Supplemental Digital Content is available in the text
Abstract
Previous studies have yielded controversial results related to the contribution of interleukin 10 (IL-10) gene polymorphisms (IL-10 -592C/A, IL-10 -1082G/A, and IL-10 -819C/T) in the progression of cardiovascular disease (CVD). Thus, we performed a meta-analysis to summarize this situation.
Eligible studies were retrieved by searching PubMed, Embase, Web of Science, and Cochrane Library with the last search up to July 7, 2015. Data were pooled by odds ratios (ORs) and their 95% confidence intervals (CIs). False-positive report probability (FPRP) analysis was conducted for all significant findings. Genotype-based mRNA expression analysis was also performed using data from 270 individuals with different ethnicities.
Finally, 19 studies for IL-10 -592C/A polymorphism (7284 cases and 7469 controls), 21 studies for IL-10 -1082G/A polymorphism (8263 cases and 5765 controls), and 12 studies for IL-10 -819C/T polymorphism (4502 cases and 3190 controls) were included in the meta-analyses. With respect to IL-10 -819C/T polymorphism, statistically significant decreased CVD risk was found when all studies were pooled into the meta-analysis (T vs C: OR = 0.91, 95% CI = 0.84–0.98; TT + TC vs CC: OR = 0.90, 95% CI = 0.81–1.00). Subgroup analyses stratified by disease subtype suggested the -819C/T polymorphism was significantly associated with a decreased CAD risk (T vs C: OR = 0.90, 95% CI = 0.83–0.97; TT vs CC: OR = 0.81, 95% CI = 0.66–1.00; TT vs TC + CC: OR = 0.82, 95% CI = 0.69–0.98; TT + TC vs CC: OR = 0.89, 95% CI = 0.80–0.99), which was noteworthy finding as evaluated by FPRP. However, with regard to IL-10 -592C/A and IL-10 -1082G/A polymorphisms, no significant association with CVD risk was observed in the overall and subgroup analyses.
In conventional meta-analyses, the results suggested that IL-10 -819C/T polymorphism was associated with decreased risk of CVD, especially CAD outcome, whereas IL-10 -592C/A and IL-10 -1082G/A polymorphisms might have no influence on the susceptibility of CVD. However, trial sequential analysis does not allow us to draw any solid conclusion for the association between IL-10 -592C/A or IL-10 -1082G/A polymorphism and CVD risk. Further large and well-designed studies are still needed.
INTRODUCTION
Cardiovascular diseases (CVDs) such as coronary artery disease (CAD) and stroke are the leading cause of death worldwide and represent a public health challenge in both industrialized and developing countries.1,2 A number of clinical risk factors for CVD have been identified for decades, involving obesity, dyslipidemia, hypertension, diabetes, and a sedentary lifestyle. Nevertheless, the molecular basis of CVD is complex and linked to a broad range of biological pathways, including lipid and glucose metabolism, vascular repair, and angiogenesis.3 Apart from these, more and more evidence showed that inflammatory molecules might take part in the pathogenesis of CVD as well.4,5
Inflammation has been shown to involve in the manifestation and development of arterial thrombotic diseases.6,7 Interleukins, a group of cytokines, were recognized as crucial agents involved in the host inflammatory response.8 Interleukin 10 (IL-10), secreted by Th2 cells as well as by macrophages, is an important anti-inflammatory cytokine with potent deactivating properties on both macrophages and T cells.9 IL-10 exerts a negative modulator effect on the inflammatory response by inhibiting cytokine synthesis.5,10 Because of its anti-inflammatory function, IL-10 is thought to be involved in arterial thrombotic diseases and further illustrated by epidemiologic studies, which recognized an association between lower levels of plasma IL-10 and increased risk of several end points of CVD such as acute coronary disease (ACS) and ischemic stroke (IS).5,11,12 Previous studies have reported that approximately 75% of individual difference in IL-10 secretion is determined by genetic factors and controlled at transcriptional level.13IL-10 gene is located on chromosome 1, has 5 exons, and has been mapped to the junction between 1q31 and 1q32.14 Three single-nucleotide polymorphisms (SNPs) (G-1082A, C-819T, and C-592A) in the promoter region of IL-10 were found to be associated with transcription activity of IL-10 gene and levels of plasma IL-10.13,15 Owing to their important roles, they were extensively studied and anticipated to be involved in arterial thrombotic diseases. This is supported by several studies that observe an increased risk of CVD in IL-10 -1082 A allele carriers.4,16,17 However, such associations could not be confirmed in other studies.10,14,18,19 The associations between IL-10 -592C/A and IL-10 -819C/T polymorphisms and CVD were not conclusive as well.10,13–17,19–32 Therefore, we performed this systematic review with meta-analysis and trial sequential analysis (TSA) of all the published case–control studies in the hope of providing more precise evidence.
METHOD
Search Strategy and Identification of Relevant Studies
We carried out a comprehensive search of electronic databases including PubMed, Embase, Web of Science, and Cochrane Library to identify relevant publications reporting on the association between the IL-10 polymorphisms and CVD risk, with the last search update on July 7, 2015. The following keywords and medical subject headings were employed: (“interleukin 10” or “interleukin-10” or “IL-10” or “IL 10”), (“acute coronary syndrome” or “myocardial infarction” or “coronary artery disease” or “coronary heart disease” or “ischemic heart disease” or “cardiovascular disease” or “cardiovascular” or “stroke” or “myocardial ischaemia” or “myocardial ischemia” or “cerebral ischemia” or “cerebral ischaemia” or “cerebral infarction” or “brain infarction”), and (“polymorphism” or “variation” or “variant” or “allele” or “mutation” or “SNP”). Additional relevant publications were identified by a manual search of bibliographies of retrieved studies and recent reviews. Ethical approval and informed consent were not necessary because our analyses were based on data from previously published studies.
Studies were included that met the following criteria: investigation of the association between IL-10 -592C/A, IL-10 -1082G/A, or IL-10 -819C/T polymorphism and CVD among unrelated subjects; case–control design; sufficient information provided to calculate odds ratio (OR) and the corresponding confidence interval (CI). In addition, exclusion criteria were as follows: meeting abstracts, case reports, reviews, or editorials; overlapping data; studies were published in languages other than English and Chinese. The articles with the largest dataset were chosen when there were multiple publications from the same population. Two investigators selected the studies according to the above criteria, and disagreements were resolved by consensus.
