Abstract
Comparative genomics of the bacterial thiamin pyrimidine synthase (thiC) revealed a paralog of thiC (bzaF) clustered with anaerobic vitamin B12 biosynthetic genes. Here we demonstrate that BzaF is a radical S-adenosylmethionine enzyme that catalyzes the remarkable conversion of aminoimidazole ribotide (AIR) to 5-hydroxybenzimidazole (5-HBI). We identify the origin of key product atoms and propose a reaction mechanism. These studies represent the first step in solving a long-standing problem in anaerobic vitamin B12 assembly and reveal an unanticipated intersection of thiamin and vitamin B12 biosynthesis.
Graphical Abstract
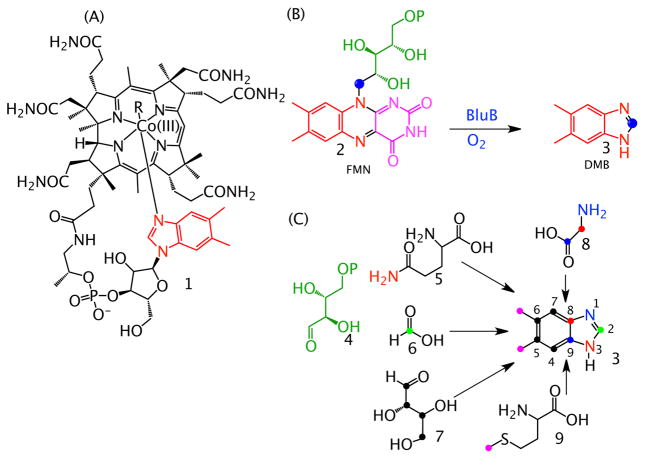
The formation of dimethylbenzimidazole (DMB, 3), the axial ligand of vitamin B12 (cobalamin), 1, (Figure 1A) is the last remaining major unsolved puzzle in B12 biosynthesis. In aerobic organisms, DMB is biosynthesized from flavin mononucleotide (FMN, 2) in a remarkable reaction catalyzed by the BluB gene product (Figure 1B). This oxygen requiring DMB synthase has been biochemically and structurally characterized, but its mechanism is not yet known.1,2 The anaerobic pathway to DMB involves different chemistry and labeling studies demonstrate that FMN is not the precursor (Figure 1C).3–6 The anaerobic DMB synthase has not been yet identified.
Figure 1.
The oxygen-dependent and oxygen-independent biosynthesis of the DMB ligand of vitamin B12. (A) The structure of vitamin B12 1 showing the DMB ligand in red. (B) Oxygen-dependent DMB formation from FMN 2. (C) Labeling studies showing the oxygen-independent biosynthetic origin of the atoms of DMB in E. limosum. 5-Hydroxybenzimidazole (HBI) and 5-hydroxy-6-methylbenzimidazole have been identified as precursors.11
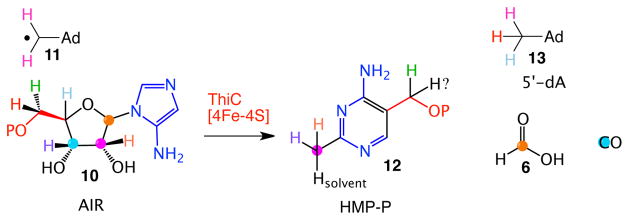
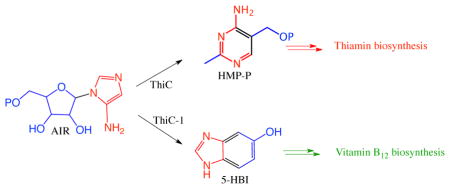
Our approach to elucidating the anaerobic biosynthesis of DMB was indirect and derived from our studies on the bacterial thiamin pyrimidine synthase (ThiC). This radical SAM enzyme catalyzes a remarkable rearrangement of aminoimidazole ribotide (AIR, 10) to the thiamin pyrimidine (12, Figure 2).7–10
Figure 2.
The remarkable, ThiC-catalyzed, conversion of AIR 10 to the thiamin pyrimidine 12.
