNet classifiers. The relationships of the SVM classifier score and reported GWAS-associated genes were determined. The graph shows differences in the probability density of genes reported as GWAS associated in comparison with other genes in the network. Genes with high SVM scores were highly significantly enriched in reported GWAS-associated genes for all three diseases.
(C) Prediction of reported IBD eQTL genes by ImmuNet IBD classifier. The relationship of the SVM classifier scores and reported eQTL genes was determined. The graph shows a significant difference in the probability density of genes identified by eQTL analysis in comparison with other genes in the network. See also Figure S2 and Table S3.
In order to evaluate further the usefulness of this approach for generating disease gene predictions, we used a curated NHGRI catalog of GWAS-identified causal genes (Welter et al., 2014) to test whether GWAS-identified genes were enriched among the highest-confidence ImmuNet disease-associated predictions. We evaluated ImmuNet gene predictions for IBD, RA, and CVID. As shown graphically in Figure 5B, we found that each set of the GWAS genes was highly enriched among the highest ranked predictions for its respective classifier (CVID p < 7.5 × 10−5; IBD p < 1.3 × 10−51; RA p < 4.1 × 10−59 by PAGE rank-based enrichment test [Kim and Volsky, 2005]). We also studied whether the ImmuNet disease-associated gene classifier was useful for predicting expression quantitative trait loci (eQTL) genes. We focused on the complex multifactorial disease IBD (Neurath and Finotto, 2009). We compared the Immu-Net classifier-generated IBD gene list to previously identified IBD eQTLs (Kabakchiev and Silverberg, 2013). Among the highest ranked ImmuNet IBD classifier predictions, reported eQTL genes were significantly enriched (p < 0.0001, PAGE rank-based enrichment test, Figure 5C). These results indicate that ImmuNet provides a useful computational approach for identification of candidate disease-associated genes based on GWAS data that can complement eQTL data and be especially useful for prioritizing potential targets when eQTL data are not available.
These three independent assessments (hold-out cross-validation, GWAS prediction, and eQTL prediction) indicate that ImmuNet is a powerful functional-genomics-based framework to facilitate understanding the genetics of immune diseases. The user-friendly interface to apply the SVM classifier prediction engine to any immune-related disease of interest should facilitate wide application of the approaches described.
DISCUSSION
Computational analyses of large collections of experimental data harbor great potential for leveraging the relevant information in public data to make novel biological inferences far beyond those generated in each dataset’s original analysis and publication. Here, we introduced a probabilistic framework that can effectively utilize the global research output of the biomedical community to address targeted immunology research questions by identifying immunologically relevant signals hidden in diverse human large-scale data. To enable any biomedical researcher to easily explore and utilize these large data collections, we made ImmuNet and its associated analysis tools publicly available for the immune research community through an intuitive user-interactive website at http://immunet.princeton.edu/.
Through rigorous and systematic evaluations, we demonstrated that ImmuNet was able to accurately identify members of immune pathways as well as genes involved in immune diseases. The immune-specific aspect of the probabilistic integration was important for these tasks, because ImmuNet substantially outperformed non-immune-focused probabilistic integration or physical interaction networks. Further, we demonstrated that ImmuNet, which is trained with immunological pathways, extracts insight relevant to immunology from datasets not generated for immunological purposes. For example, in our illustration of the use of ImmuNet to study responses to influenza A virus, ImmuNet utilized datasets that would not initially seem to contain relevant immunological information, such as gene expression studies of medulloblastoma metastasis or prostate cancer cell lines exposed to DNA-methylation inhibitors. Overall, we found that integration focused only on datasets collected in studies directly relevant to immunology was less accurate than ImmuNet’s integration of the entire data compendium. This finding demonstrates that immunologically relevant insight is contained in non-immunological public data, and it can be extracted by immunologically focused probabilistic integration.
The resulting immune-specific functional networks aid in the interpretability of immune disease genome-wide association studies by leveraging functional genomics synergistically with quantitative genetics. We demonstrated the usefulness of Immu-Net in prioritizing GWAS-reported genes and in predicting disease-associated eQTL-confirmed genes. Major challenges still exist in identifying disease-linked loci due to biases in verification of GWAS-implicated loci linked to intergenic SNPs, especially with regard to bias for the nearest gene. Our customizable, web-accessible engine, which prioritizes genes in such loci based on functional genomic information, complements the use of physical distance and improves prioritization for experimental validation.
