Highlight
Assessing gall numbers in 331 cultivars of a rice diversity panel has identified two resistant landraces, 11 quantitative trait loci and good candidate genes for resistance to Meloidogyne graminicola.
Key words: GWAS, lectin, Mla, Nematode, resistance, rice diversity panel.
Abstract
The root-knot nematode Meloidogyne graminicola is one of the most serious nematode pests worldwide and represents a major constraint on rice production. While variation in the susceptibility of Asian rice (Oryza sativa) exists, so far no strong and reliable resistance has been reported. Quantitative trait loci for partial resistance have been reported but no underlying genes have been tagged or cloned. Here, 332 accessions of the Rice Diversity Panel 1 were assessed for gall formation, revealing large variation across all subpopulations of rice and higher susceptibility in temperate japonica accessions. Accessions Khao Pahk Maw and LD 24 appeared to be resistant, which was confirmed in large pot experiments where no galls were observed. Detailed observations on these two accessions revealed no nematodes inside the roots 2 days after inoculation and very few females after 17 days (5 in Khao Pahk Maw and <1 in LD 24, in comparison with >100 in the susceptible controls). These two cultivars appear ideal donors for breeding root-knot nematode resistance. A genome-wide association study revealed 11 quantitative trait loci, two of which are close to epistatic loci detected in the Bala x Azucena population. The discussion highlights a small number of candidate genes worth exploring further, in particular many genes with lectin domains and genes on chromosome 11 with homology to the Hordeum Mla locus.
Introduction
The genus Meloidogyne (root-knot nematodes) has been identified as the most scientifically and economically important group of nematodes (Jones et al., 2013). Within this genus, the rice root-knot nematode, Meloidogyne graminicola (Golden and Birchfield, 1965), is considered one of the most important pests of Asian rice (Oryza sativa L.) (Plowright and Bridge, 1990; De Waele and Elsen, 2007; Win et al., 2011; Kyndt et al., 2012; Ji et al., 2013; Jones et al., 2013).
M. graminicola is widely distributed, especially in South and Southeast Asia where it occurs in every rice-producing country surveyed so far (Jain et al., 2012). It has been clearly shown to cause substantial damage to the rice root system (Plowright and Bridge, 1990). It can cause substantial yield losses to upland, lowland, and deepwater rice as well as in rice nurseries (Bridge et al., 2005; Pokharel et al., 2010; Win et al., 2011). It is now considered one of the biotic causal agents of yield failure in tropical aerobic rice (Kreye et al., 2009; De Waele et al., 2013). Currently, M. graminicola infestation of rice-based agrosystems is only a serious problem in the tropics (Jones et al., 2013) but the geographic areas affected might be expected to expand owing to climate change (Bebber et al., 2013).
Second-stage juveniles (J2) invade rice roots just behind the root tips and migrate intercellularly towards the central cylinder. At this site, the J2 puncture selected vascular cells with their stylet and inject pharyngeal secretions, which ultimately leads to the reorganization of the plant cells into typical feeding structures called giant cells. These giant cells are imbedded in galls induced upon penetration of the roots by the J2. The J2 will start to swell and after three moults develops into a swollen female. For the remainder of its sedentary life, this female will feed on the giant cells (Gheysen and Mitchum, 2011). Although gall number is not always correlated with the level of ability of the plant to support nematode reproduction (susceptibility), it usually is a good indicator of the infection level (Dong et al., 2007; Wang et al., 2012). M. graminicola infection of rice plants may result in seedling mortality or reduced vigour, and a substantial reduction in vegetative and yield-contributing traits, yield, and seed quality (De Waele et al., 2013; Patil and Gaur, 2014; Win et al., in press). In an M. graminicola-infested rainfed lowland rice field in Bangledesh, and a similarly infested rainfed upland rice field in Thailand, nematicide application resulted in yield increases of 16–20% (or 1 t/ha) and 12–33%, respectively (Arayarungsarit, 1987; Sharma-Poudyal et al., 2004).
The options to manage M. graminicola population densities below damage threshold levels are limited at the moment. Crop rotation, flooding, and the use of nematicides are practices that are often used to manage plant-parasitic nematodes in infested fields (Bridge et al., 2005) but each of these practices has a number of important drawbacks (De Waele et al., 2013). In this context, the identification and use of rice genotypes with resistance to M. graminicola offers an interesting alternative to limit the yield losses caused by this nematode species.
Accessions with good resistance to M. graminicola, showing consistently low nematode reproduction compared with susceptible reference genotypes, have been found in O. longistaminata A. Chev. & Roehrich and O. glaberrima Steud. (Plowright et al., 1999; Soriano et al., 1999; Cabasan et al., 2012) but so far introgression of this resistance into Asian rice has not been very successful because the interspecific progenies do not express the same degree of resistance observed in African rice. In Asian rice, some resistance to M. graminicola has been reported (Jena and Rao, 1976; Yik and Birchfield, 1979; Sharma-Poudyal et al., 2004; Prasad et al., 2006); however, according to Bridge et al. (2005), only a few of these accessions are truly resistant and the majority are in fact susceptible to M. graminicola (Cabasan MTN, Lahari Z, Galeng J, Price AH, Gheysen G, and De Waele D. Re-examination of the resistance to M. graminicola infection within Asian rice genotypes. In preparation).
A comparative transcriptomics study of the site of M. graminicola infection (galls versus root tips) after 3 days in rice revealed 156 differentially expressed genes (Kyndt et al., 2012). Key metabolic pathways, hormone homeostasis, and epigenetic processes are affected during M. graminicola-induced giant cell development in rice (Ji et al., 2013).
Applying plant molecular genetics has resulted in the identification of several nematode resistance (R) genes or quantitative trait loci (QTLs) for resistance to sedentary endoparasitic nematodes, and some genes mapped to chromosomal locations or linkage maps (Messeguer et al., 1991; Ganal et al., 1995). A few have been cloned (Veremis and Roberts, 2000; Thurau et al., 2010). One of the best characterized and commercially used root-knot nematode resistance genes is Mi-1.2, whch is found in the wild relative of tomato (Lycopersicon peruvianum complex) and confers resistance to several Meloidogyne species (Veremis and Roberts, 2000). In African rice, the major gene Hsa-1 Og confers resistance to the cyst nematode Heterodera sacchari and has been mapped on chromosome 11 of an O. sativa x O. glaberrima interspecific cross (Lorieux et al., 2003). In Asian rice, QTLs for partial resistance to M. graminicola were identified on chromosomes 1, 2, 6, 7, 9, and 11 using F6 recombinant inbred lines of a Bala x Azucena O. sativa mapping population (Shrestha et al., 2007).
The variation in landraces provides an important reservoir of genetic diversity and potential sources of beneficial alleles for rice breeding and improvement (Zhang et al., 2009). Thus germplasm collections are invaluable resources for screening more rice accessions for M. graminicola resistance and offer opportunities for plant breeders to apply conventional selection techniques if resistance is found, and even marker-assisted selection if the resistance can be mapped to the genome. Genome-wide association (GWA) studies with a global collection of 413 diverse rice (O. sativa) accessions including landraces and elite cultivars from 82 countries, known as the Rice Diversity Panel 1 (RDP1), have recently been conducted (Zhao et al., 2011). The authors used 44100 single nucleotide polymorphisms (SNPs) (44K chip) as markers and systematically phenotyped 34 morphological, developmental, and agronomic traits over two consecutive field seasons. GWA analysis of the same RPD1 panel has been used to study the genetic architecture of rice aluminium tolerance (Famoso et al., 2011) and QTLs for grain ionome composition (Norton et al., 2014). In our study, we examined the root galling of 332 accessions of the RDP1 panel 2 weeks after inoculation with J2 of M. graminicola to evaluate the host response of these genotypes, to identify resistant genotypes, and to highlight potential QTLs and candidate genes worthy of further characterisation. Additional experiments with a small number of accessions were carried out to examine if observations were reproducible across differing experimental conditions (substrate, pot size, plant age, and laboratory).