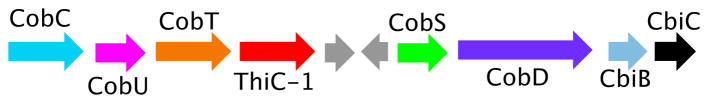
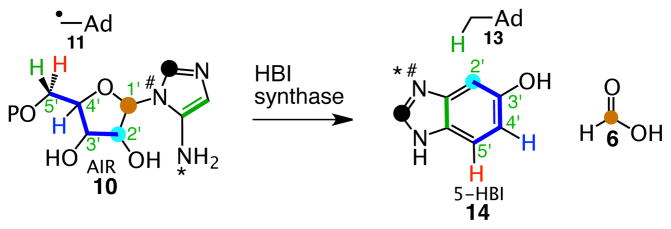
While looking for a ThiC paralog, to facilitate our mechanistic studies on this complex reaction, we noticed that some anaerobes in the SEED database (http://theseed.uchicago.edu/FIG/) had a thiC paralog (bzaF) clustered with vitamin B12 biosynthetic genes (Figure 3). This suggested the possibility that BzaF might catalyze the conversion of AIR to a DMB precursor. Previous in vivo labeling studies, using 4-aminoimidazole, eliminated this heterocycle as a DMB precursor.12 In this communication we report the successful reconstitution of the BzaF-catalyzed conversion of AIR to 5-hydroxybenzimidazole (HBI), we identify the hydrogen atom initially abstracted by the 5′-deoxyadenosyl radical and the origin, in AIR, of key HBI atoms. These results are integrated into a mechanistic hypothesis for the BzaF (HBI synthase) catalyzed reaction.
Figure 3.
BzaF gene cluster in D. acetoxidans. CobCUT, CobSD and CbiBC have established functions in vitamin B12 biosynthesis. BzaF has 38% sequence identity and 53.6% sequence similarity to the D. acetoxidans ThiC.
The bzaF gene from Desulfuromonas. acetoxidans was synthesized with codon optimization (for gene details - Figure S33), cloned in the THT vector and co-expressed in the presence of a plasmid encoding the suf operon for in vivo [4Fe-4S] reconstitution in E.coli BL21 (DE3) (See supporting information - page 13). The enzyme was then purified under anaerobic conditions using Ni-NTA chromatography (see SI for experimental details). The enzyme yield was 3 mg/L of culture. BzaF has a molecular mass of 48 kDa, and contains 3 irons and 2.6 sulfides/monomer. The UV-Visible spectrum of the purified protein showed the long wavelength absorption characteristic of the [4Fe-4S]+2 cluster (Figure 4B).
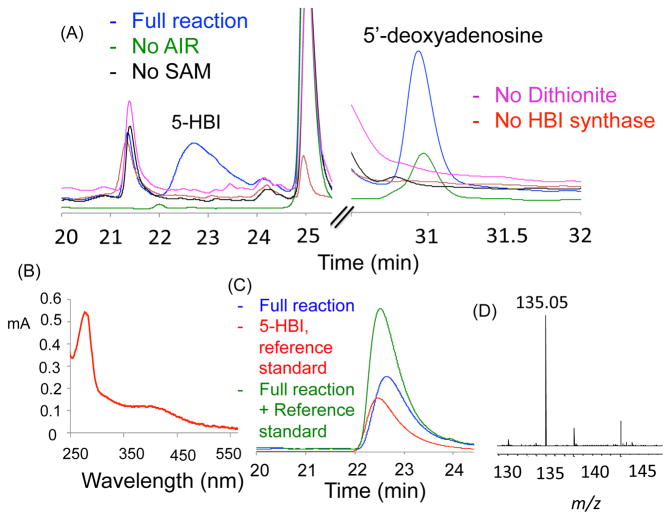
Figure 4.
Characterization of the product of the BzaF catalyzed reaction. (A) HPLC (254 nm) analysis of the HBI synthase reaction mixture showing a new product eluting at 22.7 min and 5′-dA eluting at 31 min (blue trace). This product was not formed in reaction mixtures lacking dithionite (red trace). (See Figure S31 for all controls). (B) UV-Vis spectrum of HBI synthase as isolated. (C) Product identification by HPLC analysis (red trace – HBI, blue trace – full enzymatic reaction, green trace - co-injection of HBI and the reaction product). (D) MS analysis of the 22.7 min enzymatic product showing [M+H]+ = 135.05 Da, consistent with HBI formation.
