Abstract
Histiocytoid cardiomyopathy (Histiocytoid CM) is a rare form of cardiomyopathy observed predominantly in newborn females that is fatal unless treated early in life. We have performed whole exome sequencing on five parent-proband trios and identified nuclear-encoded mitochondrial protein mutations in three cases. Two probands had de novo non-sense mutations in the second exon of the X-linked nuclear gene NDUFB11, which has not previously been implicated in any disease, despite evidence that deficiency for other mitochondrial electron transport complex I members leads to cardiomyopathy. A third proband was doubly heterozygous for inherited rare variants in additional components of complex I, NDUFAF2 and NDUFB9, confirming that Histiocytoid CM is genetically heterogeneous. In a fourth case, the proband with Histiocytoid CM inherited a mitochondrial mutation from her heteroplasmic mother, as did her brother who presented with cardiac arrhythmia. Strong candidate recessive or compound heterozygous variants were not found for this individual or for the fifth case. Although NDUFB11 has not been implicated before in cardiac pathology, morpholino-mediated knockdown of Ndufb11 in zebrafish embryos generated defective cardiac tissue with looping defects, which confirms the causative role of NDUFB11 in cardiac pathology. Therefore, the NDUFB11 mutation represents a genetic basis of this heterogeneous disease.
Keywords: histiocytoid cardiomyopathy, NDUFB11, zebrafish, morpholinos, whole exome sequencing, de novo mutation
INTRODUCTION
Infantile histiocytoid cardiomyopathy (Histiocytoid CM, MIM 500000) is a rare, but distinctive arrhythmogenic disorder characterized by incessant ventricular tachycardia, cardiomegaly, and sudden death within the first two years of life if left untreated. Approximately 100 Histiocytoid CM cases have been reported in the literature [Bove et al., 1973; Ferrans et al., 1976; Shehata et al., 1998; Shehata et al., 2011; Ferrans et al., 1976; Gelb et al., 1993; Zimmermann et al., 1982; Malhotra et al., 1994; MacMahon et al., 1971; Andreu et al., 2000; Vallance et al., 2000; Ruszkiewicz et al., 1995; Prahlow et al., 1993; Heifetz et al., 1995], but the prevalence is likely to be higher since many cases of Histiocytoid CM may have been misdiagnosed as Sudden Infant Death Syndrome [Grech et al., 2000] [MIM 272120]. The disease was first confused with rhabdomyoma, benign tumors of the myocardium, and was not recognized as a separate pathologic entity until 1962 by Voth [Voth., 1962]. After observing familial tendency, the first author created an Histiocytoid CM registry in order to collect cases and perform analyses with the objective of identifying the causative gene(s). The registry can be found at spponline.org.
Although several disease mechanisms have been proposed, the etiology of Histiocytoid CM remains unknown. An early report [Andreu et al., 2000] of a missense mutation in Cytochrome b (MTCYB [MIM 516020]), a mitochondrion-encoded protein from complex III of the electron transport chain (ETC), suggested a mitochondrial pathology. However, analysis of additional 27 cases from the Histiocytoid CM registry through collaborative work with two institutions failed to verify this gene as a causal gene for Histiocytoid CM. [Andreu et al., 2000] A second mitochondrial mutation was found at A8344G within the MT-TK gene that encodes tRNALYS [MIM 590060] in the mtDNA, and has previously been implicated in myoclonic epilepsy associated with ragged-red fibers syndrome [Vallance et al., 2004] (MERRF [MIM 545000]).
Whole transcriptome DASL profiling of heart tissue from 12 cases and 12 age-matched controls [Shehata et al., 2011] failed to provide evidence that differential gene expression of these putative causal genes for Histiocytoid CM from case studies may have a broader role in pathology. That study did identify seven differentially expressed genes in Histiocytoid CM that relate to interleukin signaling, but a causative role in cardiovascular pathology has not been demonstrated.
Some physiological characteristics of the disease also support mitochondrial dysfunction in the etiology. Most cases of Histiocytoid CM show accumulation of aberrantly-shaped and excessive numbers of mitochondria in the cardiomyocytes. Lipids in small vacuoles are commonly observed to fill the intracellular space, generating a foamy cytoplasm that has been considered to result from failure of energy generation. Unlike other mitochondrial diseases, however, Histiocytoid CM does not affect all cardiomyocytes equally. The observation that lesions tend to be clustered in the vicinity of the Purkinje fibers has also prompted the argument that the disease may be related to defective activity of this sub-type of cardiomyocyte that coordinate the cardiac action potential [Brunton et al., 1977].
MATERIALS AND METHODS
The material, collected over the last decade, includes over 100 cases, the majority of which are from autopsies, as well as over 20 cases collected from children who were diagnosed early and treated either with surgical excision or ablation of the arrhythmogenic foci. In cases where there is extensive involvement of Histiocytoid CM a cardiac transplant was performed. We estimate the sibling recurrence rate to be approximately 5%, but note that this is likely to be downward biased due to the incidence of recurrent miscarriages in some families.