Materials and Methods
Host response of 332 rice accessions to M. graminicola infection 2 weeks after nematode inoculation
This experiment was carried out in a growth chamber at the University of Aberdeen. The 332 cultivars screened were a subset of the 413 rice O. sativa accessions that have been genotyped with 44100 SNPs as previously described (Zhao et al., 2011) (Supplementary Table 1). The 332 cultivars mostly belonged to one of five recognized subpopulations of rice, with 12 aromatic, 57 aus, 64 indica, 78 tropical japonica, and 88 temperate japonica, with the rest (32) an admix between those subpopulations. The seeds of the rice accessions were obtained from the National Rice Research Centre, USA, and bulked in Aberdeen, UK. The nematode screening method was adopted from Shrestha et al., (2007). The accessions were assessed in batches of 40 using 10 temporally separated runs. Each run consisted of three plug trays (LBS, Colne, UK) of 84 wells each [36.5×36.5×50mm (length × width × height)] with each tray containing two replicates of 40 accessions from the RDP1 plus Azucena (tropical japonica rice variety) and Bala (indica rice variety) as reference genotypes. All genotypes were replicated six times, making a total of 252 plants per screening run. Within the three plug trays, a randomized complete block design was used where a block was one half of a tray. The trays were filled with sand and sown directly with two seeds per plug, which were then thinned to one seed per plug after 1 week. Every 2 weeks, another batch of 40 RDP1 accessions plus Azucena and Bala reference plants were sown in a separate screening run. J2 were collected from the roots of 50 mature rice plants as described in Shrestha et al. (2007), except that where cut galls from these stock plant roots were incubated for 7 days at 30°C in Shrestha et al., (2007), we incubated them for only 3 days at 28°C.
The M. graminicola inoculum used was originally obtained from CABI (Egham, UK) and has been maintained on rice for 10 years in Aberdeen. The plants were inoculated with 200 J2 per plant 2 weeks after planting. Two weeks after inoculation, the roots were carefully removed and washed with tap water, and the nematode galls counted. An incubation period of 2 weeks was chosen to terminate the experiment near the end of one nematode life cycle to avoid secondary galling. The life cycle of M. graminicola is completed in 19 days at 22–29°C in well-drained soil (Pokharel et al., 2010; Kyndt et al., 2012; Ji et al., 2013). The temperature in the growth chamber ranged from 28°C (day) to 25°C (night), with 50–70% humidity, and light of 350 µmol m−2 s−1 photosynthetically active radiation at plant height for 12h d−1. The plants were watered daily to field capacity and fertilized twice a week with Yoshida’s nutrient solution (Yoshida et al., 1976). One of the runs included 30 randomly selected plants of RDP1 assessed for a second time to confirm the repeatability of the method.
Host response of 10 rice accessions to M. graminicola infection 5 weeks after nematode inoculation
This experiment was carried out in a greenhouse, also at the University of Aberdeen. After the first screening with the 332 accessions, 10 rice accession were selected based on their root galling: five with the lowest nematode gall numbers (LD 24, Khao Pahk Maw, DV85, Gerdeh, and Ku115) and five with the highest gall numbers (Lusitano Zerawchanica, Karatalski, Taipei 309, Ta Mao Tsao, and Benllok) (Table 1). Seeds were sown in individual 13-cm diameter pots in the same sandy soil as used in the first experiment. Two weeks after planting, each seedling was inoculated with 100 J2 of M. graminicola. Each accession was replicated six times. The experiment was carried out in a greenhouse in July–September 2012 without supplementary light. The minimum temperature was 25°C. The plants were watered daily to field capacity and fertilized once a week with full Yoshida’s nutrient solution (Yoshida et al., 1976). Gall numbers were counted 5 weeks after inoculation with J2.
Table 1.
The 10 rice cultivars used in the 5-week inoculation experiment
| Accession number | Accession name | Origin | Subpopulation |
|---|---|---|---|
| 298 | LD 24 | Sri Lanka | indica |
| 330 | Khao Pahk Maw | Thailand | aus |
| 49 | DV85 | Bangladesh | aus |
| 55 | Gerdeh | Iran | aromatic |
| 96 | KU115 | Thailand | tropical japonica |
| 289 | Lusitano | Portugal | temperate japonica |
| 287 | Zerawchanica Karatalski | Poland | temperate japonica |
| 158 | Taipei 309 | Taiwan | temperate japonica |
| 155 | Ta Mao Tsao | China | temperate japonica |
| 180 | Benllok | Peru | temperate japonica |
Comparison of M. graminicola development on LD 24 and Khao Pahk Maw, with reference rice genotypes
These experiments were carried out at Ghent University. The M. graminicola culture was provided by Prof. Dirk De Waele (University of Leuven, Belgium) and was originally isolated from rice in the Philippines. M. graminicola was maintained on O. sativa cv. Nipponbare in potting soil or on the grass Echinochloa crusgalli. Three-month-old infected plants were used to extract J2 of M. graminicola using a modified Baermann method (Luc et al., 2005). Seventeen-day-old rice seedlings in SAP medium (Reversat et al., 1999) were inoculated with ±200 freshly harvested J2 per plant as previously described (Nahar et al., 2011). Eight plants were analysed per genotype. Rice (O. sativa) genotypes CO39 (ssp. indica) and Taichung65 (ssp. japonica) were used as susceptible references and TOG5674 (O. glaberrima from the International Rice Research Institute) as a resistant reference genotype (Cabasan et al., 2012). The infection level of the plants and the developmental stages of the nematodes were evaluated 2 days after inoculation (DAI) and 17 DAI using acid fuchsin staining (Nahar et al., 2011). The root and shoot lengths were also recorded.
Statistical analyses
Collected experimental data of the RDP1 screen were corrected for block effects (within each run) using Minitab 16 (Minitab Inc, USA). To account for differences between runs, the mean gall numbers of the six replicates of each accession in each run was divided by the combined mean of the check genotypes Azucena and Bala in that particular screening. Because the relative gall number across all RDP1 accessions tested was not normally distributed (Fig. 1), it was square root transformed before further analysis. One-way ANOVA was carried out using Minitab 16 to compare relative galls numbers between rice subpopulations as given in Zhao et al. (2011). Pairwise comparisons among subpopulation means were achieved with Tukey’s method at a 5% level of significance. Correlation analyses using Minitab were conducted between relative means of nematode gall numbers from 30 accessions that were included in two separate runs (to assess reproducibility between runs), and between the relative means of gall number from the 10 accessions assessed in the 332-cultivar screen and the absolute number of galls in the 5-week inoculation experiment. In the analysis of nematode development, ANOVA was performed using the software SPSS (Version: SPSS Statistics 22). The means of the control and treated groups were compared by Duncan’s multiple mean comparison test at the 5% level of significance.