As genomic data collections continue to grow, our immune-specific probabilistic framework will be updated to continue to provide a flexible and intuitive approach for exploring these data to make hypotheses about immune biology and the molecular basis for immune diseases. In its current release, ImmuNet might have less relevance for some aspects of immunology, such as cell-cell interaction over space and time via the action of cytokines, because these relationships might not be well captured by public data. However, the applicability of Immu-Net to wide-ranging areas of immunology should grow with incorporation of continually increasing public big data. Our framework enables biomedical researchers to mine these data from an overall immunology-relevant perspective as well as from the perspective of specific immune processes. In addition, our framework makes it easy for the researcher to utilize SVM machine learning to predict genes associated with any specific disease or condition based on an ImmuNet-generated network. By enabling immune researchers from diverse backgrounds to intuitively leverage these valuable but noisy and heterogeneous data collections, ImmuNet has the potential to accelerate discovery in immunology.
COMPUTATIONAL METHODS
Immune Functional Relationship Networks
Integrated functional relationship networks summarize heterogeneous collections of genome-scale data into a concise graph representation. In this graph, molecular entities (genes, proteins) are nodes. The edge weights between nodes represent the probability that these molecular entities function together within a biological process. ImmuNet functional networks are generated by Bayesian data integration, which assesses the conditional probability that individual data sources (e.g., microarray experiments, protein-protein interaction data, etc.) contain evidence for gene relationships based on a training set of positive and negative examples (Pearl, 1988; Troyanskaya et al., 2003). Intuitively, this process assesses the accuracy and coverage of each data source, automatically down-weighting noisy datasets and experiments that are simply not relevant to the immune processes used for training the network. Bayesian inference then predicts the pair-wise posterior probabilities of functional relationships between all genes based on these per-dataset weights and behavior of these genes in the corresponding datasets. By choosing the training examples appropriately, we can generate networks that are specific to particular immunological pathways. This context specificity improves accuracy (Huttenhower et al., 2009) and provides users an opportunity to “summarize” the heterogeneous data collections focusing on specific areas of interest. To train networks to identify novel relationships in immunological research areas, we used 15 curated, immune-related KEGG pathways (Ogata et al., 1999) and a massive data compendium to generate 15 immune context networks. We also generated a context-averaged summary network (Immune Global), based on the 15 specific networks.
A network can be trained with any available set of annotated immunological relationships among molecular entities. We compared the predictive capacity of networks trained with immune relationships from Gene Ontology (Harris et al., 2004), Reactome (Croft et al., 2011), and KEGG. Networks trained on KEGG outperformed those based on the other training sets (Figure S2B). The description of the training examples, data sources, and generation of Immune Global are provided in the Supplemental Computational Methods.
Evaluation of ImmuNet Functional Relationships
We evaluated the ability of ImmuNet to capture immune-process-specific interactions by conducting a 3-fold cross-validation for each of the 15 KEGG immune pathways. The total set of molecular entities present in the reference pathway was randomly portioned into three sets. In each of the three cross-validation runs, a functional relationship network was generated using a training set limited to molecular entities present in two of the three subsets. The held-out third of the training set was used for evaluating the performance of the network generated using the other two-thirds (as measured by the area under the receiver operator characteristic curve [AUC]). Additionally, for each cross-validation, a context-averaged Immune Global network was generated and evaluated using the 15 networks of the corresponding cross-validation run (see Supplemental Computational Methods).
Gene Signature Prediction for Virus Infection
NC/99 Gene Response Signature
We identified genes characteristic of the early transcriptional response to H1N1 influenza A/New Caledonia/20/1999 (NC/99) infection in monocyte-derived dendritic cells (Zaslavsky et al., 2013). In that study, microarray profiling was used to select 183 genes that were differentially upregulated at the 4 hr post-infection time point. These genes were used as the input seed set for further analysis with ImmuNet.
SVM Classifier
SVM (Support Vector Machine) is a supervised machine learning method that uses a training set of examples belonging to two classes and a collection of relevant data/features (ImmuNet networks) to build a classification scheme that assigns each new example to one of these classes (Cortes and Vapnik, 1995). In our case, the two categories represent genes that either are related to a specific immune response/disease (positive examples) or are unrelated to this group (negative examples). In our previous work, we have demonstrated that functional networks can be used as input to machine learning methods, such as SVM, to accurately predict gene knockout phenotype, biological process membership, and disease association (Guan et al., 2010; Wong et al., 2012). Intuitively, gene-gene interaction probabilities derived by functional networks provide an accurate, integrative summary of high-throughput data, allowing machine learning methods to identify predictive gene connections related to the trait or disease of interest.