In order to overcome the ambiguity of candidate gene studies, we have initiated whole exome sequencing [Ng et al., 2009] of Histiocytoid CM cases and here report results from five trios consisting of an affected proband and both biological parents (with one exception, Figure 1, HC4) who provided informed consent for the study. Each individual exome was sequenced to an average depth of 172× using Illumina TruSeq technology (Table SI). In addition to cataloguing all instances of recessive and compound heterozygous inheritance in each proband, custom perl scripts were written to identify all heterozygous mutations where both parents were homozygous for the HG19 reference allele. Synonymous substitutions were annotated using the SeattleSeq database [http://snp.gs.washington.edu/SeattleSeqAnnotation138/]. The results are summarized in Table I, which lists de novo and inherited rare variants (MAF<1%), and compound heterozygotes where at least one allele is predicted deleterious by the MutationTaster algorithm [Schwarz et al., 2010].
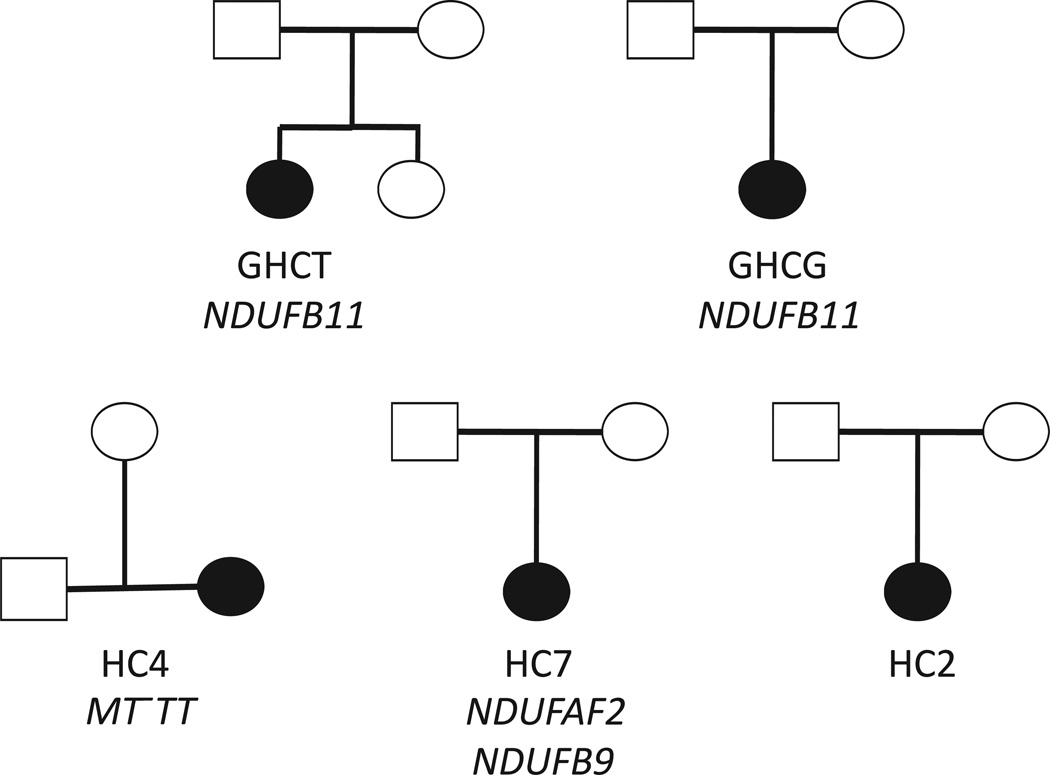
Figure 1.
Patient pedigrees and mutations. Probands are indicated as filled circles (all are females). For HC4, the father was unavailable and is denoted in gray. Implicated causal mutations are shown underneath each trio.
Table I.
Summary of Exome Sequencing Results#
| Individual | De novo non-sense |
De novo mis-sense |
RHD non-syn1 |
RHD syn2 | DCH3 | Candidate mutation |
|---|---|---|---|---|---|---|
| GHC-T | 2 | 0 | 15 | 17 | 1 | NDUFB11 |
| GHC-G | 1 | 0 | 15 | 10 | 2 | NDUFB11 |
| HC2 | 0 | 0 | 9 | 12 | 1 | |
| HC4 | n/a4 | n/a | n/a | n/a | n/a | MT-TT |
| HC7 | 0 | 1 | 19 | 16 | 7 | NDUFAF2/B9 |
See Supplemental Table S3 for a description of the rare or de novo variants at each locus.
Rare homozygous non-synonymous substitutions inherited from carrier parents
Rare homozygous synonymous substitutions inherited from carrier parents
Deleterious compound heterozygotes, at least one allele predicted deleterious by Mutation Taster
Data not available since one parent missing
Sample Acquisition and DNA Isolation
Whole blood samples were obtained from patients and first degree relatives (parents and living siblings) after informed written consent was received under the research protocol and approved by the Emory University Institutional Review Board (IRB). This research is in compliance with the Helsinki Declaration for conduct of research utilizing human subjects. All the cases in this study have been confirmed by Dr Bahig Shehata as Histiocytoid CM. For isolation of DNA, whole blood was stored at 4°C until DNA extraction using the Mag-Bind SQ Blood DNA Isolation Kit (Omega Biotek, Atlanta, GA) according to the manufacturer’s instructions. The concentration of DNA was confirmed on a Nanodrop spectrophotometer, and the RNA quantity values were greater than 1.9.