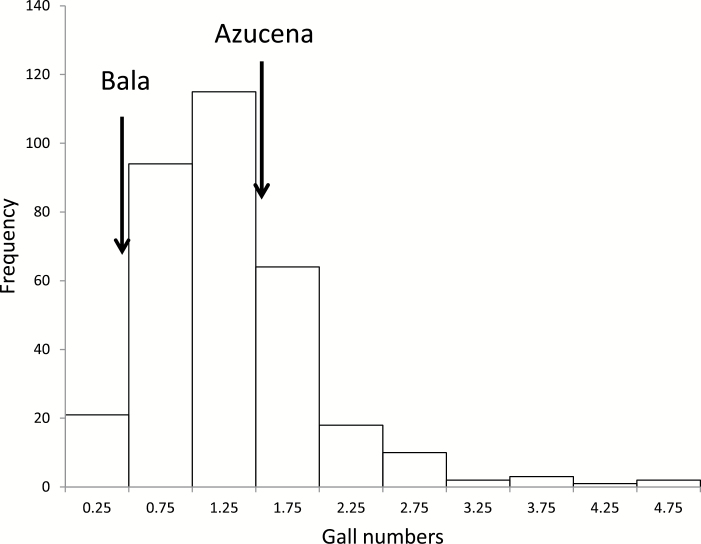
Fig. 1.
Frequency distribution of gall numbers in 332 RDP1 accessions relative to the check genotypes Azucena and Bala.
GWA analysis was performed on all the 332 accessions and also for each of the four subpopulations indica, aus, tropical japonica, and temperate japonica according to Zhao et al. (2011). Briefly, a linear mixed effects model (LMM) was used to model the association between each SNP and the phenotype (mean relative gall number) whilst accounting for population structure and potential relatedness between accessions using efficient mixed model analysis (EMMA) (Yu et al., 2006). For the fixed effects, population structure (Price et al., 2006) was included as the first four principal components of a principal components analysis of all SNPs across accessions (Zhao et al., 2010). Relatedness was incorporated into models as a random effect, estimated using a kinship matrix (Zhao et al., 2011) that measured the genetic similarity between individuals as the proportion of times a given pair of accessions had the same genotype across all SNPs (IBS values). For models of each of the four subpopulations, only random effects were incorporated.
An association between a SNP and phenotype was considered significant if the minor allele frequency (MAF) was >5% and the P-value from LMM <1×10–4. This threshold has been used previously based on the upper-limit false discovery rate, determined from the candidate genes in the same approach as used previously (Li et al., 2010; Zhao et al., 2010; Famoso et al., 2011; Zhao et al., 2011; Norton et al., 2014). For each SNP considered significant with this method, we determined if they were also significant after applying a Benjamini–Hochberg correction. All reporting of SNP position used Pseudomolecules version 7 from the Rice Genome Annotation Project (RGAP) (http://rice.plantbiology.msu.edu/index.shtml).
Candidate gene selection
In this study, QTLs were considered to be within 200kb of a significant SNP. The 200-kb window was selected to fall within the estimated window of linkage disequilibrium (LD) decay in rice (~50–500kb) (Mather et al., 2007; Rakshit et al., 2007; McNally et al., 2009). This is close to the average for different subpopulations in this panel (100kb for indica, 200kb for aus and temperate japonica, and 300kb for tropical japonica) identified by Zhao et al. (2011) and is the approach for listing candidate genes used for this population by those authors. If more than one significant SNP was located within 200kb of another, they were considered to be one QTL. Genes within 200kb of the chosen significant SNP were selected using rice Pseudomolecule version 7. All genes described as transposon-related or retrotransposon-related were not selected. To identify which of these genes are good candidates for resistance to nematodes, we examined further information from the RGAP, such as gene ontology classification, PFAM hits, and Interpro hits, which provide insight into gene function and known gene orthology and homology.
Results
Host response of 332 rice accessions to M. graminicola infection 2 weeks after nematode inoculation
Figure 1 shows the frequency distribution of the relative gall numbers, while all values are given in Supplementary Table 1. As an example, the results for some accessions of the RDP1 in one run that show low, medium, and high galls numbers are presented in Fig. 2. Within most runs there was highly significant variation due to genotype (significant in 9 out of 10 runs), indicating genetic variation for gall numbers is readily detectable. The result of the repeat run with the 30 randomly selected rice accessions revealed a strong correlation with the initial assessment (r = 0.730, P < 0.001) (data not shown), indicating the method is highly repeatable.
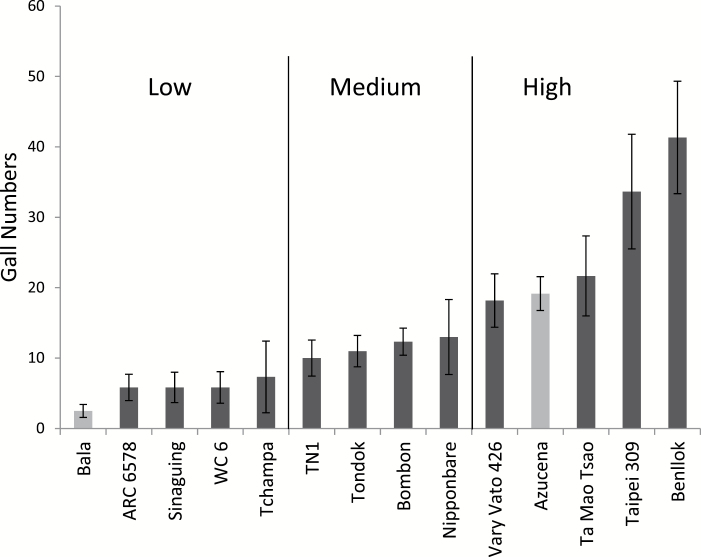
Fig. 2.
Plot of example accessions in one run, including reference cultivars Azucena and Bala and 12 RDP1 accessions grouped as four low, four medium, and four high. Bar is standard error.
The mean number of galls relative to Azucena and Bala was 1.3 while the highest values obtained were over four times the average of the reference genotypes. The three highest scoring accessions were all temperate japonicas: Ta Mao Tsao from China (4.6 relative galls), Taipei 309 from Taiwan (4.5), and Benllok from Peru (5). LD 24, an indica accession from Sri Lanka, showed no galls in any of the six replicates whereas another accession from Thailand, Khao Pahk Maw from the aus subpopulation, had just one gall in one of the replicates with zero galls in the other five replicates. There was a significant difference (P < 0.001) in gall number between rice subpopulations that explained 21% of the variation in relative gall number (Fig. 3). The temperate japonica had the highest number of galls, followed by tropical japonica, aromatic, indica, and aus. A Tukey’s test indicated that the number of galls in the temperate japonicas was significantly higher than in the other subpopulations.
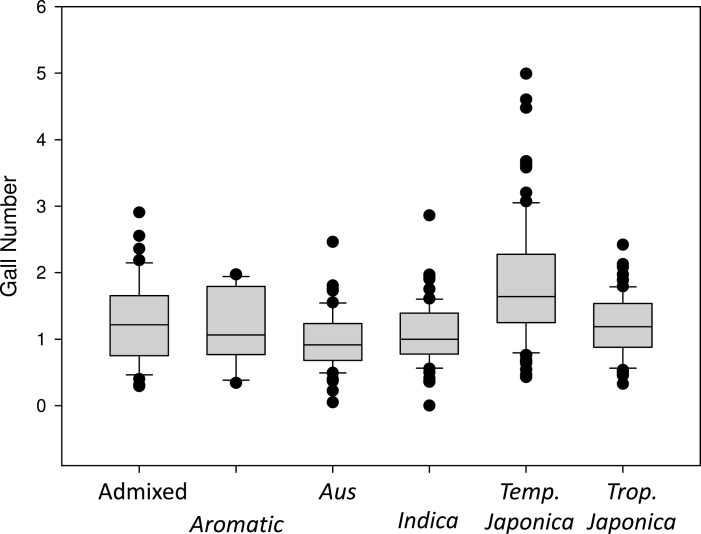
Fig. 3.