Anaerobic incubation of AIR and SAM with dithionite-reduced BzaF (RT, 60 min) resulted in the formation of 5′-deoxyadenosine (5′-dA) and a new product as determined by HPLC analysis (Figure 4A). This product had an [M+H]+ of 135.05 Da and co-migrated with an authentic standard of HBI (Figure 4C and D). This identification was confirmed by scale-up of the reaction followed by NMR analysis of the purified product. The ratios of HBI and 5′-dA to BzaF were 0.7 (HBI:BzaF) and 1.1 (5′dA:BzaF) respectively. The corresponding ratios for ThiC are 0.4 (HMP-P: C.c.ThiC) and 0.6 (5′dA: C.c.ThiC).
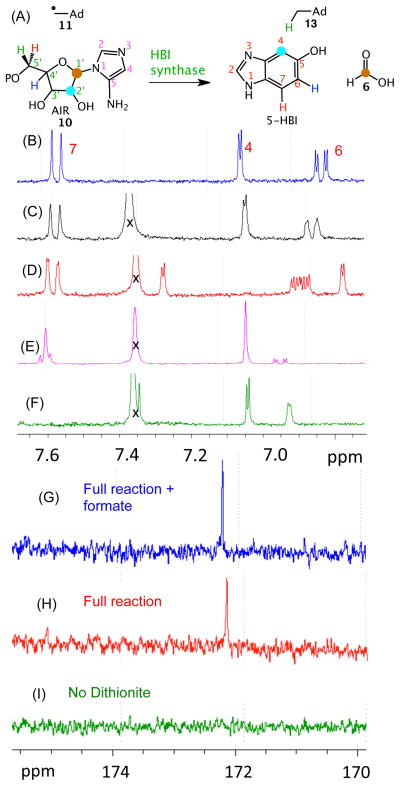
As a first step in elucidating the mechanism of HBI synthase, we have determined the origin of key atoms of HBI. Our strategy involved the conversion of a set of 2H- and 13C-AIR isotopologs to HBI followed by the identification of the site of labeling by NMR and MS analysis. The results of these experiments are shown in Figure 5 and in Table S1 (See also Figures S20–S29). The key findings, summarized in Figure 5A are as follows: 1) The proS hydrogen at C5 of AIR is abstracted by the adenosyl radical (Table S1, lines 2–4). 2) The C7 hydrogen of HBI is derived from the C5′ proR hydrogen of AIR (Table S1, line 3 and Figure 5F). 3) The C6 hydrogen of HBI is derived from the C4′ hydrogen of AIR (Table S1, line 5 and Figure 5E). 4) The C1′ carbon of AIR is lost as formate (Table S1, lines 6–9 and Figure 5GHI). 5) The C5 amino group nitrogen of AIR is partially retained in the product (Table S1, line 10, 62% retention, see legend to Table S1). 6) The C4 carbon of HBI is derived from the C2′ carbon of AIR (Figure 5D). From these experiments, we can deduce the origin of all of the atoms of HBI as shown in Figure 6.
Figure 5.
Labeling studies to determine the origin of key HBI atoms. (A) Summary of the labeling studies. (B) Partial 1H NMR for authentic HBI. (C)–(F): 1H NMR of HBI purified from enzymatic reactions using (C) unlabeled AIR (D) 2′-13C-AIR (E) 4-2H-AIR and (F) 5′,5′-2H2-AIR. (G) – (I): 13C DEPT 90 analysis of the reaction mixture. (G) HBI synthase reaction with 1′,2′,3′,4′,5′-13C5-AIR spiked with formate (10 mM) after 2hrs. (H) HBI synthase reaction with 1′,2′,3′,4′,5′-13C5-AIR. (I) No Dithionite – control. X is an unknown impurity. The doublet at 7.6 and the doublet of doublets at 7.05 in spectrum E are due to a trace of unlabeled AIR in the sample (See Figure S30 for 13C-NMR).
Figure 6.
Origin of all of the atoms of HBI based on the labeling studies shown in Figure 5 and in Table S1.