Exome Sequencing and Variant Prediction
To screen for causal mutations for Histiocytoid CM, the isolated DNA was enriched for 51MB of exonic DNA using Agilent SureSelect Solution probes (Agilent Technologies, Santa Clara, CA). Using the resulting exonic DNA, exome data was generated at Vanderbilt University (Nashville, TN) using high-throughput sequencing on an Illumina HiSeq2000, according to standardized operating procedures. The BWA short read alignment tool [Li et al., 2009] was used to map each mate-pair of reads (length 2×102 nt or 2×77 nt) to the human genome (GRCh37). Samtools [Li et al., 2009] was then used to sort and index the bam files, and subsequently for generation of a pileup which was ported to VarScan [Koboldt et al., 2009] for variant calling. Calls with less than 10% of support from one strand were subsequently removed from further consideration. A total of 273,184 SNPs and 38,321 short indels were identified. Mitochondrial exons were included in the analysis, as they are pulled down passively with the Agilent SureSelect protocol, and the variants reported here had an average read depth of 40×.
Gene Annotation
Variants were analyzed against the RefSeq hg18 gene definitions including 18,933 genes. Potential causal mutations were then annotated with the software package SeattleSeq [SeattleSeq]. A list of rare homozygous deleterious (RHD) mutations was obtained after excluding SNP/Indel calls not within the CDS regions or with less than 20× depth. SNPs were reported as RHD if they had a minor allele frequency (MAF) of less than 0.01 or were not present in dbSNP137. The calls were then manually inspected using the Integrative Genome Viewer (IGV) [Thorvaldsdottir et al., 2013].
MitoMap [Brandon et al., 2005] is a comprehensive database of human mitochondrial DNA from adult human tissues and serves as a universal reference of human mitochondrial DNA variation including tRNA/rRNA mutations, as well as coding and control region mutations. MitoMap was used to identify causal mitochondrial mutations in the disease and first-degree relative samples. We specifically focused on SNPs where the affected patient was heterozygous and the patients’ parents were homozygous for the reference allele.
Zebrafish Husbandry and Morpholino Injection
Zebrafish (Danio rerio) used in this study were housed at the CSIR-Institute of Genomics and Integrative Biology following standard husbandry practices [Westerfield et al., 2000]. All experiments were performed in strict accordance with the recommendations and guidelines laid down by the CSIR Institute of Genomics and Integrative Biology, India, and the protocol was approved by the Institutional Animal Ethics Committee (IAEC) of the Institute. All efforts were made to minimize animal suffering.
Wildtype and transgenic zebrafish embryos were obtained by pair wise mating of adult. Tg(cmlc2:mRFP) that expresses red fluorescent protein in the heart. Alternatively, the Tg(fli1:EGFP; gata1a:dsRed) zebrafish line that expresses green fluorescent protein (GFP) in endothelial cells and red fluorescent protein in blood cells was used [Lalwani et al., 2012].
Morpholino (MO) oligonucleotides (Gene Tools, USA) were dissolved in nuclease free water (Ambion, USA) at a concentration of 1mM according to the protocols recommended by Gene Tools. 1mM stocks of MO oligos were stored at −80°C. Working aliquots of MO oligos were prepared and stored at 4°C. The ndufb11 MO oligo sequences are GTTTCGAGACAGCTACCGCTTCGAG and AGACGTGAGAGCATTCTCCCGACTT. Microinjection glass capillary (World Precision) micropipettes were pulled using Sutter Instrument (USA) and clipped appropriately to deliver 1–3 nl solution into 1–2 cell zebrafish embryos.
RESULTS
In two of the Histiocytoid CM cases, GHCT and GHCG, distinct non-sense de novo mutations were detected in exon 2 of the NDUFB11 gene [MIM 300403, Refseq accession number NC_000023.10]. Given an average of one non-sense mutation per individual, the probability that two of the five cases would have disruptions in the same gene is less than 1 in 4,500. Including a prior expectation of disrupted mitochondrial function, the odds of this observation are considerably smaller. Furthermore, the Exome Variant Server lists just 4 mis-sense rare variants in the gene, only two of which are possibly deleterious, fewer than any other NDUF gene in the database and implying a high level of purifying selection. NDUFB11 encodes the NADH-ubiquinone oxidoreductase ESSS subunit component of complex 1, and the two truncating mutations are likely to have a deleterious effect on the stability of the complex, compromising energy production. One of the two de novo mutations is an A→C transversion that changes a tyrosine at codon 108 to a premature stop, while the other is a C→T transition that changes a tryptophan at codon 85 to a premature stop. Both were confirmed by Sanger sequencing (Supplemental Figure S1). Since there is only a single intron in the gene, it is unlikely that alternative splicing would rescue protein function bypassing the premature stop codons, but no attempt was made to exclude this possibility since RNA was not available for sequencing.