Data Extraction
Data were extracted from all eligible studies by primary investigators using a standardized extraction form. Extracted forms were reviewed by co-authors and a research assistant to ensure accuracy with dissent settled by consensus. The following information was collected: first author's name, publication year, country and ethnicity of population, outcome, matching status, genotyping methods, number of cases and controls, genotype distributions in cases and controls, and the Hardy-Weinberg Equilibrium (HWE) in controls (P). If these were not possible, the authors of the publications were contacted via E-mail for more detailed data.
Genotype-based mRNA Expression Analysis
The genotypes data for IL-10 -592C/A, IL-10 -1082G/A, and IL-10 -819C/T polymorphisms were available from the HapMap (http://hapmap.ncbi.nlm.nih.gov/) for 270 subjects with 3 different ethnicities and their corresponding mRNA expression levels data were available from SNPexp (http://app3.titan.uio.no/biotools/tool.php?app=snpexp) as described previously.33,34
Quality Assessment
The methodological quality of the included studies was accessed by 2 authors respectively according to the Newcastle Ottawa Scale (NOS) (www.ohri.ca/programs/clinical_epidemiology/oxford.asp).35 The NOS criteria consist of 3 aspects: selection, comparability, and exposure. Scores ranged from 0 stars (worst) to 9 stars (best) and a score ≥7 indicated that a study was of high quality. Dissent was settled as described above.
Statistical Analyses
We initially assessed HWE among control subjects by χ2 test and P < 0.05 was considered as significant disequilibrium. The pooled ORs with their 95% CIs were calculated to evaluate the strength of the association between the IL-10 gene polymorphisms and CVD risk based on 5 genetic comparison models: allele model, homozygous model, heterozygous model, dominant model, and recessive model. Statistical heterogeneity between eligible studies was evaluated by using the Cochran's Q statistic and I2 test.36P < 0.1 and I2 exceeding 50% indicated substantial heterogeneity across studies, then a random-effects model was chosen to perform meta-analysis; otherwise, the fixed-effects model was selected. Predefined subgroup analyses were conducted a priori according to ethnicity (Asian, white, or mixed), disease subtype (CAD or stroke), and quality score (low quality: score <7; high quality: score ≥7). A power calculation was performed using Power and Sample Size Calculation version 3.1.2 (http://biostat.mc.vanderbilt.edu/twiki/bin/view/Main/PowerSampleSize). Sensitivity analyses were performed to look at more narrowly drawn subsets of the studies by removing an individual study or by removing studies with similar feature to assess their influence separately. Begg funnel plot and Egger regression test were used to search for publication bias and a P value >0.05 suggests no significant publication bias has been detected.37 The fail-safe number (Nfs) set at a significance of 0.05 was also calculated to inspect publication bias, according to the formula Nfs0.05 = (∑Z/1.64)2-k, where k is the number of studies included. If the Nfs was less than the number of observed studies for a polymorphism, we deemed that there exists a significant publication bias.
For each statistically significant association identified, we estimated the false-positive reporting probability (FPRP).38 The FPRP value is determined by the P value, the prior probability for the association, and statistical power. We set 0.2 as an FPRP threshold and assigned a prior probability of 0.1 to detect an OR of 1.50/0.67 for alleles with a risk/protective effect. Only the results with FPRP values <0.2 were referred as noteworthy.
All P values were 2-sided. All above statistical analyses were performed using STATA software version 12.0 (STATA Corporation, College Station, TX).
TSA
Meta-analyses may result in type I errors owing to an increased risk of random error when sparse data are analyzed and to repeated significance testing when a cumulative meta-analysis is updated with new trials. TSA has been introduced to control the risk of type I error by estimating the required information size and an adjusted threshold for statistical significance. 39–41 A required information size was calculated with the adjustment by diversity (D2) between trials. We performed TSA with a desire to maintain an overall 5% risk of a type I error and 20% of the type II error (a power of 80%).42 As for IL-10 -592C/A, IL-10 -1082G/A, and IL-10 -819C/T polymorphisms, the required information size was calculated based on a relative risk increase of 6%, a relative risk reduction of 2%, a relative risk reduction of 10%, respectively, with low-risk bias (taking the data of dominant model for example). For IL-10 -819C/T polymorphism and CAD, we observed an 11% relative risk reduction. The control event proportion was calculated from the actual meta-analyses.
When the cumulative Z-curve crosses the trial sequential monitoring boundary, a sufficient level of evidence may have been reached and further trials are unnecessary. If the Z curve does not cross any of the boundaries and the required information size has not been reached, evidence to reach a conclusion is insufficient.43 We used software Trial Sequential Analysis (version 0.9, http://www.ctu.dk/tsa/) and provided the 95% CIs adjusted for sparse data or repetitive testing, which we describe as the TSA-adjusted 95% CIs.
RESULTS
The Main Characteristics of Included Studies
The process of literature retrieval and exclusion was shown in Figure 1. The initial comprehensive search identified a total of 1087 potentially relevant articles, 227 articles were excluded for duplication, and 789 additional articles were excluded for their unmatched titles or abstracts. After reading the full text of the remaining 71 articles, 44 articles were removed due to reviews, meeting abstract, studies with insufficient data, and so on. Since 1 article included 2 populations, both of them were considered as an independent study.27 Finally, 27 articles including 28 case–control studies, involving a total sample size of 20875, were included in our meta-analysis.4,10,13–32,44–48 Detailed characteristics and genotype distributions of included studies were summarized in Tables 1 and 2, respectively.4,10,13–32,44–48 The 28 studies concerned IL-10 -592C/A polymorphism, IL-10 -819C/T polymorphism, IL-10 -1082G/A polymorphism, respectively. These 3 polymorphisms were found to occur in frequencies consistent with HWE in the control populations of the vast majority of the published studies. There were 10 studies based on Asian population,4,13,16,17,20,23,25,28,32,48 16 studies conducted in white population,10,14,18,19,21,22,26,27,29–31,44–47 and 2 studies from mixed population.15,24 Among these included studies, cases were generally recruited in referral centers with documented CAD or stroke, and the controls were without any direct evidence of overt disease. The number of cases among all selected studies varied from 86 to 1791, whereas the numbers of controls varied from 48 to 2089. All the studies included met quality criteria ranging from 4 to 9 (Supplemental Table 1).
FIGURE 1.