We leveraged ImmuNet networks as input to the SVM classifier to predict genes that show network properties similar to the set of genes that characterizes the early transcriptional response to NC/99 infection. Genes that were differentially upregulated in response to NC/99 infection (183 genes, as above) comprised the positive examples for our training set. Training negatives used the same negative set for disease SVM classifiers, excluding genes that have been identified as differentially regulated within the NC/99 transcriptional response at any time point. The network edge probabilities in the Immune Global functional network were used in the feature vector for supervised training of a linear SVM model. The SVM classifier was evaluated using 3-fold cross-validation. Seven cost parameters were tested for each classifier (C = 10n: −3 ≥ n ≥ 3), and the best-performing parameter for each disease classifier was used. The resulting SVM scores for each gene were then converted to probabilities by a sigmoid transformation (Platt, 1999).
Disease Prediction and Evaluation
To train SVM disease prediction classifiers, we utilized the ontological structure of Disease Ontology (Schriml et al., 2012) with the annotated genes in OMIM (Hamosh et al., 2005). OMIM-annotated genes were associated to their corresponding Disease Ontology terms, and the ontology structure was used to aggregate genes annotated to any branch of each disease subgroup studied. Using Disease Ontology terms, we identified nine immune disease subgroups that had six or more associated positive genes for training (see Table S3). Training negatives were selected randomly from genes associated with some OMIM disease term, excluding immune disease-annotated genes. Because there are GWAS data available for CVID, which is annotated in Disease Ontology as a child term of one of the nine disease categories, we trained a separate classifier for CVID (Table S3). The network edge probabilities in the Immune Global functional network were used in the feature vector for supervised training of a linear SVM model. Each SVM classifier was generated and evaluated using cross-validation, with cost parameters and conversion to probability of disease association as described above. Disease-associated genes predicted by these classifiers were evaluated against GWAS and eQTL catalogs, as described in the Results.
Supplementary Material
Highlights.
Interactive web-accessible immunology resource leverages 38,088 experiments
Powerful computational methods generate big-data-driven hypotheses for immunology
Predicts new immune pathway interactions, mechanisms, and disease-associated genes
Flexible, user-friendly platform addresses diverse immunological research questions
Acknowledgments
We thank Drs. Judy Cho and Gareth John for helpful discussions or manuscript comments, Nada Marjanovic for technical support, and the Icahn School of Medicine qPCR Core Facility. Supported by NIH Contract HHSN272201000054C and Grant 1U19AI117873. O.G.T. is a Senior Fellow of CIFAR. M.F. was supported by T32 MH096678.
Footnotes
AUTHOR CONTRIBUTIONS
Computational Experiments, D.G., C.Y.P., E.Z., and M.F.; Wetlab Experiments, M.F., B.M.H., and R.A.A.; Website and System Interface, D.G., A.K.W., and A.T.; Manuscript, D.G., C.Y.P., E.Z., M.F., O.G.T., S.H.K., R.A.A., A.G.-S., and S.C.S.; Conception and Oversight of Execution, E.Z., S.C.S., and O.G.T.
Supplemental Information includes two figures, three tables, and Supplemental Computational Methods and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2015.08.014.
References
- Albert ML, Sauter B, Bhardwaj N. Dendritic cells acquire antigen from apoptotic cells and induce class I-restricted CTLs. Nature. 1998;392:86–89. doi: 10.1038/32183. [DOI] [PubMed] [Google Scholar]
- Alexeyenko A, Sonnhammer EL. Global networks of functional coupling in eukaryotes from comprehensive data integration. Genome Res. 2009;19:1107–1116. doi: 10.1101/gr.087528.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Andorf S, Gomes L, Dunn P, Schaefer H, Pontius J, Berger P, Desborough V, Smith T, Campbell J, et al. ImmPort: disseminating data to the public for the future of immunology. Immunol Res. 2014;58:234–239. doi: 10.1007/s12026-014-8516-1. [DOI] [PubMed] [Google Scholar]
