Abstract
Programmed cell death 4 (PDCD4) is a novel tumor suppressor, which is involved in the initiation and progression of cancers. However, the role of PDCD4 in hepatocellular carcinoma (HCC) has not been reported. The aim of this study was to investigate the molecular mechanism and clinical significance of PDCD4 inactivation in HCC.
The mRNA levels of PDCD4 in HCC tissues and adjacent nontumor tissues were analyzed by quantitative real-time polymerase chain reaction (qRT-PCR). Bisulfite sequencing PCR was performed to determine the methylation status of PDCD4 promoter. Furthermore, the mRNA expression level and the methylated level of PDCD4 were analyzed with the clinical and pathological characteristics.
qRT-PCR analysis showed that PDCD4 mRNA levels in tumor tissues were significantly decreased compared with that in adjacent nontumor tissues. The methylation rate of PDCD4 promoter was significantly higher in HCC tissues than that in adjacent nontumor tissues. PDCD4 mRNA levels and promoter methylation levels were both statistically correlated with metastasis and the degree of differentiation in HCC. In addition, the correlation between PDCD4 hypermethylation, mRNA levels, and overall survival (OS) was statistically significant.
Our results indicated that PDCD4 may be a novel candidate of tumor suppressor gene in HCC, and that promoter hypermethylation is an important mechanism for its downregulation and is also a good predictor of OS for HCC.
INTRODUCTION
Liver cancer is 1 of the most common malignant tumors and the second leading cause of cancer-related death in the world.1 In 2008, about 750,000 liver cancers were reported worldwide, with about 700,000 deaths. Hepatocellular carcinoma (HCC) is the main type of liver cancer, accounting for over 90% of cases worldwide. The highest incidence rates of HCC in the world are in Asia and Africa. Approximately 75% of HCC occurs in Asia, with China accounting for more than 50% of the world's burden. In contrast, the lowest incidence rates in the world occur in countries of Northern Europe, Middle East, Oceania, and North and South America. The disease is associated with poor prognosis because of the difficulty of early detection and recurrence after resection.2 Due to the asymptomatic nature of early HCC and lack of effective screening strategies, 80% of patients present with advanced HCC at the time of diagnosis. Therefore, a better understanding of the molecular mechanisms involved in the initiation and progression of HCC will help us to develop strategies for better disease detection. As for many other tumors, the pathogenesis of HCC is also believed to be a multistep process resulting from aberrant oncogene activation and tumor suppressor gene (TSG) inactivations. Studies have shown that oncogenes are activated mainly through genetic alterations, whereas aberrant TSG inactivation can result from both genetic and epigenetic alterations.3 Epigenetic alterations, particularly promoter hypermethylation of TSGs with consequent gene-silencing, play important roles in the carcinogenesis of HCC.3,4 Furthermore, aberrant hypermethylation of TSGs can also serve as a novel biomarker for detecting cancer as well as predicting prognosis.5
Programmed cell death 4 (PDCD4) is a novel TSG, which has been reported to suppress tumor genesis by regulating tumor cell apoptosis, cell cycle, and cell proliferation.6 The PDCD4 gene has been found to be downregulated or lost in various tumors, such as human glioma,7 lung cancer,8 breast cancer,9 and HCCs.10 Recent studies have shown that the expression of PDCD4 is frequently silenced or reduced as a result of hypermethylation in human gliomas.7 However, to date, there are no studies that investigated the relationship between the methylation status of the PDCD4 promoter and its expression in HCC.
In this study, we compared the methylation status of the PDCD4 gene promoter as well as the expression of PDCD4 mRNA levels in 56 paired samples of HCC and adjacent nontumor tissues. We also investigated the potential correlation of methylation of the PDCD4 gene with clinical and pathological features of HCC.