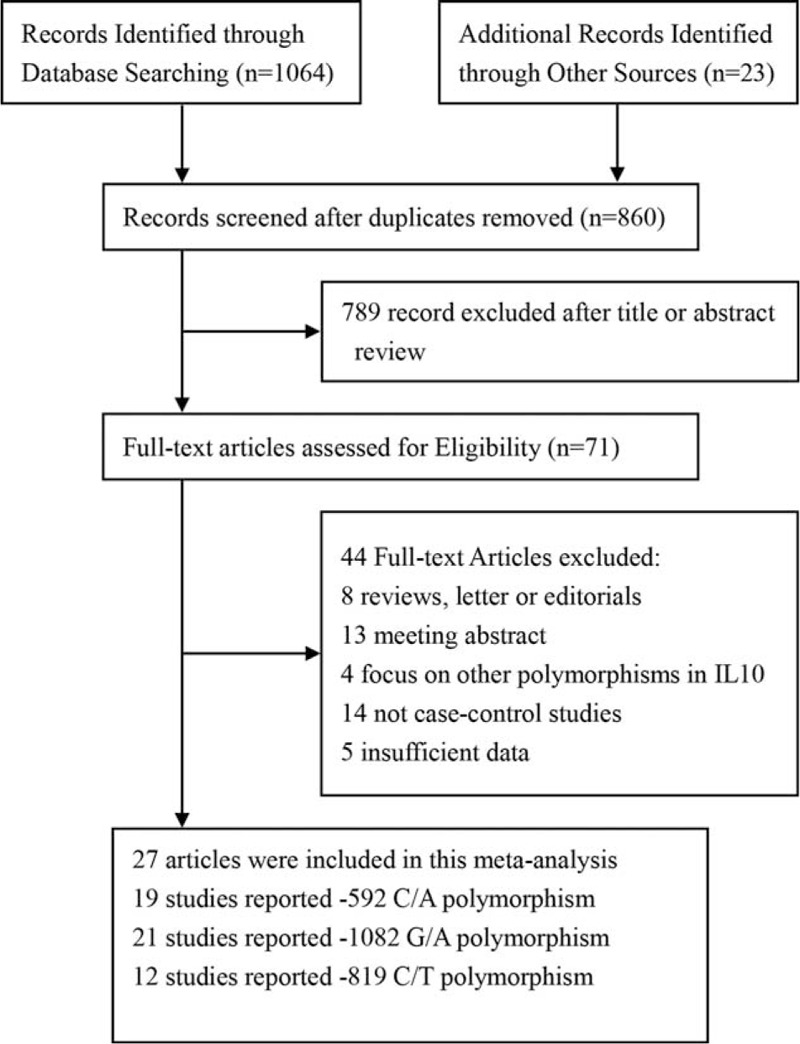
Flow chart of the search strategy and study selection. The terms “N” in the boxes represent the number of corresponding studies.
TABLE 1.
Characteristics of the Eligible Studies Included in This Meta-analysis
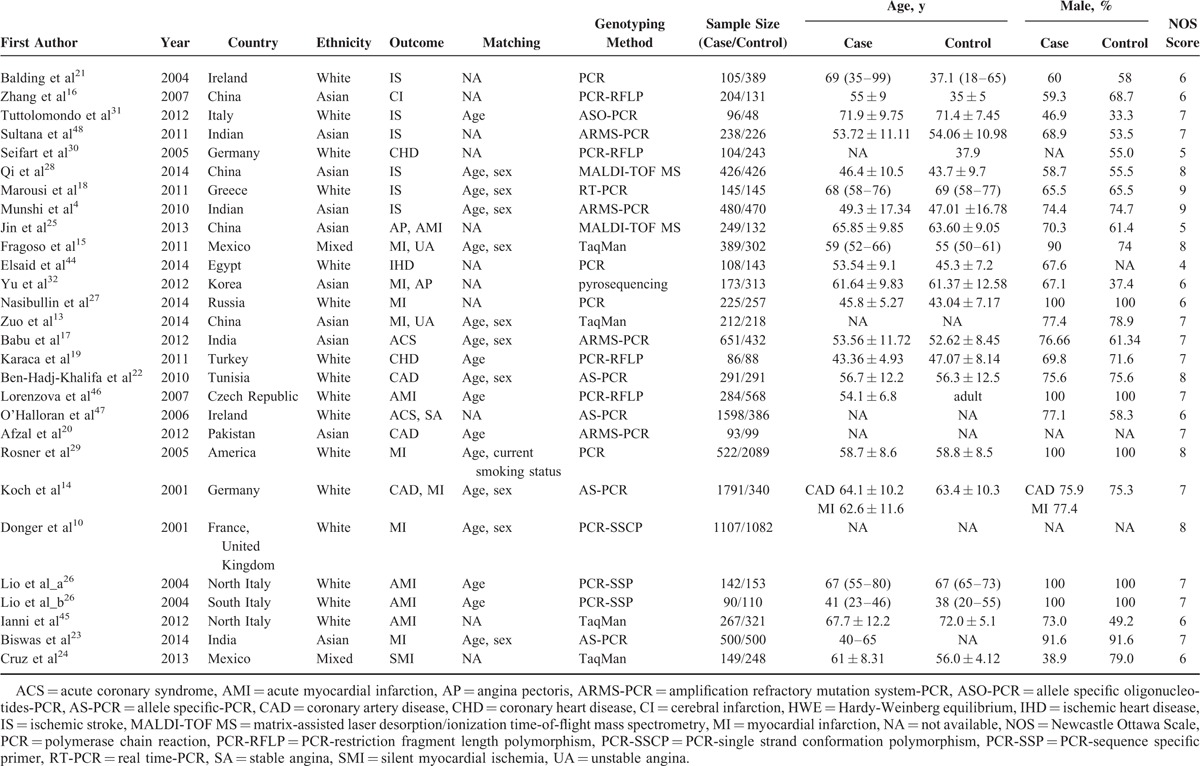
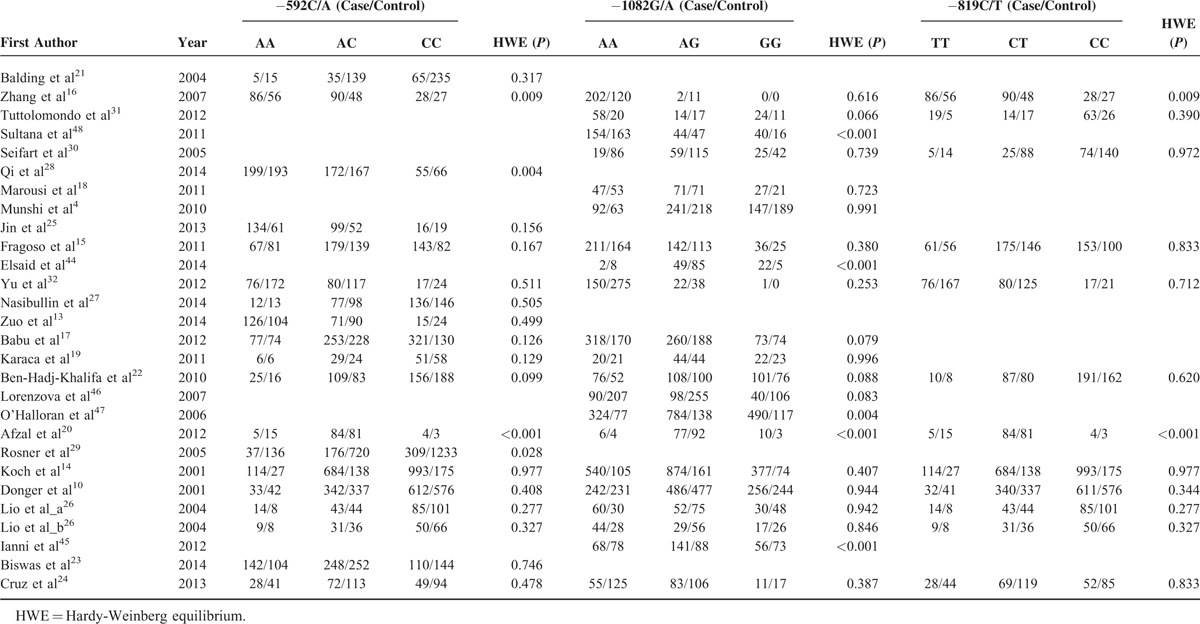
TABLE 2.