- Brazma A, Parkinson H, Sarkans U, Shojatalab M, Vilo J, Abeygunawardena N, Holloway E, Kapushesky M, Kemmeren P, Lara GG, et al. ArrayExpress–a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003;31:68–71. doi: 10.1093/nar/gkg091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs RC, Shults KE, Flye LA, McClintock-Treep SA, Jagasia MH, Goodman SA, Boulos FI, Jacobberger JW, Stelzer GT, Head DR. Dysregulated human myeloid nuclear differentiation antigen expression in myelodysplastic syndromes: evidence for a role in apoptosis. Cancer Res. 2006;66:4645–4651. doi: 10.1158/0008-5472.CAN-06-0229. [DOI] [PubMed] [Google Scholar]
- Connolly DJ, Bowie AG. The emerging role of human PYHIN proteins in innate immunity: implications for health and disease. Biochem Pharmacol. 2014;92:405–414. doi: 10.1016/j.bcp.2014.08.031. [DOI] [PubMed] [Google Scholar]
- Cortes C, Vapnik V. Support-Vector Networks. Mach Learn. 1995;20:273–297. [Google Scholar]
- Coulombe F, Jaworska J, Verway M, Tzelepis F, Massoud A, Gillard J, Wong G, Kobinger G, Xing Z, Couture C, et al. Targeted prostaglandin E2 inhibition enhances antiviral immunity through induction of type I interferon and apoptosis in macrophages. Immunity. 2014;40:554–568. doi: 10.1016/j.immuni.2014.02.013. [DOI] [PubMed] [Google Scholar]
- Croft D, O’Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 2004;21:137–148. doi: 10.1016/j.immuni.2004.07.017. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sastre A. Induction and evasion of type I interferon responses by influenza viruses. Virus Res. 2011;162:12–18. doi: 10.1016/j.virusres.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisser S. Predictive Inference. CRC Press; 1993. [Google Scholar]
- Guan Y, Ackert-Bicknell CL, Kell B, Troyanskaya OG, Hibbs MA. Functional genomics complements quantitative genetics in identifying disease-gene associations. PLoS Comput Biol. 2010;6:e1000991. doi: 10.1371/journal.pcbi.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann B, Albrecht R, Marjanovic N, Patil S, Fribourg M, Sealfon S. Cell death in pandemic and seasonal influenza viruses (VIR2P.1027) J Immunol. 2014;192(1 Supplement):16. [Google Scholar]
- Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. 2. Springer; 2011. [Google Scholar]
- Hebbring SJ. The challenges, advantages and future of phenome-wide association studies. Immunology. 2014;141:157–165. doi: 10.1111/imm.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng TS, Painter MW Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- Hibbs MA, Hess DC, Myers CL, Huttenhower C, Li K, Troyanskaya OG. Exploring the functional landscape of gene expression: directed search of large microarray compendia. Bioinformatics. 2007;23:2692–2699. doi: 10.1093/bioinformatics/btm403. [DOI] [PubMed] [Google Scholar]
- Hoffman MM, Buske OJ, Wang J, Weng Z, Bilmes JA, Noble WS. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat Methods. 2012;9:473–476. doi: 10.1038/nmeth.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann WK, de Vos S, Komor M, Hoelzer D, Wachsman W, Koeffler HP. Characterization of gene expression of CD34+ cells from normal and myelodysplastic bone marrow. Blood. 2002;100:3553–3560. doi: 10.1182/blood.V100.10.3553. [DOI] [PubMed] [Google Scholar]
- Huang Q, Liu D, Majewski P, Schulte LC, Korn JM, Young RA, Lander ES, Hacohen N. The plasticity of dendritic cell responses to pathogens and their components. Science. 2001;294:870–875. doi: 10.1126/science.294.5543.870. [DOI] [PubMed] [Google Scholar]
- Huttenhower C, Hibbs M, Myers C, Troyanskaya OG. A scalable method for integration and functional analysis of multiple microarray datasets. Bioinformatics. 2006;22:2890–2897. doi: 10.1093/bioinformatics/btl492. [DOI] [PubMed] [Google Scholar]
- Huttenhower C, Haley EM, Hibbs MA, Dumeaux V, Barrett DR, Coller HA, Troyanskaya OG. Exploring the human genome with functional maps. Genome Res. 2009;19:1093–1106. doi: 10.1101/gr.082214.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabakchiev B, Silverberg MS. Expression quantitative trait loci analysis identifies associations between genotype and gene expression in human intestine. Gastroenterology. 2013;144:1488–1496. 1496.e1–1496.e3. doi: 10.1053/j.gastro.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Volsky DJ. PAGE: parametric analysis of gene set enrichment. BMC Bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerner I, Kochs G, Kalinke U, Weiss S, Staeheli P. Protective role of beta interferon in host defense against influenza A virus. J Virol. 2007;81:2025–2030. doi: 10.1128/JVI.01718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R, Laur S, Adler P, Vilo J. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics. 2012;28:573–580. doi: 10.1093/bioinformatics/btr709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004;14:1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Blom UM, Wang PI, Shim JE, Marcotte EM. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011;21:1109–1121. doi: 10.1101/gr.118992.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok CKP, Lee DCW, Cheung CY, Peiris M, Lau ASY. Differential onset of apoptosis in influenza A virus H5N1- and H1N1-infected human blood macrophages. J Gen Virol. 2007;88:1275–1280. doi: 10.1099/vir.0.82423-0. [DOI] [PubMed] [Google Scholar]
- Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeroff ME, Barabino SML, Li Y, Keller W, Krug RM. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3’end formation of cellular pre-mRNAs. Mol Cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- Neurath MF, Finotto S. Translating inflammatory bowel disease research into clinical medicine. Immunity. 2009;31:357–361. doi: 10.1016/j.immuni.2009.08.016. [DOI] [PubMed] [Google Scholar]
- Noah DL, Twu KY, Krug RM. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAS. Virology. 2003;307:386–395. doi: 10.1016/s0042-6822(02)00127-7. [DOI] [PubMed] [Google Scholar]
- Noble WS. What is a support vector machine? Nat Biotechnol. 2006;24:1565–1567. doi: 10.1038/nbt1206-1565. [DOI] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MS, García-Sastre A, Cros JF, Basler CF, Palese P. Newcastle disease virus V protein is a determinant of host range restriction. J Virol. 2003a;77:9522–9532. doi: 10.1128/JVI.77.17.9522-9532.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MS, Shaw ML, Muñoz-Jordan J, Cros JF, Nakaya T, Bouvier N, Palese P, García-Sastre A, Basler CF. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J Virol. 2003b;77:1501–1511. doi: 10.1128/JVI.77.2.1501-1511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Krishnan A, Zhu Q, Wong AK, Lee YS, Troyanskaya OG. Tissue-aware data integration approach for the inference of pathway interactions in metazoan organisms. Bioinformatics. 2015;31:1093–1101. doi: 10.1093/bioinformatics/btu786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl J. Probabilistic Reasoning in Intelligent Systems. Morgan Kaufmann; 1988. [Google Scholar]
- Platt JC. Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. In: Smola AJ, Bartlett P, Schölkopf B, Schuurmans D, editors. Advances in Large Margin Classifiers. MIT Press; 1999. [Google Scholar]
- Pradhan A, Mijovic A, Mills K, Cumber P, Westwood N, Mufti GJ, Rassool FV. Differentially expressed genes in adult familial myelodys-plastic syndromes. Leukemia. 2004;18:449–459. doi: 10.1038/sj.leu.2403265. [DOI] [PubMed] [Google Scholar]
- Schattgen SA, Fitzgerald KA. The PYHIN protein family as mediators of host defenses. Immunol Rev. 2011;243:109–118. doi: 10.1111/j.1600-065X.2011.01053.x. [DOI] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schriml LM, Arze C, Nadendla S, Chang YWW, Mazaitis M, Felix V, Feng G, Kibbe WA. Disease ontology: A backbone for disease semantic integration. Nucleic Acids Res. 2012;40:D940–D960. doi: 10.1093/nar/gkr972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- Taşan M, Drabkin HJ, Beaver JE, Chua HN, Dunham J, Tian W, Blake JA, Roth FP. A Resource of Quantitative Functional Annotation for Homo sapiens Genes. G3 (Bethesda) 2012;2:223–233. doi: 10.1534/g3.111.000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanskaya OG, Dolinski K, Owen AB, Altman RB, Botstein D. A Bayesian framework for combining heterogeneous data sources for gene function prediction (in Saccharomyces cerevisiae) Proc Natl Acad Sci USA. 2003;100:8348–8353. doi: 10.1073/pnas.0832373100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AK, Park CY, Greene CS, Bongo LA, Guan Y, Troyanskaya OG. IMP: a multi-species functional genomics portal for integration, visualization and prediction of protein functions and networks. Nucleic Acids Res. 2012;40:W484–W490. doi: 10.1093/nar/gks458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslavsky E, Hershberg U, Seto J, Pham AM, Marquez S, Duke JL, Wetmur JG, Tenoever BR, Sealfon SC, Kleinstein SH. Antiviral response dictated by choreographed cascade of transcription factors. J Immunol. 2010;184:2908–2917. doi: 10.4049/jimmunol.0903453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslavsky E, Nudelman G, Marquez S, Hershberg U, Hartmann BM, Thakar J, Sealfon SC, Kleinstein SH. Reconstruction of regulatory networks through temporal enrichment profiling and its application to H1N1 influenza viral infection. BMC Bioinformatics. 2013;14(Suppl 6):S1. doi: 10.1186/1471-2105-14-S6-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.