MATERIALS AND METHODS
Patients and Tissue Specimens
Specimens of cancer tissues and adjacent nontumor tissues were obtained from 56 patients without cirrhosis who had undergone curative hepatectomy for primary HCC at the Fifth Affiliated Hospital of Sun Yat-Sen University, Guangdong, China, during the period between January 2012 and December 2014. The primary HCC patients were diagnosed based on formalin-fixed paraffin-embedded findings, according to World Health Organization system, by 2 pathologists in our hospital who were blinded to patient data, separately. All the tumor samples were represented by bigger surgical specimens. Before DNA extraction, we predisposed some tumor specimen of all the samples to pathological detection by ourselves to confirm whether it is the tumor specimen or not again. The cases we chose in the study had only 1 neoplasm. The tissues that we used as normal controls are >5 cm distant from the margins of the neoplasm, and had no alteration like inflammation secondary to viral infections (cirrhosis), ischemia, and/or compression by the neoplasm according to pathological results. All patients had no prior chemotherapy or radiotherapy before surgery. Clinical data of the patients, including gender, age, initial presentation, postoperative irradiation, chemotherapy, follow-up, and outcome, were obtained from the hospital database. The study was approved by the human ethics committee of Sun Yat-Sen University, and the informed consent was obtained from all subjects studied.
RNA Isolation and Real-Time Quantitative PCR
Total RNA was extracted from 56 frozen HCC tissues and adjacent nontumor tissues by using trizol (Invitrogen, Carlsbad, CA, USA) reagents according to the manufacturer's instructions. And 1 μg RNA was used as a template for cDNA synthesis using Reverse-Transcribe Kit (Promega Co., Madison, WI) in accordance with the manufacturer's instructions. The real-time quantitative polymerase chain reaction (PCR) analysis was performed in a final volume of 20 μL with SYBR Premix Ex Taq Kit (TaKaRa, Dalian, Liaoning, China). The following qPCR conditions were used: preheating of the mixture at 95°C for 2 min and 40 cycles of 95°C for 10 s, followed by a final extension at 60°C for 10 s. The primers used for the amplification of PDCD4 gene were 5′-GAAGGTTGCTGGATAGGC-3′ (sense) and 5′-ATAAACACAGTTCTCCTGGTCATCA-3′ (anti-sense). All quantifications were performed with human GAPDH as an internal standard. The primers used for the amplification of GAPDH gene were 5′-ACAGCGACACCCACTCCTCC-3′ (sense), 5′-TAGCCAAATTCGTTGTCATACCAG-3′ (antisense). Real-time PCR was carried out on an ABI PRISM 7500 Sequence Detection System (Applied Biosystems, NJ). All qRT-PCR reactions were performed in triplicate.
Bisulfite DNA and Bisulfite Sequencing PCR
Genomic DNA was isolated from HCC tissues and adjacent nontumor liver tissues by use of the Universal Genomic DNA Extraction Kit Ver.3.0 (TaKaRa) as recommended by the manufacturer. The quality and integrity of DNA were determined by electrophoresis on 1% agarose gel. Bisulfite modification of DNA was performed using an Epitect Bisulfite Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. In a brief, 1 μg of purified genomic DNA was treated with NaOH, and subsequently subjected to sodium bisulfite treatment. Then, the mixture was desulfonated, and the DNA was purified to a final volume of 20 μL. The bisulfite-modified DNA was stored at −20°C for further analysis.
The position of CpG island within PDCD4 promoter was predicted and primers for bisulfite genomic sequencing PCR (BSP) were designed by methyl-primer-express software. The PCR was done using the following primers: 5′-GGATAGGTTGATTATTTGGAGT-3′ (sense) and 5′-TTATTTTTATTTTCTTCTACCCAAT-3′ (anti-sense). For bisulfite sequencing, the PCR products were purified and cloned into pGEM T-Easy vector (Promega). Five to 10 clones of each sample were sequenced. And the percentage of methylated CpG dinucleotides was calculated to evaluate the methylation level of PDCD4 gene.