Genotype Distribution Among Studies Included in the Meta-analysis
Association Between the IL-10 Polymorphisms and CVD Risk
Data on IL-10 -592C/A polymorphism were obtained from 19 studies including 7284 CVD patients and 7469 controls.10,13–17,19–29,32 In overall comparison, there was no obvious evidence of an association between IL-10 -592C/A polymorphism and the incidence of CVD under all genetic models (A vs C: OR = 1.03, 95% CI = 0.90–1.17; AA vs CC: OR = 1.08, 95% CI = 0.82–1.41; AC vs CC: OR = 1.04, 95% CI = 0.88–1.22; AA vs AC + CC: OR = 1.00, 95% CI = 0.83–1.20; AA +AC vs CC: OR = 1.06, 95% CI = 0.88–1.26) (Figure 2A and Table 3). Similar results were identified in subgroup analysis in light of ethnicity, disease subtype, and quality score. Significant between-study heterogeneity was observed under all 5 genetic models. In the subgroup analysis, heterogeneity vanished in stroke studies as well as dramatically decreased in white subgroup (Table 3).
FIGURE 2.
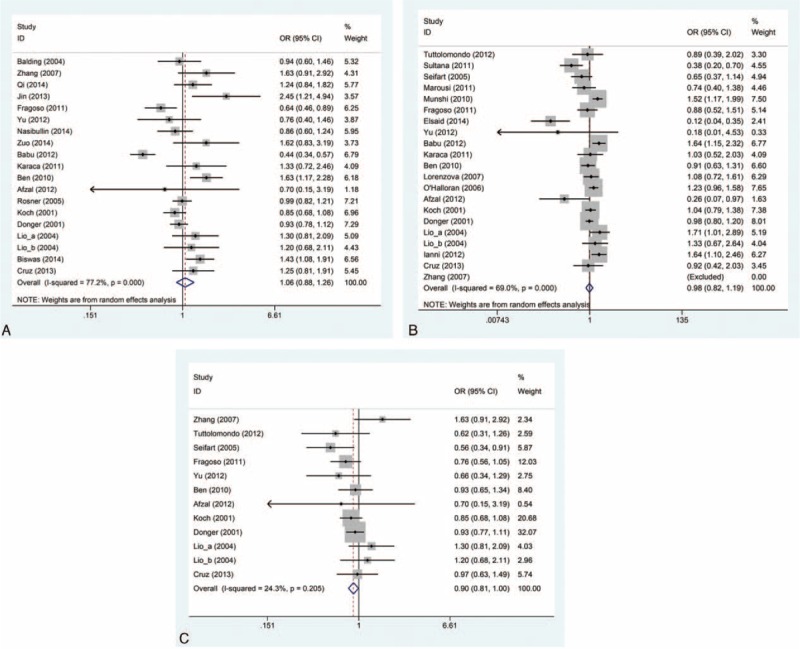
Forest plot of the risk of CVD associated with the IL-10 gene polymorphisms under dominant genetic model. (A) IL-10 -592C/A polymorphism. (B) IL-10 -1082G/A polymorphism. (C) IL-10 -819C/T polymorphism. The solid diamonds and horizontal lines correspond to the study-specific ORs and 95% CIs. The gray areas reflect the study-specific weight. The hollow diamonds represent the pooled ORs and 95% CIs of the overall population. The vertical solid lines show the OR of 1 and the vertical dashed lines indicate the corresponding pooled OR. CI = confidence interval, CVD = cardiovascular disease, OR = odds ratio.
TABLE 3.