A complete list of de novo, rare homozygous recessive, and deleterious compound heterozygous variants in each of the four trio probands and one additional case with missing paternal data is provided in Table SIII, while all discovered polymorphisms are listed in Table SIV. For one case we have been unable to highlight a likely causal genotype. The proband from trio HC7 was doubly heterozygous for mutations in two different genes, in NDUFAF2 and NDUFB9, both inherited from singly heterozygous parents, and both predicted by MutationTaster [Schwarz et al., 2010] to be deleterious. Although activity at each locus is presumably provided by the alternate allele, we postulate (in light of the NDUFB11 results) that compound loss of function of these two components of the same electron transport chain is sufficient to promote Histiocytoid CM. Furthermore, NDUFAF2 and NDUFB9 have both been implicated previously in mitochondrial complex I deficiency [Schlehe et al, 2013; Haack et al, 2012].
The fourth family, HC4, included a female infant who died of Histiocytoid CM and her brother who, like his mother, has symptoms of arrhythmia and tachycardia. The mother is heteroplasmic for A15924G in the mtDNA, affecting MT-TT (tRNATHR), and both children seem to be homoplasmic for the same mutation, which was previously reported to cause two cases of lethal infantile mitochondrial myopathy (LIMM [MIM 551000]) [Yoon et al., 1991; Brown et al., 1992]. However, according to dbSNP [Sherry et al., 2001] the variant is at a prevalence of 2.3% in humans so would appear to have variable penetrance and expressivity and cannot reliably be considered the cause of Histiocytoid CM in this proband. Table SIII includes evaluation of the potential deleteriousness of each site using the AACDS classification scheme [Preeprem et al., 2013]. A handful of variants were found that are simultaneously highly likely to be deleterious and in genes associated with diseases or traits (AACDS categories 2B and 3B). Proband GHC-G is also homozygous for a non-synonymous substitution in a gene (RAI14) previously linked to linked left