Follow-Up
In this prospective study, patients were followed up for a period of nearly to 3 years. The patients have not received radiotherapy or chemotherapy before surgery. Patients were given an examination every 6 months for 3 years with liver CT examination, alpha-fetal-protein (AFP) determination, etc. The overall survival (OS), defined as the time from the date of surgery to the date of death or last contact if the patient was still alive, ranged from 4 to 32 months (median, 22 months). Among the 56 HCC patients included in this study, 21 patients were alive until the end of the follow-up period, and 35 patients died.
Statistical Analysis
All statistical analyses of the data were performed by using SPSS version 16.0 (Chicago, IL). The difference of PDCD4 methylation status between HCC tissues and adjacent nontumor tissue was examined by Student t test. The relationship between PDCD4 methylation status, mRNA level expression, and clinical and pathological parameters were examined by multivariate analysis of variance (MANOVA). Survival curves were estimated by the Kaplan–Meier method, and the differences in OS between subgroups were compared by the log-rank test. A P value <0.05 as considered statistically significant.
RESULTS
PDCD4 Hypermethylation in HCC Tissues
To determine the methylation status of PCDC4 in HCC tissues, we designed and performed BSP analysis for PCDC methylation within the region which included 27 CpGs (Figure 1A). The results showed that the CpG sites were highly methylated in tumor samples, and the methylation level varied from 18.5% to 79.3%, with a mean ratio of 59.1% (Figure 1B and C). In contrast, the methylation level observed in the adjacent nontumor tissues ranged from 16.3% to 66.7%, with a mean of and 36.9% (Figure 1B and C). Methylation levels >23.0% were considered as hypermethylation. PDCD4 was hypermethylated in 47 (83.9%) of 56 tumors.
FIGURE 1.
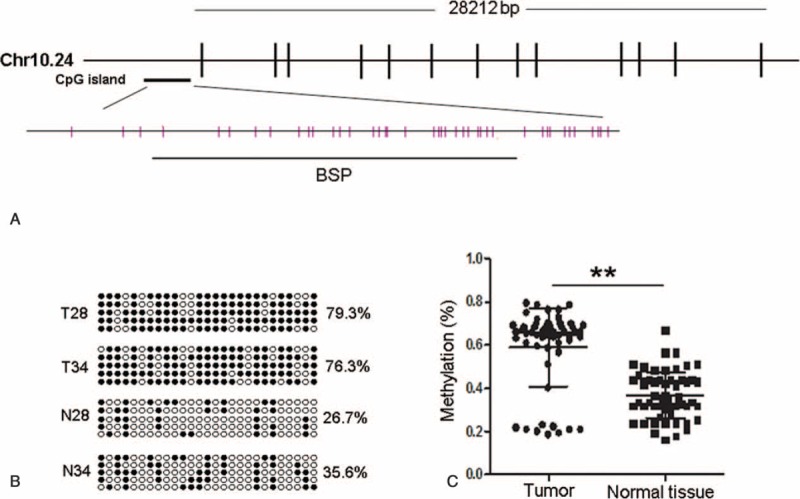
Determination of PDCD4 methylation status. (A) Schematic diagram of CpG dinucleotides in the promoter of the PDCD4 gene. PDCD4 is located on chromosome 10, and its promoter region contains a CpG island. CpG sites are shown as pink bars. The bold black line shows the region tested by BSP. (B) Representative results of the BSP analysis for PDCD4 in selected 2 pairs of samples, respectively. For each sample, at least 5 separate clones were sequenced. Black circles represent methylated CpG, and white circles represent unmethylated CpG. For each row of circles sequence, results for an individual clone of the bisulfite-PCR product are given. The methylation level is given as a percentage on the right of each bisulfite result. (C) The methylation status of PDCD4 promoter was compared between HCC tissues and adjacent nontumor tissues. BSP = bisulfite sequencing PCR, HCC = hepatocellular carcinoma, N = adjacent nontumor tissue, PCR = polymerase chain reaction, T = tumor tissue. ∗∗P < 0.001.