Meta-analysis of the Association Between 3 IL-10 Gene Polymorphisms and CVD risk
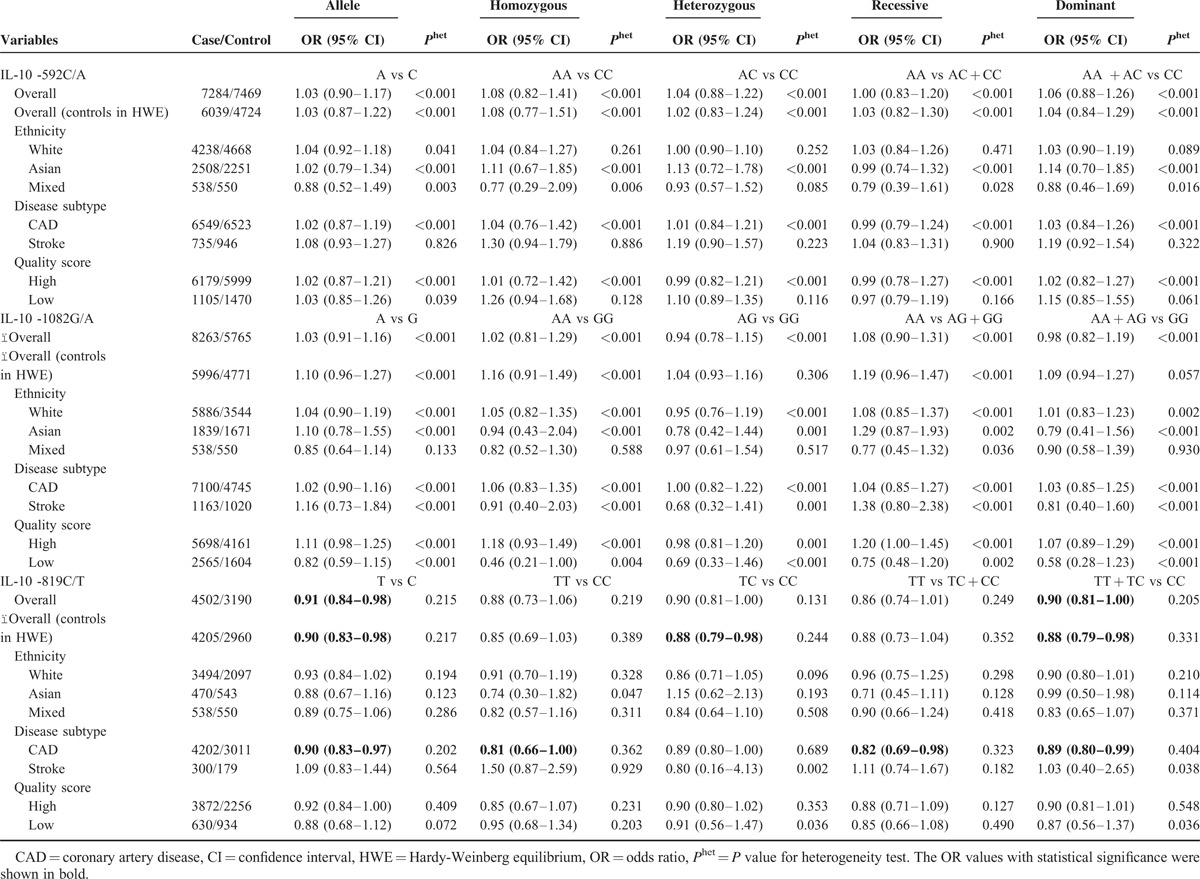
TSA showed that 39.6% (14753) of the required information size of 37,263 subjects were accrued. The cumulative Z-curve has not crossed the conventional boundary before reaching the required information size, suggesting that there was insufficient evidence to show a 6% relative risk increase, and further studies are necessary (Supplemental Figure 1). The TSA-adjusted 95% CI was 0.78 to 1.44.
Twenty-one studies had data on IL-10 -1082G/A polymorphism, with 8263 CVD patients and 5765 controls.4,10,14–20,22,24,26,30–32,44–48 Likewise, we failed to confirm the association between IL-10 -1082G/A polymorphism and CVD risk under all genetic models (A vs G: OR = 1.03, 95% CI = 0.91–1.16; AA vs GG: OR = 1.02, 95% CI = 0.81–1.29; AG vs GG: OR = 0.94, 95% CI = 0.78–1.15; AA vs AG + GG: OR = 1.08, 95% CI = 0.90–1.31; AA + AG vs GG: OR = 0.98, 95% CI = 0.82–1.19) (Figure 2B and Table 3). In the subgroup according to ethnicity, disease subtype and quality score, similar trends with overall results were observed. Substantial heterogeneities were noticed under all 5 genetic models. Following subgroup analysis, heterogeneities almost disappeared in mixed population subgroup (Table 3).
TSA showed that 13,693 of the required information size of 89,658 subjects were accrued. The required information size is far from reached and the conventional boundary has not been crossed, leaving the meta-analysis inconclusive of a 2% relative risk reduction (Supplemental Figure 2). The TSA-adjusted 95% CI was 0.46 to 2.11.
Twelve studies provided data on IL-10 −819C/T polymorphism consisting of 4502 CVD cases and 3190 controls.10,14–16,20,22,24,26,30–32 Overall, the pooled results revealed a significant association between IL-10 -819C/T polymorphism and decreased CVD risk under allelic comparison and dominant model (T vs C: OR = 0.91, 95% CI = 0.84–0.98; TT + TC vs CC: OR = 0.90, 95% CI = 0.81–1.00) (Figure 2C and Table 3). If we set α = 0.05, based on the data set for -819 T allele, we have a 77.2% power to detect an OR of 0.91. Similar results were found when the meta-analysis was restricted to studies whose controls were in agreement with HWE. In the subgroup analysis stratified by disease subtype, the statistically significant association were found for CAD under all genetic models except heterozygote comparison (T vs C: OR = 0.90, 95% CI = 0.83–0.97; TT vs CC: OR = 0.81, 95% CI = 0.66–1.00; TT vs TC + CC: OR = 0.82, 95% CI = 0.69–0.98; TT + TC vs CC: OR = 0.89, 95% CI = 0.80–0.99) (Table 3). No significant heterogeneity was detected under any genetic models. Meanwhile, no obvious heterogeneity was observed in the vast majority of subgroups.
Using the TSA, the required information size is 4495 subjects to demonstrate the issue. Until now, the cumulative Z-curve has crossed the conventional boundary and the required information size has been reached, confirming that IL-10 -819 C/T polymorphism is associated with decreased risk of CVD and further relevant trials are unnecessary (Figure 3). The TSA adjusted 95% CI was 0.80 to 1.00. Additionally, the TSA of 10 studies reporting CAD showed that sufficient evidence was established to show a relative risk reduction of 11%, the cumulative Z-curve has crossed the conventional boundary and the required information size has been reached (Figure 4). The TSA adjusted 95% CI was 0.78 to 1.01. The FPRP values for all significant findings are shown in Supplemental Table 2. For a prior probability of 0.1 and OR of 0.67, the FPRP analyses suggested that all significant associations were deserving of attention.
FIGURE 3.
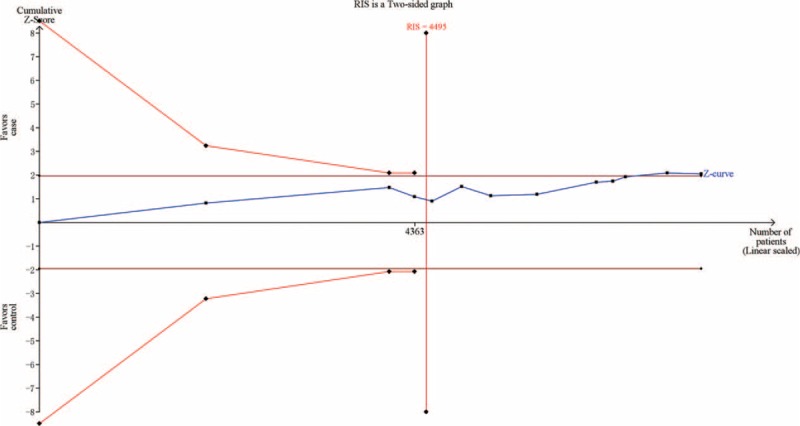
Trial sequential analysis of 12 studies reporting IL-10 -819C/T polymorphism. The required information size was calculated using α = 0.05 (2-sided), β = 0.20 (power 80%), D2 = 39%, a relative risk reduction of 10% and an event proportion of 53.5% in the control arm. The blue cumulative Z-curve was constructed using a fixed-effects model.