ventricular mass, but which is predicted to be benign, while HC4 is homozygous for a recessive variant thought to contribute to colorectal cancer (KMT2C Leucine to Phenylalanine substitution) and for a predicted benign mutation in COL17A1, a hemidesmosomal component gene that has also been repeatedly linked to epidermolysis bullosa [Vanotti et al., 2013].
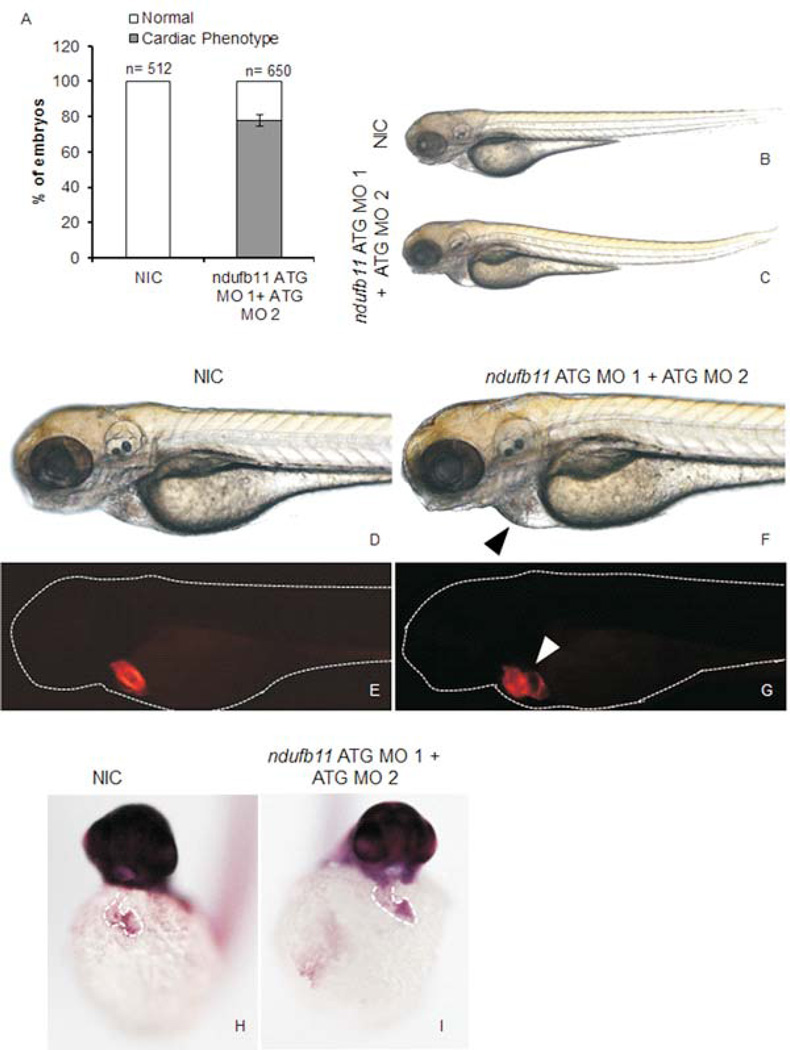
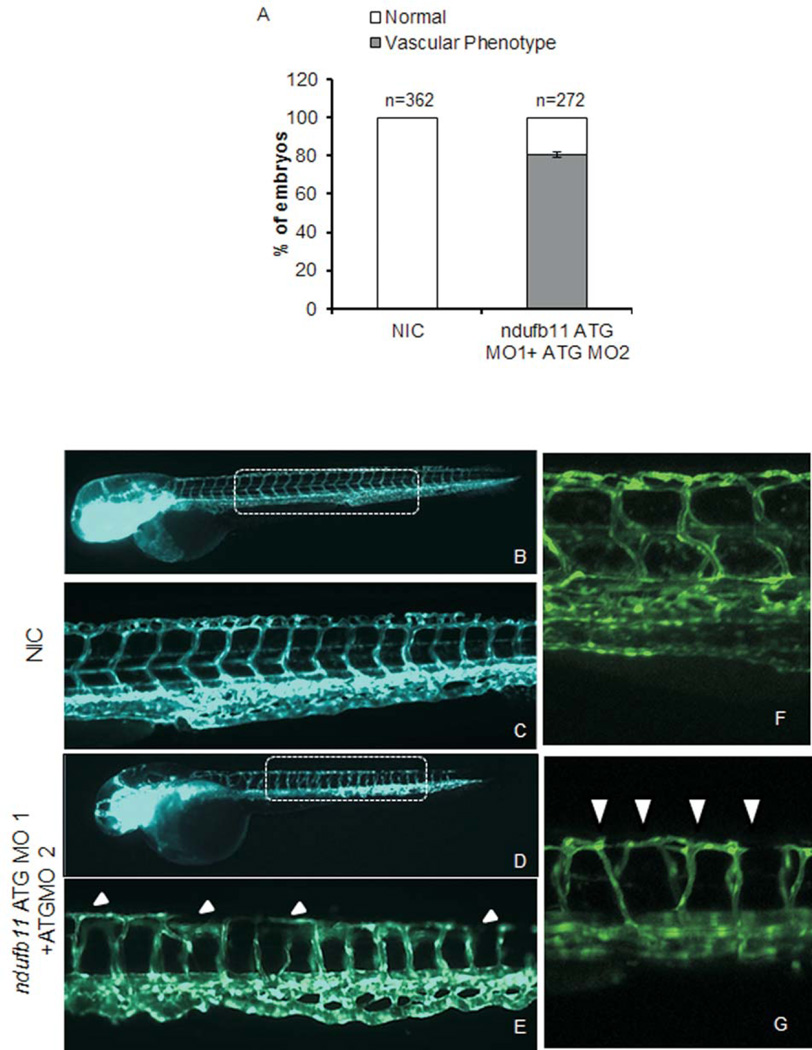
In order to examine the role of NDUFB11 in cardiac development and function, we have employed the zebrafish vertebrate model, which is well established for the study of cardiac dysfunction [Liechke et al., 2007]. Danio rerio ndufb11 is known to be expressed ubiquitously during embryogenesis [Thisse et al., 2001]. Morpholino (MO) mediated transient translational suppression of ndufb11 in zebrafish embryos was performed with two different morpholinos injected into one-cell zebrafish embryos at a final dose of 0.4mM (Figure 2A and Supplemental Figure S2). Knockdown of ndufb11 in zebrafish embryos carrying the heart marker Tg{cmlc2:mRFP} displayed edema and abnormal heart structure in approximately 80% of injected animals. Detailed analysis revealed heart structure defects such as loss of the S-shaped heart at 3 days post fertilization (dpf), resulting in a linear heart tube consistent with defective cardiac looping (Figure 2 E,G; Supplemental Movies 1,2). This linear heart tube defect was confirmed by whole mount in situ hybridization of the gata5 transcript, which is expressed in heart tissue along with the gut and pharynx (Figure 2H,I). Suppression of ndufb11 expression in Tg {fli1: EGFP: gata1: dsRED} zebrafish embryos also revealed defects in angiogenic vessels (Figure 3). We did not observe any sign of apoptosis associated with ndufb11 morpholino injections compared to non-injected zebrafish embryos as determined by whole mount TUNEL assay (Supplemental Figure S3). In summary, knockdown of ndufb11 in developing zebrafish embryos results in abnormal heart structure suggesting its putative role in cardiac development in zebrafish embryos.
Figure 2.