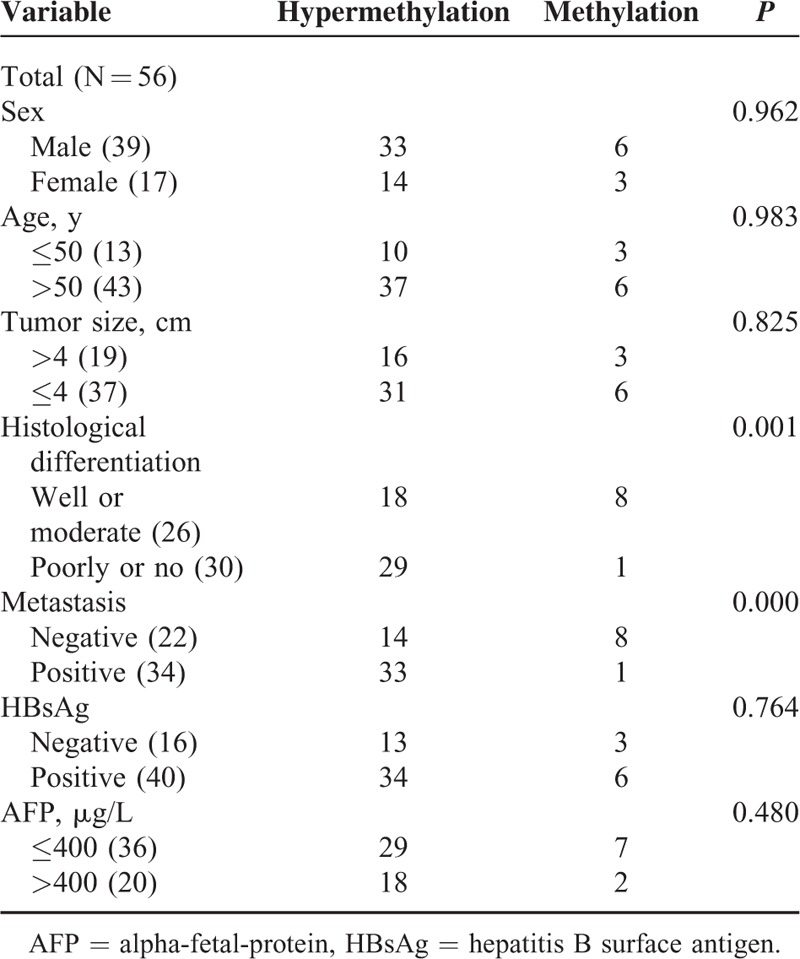
Comparisons of PDCD4 Hypermethylation With Clinical and Pathological Characteristics of Patients
We compared PDCD4 methylation level in HCC samples with different clinical and pathological characteristics including gender, age, tumor size, hepatitis B surface antigen (HBsAg), AFP, metastasis, and differentiation. The results showed that PDCD4 hypermethylation was statistically correlated with metastasis and the degree of differentiation (Table 1).
TABLE 1.