FIGURE 4.
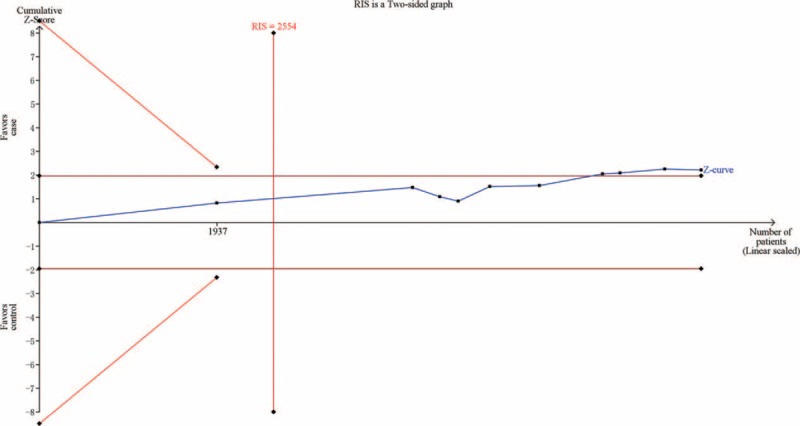
Trial sequential analysis of 10 studies reporting the association between IL-10 -819C/T polymorphism and CAD. The required information size was calculated using α = 0.05 (two sided), β = 0.20 (power 80%), D2 = 8%, a relative risk reduction of 11% and an event proportion of 52.54% in the control arm. The blue cumulative Z-curve was constructed using a fixed-effects model. CAD = coronary artery disease.
The Correlation Between the mRNA Expression and Genotypes
The correlation between IL10 mRNA expressions levels by the genotypes were explored for all population (Supplemental Figure 3). No significant alteration in the mRNA expression levels was found for the 3 variants.
Sensitivity Analysis and Publication Bias
Sensitivity analyses were performed to assess the influence of each individual study on the pooled OR in each comparison in the polymorphisms of IL-10 -592C/A, IL-10 -819C/T, and IL-10 -1082G/A. The recalculated ORs were not significantly influenced, which suggested our results were robust and reliable. Begg funnel plot and Egger test were performed to evaluate the potential publication bias of literatures. The shapes of the funnel plots showed no evidence of obvious asymmetry (Figure 5). The Egger test results did not support the existence of publication bias. The Nfs0.05 values for IL-10 -592C/A, IL-10 -1082G/A, and IL-10 -819C/T polymorphisms were 242, 208, 51, respectively, which were consistently greater than the number of studies included in this meta-analysis.
FIGURE 5.
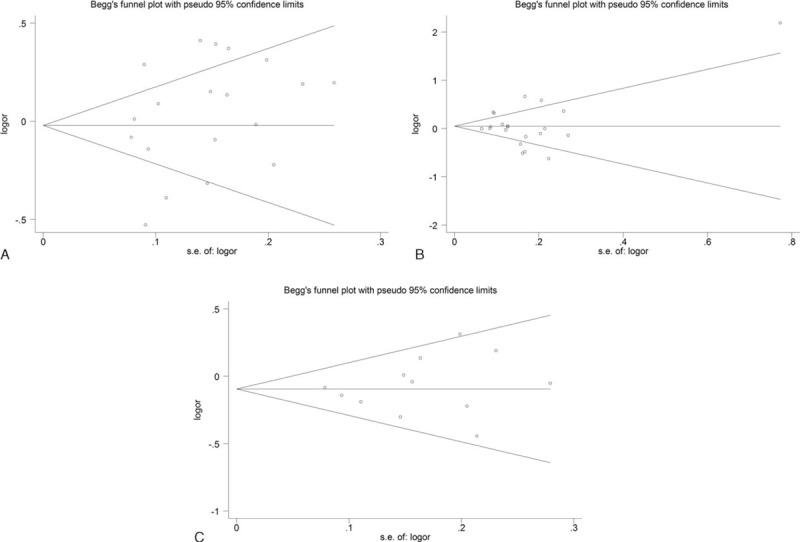
Begg funnel plot of publication bias in the meta-analysis of the association of IL-10 gene polymorphisms with CVD risk under allele genetic model. (A) IL-10 -592C/A polymorphism. (B) IL-10 -1082G/A polymorphism. (C) IL-10 -819C/T polymorphism. Each circle represents a separate study for the indicated association. CVD = cardiovascular disease, logor = natural logarithm of OR; s.e. = standard error.
DISCUSSION
It is now accepted that inflammation play a significant role in the pathophysiology of CVD.4,5 IL-10 is a potent anti-inflammatory cytokine with multiple functions taking part in inflammation reaction as well as the development of CVD.4,11,12 Recently, the associations between 3 IL-10 gene polymorphisms and the risk of CVD have been intensively investigated; however, the results are inconsistent. Thus, we conducted a systematic review with meta-analysis and TSA to obtain a more precise conclusion.
Although data from some individual studies suggested a relationship, the overall result of the present meta-analysis argued against an association of IL-10 -592C/A or IL-10 -1082G/A polymorphism with CVD risk in all genetic models. We also performed genotype-based mRNA expression analysis using the data from 270 individual. The biological results are in accordance with the observed association. Moreover, further sub-analysis of either gene polymorphism based on ethnicity, disease subtype, or quality score did not suggest a significantly different result. There are 3 potential reasons for the results. First, because of the complex nature of CVD, it is unlikely that a SNP in a single gene would be associated with an increased risk of CVD, without a contribution from other polymorphic susceptibility genes. Second, IL-10 -592C/A or IL-10 -1082G/A polymorphism itself might exhibit null contribution to the susceptibility of CVD. Third, other factors, such as age, medical treatment, and nutrient status, can also influence the risk of CVD. However, TSA did not allow us to draw any solid conclusion on the association between IL-10 -592C/A or IL-10 -1082G/A polymorphism and CVD risk. Thus, these issues need to be further studied.