Box plot of relative gall numbers in the different subgroups of rice. Temperate japonicas were significantly higher.
Host response of 10 rice accessions to M. graminicola infection 5 weeks after nematode inoculation
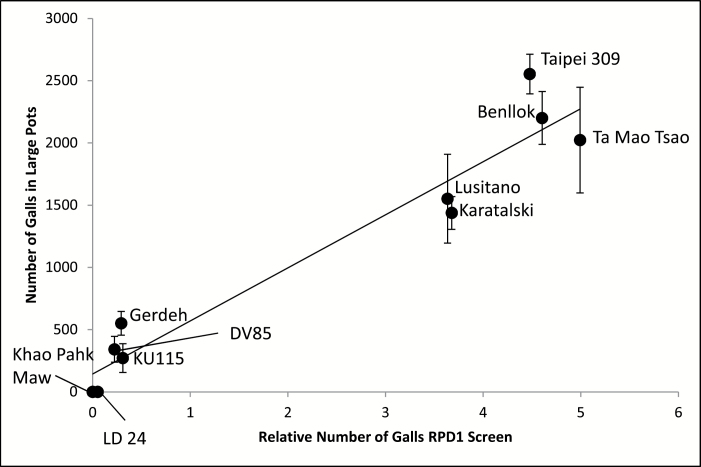
The number of galls observed in the 10 selected accessions of the RDP1 5 weeks after inoculation was highly significantly different between accessions (P < 0.001, R2 = 79%) and correlated very strongly (r = 0.970, P < 0.001) with the nematode gall numbers in the initial 2-week nematode screening of 332 diverse cultivars (Fig. 4). The indica LD 24 and aus Khao Pahk Maw had zero galls in the six replicates of this experiment, confirming the same result in the initial 2-week nematode screening of the RDP1.
Fig. 4.
Plot of mean and standard error of gall numbers in 10 RDP1 accessions inoculated for 5 weeks in large pots versus the relative gall numbers obtained after 2 weeks when RDP1 was screened. Note no galls were detected in Khao Pahk Maw and LD 24. Correlation r = 0.967.
Comparison of M. graminicola development on LD 24 and Khao Pahk Maw with that on reference rice genotypes
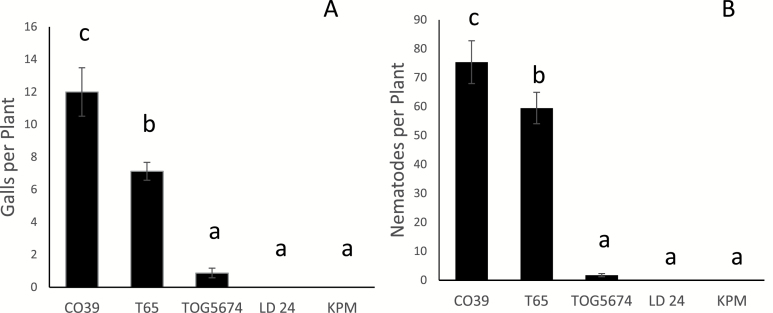
Two M. graminicola-susceptible O. sativa genotypes, CO39 and Taichung65 (T65), and one M. graminicola-resistant African rice, O. glaberrima, genotype TOG5674 were used as controls. LD 24 and Khao Pahk Maw showed nematode infection-resistance comparable to that of TOG5674. At 2 DAI, on average less than two J2 of M. graminicola had penetrated the TOG5674 roots and no J2 had penetrated the roots of LD 24 and Khao Pahk Maw (Fig. 5 A, B). The susceptible rice genotypes CO39 and T65 had 7–12 galls per plant, with 59–75 J2 in their roots (Figs 5 A, B).
Fig. 5.
Responses of rice genotypes to root knot nematode M. graminicola 2 DAI. Two susceptible genotypes, CO39 and T65, and one resistant genotype, TOG5674, were used as control treatments to analyse the susceptibility of the rice accessions LD 24 and Khao Pahk Maw. Each treatment was a pool of eight individual plants. The number of inoculated nematodes per plant was ±200 J2. The response of each genotype was evaluated in terms of (A) number of galls per plant and (B) number of nematodes (J2) per plant. Each bar with standard error (±SE) represents the average number of galls or nematodes. Letter(s) on error bars are based on statistical analysis (Duncan’s multiple range tests after one-way ANOVA). Genotypes having different letters are statistically significant (P = 0.05). The experiment was done twice with similar results.
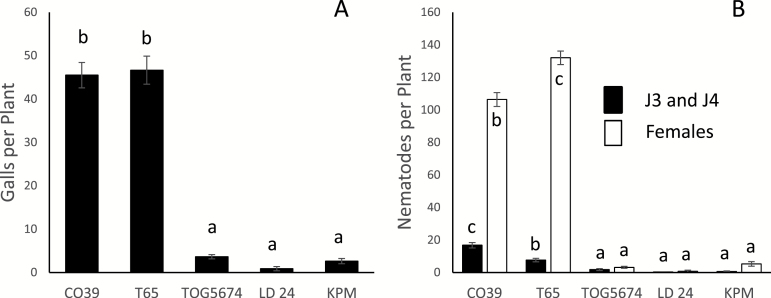
At 17 DAI, the trend of nematode-infection level in resistant and susceptible rice genotypes was similar to that at 2 DAI. The number of galls and developed females were comparable in the resistant rice genotype TOG5674 and Khao Pahk Maw (about five females per plant) and this was significantly lower than in the susceptible rice genotypes, which had on average >100 females per plant (Fig. 6). Most remarkably, in LD 24 we observed on average less than one gall and one female per plant (Fig. 6).
Fig. 6.
Responses of susceptible CO39 and T65, and resistant TOG5674, LD 24, and Khao Pahk Maw rice genotypes to M. graminicola 17 DAI with ±200 nematodes. Response of each genotype is evaluated in terms of (A) number of galls per plant, (B) number of nematodes (J3 and J4, and females) per plant. Each bar with standard error (±SE) represents the average number of galls or nematodes recorded on eight plants for each genotype. Letter(s) on error bars are based on statistical analysis (Duncan’s multiple range tests after one-way ANOVA). Genotypes having different letters are statistically significant (P = 0.05). The experiment was done twice with similar results.
GWA study
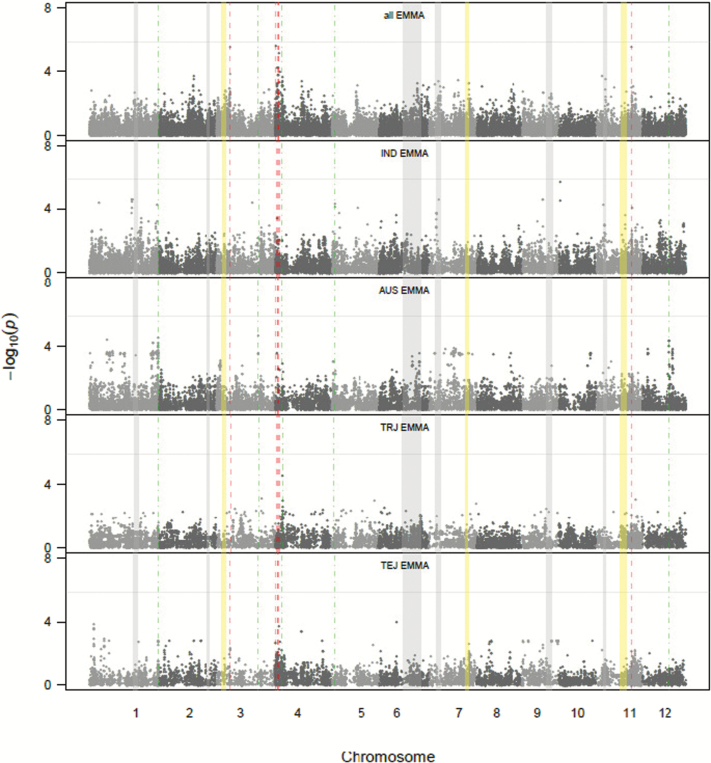
The result of the GWA scans is summarized in Fig. 7 and Table 2. The qq plots are presented in Supplementary Fig. 1. A total of 18 significant SNPs were associated with nematode gall numbers using the EMMA, judged to represent 11 QTLs (Table 2). Five of the SNPs were not significant after a Benjamini–Hochberg correction, which represents four of the 11 QTLs. This gives stronger confidence for the seven QTLs that were detected after this correction, but it does not mean the others are not true QTLs. When analysing the population as a whole, we identified eight SNPs as associated with gall number, appearing to represent five QTLs, on chromosomes 3, 4 (three QTLs), and 11. These are identified with red dotted lines in Fig. 7. In the separate subpopulation analysis, we detected seven significant SNPs representing three QTLs on chromosome 1, 4, and 12 in the aus subpopulation, two SNPs from one QTL on chromosome 5 in indica, and one SNP on chromosome 4 in the tropical japonica. No SNP was observed in temperate japonica.