Correlation of PDCD4 Hypermethylation With Clinicopathological Characteristics
Correlation of PDCD4 Hypermethylation With Its Expression Levels
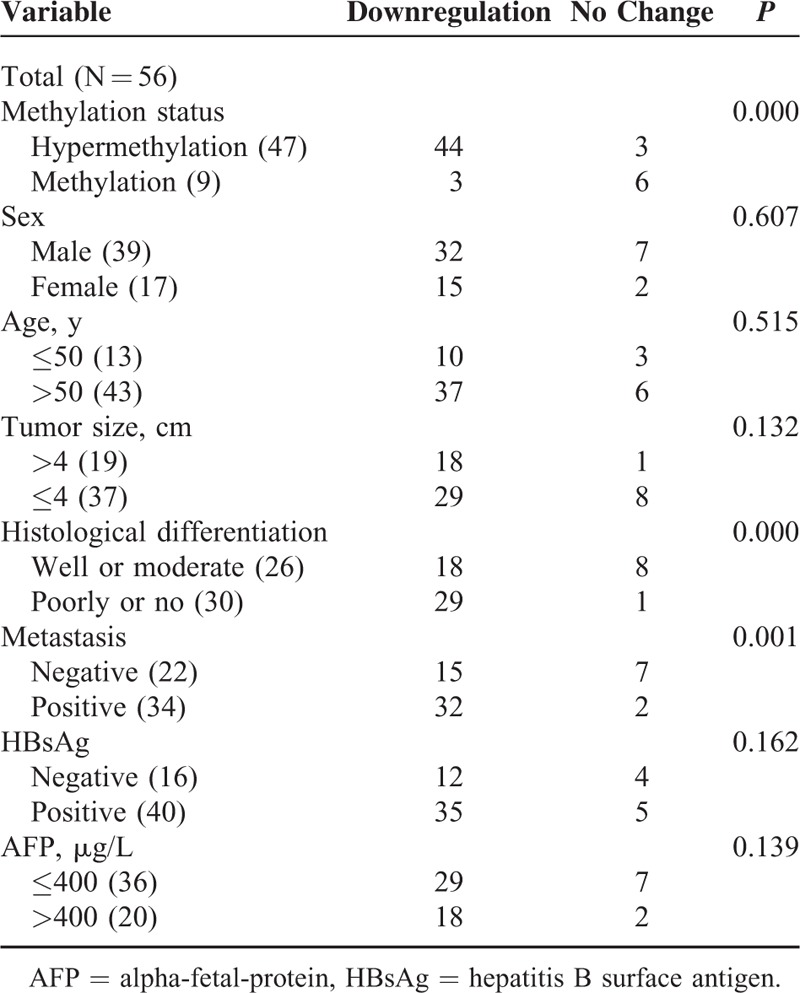
To determine whether the reduced expression of PDCD4 correlates to its hypermethylation in HCC, qRT-PCR was performed to analyze the mRNA level expression of PCDC4. As shown in Figure 2, the PDCD4 mRNA levels were decreased in HCC tissues by 65.0% (0.4434/1.2655; the PDCD4 mRNA level in 1 nontumor liver tissue was designated as 1 in comparison of other tumor or nontumor samples with it). Furthermore, we analyzed the relationship between methylation data and qRT-PCR findings. The PDCD4 mRNA levels <0.8343 were considered as downregulation. The results revealed that downregulation of PDCD4 mRNA expression in 44 (93.6%) of the 47 tumors harboring PDCD4 hypermethylation. In contrast, no changed PDCD4 mRNA level was observed in 6/9 (66.7%) of low-level methylated tumors and reduced expression was only observed in 3/9 (33.3%) (Table 2). There was a significant association between PDCD4 hypermethylation and its mRNA level expression (P < 0.000, Table 2).
FIGURE 2.