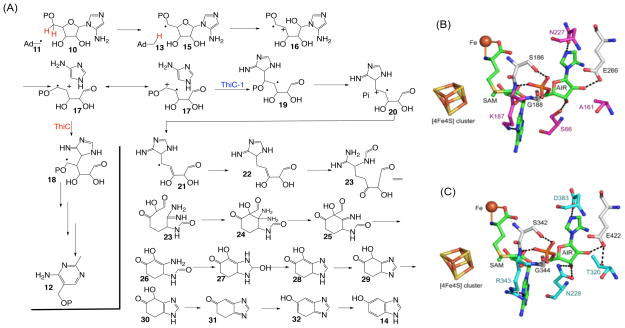
These labeling studies allow us to propose a mechanistic hypothesis for the HBI-synthase-catalyzed reaction (Figure 7A). As expected from the sequence similarity with ThiC, HBI formation is initiated by the abstraction of the C5′ proS hydrogen of AIR by the 5′-deoxyadenosyl radical to give 15. C-O bond cleavage followed by N-glycosyl bond cleavage gives 17. At this point, the chemistry diverges from the proposed ThiC pathway (See Figure S32). Electrophilic addition of the ribose C4′ to the aminoimidazole leads to the thiamin pyrimidine while electrophilic addition of the ribose C5′ gives 19 which leads to DHBI. Loss of phosphate followed by deprotonation gives 21, which is quenched by hydrogen atom donation from an unknown source. A ring opening/closing sequence gives 24. Loss of ammonia from 24 gives imine 25 in which 38% of the imine nitrogen is derived from N1 of AIR and the remainder is derived from the C5 amino group. This is consistent with the partial scrambling of the two amidine nitrogens in 23 by C-C bond rotation. Loss of formate, ring closure and a series of dehydrations and tautomerizations complete the formation of HBI 14.
Figure 7.
(A) Mechanistic proposal for the radical-triggered conversion of AIR to HBI catalyzed by HBI synthase. (B) Homology model for HBI synthase. (C) X-ray structure of the ThiC active site. The [4Fe-4S] cluster, additional iron, SAM, AIR, and key active site residues are shown. SAM was generated by adding a methyl group in the S-configuration to SAH. The additional iron is also chelated to two conserved side chains (His270 and His334) in HBI synthase and His426 and His490 in ThiC.
A homology model for HBI synthase was constructed using the structure of the Arabidopsis thaliana ThiC/AIR/SAH/Fe complex as a template.13 HBI synthase lacks the chloroplast-targeting sequence specific to the Arabidopsis ThiC as well as the N-terminal ThiC domain.7,13,14 This N-terminal domain is also missing from ThiCs from anaerobic organisms. HBI synthase is predicted to have the noncanonical active site architecture recently reported for ThiC with an additional iron chelating two conserved histidine residues and the amino and carboxylate groups of SAM.13 The homology model was consistent with the observed abstraction of the 5’-ProS hydrogen atom of the substrate and showed remarkable conservation of active site residues with only four differences out of 21 residues in the first shell when compared to ThiC (Figure 7B – 7C). Three of the differences involve contacts with AIR. In ThiC, Asn228 hydrogen bonds to AIR O3′ and is replaced by Ser66 in HBI synthase. Thr320 hydrogen bonds to Glu422, which in turn hydrogen bonds to AIR O2′, and is replaced by Ala161 in HBI synthase; Glu244 is equivalent to Glu422 and still hydrogen bonds to AIR O2′. Asp383 (if protonated) hydrogen bonds to AIR N3 (2.8 Å) and is replaced by Asn227 in HBI synthase. The fourth difference is near SAM, where Arg343 hydrogen bonds to N7 of SAM and is replaced by Lys187, which is also predicted to hydrogen bond to N7, in HB synthase. BLAST searches15 starting with either D. acetoxidans HBI synthase or A. thaliana ThiC suggest that these four differences alone distinguish the two paralogs.