As for IL-10 -819C/T polymorphism, our result showed that the individuals who carry the T allele have 10% decreased risk of CVD compared with the CC homozygote carriers, and a significantly decreased risk of CVD was also found in allele model. This may be because of the fact that IL-10 -819 T allele potently alters the IL-10 gene activity resulting in a marked increase of plasma IL-10 concentration. Moreover, TSA provided firm evidence of -819C/T polymorphism associated with decreased risk of CVD. In the subgroup analysis stratified by disease subtype, we discovered that -819C/T polymorphism had a significant correlation with CAD in all genetic models except heterozygous model. The result of TSA suggested evidence was sufficient enough for this relationship. However, in stroke subgroup, the data were obtained only from two studies. So, the findings in this subgroup should be interpreted with caution.
To make the conclusion more credible, we performed the FPRP analysis, publication bias analysis and sensitivity analysis. All significant associations passed the FPRP analyses, indicating that these associations were robust. Funnel plots suggested that no obvious publication bias was detected. The Nfs0.05 for 3 polymorphisms were greater than the number of studies included in this meta-analysis, also indicating a low probability of publication bias. The sensitivity analysis revealed that the results are robust and no single study could alter the pooled ORs obviously.
For meta-analysis, the existence of heterogeneity among the available studies affects the reliability of the results in a large extent. Thus, we defined a limited number of potential heterogeneity factors before performing our meta-analysis. As for IL-10 -592C/A polymorphism, results from subgroup analysis suggested that the ethnicity and disease subtype might be the sources of heterogeneity. Regarding IL-10 -1082G/A polymorphism, the ethnicity might contribute to the between-study heterogeneity.
As far as we know, this is the first comprehensive meta-analysis exploring the association between 3 IL-10 gene polymorphisms and CVD risk up to now. Previous meta-analysis mainly focused on IL-10 -1082G/A polymorphism and stroke risk,49–52 whereas our meta-analysis included more studies concerning 3 well-characterized polymorphisms in the IL-10 gene and two CVD outcomes (CAD and stroke). Our meta-analysis also has some advantages. First, the search and selection studies were conducted strictly. Second, no evidence of publication bias was found by Begg funnel plot and Egger test. Third, TSA was performed, which could reduce the type I error rate. In addition, we performed false-positive report probability analysis to preclude false association resulting from multiple calculations.
Despite the clear strengths of this meta-analysis, including the large sample size and the implementation of TSA, several limitations should be addressed. First, the included studies were published in English and Chinese, whereas studies published in other languages were ignored. Second, there was significant heterogeneity in some of the pooled analysis, which may have affected the meta-analysis results even though we adopted the random-effects model. Third, several studies deviate from HWE expectations. Though, when the analysis was restricted to the studies in HWE, the pooled results did not alter significantly. Fourth, 2 studies in our meta-analysis included population with Latinos, and we did not find studies performed in other mixed ethnicities, so it is hard to make a definite conclusion about the population-specific genetic differences between the 3 polymorphisms and CVD risk; further studies should pay attention to the ethnic-specific effects on CVD susceptibility.
In conventional meta-analyses, IL-10 -592C/A and IL-10 -1082G/A polymorphisms were not likely to exert any influence on the susceptibility of CVD, whereas the IL-10 -819C/T polymorphism might be a protective factor for CVD, especially for CAD outcome, suggesting potential implications for genotyping the IL-10 -819C/T polymorphism in CAD risk appraisal. After TSA adjustment for sparse data and multiple updating in cumulative meta-analysis, it seems unsure that IL-10 -592C/A and IL-10 -1082G/A polymorphisms were not associated with the risk of CVD. Considering our main limitations, larger well-designed studies are necessary. Moreover, other IL polymorphisms and gene–gene interactions should also be considered in future studies.
Supplementary Material
Footnotes
Abbreviations: CAD = coronary artery disease, CI = confidential interval, CVD = cardiovascular disease, FPRP = False-positive report probability, HWE = Hardy-Weinberg equilibrium, IL-10 = interleukin 10, IS = ischemic stroke, OR = odds ratio, PCR = polymerase chain reaction, SNP = single-nucleotide polymorphism, TSA = trial sequential analysis.
The authors have no conflicts of interest to disclose.
This work was supported in part by the National Natural Science Foundation of China (No. 81470474).
REFERENCES
- 1.Moran AE, Roth GA, Narula J, et al. 1990–2010 global cardiovascular disease atlas. Glob Heart 2014; 9:3–16. [DOI] [PubMed] [Google Scholar]
- 2.Roger VL, Go AS, Lloyd-Jones DM, et al. Heart disease and stroke statistics–2012 update: a report from the American Heart Association. Circulation 2012; 125:e2–e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaess BM, Preis SR, Lieb W, et al. Circulating brain-derived neurotrophic factor concentrations and the risk of cardiovascular disease in the community. J Am Heart Assoc 2015; 4:e001544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munshi A, Rajeshwar K, Kaul S, et al. Interleukin-10-1082 promoter polymorphism and ischemic stroke risk in a South Indian population. Cytokine 2010; 52:221–224. [DOI] [PubMed] [Google Scholar]
- 5.Anguera I, Miranda-Guardiola F, Bosch X, et al. Elevation of serum levels of the anti-inflammatory cytokine interleukin-10 and decreased risk of coronary events in patients with unstable angina. Am Heart J 2002; 144:811–817. [DOI] [PubMed] [Google Scholar]
- 6.Meuwissen M, van der Wal AC, Siebes M, et al. Role of plaque inflammation in acute and recurrent coronary syndromes. Neth Heart J 2004; 12:106–109. [PMC free article] [PubMed] [Google Scholar]
- 7.Marousi S, Antonacopoulou A, Kalofonos H, et al. Functional inflammatory genotypes in ischemic stroke: could we use them to predict age of onset and long-term outcome? Stroke Res Treat 2011; 2011:792923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slattery ML, Herrick JS, Torres-Mejia G, et al. Genetic variants in interleukin genes are associated with breast cancer risk and survival in a genetically admixed population: the Breast Cancer Health Disparities Study. Carcinogenesis 2014; 35:1750–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mallat Z, Besnard S, Duriez M, et al. Protective role of interleukin-10 in atherosclerosis. Circ Res 1999; 85:e17–e24. [DOI] [PubMed] [Google Scholar]