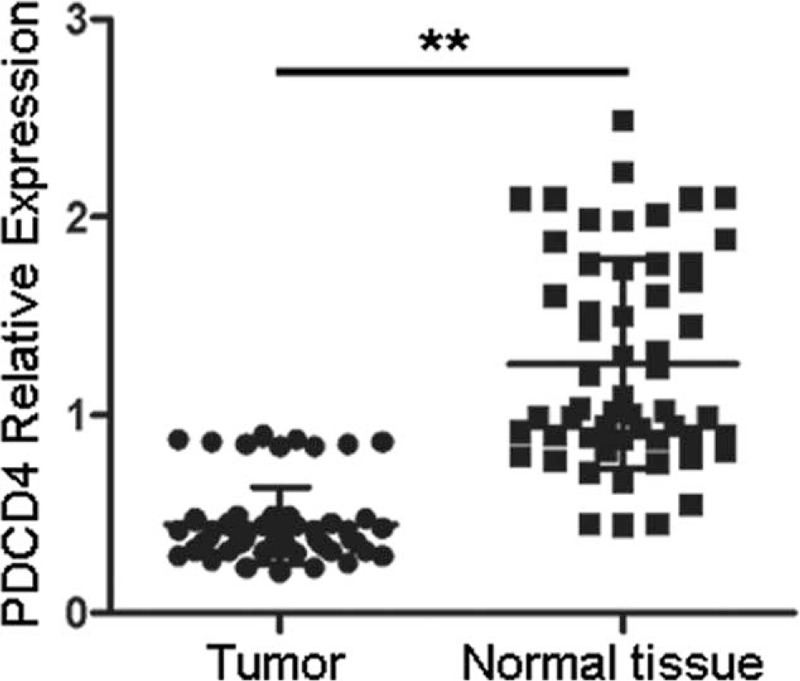
Determination of PDCD4 mRNA expression levels. qRT-PCR was performed to analyze the mRNA level expression of PCDC4 in 56 HCC tissues and the adjacent nontumor tissues. The PDCD4 mRNA level in 1 nontumor liver tissue was designated as 1, and other samples were compared with it. HCC = hepatocellular carcinoma, qRT-PCR = quantitative real-time polymerase chain reaction. ∗∗P < 0.001.
TABLE 2.
Correlation of PDCD4 Downregulation With Its Hypermethylation and Clinicopathological Characteristics
Comparisons of PDCD4 mRNA Levels With Clinical and Pathological Characteristics of Patients
We also compared PDCD4 mRNA level expression in HCC samples with different clinical and pathological characteristics, and found that PDCD4 mRNA level was also statistically correlated with metastasis and the degree of differentiation (Table 2).
Clinical Outcome of the Patients
To investigate the prognostic value of PDCD4 hypermethylation status and PDCD4 mRNA level expression for HCC, we assessed the relationship between PDCD4 methylation status or its mRNA level and survival duration by using the Kaplan–Meier analysis. Patients with hypermethylated PDCD4 promoter in HCC tissues revealed shorter OS compared with patients without hypermethylated PDCD4 promoter (P = 0.0213, Figure 3A). The correlation of PDCD4 mRNA level and OS was also showed to be markedly significant, and statistically reduced OS was observed in HCC patients having PDCD4 downexpression (P = 0.0234, Figure 3B).
FIGURE 3.
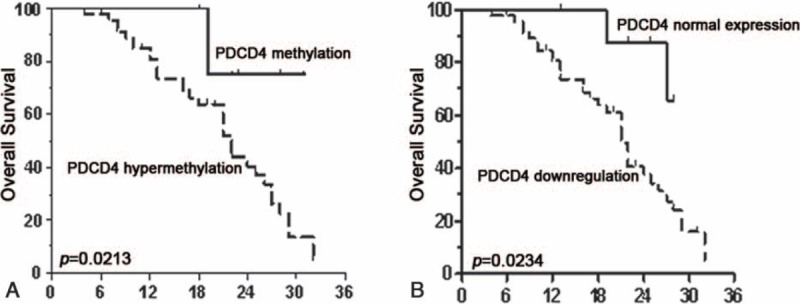
Kaplan–Meier analysis of overall survival (OS) in relation to PDCD4 methylation status and PDCD4 mRNA expression levels. (A) The correlation between PDCD4 methylation status in tumor tissues and the OS of the HCC patients. The patients with PDCD4 hypermethylation had a shorter OS than those with nontumor levels. (B) The correlation between PDCD4 mRNA expression levels in the tumor tissues and the OS of the HCC patients. The patients with PDCD4 downregulation had a poor outcome. HCC = hepatocellular carcinoma.