Conserved active sites suggest similar catalytic mechanisms for most enzymatic reactions. This is clearly not the case for ThiC and BzaF. These proteins reveal unique features of enzymatic catalysis of reactions that proceed via high-energy intermediates. For such reactions, it is likely that the high-energy substrate radical 15 is converted to HMP-P 12 or to HBI 14 in a complex multistep sequence in which several steps along the reaction coordinate do not require catalytic assistance from the enzyme. This allows for similar active sites to produce different products. This catalytic motif has previously been observed in terpene cyclases where similar active sites can catalyze the conversion of high-energy carbocations to different products. For example, the D100E mutant of trichodiene synthase generates six terpenes16 and native baruol synthase produces a library of 23 terpenes.17
Our studies on the identification and reconstitution of the vitamin B12 HBI synthase represent the first steps in solving a long-standing problem in anaerobic vitamin B12 biosynthesis. The biosynthesis involves an unanticipated intersection of thiamin and vitamin B12 biosynthesis and uses chemistry that exceeds in complexity any of the enzymatic reactions used in the assembly of the coronoids of vitamin B12. Mechanistic and structural studies are in progress to elucidate the details of this remarkable reaction.
Supplementary Material
Acknowledgments
This research was supported by the Robert A. Welch Foundation (A-0034 to TPB) and the National Institutes of Health (DK44083 to TPB; DK067081 to SEE).
Footnotes
ASSOCIATED CONTENT
Supporting Information Available: Detailed experimental procedures are provided in the supporting information. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Taga ME, Larsen NA, Howard-Jones AR, Walsh CT, Walker GC. Nature (London, U K) 2007;446:449. doi: 10.1038/nature05611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cooper AJL, Pinto JT. Chemtracts. 2006;19:474. [Google Scholar]
- 3.Scott AI. J Org Chem. 2003;68:2529. doi: 10.1021/jo020728t. [DOI] [PubMed] [Google Scholar]
- 4.Munder M, Vogt JRA, Vogler B, Renz P. Eur J Biochem. 1992;204:679. doi: 10.1111/j.1432-1033.1992.tb16681.x. [DOI] [PubMed] [Google Scholar]
- 5.Vogt JRA, Lamm-Kolonko L, Renz P. Eur J Biochem. 1988;174:637. doi: 10.1111/j.1432-1033.1988.tb14145.x. [DOI] [PubMed] [Google Scholar]
- 6.Vogt JRA, Renz P. Eur J Biochem. 1988;171:655. doi: 10.1111/j.1432-1033.1988.tb13836.x. [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee A, Li Y, Zhang Y, Grove TL, Lee M, Krebs C, Booker SJ, Begley TP, Ealick SE. Nat Chem Biol. 2008;4:758. doi: 10.1038/nchembio.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterjee A, Hazra AB, Abdelwahed S, Hilmey DG, Begley TP. Angew Chem, Int Ed. 2010;49:8653. doi: 10.1002/anie.201003419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawhorn BG, Mehl RA, Begley TP. Org Biomol Chem. 2004;2:2538. doi: 10.1039/B405429F. [DOI] [PubMed] [Google Scholar]
- 10.Yamada K, Kumaoka H. J Nutr Sci Vitaminol (Tokyo) 1983;29:389. doi: 10.3177/jnsv.29.389. [DOI] [PubMed] [Google Scholar]
- 11.Renz P, Endres B, Kurz B, Marquardt J. Eur J Biochem. 1993;217:1117. doi: 10.1111/j.1432-1033.1993.tb18344.x. [DOI] [PubMed] [Google Scholar]
- 12.Endres B, Weurfel A, Volgler B, Renz P. Biol Chem Hoppe-Seyler. 1995;376:595. doi: 10.1515/bchm3.1995.376.10.595. [DOI] [PubMed] [Google Scholar]
- 13.Fenwick MK, Mehta AP, Zhang Y, Abdelwahed SH, Begley TP, Ealick SE. Nature Commun. 2015;6:6480. doi: 10.1038/ncomms7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coquille S, Roux C, Mehta A, Begley TP, Fitzpatrick TB, Thore S. J Struct Biol. 184:438. doi: 10.1016/j.jsb.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J Mol Biol. 1990;215:403. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 16.Cane DE, Xue Q, Fitzsimons BC. Biochemistry. 1996;35:12369. doi: 10.1021/bi961344y. [DOI] [PubMed] [Google Scholar]
- 17.Lodeiro S, Xiong Q, Wilson WK, Kolesnikova MD, Onak CS, Matsuda SPT. J Am Chem Soc. 2007;129:1121. doi: 10.1021/ja073133u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.