Fig. 7.
GWA of nematode gall numbers across and within subpopulations. P-values from the mixed model for all cultivars in top panel (all), and for tropical japonica (TRJ), temperate japonica (TEJ), indica (IND), and aus (AUS) subpopulations individually in subsequent panels. The aromatic subpopulation was not included owing to its small sample size. X-axis indicates the SNP location along the 12 chromosomes y-axis is the −log 10 (P value). Red dotted lines indicate where the associated SNPs in the ‘all’ analysis were detected while green were detected in one of the subpopulation analyses. Grey shading indicates QTLs detected in [33] while those in yellow are biallelic epistatic interactions also detected in the [33] study but not reported.
Table 2.
Eighteen SNPs with significance association with relative nematode gall numbers using EMMA. SNPs in bold were significant after Benjamini–Hochberg correction
| Suspected QTLs | SNP id | Analysis | Chromosome | Position (bp) | P-value | MAF (%) |
|---|---|---|---|---|---|---|
| 1 | dd1002102 | AUS | 1 | 42554085 | 2.81E−05 | 8.6 |
| 1 | id1027818 | AUS | 1 | 42581030 | 7.57E−05 | 5.6 |
| 1 | id1027825 | AUS | 1 | 42582856 | 6.37E−05 | 5.5 |
| 3.1 | id3004493 | All | 3 | 8544211 | 2.75E−06 | 45 |
| 3.1 | id3004510 | All | 3 | 8573136 | 3.28E−06 | 45 |
| 3.1 | id3004522 | All | 3 | 8595237 | 3.28E−06 | 45 |
| 3.2 | id3011085 | AUS | 3 | 8596299 | 2.18E−05 | 5.4 |
| 4.1 | id4000274 | All | 4 | 474253 | 2.27E−06 | 8.6 |
| 4.2 | id4000769 | All | 4 | 1405412 | 6.59E−05 | 8.4 |
| 4.3 | id4000913 | All | 4 | 1876908 | 6.81E−05 | 12 |
| 4.4 | id4001092 | All | 4 | 2457675 | 8.00E−06 | 17 |
| 4.5 | id4001733 | TRJ | 4 | 4276025 | 3.06E−05 | 14 |
| 5 | id5001000 | IND | 5 | 1572494 | 7.78E−05 | 7.7 |
| 5 | id5001019 | IND | 5 | 1656600 | 5.18E−05 | 6.8 |
| 11 | id11008353 | All | 11 | 22245292 | 2.65E−06 | 5.2 |
| 12 | id12005724 | AUS | 12 | 16909682 | 9.95E−05 | 5 |
| 12 | id12005764 | AUS | 12 | 17074099 | 4.50E−05 | 11 |
| 12 | id12005794 | AUS | 12 | 17282922 | 4.51E−05 | 6.8 |
None of the QTLs detected by the GWA study are co-located with main effect QTLs reported for the same trait in the Bala x Azucena mapping population (Shrestha et al., 2007) (grey shading in Fig. 7). However, Shrestha et al. (2007) did not report biallelic epistatic interactions. Subsequent analysis of that data using the programme QTLMapper version 1 (Wang et al., 1999a; Wang et al., 1999b) revealed two epistatic interactions in the Bala x Azucena mapping population, both involving a locus on chromosome 11 at around 13–18 Mbp, one with a locus on chromosome 3 at around 4 Mbp (explaining 23% of the genetic variation) and another with a locus on chromosome 7 around 24 Mbp (explaining 18% of the genetic variation) (yellow shading in Fig. 7). These two epistatic interactions were confirmed to be both strong and highly significant by a three-way ANOVA of the gall numbers in the 4-week experiment reported in Shrestha et al. (2007) with markers RG409, RG650, and a1245y as factors. This model explains 30% of the variation for gall numbers, and significant interactions were detected between RG409 and a1245y (P = 0.001) and between RG650 and a1245y (P < 0.001). These interactions are presented graphically in Supplementary Fig. 2. The epistatic loci on chromosomes 3 and 11 are within a few megabase pairs of QTLs detected by the GWA study and may well be within expected confidence intervals of biallelic linkage mapping.
A total of 493 positional candidate genes were selected within 200kb of the identified significant SNPs/genomic regions representing 11 suspected QTLs for nematode resistance (see Supplementary Table 2). These included genes without functional annotation (191 expressed proteins and 34 hypothetical proteins). Of these 493 genes, 16 were previously identified as differentially regulated after 3 or 7 days in rice roots infected with M. graminicola (Kyndt et al., 2012) (15 up-regulated and one down-regulated) whereas 51 were identified as up-regulated and 6 down-regulated in the giant cells of M. graminicola-infected rice by Ji et al. (2013) (indicated in Supplementary Table 2). Five genes were differentially regulated in both studies. These genes may be good candidate genes for nematode resistance.
Discussion
Host response of the rice cultivars examined to M. graminicola infection
The present study screened 332 diverse rice accessions from the RDP1 for gall development of M. graminicola. We found significant variation in the degree of susceptibility of O. sativa accessions to infection. Thirty randomly selected rice accessions from the RDP1 were used to repeat the 2-week experiment, confirming the repeatability of the method, while the method was further evaluated using a longer pot experiment for 5 weeks after inoculation using 10 selected accessions of the RDP1. Our results provide robust evidence that this is a good method to screen for rice–nematode interactions. The mean gall number ranged from 0 galls per plant in cultivar LD 24 (in a run with a mean number of galls in Azucena and Bala of 14.2 and 1.3 respectively) to 41 galls per plant in Benllok (in a run with a mean number of galls in Azucena and Bala of 19.2 and 2.5 respectively) when assessed 2 weeks after inoculation. In the larger, longer pot experiments, Benllok had over 2000 galls whereas LD 24 still had 0 galls per plant (Fig. 4). The correlation coefficient reflected a strong significant positive relationship between gall numbers per accession in the two experiments (2 and 5 weeks after inoculation). The differential susceptibility of O. sativa accessions to M. graminicola revealed by the results of our experiment supports the findings reported by other authors (Plowright et al., 1999; Soriano et al.; 1999, Shrestha et al., 2007) that accessions differ in their reaction to M. graminicola. However, the remarkable result of the present data is that they suggest for the first time a complete resistance to infection of M. graminicola of two O. sativa accessions, LD 24 (an indica from Sri Lanka) and Khao Pahk Maw (an aus from Thailand). These accessions had no nematode galls in the six replicates of the 5-week experiment (Fig. 4).