Microinjection of ndufb11morpholino (MO) in zebrafish embryos leads to cardiac tissue defects. (A) Bar graph showing cardiac defect phenotype in ndufb11ATG MO 1+ATG MO 2 injected Tg(cmlc2: mRFP) zebrafish embryos at 3 days post fertilization (dpf). Data is represented as mean percentage ± SD (standard deviation) collected over 7 independent experiments and n is number of embryos analyzed. (B–G) Representative images of cardiac defects in zebrafish embryos at 3 dpf. (B,D,E) Non-injected control embryos (NIC) with normal cardiac development and (C,F,G) ndufb11 MO injected embryos displaying defect in cardiac structure. Arrowheads indicate regions with cardiac tissue defect. (H, I) Expression of gata5 transcript in 48 hpf zebrafish embryos. (B,C) 2.5× magnification, (D–G) 5× magnification.
Figure 3.
Microinjection of ndufb11morpholino (MO) in zebrafish embryos leads to vasculature defects. (A) Bar graph showing embryos with normal and defective inter-segmental vessel in non-injected control (NIC) and 400 µM ndufb11ATG MO 1+ ATG MO 2 morpholino injected embryos at 72 hpf. (B–G) Representative images of 72 hpfTg(fli1:EGFP, gata1a: dsRed) zebrafish embryos. (B,C,F) Non-injected control embryos (NIC) with normal vasculature and (D,G,H) ndufb11 MO injected embryos displaying vasculature defect. (B–E) 2.5× magnification. (F,G) 20× magnification. Images are arranged in a lateral view and inset displaying intersegmental vessels from the trunk region. Arrowheads indicate regions with vascular defects.
DISCUSSION
A causal role for truncation of NDUFB11 in the etiology of Histiocytoid CM helps to explain the female-bias of the disease, which presents in girls in 75% of cases. Whereas most complex I deficiencies are thought to be inherited in a Mendelian recessive manner, these two de novo mutations establish a dominant haploinsufficient phenotype. The gene is located on the short arm of the X-chromosome at interval Xp11.23. It is tempting to speculate that similar mutations occurring in males are embryonic lethal and cause miscarriage, since there would be no residual protein activity. By contrast, male bias for mitochondrial encephalomyopathy was associated with recessive X-linked NDUFA1 mutations [Fernandez-Moreira et al., 2007].
Our data also confirm that Histiocytoid CM is genetically heterogeneous since NDUFB11 did not carry mutations in the other three probands. Thus, some cases are likely due to autosomal mutations that may segregate in a recessive manner, some of which may also be modified in male and female backgrounds.
Many previous studies have implicated 16 different NDUF genes in respiratory chain complex I deficiency, which presents with highly variable morbidity due to exercise intolerance and muscle wasting, and may also result in neonatal mortality [Von Kleist-Retzow JC et al., 1998; Calvo et al., 2012; Swalwell et al., 2011]. Typical features include lactic acidosis, neuropathy, and neurodegeneration, and in approximately one quarter of cases, cardiomyopathy. This raises two questions: why has NDUFB11 escaped identification as a disease gene until now, and why did these two cases present with such a specific and severe cardiac defect?
A possible explanation for the severe defect is that NDUFB11 may be particularly critical in heart tissue. Intriguingly, the BioGPSserver [Wu et al., 2009] reports three-fold elevated expression of NDUFB11 transcripts in the heart over most other tissues, a feature it shares with another cardiomyopathy-associated family member NDUFS2 [MIM 602985]. Most other NDUF genes do not have elevated cardiac expression. However, the correlation is not complete as the Leigh Syndrome [MIM 25600] associated NDUFS7 [MIM 601825] and NDUFS8 [MIM 602141] also show relatively high expression in the heart. We consider it more likely that the Histiocytoid CM phenotype arises as a result of modification of the primary defect in mitochondrial electron transport by the genetic background, either additional rare variants such as those mentioned in the text, or unknown common variant contributions to mitochondrial function.
One further interesting finding arose from the comparison of exome sequence data derived from cardiac tissue and whole blood for the proband GHCG with the de novo Trp85 non-sense mutation. A second non-sense mutation was detected in Cytochrome b, but only at a frequency of 20% and only in the cardiac tissue, possibly indicating clonal selection of a somatic mutation in the diseased heart. This individual also showed a Val→Met mis-sense substitution in ACSS1 [MIM 614355], which encodes a mitochondrial acetyl-coA synthetase. Since the variant is common in all human populations is not causal for Histiocytoid CM, but may have modified the phenotype as well. Whole transcriptome expression profiling of 12 cases and 12 age-matched controls [Shehata et al., 2011] detected significant differential expression of 1356 probes enriched for cell death as well as cardiac development and function. Consequently, there are many possible avenues through which the expressivity of defects in complex I activity might be regulated, giving rise to diseases as diverse as Histiocytoid CM and Leigh syndrome.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge help received from the zebrafish facility staff at CSIR-IGIB, sequencing performed at the genomics core facility of Vanderbilt University directed by Travis Clark, and the assistance of Mark Bouzyk and Dalia Arafat. Thanawadee Preeprem completed the AACDS analyses reported in Supplemental Table SIII.