DISCUSSION
Epigenetic alterations are important for human carcinogenesis, which contribute to the initiation of cancers and also trigger genetic events leading to tumor development.11 To date, DNA methylation is the most commonly studied epigenetic mechanism, which is involved in the tumorgenesis of nearly all types of cancer, including HCC.12 Hypermethylation of many TSGs such as RASSF1A, APC, and GPX3 has been found in many HCC.13,14 The silencing of TSGs by hypermethylation is 1 of the major molecular alterations in the process of HCC carcinogenesis and also a potential diagnostic and prognostic marker.15
The PDCD4 gene was originally identified as being differentially upregulated during the procession of apoptosis. The human PDCD4 gene was localized to chromosome 10q24. Experimental models have shown that PDCD4 functions as a tumor suppressor, and play a vital role in inhibiting tumor development in vitro and vivo.16–19 PDCD4 has been found to be downregulated or frequently lost in many tumors such as lung cancer, breast cancer, and gliomas, and is a promising biomarker for better outcomes in these tumors.7–9 Yang et al20 indicated that miR-21 post-transcriptionally regulates PDCD4 expression in lung cancer. Dorrello et al21 found that S6K1 and β-TRCP may control the levels of PDCD4 protein through an ubiquitination-dependent mechanism. Gao et al showed that PDCD4 gene silencing in gliomas is associated with 5′CpG island methylation.7 Recent studies revealed that PDCD4 could suppress metastastic potential of human HCC cells, and it is reduced in the HCC tissues.10 However, the mechanisms of PDCD4 silencing in HCC are not well known.
In this study, the result of qRT-PCR analysis showed that PDCD4 mRNA levels were significantly downregulated in HCC tissues. In our methylation analysis, HCC samples were significantly hypermethylated with a mean methylation level of 59.1%, compared with adjacent nontumor samples (a mean of 36.9%). Furthermore, a statistically significant correlation between PDCD4 downregulation and PDCD4 hypermethylation in HCC was observed, which suggests that DNA hypermethylation may be 1 important mechanism for PDCD4 downregulation in HCC. In addition, we analyzed the correlation between PDCD4 methylation status and the clinical and pathological features. The data revealed that there was a statistically correlation of tumor metastasis and differentiation with PDCD4 hypermethylation. Moreover, Kaplan–Meier analysis showed that PDCD4 methylation status was inversely correlated with OS of the HCC patients. These results indicated that PDCD4 hypermethylation may play an important role in the progression and metastasis of HCC. In the present study, we found a predominance of poorly undifferentiated neoplasms in all the tumor samples studied. In China, chronic HBV infection is 1 major risk factor. Due to the asymptomatic nature of early HCC and lack of effective screening strategies, 80% of patients present with advanced HCC at the time of diagnosis. The standard treatments for early stage HCC are surgical resection and liver transplantation. However, most people cannot receive transplantation because of a lack of livers. There are some factors that interfere in OS and/or may be associated with the molecular findings. We analyzed the relationship between these points and the molecular findings just to explain that this molecule may be a biomarker for the prognosis and prediction of HCC. And whether it can be extrapolated to other populations will be needed to verify in larger samples. Thus, PDCD4 hypermethylation would provide a potential biomarker of prognosis and clinical outcome in HCC patients.
In summary, our study provides evidence that PDCD4 may function as a candidate TSG in HCC, and its downregulation may be associated with promoter hypermethylation. PDCD4 hypermethylation can be a novel biomarker of prognosis and clinical outcome for HCC patients.
ACKNOWLEDGMENTS
We thank Beibei Zhang in the 5th Affiliated Hospital of Sun Yat-Sen University for the help with BSP assays. XD is responsible for the experiments and paper writing; XC, MG, and XC are responsible for data statistic; KL is responsible for the whole design and paper writing.
Footnotes
Abbreviations: AFP = alpha-fetal-protein, BSP = bisulfite sequencing PCR, HCC = epatocellular carcinoma, OS = verall survival, PDCD4 = rogrammed cell death 4 = qRT-PCR, uantitative real-time polymerase chain reaction.
The authors have no funding and conflicts of interest to disclose.