This resistance of LD 24 and Khao Pahk Maw was confirmed by an independent study on the same cultivars in a different laboratory using a different nematode population. Analysis of the infected rice plants at 2 DAI revealed the absence of penetrated nematodes in the resistant accessions, suggesting that the resistance acts at a very early step during the infection by preventing or delaying penetration. This indicates a different resistance mechanism than in the case of Mi-1.2, where the nematodes enter the roots but induce a rapid necrosis when attempting to induce giant cells (Williamson and Hussey, 1996). Analysis of LD 24 and Khao Pahk Maw roots at 17 DAI showed that very few nematodes had entered the root later and developed into females.
The very low nematode infection was not due to poor root growth because both Khao Pahk Maw and LD 24 had more roots than the susceptible genotypes CO39 and T65 (data not shown). Nevertheless, it is important to know how LD 24 and Khao Pahk Maw perform in the field and how nematode reproduction in these accessions compares to that in susceptible accessions. Experiments in raised beds in the Philippines have already confirmed the resistance in field-like conditions (Cabasan et al., in preparation).
The fact that reports of resistance to root-knot nematodes in O. sativa are rare and irreproducible (Cabasan et al., in preparation) was a major motivation to further analyse the accessions that showed no galls in the RDP1 screening. Shrestha et al. (2007) reported QTLs for partial resistance to M. graminicola in O. sativa and resistance in the O. glaberrima accession CG14. Lorieux et al. (2003) reported resistance to Meloidogyne spp. and the cyst nematode Heterodera sacchari in varieties of the African rice cultivated species O. glaberrima. Effort has been made in the past to transfer the Hsa-1 Og gene from rice O. glaberrima chromosome 11 to O. sativa through interspecific crossing but the interspecific progenies tested so far have not been able to express the same degree of resistance (Plowright et al., 1999; Lorieux et al., 2003).
Phenotyping of the diversity panel has provided valuable information about the range and distribution of nematode susceptibility in O. sativa and offered new insights into the evolution of the susceptibility of the rice subspecies to nematode establishment (i.e. ability to form galls). The mean number of nematode galls in the subspecies japonica was greater than 1.5 times that of indica (P < 0.05) and the relative number of nematode galls in the five subpopulations indicates that temperate japonicas had most galls on average (temperate japonica > tropical japonica ≈ aromatic ≈ indica ≈ aus), which may reflect the fact that the nematode is not a pest of temperate rice production so no selection has been operating to introduce resistance. It has been suggested that the range of most crop pests and pathogens are moving in a poleward direction under the influence of global warming at an average rate of 2.7 km per year, with wide variation between types and individual pests/pathogens (Bebber et al., 2013). It may therefore be expected that the root-knot nematode will become an increasingly important pest in temperate rice regions as the planet warms. The high gall numbers observed in the temperate japonicas should be a cause of concern in this context.
Genetic loci affecting galling
In the interest of brevity, we are highlighting only two of the most noteworthy functional links between genes under QTLs and nematode resistance. First it is noteworthy that six of the identified 11 QTL regions contain genes annotated as containing lectin domains (See Supplementary Table 2). There is one gene in QTL 1 (annotated as S-locus lectin protein kinase), one in QTL 3 (annotated as lectin-like receptor kinase), two in QTL 4.1 (both legume lectins beta domain containing protein), three in QTL 4.2 (one annotated as above and two jacalin-like lectin domain containing protein), two in QTL 4.4 (both the legume lectin type), and one in QTL 5 (lectin protein kinase). These 10 genes represent only 8% of those genes in rice annotated with ‘lectin’ because there are 125 of them. But given that these 11 regions together are only 0.55% of the genome [(0.2×11)/400 Mb], this is more ‘lectin’ genes than chance would suggest. Doing similar analysis for genes with common annotations reveals that these lists contain 3.75% of the 160 ‘peroxidase’ genes, 2.6% of the 380 ‘transferase’ genes, 1.5% of the 470 ‘synthase’ genes, 1.0% of the 192 ‘ubiquitin’ genes, and 0% of the 221 ‘reductase’ genes. Also noteworthy here is that in the QTL 4.1, three additional genes have a lectin legB domain despite not being annotated as lectin genes (LOC_04g01860, 01874, and 01890). Lectins are recognized as proteins associated with plant resistance to nematodes and transgenic alteration of lectin expression has been shown to confer nematode resistance (Fuller et al., 2008).
The second observation worth highlighting is that QTL 11.1 contains clusters of both mla1-like genes and highly homologous stripe rust-resistance proteins. In the gene list are seven stripe rust-resistance proteins surrounding two genes annotated as mla or mla1 putative. Mla is a powdery mildew resistance gene of barley (Hordeum vulgare L.) (Wei et al., 1999). It has very strong homology to stripe rust-resistance genes [e.g. 64% amino acid identity between LOC_Os11g37790 (mla putative) and LOC_Os11g37774 (rust stripe resistance protein Yr10)]. Two of these stripe rust genes were shown to be down-regulated in giant cells (LOC_Os11g37774 and LOC_Os11g37860) (Ji et al., 2013) while one (LOC_Os11g37850) is up-regulated in infected rice roots after 7 days (Kyndt et al., 2012). Because this locus may co-localize with the epistatic QTL identified in the Bala x Azucena mapping population, it is relevant to examine the Bala and Azucena genome sequences (Fastq data have been deposited in the NCBI Short Read Archive Acc_ID SRA050654.1). This reveals that the stripe rust genes LOC_Os11g37759, Os11g37774, and Os11g37780 appear to be missing in Bala while the two mla1 genes are missing in both Azucena and Bala, suggestive of major chromosomal re-arrangement in this region. Seifi et al. (2011) have provided strong genetic evidence in tomato that the nematode- and insect-resistance gene Mi-1 co-segregates or is causally related to mildew resistance.
The two cultivars LD 24 and Khao Pahk Maw are remarkable because they hardly form galls compared to all the other accessions assessed. Because this phenotype is very rare (in the RDP1 and across O. sativa), GWA studies will not detect it (because alleles with frequency below 5% will not be considered). If these two accessions are resistant because of the presence of the same single and very rare gene (an assumption that might well not be true) then it should be possible to identify a likely position of that gene based on the SNP data. An attempt was made to do that in three steps. First, SNPs were filtered to remove all SNPs that were not common between LD 24 and Khao Pahk Maw, leaving about 20 000 SNPs. Second, SNPs were eliminated if they were found in the nearest accession to LD 24 based on principal components analysis, the Indonesian indica Seratoes Hari that had a mean gall number value of 1 (i.e. exactly on the mean of Bala and Azucena). This left 3737 SNPs. Finally, because the region in LD 24 and Khao Pahk Maw must be rare, all SNPs within this group with a MAF value below 5% were identified. There were 16 SNPs where the allele in the resistant cultivars had a low frequency. Six of these fall in a cluster between 42.31 and 42.60 Mbp on chromosome 1, five cluster between 0.95 and 1.24 Mbp on chromosome 3, and three cluster between 26.32 and 26.33 Mbp on chromosome 11, leaving two singletons at 2.47 and 18.45 Mbp on chromosome 11. Further mapping will be required to confirm if these loci really harbour resistance genes but it is noteworthy that at 26.4Mb on chromosome 11 is a resistance gene homolog (LOC_Os11g43700) annotated as RGH1A, which is associated with the Mla locus in barley (Wei et al., 2002).