Appendices/Funding
Exome sequencing was supported by a private donor to BS. KL was the recipient of a Presidential Fellowship from the Regents of the Georgia Institute of Technology. The authors acknowledge funding from the Council of Scientific and Industrial Research (CSIR), India through the BSC0122 Grant. AS acknowledges a junior research fellowship (JRF) from CSIR. MKL acknowledges fellowship funding from BSC0123 grant of CSIR-IGIB. GG is partially supported by Project 3 of NIGMS P01 GM099568 (B. Weir, U. Washington, PI). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors have no conflict of interest and nothing to disclose.
Internet Resources
Online Medelian Inheritance in Man (OMIM): http://www.omim.org/
References
- Andreu AL, Checcarelli N, Iwata S, Shanske S, DiMauro S. A missense mutation in the mitochondrial cytochrome b gene in a revisited case with histiocytoid cardiomyopathy. Pediatr Res. 2000;48:311–314. doi: 10.1203/00006450-200009000-00008. [DOI] [PubMed] [Google Scholar]
- Bove KE, Schwartz DC. Focal lipid cardiomyopathy in an infant with paroxysmal atrial tachycardia. Arch Pathol. 1973;95:26–36. [PubMed] [Google Scholar]
- Brandon MC, Lott MT, Nguyen KC, Spolim S, Navathe SB, Baldi P, Wallace DC. MITOMAP: a human mitochondrial genome database - 2004 update. Nucl. Acids Res. 2005;33:D611–D613. doi: 10.1093/nar/gki079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MD, Torroni A, Shoffner JM, Wallace DC. Mitochondrial trna(thr) mutations and lethal infantile mitochondrial myopathy. Am. J. Hum. Genet. 1992;51:446–447. [PMC free article] [PubMed] [Google Scholar]
- Brunton D, Herdson PB, Becroft DMO. Histiocytoid cardiomyopathy of infancy: an unexplained myofiber degeneration. Pathology. 1977;9:115–122. doi: 10.3109/00313027709085249. [DOI] [PubMed] [Google Scholar]
- Calvo SE, Compton AG, Hershman SG, Lim SC, Lieber DS, Tucker EJ, Laskowski A, Garone C, Liu S, Jaffe DB, Christodoulou J, Fletcher JM. Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Sci Transl Med. 2012;4:118ra10. doi: 10.1126/scitranslmed.3003310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Moreira D, Ugalde C, Smeets R, Rodenburg RJT, Lopez-Laso E, Ruiz-Falco ML, Briones P, Martin MA, Smeitink JA, Arenas J. X-linked NDUFA1 gene mutations associated with mitochondrial encephalomyopathy. Ann. Neurol. 2007;61:73–83. doi: 10.1002/ana.21036. [DOI] [PubMed] [Google Scholar]
- Ferrans VJ, McAllister HA, Haese WH. Infantile cardiomyopathy with histiocytoid change in cardiac muscle cells. Report of six patients. Circulation. 1976;53:708–719. doi: 10.1161/01.cir.53.4.708. [DOI] [PubMed] [Google Scholar]
- Gelb AB, Meter SHV, Billingham ME, Berry GJ, Rouse RV. Infantile histiocytoid cardiomyopathy – myocardial or conduction system hamartoma: what is the cell type involved? Hum Pathol. 1993;24:1226–1231. doi: 10.1016/0046-8177(93)90219-7. [DOI] [PubMed] [Google Scholar]
- Grech V, Ellul B, Montalto SA. Sudden cardiac death in infancy due to histiocytoid cardiomyopathy. Cardiol Young. 2000;10:49–51. doi: 10.1017/s1047951100006387. [DOI] [PubMed] [Google Scholar]
- Haack TB, Madignier F, Herzer M, Lamantea E, Danhauser K, Invernizzi F, Koch J, Freitag M, Drost R, Hillier I, Haberberger B, Mayr JA, Ahting U, Tiranti V, Rötig A, Iuso A, Horvath R, Tesarova M, Baric I, Uziel G, Rolinski B, Sperl W, Meitinger T, Zeviani M, Freisinger P, Prokisch H. Mutation screening of 75 candidate genes in 152 complex I deficiency cases identifies pathogenic variants in 16 genes including NDUFB9. J Med Genet. 2012;49:83–89. doi: 10.1136/jmedgenet-2011-100577. [DOI] [PubMed] [Google Scholar]
- Heifetz SA, Faught PR, Bauman M. Pathological case of the month. Histiocytoid (oncocytic) cardiomyopathy. Arch Pediatr Adolesc Med. 1995;149:464–465. doi: 10.1001/archpedi.1995.02170160118020. [DOI] [PubMed] [Google Scholar]
- Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, Weinstock GM, Wilson RK, Ding L. Varscan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalwani MK, Sharma M, Singh AR, Chauhan RK, Patowary A, Singh N, Scaria V, Sivasubbu S. Reverse genetics screen in zebrafish identifies a role of miR-142a-3p in vascular development and integrity. PLoS ONE. 2012;7:e52588. doi: 10.1371/journal.pone.0052588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and samtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat. Rev. Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- MacMahon HE. Infantile xanthomatous cardiomyopathy. Pediatrics. 1971;48:312–315. [PubMed] [Google Scholar]