REFERENCES
- 1.Kim do Y, Han KH. Epidemiology and surveillance of hepatocellular carcinoma. Liver Cancer 2012; 1:2–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villanueva A, Minguez B, Forner A, et al. Hepatocellular carcinoma: novel molecular approaches for diagnosis, prognosis, and therapy. Annu Rev Med 2010; 61:317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishida N, Goel A. Genetic and epigenetic signatures in human hepatocellular carcinoma: a systematic review. Curr Genomics 2011; 12:130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang B, Guo M, Herman JG, et al. Aberrant promoter methylation profiles of tumor suppressor genes in hepatocellular carcinoma. Am J Pathol 2003; 163:1101–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calvisi DF, Ladu S, Gorden A, et al. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest 2007; 117:2713–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cmarik JL, Min H, Hegamyer G, et al. Differentially expressed protein PDCD4 inhibits tumor promoter-induced neoplastic transformation. Proc Natl Acad Sci USA 1999; 96:14037–14042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao F, Wang X, Zhu F, et al. PDCD4 gene silencing in gliomas is associated with 5′CpG island methylation and unfavourable prognosis. J Cell Mol Med 2009; 13:4257–4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Knosel T, Kristiansen G, et al. Loss of PDCD4 expression in human lung cancer correlates with tumour progression and prognosis. J Pathol 2003; 200:640–646. [DOI] [PubMed] [Google Scholar]
- 9.Wen YH, Shi X, Chiriboga L, et al. Alterations in the expression of PDCD4 in ductal carcinoma of the breast. Oncol Rep 2007; 18:1387–1393. [PubMed] [Google Scholar]
- 10.Zhang H, Ozaki I, Mizuta T, et al. Involvement of programmed cell death 4 in transforming growth factor-beta1-induced apoptosis in human hepatocellular carcinoma. Oncogene 2006; 25:6101–6112. [DOI] [PubMed] [Google Scholar]
- 11.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol 2010; 28:1057–1068. [DOI] [PubMed] [Google Scholar]
- 12.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 2003; 33 (Suppl):245–254. [DOI] [PubMed] [Google Scholar]
- 13.Cao S, Yan B, Lu Y, et al. Methylation of promoter and expression silencing of GPX3 gene in hepatocellular carcinoma tissue. Clin Res Hepatol Gastroenterol 2015; 39:198–204. [DOI] [PubMed] [Google Scholar]
- 14.Formeister EJ, Tsuchiya M, Fujii H, et al. Comparative analysis of promoter methylation and gene expression endpoints between tumorous and non-tumorous tissues from HCV-positive patients with hepatocellular carcinoma. Mutat Res 2010; 692:26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishida N, Nagasaka T, Nishimura T, et al. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology 2008; 47:908–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jansen AP, Camalier CE, Colburn NH. Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res 2005; 65:6034–6041. [DOI] [PubMed] [Google Scholar]
- 17.Yang HS, Jansen AP, Nair R, et al. A novel transformation suppressor, PDCD4, inhibits AP-1 transactivation but not NF-kappaB or ODC transactivation. Oncogene 2001; 20:669–676. [DOI] [PubMed] [Google Scholar]
- 18.Yang HS, Knies JL, Stark C, et al. PDCD4 suppresses tumor phenotype in JB6 cells by inhibiting AP-1 transactivation. Oncogene 2003; 22:3712–3720. [DOI] [PubMed] [Google Scholar]
- 19.Hilliard A, Hilliard B, Zheng SJ, et al. Translational regulation of autoimmune inflammation and lymphoma genesis by programmed cell death 4. J Immunol 2006; 177:8095–8102. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y, Meng H, Peng Q, et al. Downregulation of microRNA-21 expression restrains non-small cell lung cancer cell proliferation and migration through upregulation of programmed cell death 4. Cancer Gene Ther 2015; 22:23–29. [DOI] [PubMed] [Google Scholar]
- 21.Dorrello NV, Peschiaroli A, Guardavaccaro D, et al. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 2006; 314:467–471. [DOI] [PubMed] [Google Scholar]