In conclusion, we have demonstrated significant variation in the degree of susceptibility of O. sativa accessions to M. graminicola infection. For the first time, two O. sativa accessions (LD 24 and Khao Pahk Maw) have been revealed to have complete resistance to infection by M. graminicola. Significant differences were seen in the number of galls among the rice subpopulations. The temperate japonicas had a higher number of galls on average than the tropical japonicas, aromatics, indicas, and aus. We have also identified candidate genes worth pursuing for nematode resistance.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. 1 qq plots for all data with naïve and mixed model results, plus naïve for each of the four subpopulations. Dotted red line is the one-to-one line.
Supplementary Fig. 2 Epistatic interaction for gall number in the Bala x Azucena mapping population. The mean number of galls 4 weeks after inoculation with M. graminicola for all six combinations of allelic combinations of markers in order of RG409 (chromosome 3), RG650 (chromosome 7), and a1245y (chromosome 11) where A is Azucena and B is Bala. Bar is standard error. Bars that do not share the same letter above are different by Tukey’s test at P < 0.05. Note this is a further analysis of data first reported in Shrestha et al. (2007). A three-way ANOVA with these markers as factors explains 30% of the variation for gall numbers, and significant interactions are detected between RG409 and a1245y (P = 0.001) and between RG650 and a1245y (P < 0.001).
Supplementary Table 1 Accession details and relative gall numbers for the Rice Diversity Panel 1.
Supplementary Table 2 List of Gene 200 kb either side of the significant SNPs.
Acknowledgements
SD received sponsorship from the Government of the Rivers State, Nigeria. We acknowledge the financial support of GOA 01GB3013 from Ghent University. The authors wish to thank Prof. Dirk De Waele for valuable advice on the manuscript.
References
- Arayarungsarit L. 1987. Yield ability of rice varieties in fields infested with the root-knot nematode. International Rice Research Newsletter 12, 14. [Google Scholar]
- Bebber DP, Ramotowski MAT, Gurr SJ. 2013. Crop pests and pathogens move polewards in a warming world. Nature Climate Change 3, 985–988. [Google Scholar]
- Bridge J, Plowright RA, Peng D. 2005. Nematode parasites of rice. In: Luc M, Sikora RA, Bridge J, eds. Plant-Parasitic Nematodes in Subtropical and Tropical Agriculture . CAB International: Wallingford, pp. 87–130. [Google Scholar]
- Cabasan MTN, Kumar A, De Waele D. 2012. Comparison of migration, penetration, development and reproduction of Meloidogyne graminicola on susceptible and resistant rice genotypes. Journal of Nematology 14, 405–415. [Google Scholar]
- De Waele D, Das K, Zhao D, Tiwari RKS, Shrivastava DK, Vera-Cruz C, Kumar A. 2013. Host response of rice genotypes to the rice root-knot (Meloidogyne graminicola) under aerobic conditions. Archives of Phytopathology and Plant Protection 46, 670–681. [Google Scholar]
- De Waele D, Elsen A. 2007. Challenges in tropical plant nematology. Annual Review of Phytopathology 45, 457–485. [DOI] [PubMed] [Google Scholar]
- Dong W, Holbrook CC, Timper P, Brenneman TB, Mullinix BG. 2007. Comparison of methods for assessing resistance to Meloidogyne arenaria in peanut. Journal of Nematology 39, 169–175. [PMC free article] [PubMed] [Google Scholar]
- Famoso AN, Zhao K, Clark RT, Tung C, Wright MH, Bustamante C, Kochian LV, McCouch SR. 2011. Genetic architecture of aluminium tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping, PLoS Genetics 7, e1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller VL, Lilley CJ, Urwin PE. 2008. Nematode resistance. New Phytologist 180, 27–44. [DOI] [PubMed] [Google Scholar]
- Ganal MW, Simon R, Brommonschenkel S, Arndt M, Phillips MS, Tanksley SD, Kumar A. 1995. Genetic mapping of a wide spectrum nematode resistance gene (Hero) against Globodera rostochiensis in tomato. Molecular Plant Microbe Interactions 8, 886–891. [DOI] [PubMed] [Google Scholar]
- Gheysen G, Mitchum MG. 2011. How nematodes manipulate plant development pathways for infection. Current Opinion in Plant Biology 14, 415–421. [DOI] [PubMed] [Google Scholar]
- Golden AM, Birchfield W. 1965. Rice root-knot nematode (Meloidogyne graminicola) as a new pest of rice. Plant Disease Reports 52, 423. [Google Scholar]
- Jain RK, Khan MR, Kuman V. 2012. Rice root-knot nematode (Meloidogyne graminicola) infestation in rice. Archives of Phytopathology and Plant Protection 45, 635–645. [Google Scholar]
- Jena RN, Rao YS. 1976. Nature of root-knot (Meloidogyne graminicola) resistance in rice (Oryza sativa). 1. Isolation of resistant genotypes. Proceedings of the Indian Academy of Science 83, 177–184. [Google Scholar]
- Ji H, Gheysen G, Denil S, Lindsey K, Topping JF, Nahar K, Haegeman A, De Vos WH, Trooskens G, Van Criekinge W, De Meyer T, Kyndt T. 2013. Transcriptional analysis through RNA sequencing of giant cells induced by Meloidogyne graminicola in rice roots. Journal of Experimental Botany 64, 3885–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JT, Haegeman A, Danchin EGJ, Gaur HS, Helder J, Jones MGK, Kikuchi T, Manzanilla-Lopez R, Palomares-Rius JE, Wesemael WML, Perry RN. 2013. Top 10 plant-parasitic nematodes in molecular plant pathology. Plant Molecular Pathology 14, 946–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreye C, Bouman BAM, Reversat G, Fernandez L, Vera Cruz C, Elazegui F, Faronilo JE, Llorca L. 2009. Biotic and abiotic causes of yield failure in tropical aerobic rice. Field Crops Research 112, 97–106. [Google Scholar]
- Kyndt T, Denil S, Haegeman A, Trooskens G, De Meyer T, Van Criekinge W, Gheysen G. 2012. Transcriptional reprogramming by root knot and migratory nematode infection in rice. New Phytologist 196, 887–900. [DOI] [PubMed] [Google Scholar]
- Li Y, Huang Y, Bergelson J, Nordborg M, Borevitz JO. 2010. Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana . Proceedings of the National Academy of Science USA 107, 21199–21024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieux M, Reversat G, Diaz SXG, Denance C, Jouvenet N, Orieux Y, Bourger N, Pando-Bahuon A, Ghesquiere A. 2003. Linkage mapping of Hsa-1Og, a resistance gene of African rice to the cyst nematode, Heterodera sacchari . Theoretical and Applied Genetics 107, 691–696. [DOI] [PubMed] [Google Scholar]
- Luc M, Bridge J, Sikora RA. 2005. Reflections on nematology in subtropical and tropical agriculture. In: Luc M, Bridge J, Sikora RA, eds. Plant Parasitic Nematodes in Subtropical and Tropical Agriculture . CAB International: Wallingford, UK, pp. 1–10. [Google Scholar]
- Mather K, Caicedo A, Polato N, Olsen K, McCouch S, Purugganan MD. 2007. The extent of linkage disequilibrium in rice (Oryza sativa L.), Genetics 177, 2223–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally KL, Childs KL, Bohnert R, Davidson RM, Zhao K, Ulat VJ, Zeller G, Clark RM, Hoen DR, Bureau TE, Stokowski R, Ballinger DG, Frazer KA, Cox DR, Padhukasahasram B, Bustamante CD, Weigel D, Mackill DJ, Bruskiewich RM, Ratsch G, Buell CR, Leung H, Leach JE. 2009. Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proceedings of the National Academy of Science USA 106, 12273–12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messeguer R, Ganal M, de Vicente MC, Young ND, Bolkan H, Tanksley SD. 1991. High resolution RFLP map around the root knot nematode resistance gene (Mi) in tomato. Theoretical and Applied Genetics 82, 529–536. [DOI] [PubMed] [Google Scholar]