- Malhotra V, Ferrans VJ, Virmani R. Infantile histiocytoid cardiomyopathy: three cases and literature review. Am Heart J. 1994;128:1009–1021. doi: 10.1016/0002-8703(94)90601-7. [DOI] [PubMed] [Google Scholar]
- Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler E, Bamshad M, Nicherson DA, Shendure J. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahlow JA, Teot LA. Histiocytoid cardiomyopathy: case report and literature review. J Forensic Sci. 1993;38:1427–1435. [PubMed] [Google Scholar]
- Preeprem T, Gibson G. An association-adjusted consensus deleterious scheme to classify homozygous Mis-sense mutations for personal genome interpretation. BioData Mining. 2013;6 doi: 10.1186/1756-0381-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruszkiewicz AR, Vernon-Roberts E. Sudden death in an infant due to histiocytoid cardiomyopathy. A light-microscopic, ultrastructural, and immunohistochemical study. Am J Forensic Med Pathol. 1995;16:74–80. doi: 10.1097/00000433-199503000-00017. [DOI] [PubMed] [Google Scholar]
- Schlehe JS, Journal MS, Taylor KP, Amodeo KD, LaVoie MJ. The mitochondrial disease associated protein Ndufaf2 is dispensable for Complex-1 assembly but critical for the regulation of oxidative stress. Neurobiol Dis. 2013;58:57–67. doi: 10.1016/j.nbd.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- SeattleSeq. http://snp.gs.washington.edu/SeattleSeqAnnotation137/ [Google Scholar]
- Shehata BM, Bouzyk M, Shulman SC, Tang W, Steelman CK, Davis GK, Moreno CS. Identification of candidate genes for histiocytoid cardiomyopathy (HC) using whole genome expression analysis: analyzing material from the HC registry. Pediatr Dev Pathol. 2011;14:370–377. doi: 10.2350/10-05-0826-OA.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata BM, Patterson K, Thomas JE, Scala-Barnett D, Dasu S, Robinson HB. Histiocytoidcardiomyopathy: three new cases and a review of the literature. Pediatr Dev Pathol. 1998;1:56–69. doi: 10.1007/s100249900007. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbsnp: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swalwell H, Kirby DM, Blakely EL, Mitchell A, Salemi R, Sugiana C, Compton AG, Tucker EJ, BX K, Lamont PJ, Turnbull DM, McFarland R, Taylor RW, Thorburn DR. Respiratory chain complex I deficiency caused by mitochondrial DNA mutations. Eur. J. Hum. Genet. 2011;19:769–775. doi: 10.1038/ejhg.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Pflumio S, Fürthauer M, Loppin B, Heyer V, Degrave A, Woehl R, Lux A, Steffan T, Charbonnier XQ, Thisse C. Expression of the zebrafish genome during embryogenesis. 2001 ZFIN Online Publication ( http://zfin.org/ZDB-PUB-010810-1). [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative genomics viewer (igv): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallance HD, Jeven G, Wallace DC, Brown MD. A case of sporadic infantile histiocytoid cardiomyopathy caused by the A8344G (MERRF) mitochondrial DNA mutation. Pediatr Cardiol. 2004;25:538–540. doi: 10.1007/s00246-003-0446-y. [DOI] [PubMed] [Google Scholar]
- Vanotti S, Chiaverini C, Charlesworth A, Bonnet N, Berbis P, Meneguzzi G, Lacour JP. Late-onset skin fragility in childhood: a case of junctional epidermolysis bullosa of late onset caused by a missense mutation in COL17A1. Br. J. Dermatol. 2013;169:714–715. doi: 10.1111/bjd.12353. [DOI] [PubMed] [Google Scholar]
- Von Kleist-Retzow JC, Cormier-Daire V, de Lonlay P, Parfait B, Chretien D, Rustin P, Feingold J, Rötig A, Munnich A. A high rate (20%–30%) of parental consanguinity in cytochrome-oxidase deficiency. Am J Hum Genet. 1998;63:428–435. doi: 10.1086/301957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth D. Uber die arachnocytose des herzmuskels. Frankfurter Z. Path. 1962;71:646–656. [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Daniorerio) 4th Edition. Eugene: University of Oregon Press; 2000. [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov J, Hodge CL, Haase J, Janes J, Huss JW, Su AI. BioGPS: An extensible and customizable portal for querying and organizing gene annotation resources. Genome Biology. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KL, Aprille JR, Ernst SG. Mitochondrial trna(thr) mutation in fatal infantile respiratory enzyme deficiency. Biochem. Biophys. Res. Commun. 1991;176:1112–1115. doi: 10.1016/0006-291x(91)90399-r. [DOI] [PubMed] [Google Scholar]
- Zimmermann A, Diem P, Cottier H. Congenital ”histiocytoid” cardiomyopathy: evidence suggesting a developmental disorder of the purkinje cell system of the heart. Virchows Arch A Pathol Anat Histol. 1982;396:87–195. doi: 10.1007/BF00431240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.