- Nahar K, Kyndt T, De Vleesschauwer D, Hofte M, Gheysen G. 2011. The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice. Plant Physiology 157, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton GJ, Douglas A, Lahner B, Lahner B, Yakubova E, Guerinot ML, Pinson SRM, Tarpley L, Eizenga GC, McGrath SP, Zhao FJ, Islam MR, Islam S, Duan G, Zhu Y, Salt DE, Meharg AA, Price AH. 2014. Genome wide association mapping of grain arsenic, copper, molybdenum and zinc in rice (Oryza sativa L.) grown at four international field sites. PLoS One 9, e89685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil J, Gaur HS. 2014. The effect of root-knot nematode, Meloidogyne graminicola, on the quality and vigour of rice seed. Nematology 16, 555–564. [Google Scholar]
- Plowright R, Bridge J. 1990. Effects of Meloidogyne graminicola (Nematoda) on the establishment, growth and yield of rice cv. IR36. Nematologica 36, 81–89. [Google Scholar]
- Plowright RA, Coyne DL, Nash P, Jones MP. 1999. Resistance to the rice nematodes Heterodera sacchari, Meloidogyne graminicola and M. incognita in Oryza glaberrima and O. glaberrima x O. sativa hybrids. Nematology 1, 745–751. [Google Scholar]
- Pokharel RR, Abawi GS, Duxbury JM, Smat CD, Wang X, Brito JA. 2010. Variability and the recognition of two races in Meloidogyne graminicola . Australasian Plant Pathology 39, 326–333. [Google Scholar]
- Prasad JS, Vijayakumar CHM, Sankar M, Varaprasad KS, Srinivasa Prasad M, Kondala Rao Y. 2006. Root-knot nematode resistance in advanced back cross populations of rice developed for water stress conditions. Nematologia Mediterranea 34, 3–8. [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics 38, 904–909. doi:10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Rakshit S, Rakshit A, Matsumura H, Takahashi Y, Hasegawa Y, Ito A, Ishii T, Miyashita NT, Terauchi R. 2007. Large-scale DNA polymorphism study of Oryza sativa and O. rufipogon reveals the origin and divergence of Asian rice. Theoretical and Applied Genetics 114, 731–743. [DOI] [PubMed] [Google Scholar]
- Reversat G, Boyer J, Sannier C, Pando-Bahuon A. 1999. Use of a mixture of sand and water-absorbent synthetic polymer as substrate for the xenic culturing of plant-parasitic nematodes in the laboratory. Nematology 1, 209–212. [Google Scholar]
- Seifi A, Kaloshian I, Vossen J, Che D, Bhattarai KK, Fan J, Naher Z, Goverse A, Tjallingii WF, Lindhout P, Visser RGF, Bai Y. 2011. Linked, if not the same, Mi-1 homologues confer resistance to tomato powdery mildew and root-knot nematodes. Molecular Plant Microbe Interactions 24, 441–450. [DOI] [PubMed] [Google Scholar]
- Sharma-Poudyal D, Pokharel RR, Shrestha SM, Khatri-Chhetri GB. 2004. Evaluation of common Nepalese rice cultivars against rice root knot nematode. Nepal Agricultural Research Journal 5, 33–36. [Google Scholar]
- Shrestha R, Uzzo F, Wilson MJ, Price AH. 2007. Physiological and genetic mapping study of tolerance to root-knot nematode in rice. New Phytologist 176, 665–672. [DOI] [PubMed] [Google Scholar]
- Soriano IR, Schmit V, Brar DS, Prot J, Reverstat G. 1999. Resistance to rice root-knot nematode Meloidogyne graminicola identified in O. longistaminata and O. glaberrima . Nematology 1, 395–398. [Google Scholar]
- Thurau T, Ye W, Cai D. 2010. Insect and nematode resistance. In: Kempken F, Jung C, eds. Genetic Modification of Plants, Biotechnology in Agriculture and Forestry , Vol. 64 Springer-Verlag: Berlin Heidelberg, pp. 177–197. [Google Scholar]
- Veremis JC, Roberts PA. 2000. Diversity of heat-stable genotype specific resistance to Meloidogyne in Maranon races of Lycopersicon peruvianum complex. Euphytica 111, 9–16. [Google Scholar]
- Wang C, Ulloa M, Mullens TR, Yu JZ, Roberts PA. 2012. QTL analysis for transgressive resistance to root-knot nematode in interspecific cotton (Gossypium spp.) progeny derived from susceptible parents. PLoS One 7, e34874. doi: 10.1371/journal.pone.0034874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DL, Zhu J, Li ZK, Paterson AH. 1999a. Mapping QTLs with epistatic effects and QTL x environment interactions by mixed linear model approaches. Theoretical and Applied Genetics 99, 1255–1264. [Google Scholar]
- Wang DL, Zhu J, Li ZK, Paterson AH. 1999b. User Manual for QTLMapper Version 1.0 . Texas A&M University: College Station, TX. [Google Scholar]
- Wei F, Wing RA, Wise RP. 2002. Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell 14, 1903–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei FS, Gobelman-Werner K, Morroll SM, Kurth J, Mao L, Wing R, Leister D, Schulze-Lefert P, Wise RP. 1999. The Mla (powdery mildew) resistance cluster is associated with three NBS-LRR gene families and suppressed recombination within a 240-kb DNA interval on chromosome 5S (1HS) of barley. Genetics 153, 1929–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson VM, Hussey RS. 1996. Nematode pathogenensis and resistance in plants. Plant Cell 8, 1735–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win PP, Kyi PP, De Waele D. 2011. Effect of agro-ecosystem on the occurrence of the rice root-knot nematode Meloidogyne graminicola on rice in Myanmar. Australasian Plant Pathology 40, 187–196. [Google Scholar]
- Win PP, Kyi PP, Maung ZTZ, Myint YY, De Waele D.(in press) Comparison of the damage potential and yield loss of the rice root-knot nematode Meloidogyne graminicola on lowland and upland rice varieties from Myanmar. Russian Journal of Nematology. [Google Scholar]
- Yik CP, Birchfield W. 1979. Host studies and reactions of cultivars to Meloidogyne graminicola . Phytopathology 69, 497–499. [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA. 1976. Laboratory Manual for Physiological Studies of Rice . IRRI: Los Banos, Philippines. [Google Scholar]
- Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckler ES. 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nature Genetics 38, 203–208. doi:10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- Zhang D, Zhang H, Wang M, Sun J, Qi Y, Wang F, Wang X, Li Z. 2009. Genetic structure and differentiation of Oryza sativa L. in China revealed by microsatellites. Theoretical and Applied Genetics 119, 1105–1117. [DOI] [PubMed] [Google Scholar]
- Zhao K, Tung CW, Eizenga GC, Wright MH, Ali ML, Price AH, Norton GJ, Islam MR, Reynolds A, Mezey J, McClung AM, Bustamante CD, McCouch SR. 2011. Genome-wide association mapping reveals rich genetic architecture of complex traits in Oryza sativa . Nature Communications 2, 467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Wright M, Kimball J, Eizenga G, McClung A, Kovach M, Tyagi W, Ali ML, Tung C, Reynolds A. 2010. Genomic diversity and introgression in O. sativa reveal the impact of domestication and breeding on the rice genome. PLoS One 5, e10780. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.