- 10.Donger C, Georges JL, Nicaud V, et al. New polymorphisms in the interleukin-10 gene - relationships to myocardial infarction. Eur J Clin Invest 2001; 31:9–14. [DOI] [PubMed] [Google Scholar]
- 11.Heeschen C, Dimmeler S, Hamm CW, et al. Serum level of the antiinflammatory cytokine interleukin-10 is an important prognostic determinant in patients with acute coronary syndromes. Circulation 2003; 107:2109–2114. [DOI] [PubMed] [Google Scholar]
- 12.Xie G, Myint PK, Zaman MJ, et al. Relationship of serum interleukin-10 and its genetic variations with ischemic stroke in a Chinese general population. PLoS One 2013; 8:e74126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuo HP, Che L, Wu XZ. Correlation between the polymorphism of C-592A in Interleukin-10 gene and genetic susceptibity to coronary heart disease. Exp Clini Cardiol 2014; 20:3440–3454. [Google Scholar]
- 14.Koch W, Kastrati A, Bottiger C, et al. Interleukin-10 and tumor necrosis factor gene polymorphisms and risk of coronary artery disease and myocardial infarction. Atherosclerosis 2001; 159:137–144. [DOI] [PubMed] [Google Scholar]
- 15.Fragoso JM, Vallejo M, Alvarez-Leon E, et al. Alleles and haplotypes of the interleukin 10 gene polymorphisms are associated with risk of developing acute coronary syndrome in Mexican patients. Cytokine 2011; 55:29–33. [DOI] [PubMed] [Google Scholar]
- 16.Zhang GZ, Pan SY, Du R, et al. The relationship between interleukin-10 gene polymorphisms and cerebral infarction [in Chinese]. Chin J Cerebrovasc Dis 2007; 4:294–297. [Google Scholar]
- 17.Babu B, Reddy BP, Priya VHS, et al. Cytokine gene polymorphisms in the susceptibility to acute coronary syndrome. Genet Test Mol Biomarkers 2012; 16:359–365. [DOI] [PubMed] [Google Scholar]
- 18.Marousi SG, Ellul J, Antonacopoulou A, et al. Functional polymorphisms of interleukin 4 and interleukin 10 may predict evolution and functional outcome of an ischaemic stroke. Eur J Neurol 2011; 18:637–643. [DOI] [PubMed] [Google Scholar]
- 19.Karaca E, Kayikcioglu M, Onay H, et al. The effect of interleukin-10 gene promoter polymorphisms on early-onset coronary artery disease. Anadolu Kardiyol Derg 2011; 11:285–289. [DOI] [PubMed] [Google Scholar]
- 20.Afzal MS, Anjum S, Farooqi ZUR, et al. Influence of IL-10 polymorphism on the development of coronary artery disease in Pakistan. Asian Biomed 2012; 6:159–165. [Google Scholar]
- 21.Balding J, Livingstone WJ, Pittock SJ, et al. The IL-6 G-174C polymorphism may be associated with ischaemic stroke in patients without a history of hypertension. Irish J Med Sci 2004; 173:200–203. [DOI] [PubMed] [Google Scholar]
- 22.Ben-Hadj-Khalifa S, Ghazouani L, Abboud N, et al. Functional interleukin-10 promoter variants in coronary artery disease patients in Tunisia. Eur Cytokine Netw 2010; 21:136–141. [DOI] [PubMed] [Google Scholar]
- 23.Biswas S, Ghoshal PK, Mandal N. Synergistic effect of anti and pro-inflammatory cytokine genes and their promoter polymorphism with ST-elevation of myocardial infarction. Gene 2014; 544:145–151. [DOI] [PubMed] [Google Scholar]
- 24.Cruz M, Fragoso JM, Alvarez-Leon E, et al. The TGF-B1 and IL-10 gene polymorphisms are associated with risk of developing silent myocardial ischemia in the diabetic patients. Immunol Lett 2013; 156:18–22. [DOI] [PubMed] [Google Scholar]
- 25.Jin H, Wang Y, Xu LX. Association of interleukin 10 gene -592C/A polymorphism with coronary artery disease [in Chinese]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2013; 30:724–728. [DOI] [PubMed] [Google Scholar]
- 26.Lio D, Candore G, Crivello A, et al. Opposite effects of interleukin 10 common gene polymorphisms in cardiovascular diseases and in successful ageing: genetic background of male centenarians is protective against coronary heart disease. J Med Genet 2004; 41:790–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nasibullin TR, Timasheva YR, Tuktarova IA, et al. Combinations of cytokine gene network polymorphic markers as potential predictors of myocardial infarction. Russian J Genet 2014; 50:987–993. [PubMed] [Google Scholar]
- 28.Qi XF, Feng TJ, Yang P, et al. Role of inflammatory parameters in the susceptibility of cerebral thrombosis. Genet Mol Res 2014; 13:6350–6355. [DOI] [PubMed] [Google Scholar]
- 29.Rosner SA, Ridker PM, Zee RYL, et al. Interaction between inflammation-related gene polymorphisms and cigarette smoking on the risk of myocardial infarction in the Physician's Health Study. Hum Genet 2005; 118:287–294. [DOI] [PubMed] [Google Scholar]
- 30.Seifart C, Dempfle A, Plagens A, et al. TNF-alpha-, TNF-beta-, IL-6-, and IL-10-promoter polymorphisms in patients with chronic obstructive pulmonary disease. Tissue Antigens 2005; 65:93–100. [DOI] [PubMed] [Google Scholar]
- 31.Tuttolomondo A, Di Raimondo D, Forte GI, et al. Single nucleotide polymorphisms (SNPs) of pro-inflammatory/anti-inflammatory and thrombotic/fibrinolytic genes in patients with acute ischemic stroke in relation to TOAST subtype. Cytokine 2012; 58:398–405. [DOI] [PubMed] [Google Scholar]
- 32.Yu GI, Cho HC, Cho YK, et al. Association of promoter region single nucleotide polymorphisms at positions -819C/T and -592C/A of interleukin 10 gene with ischemic heart disease. Inflamm Res 2012; 61:899–905. [DOI] [PubMed] [Google Scholar]
- 33.Holm K, Melum E, Franke A, et al. SNPexp - A web tool for calculating and visualizing correlation between HapMap genotypes and gene expression levels. BMC Bioinformatics 2010; 11:600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He J, Shi TY, Zhu ML, et al. Associations of Lys939Gln and Ala499Val polymorphisms of the XPC gene with cancer susceptibility: a meta-analysis. Int J Cancer 2013; 133:1765–1775. [DOI] [PubMed] [Google Scholar]
- 35.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010; 25:603–605. [DOI] [PubMed] [Google Scholar]
- 36.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002; 21:1539–1558. [DOI] [PubMed] [Google Scholar]
- 37.Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. Bmj 1997; 315:629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wacholder S, Chanock S, Garcia-Closas M, et al. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst 2004; 96:434–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wetterslev J, Thorlund K, Brok J, et al. Trial sequential analysis may establish when firm evidence is reached in cumulative meta-analysis. J Clin Epidemiol 2008; 61:64–75. [DOI] [PubMed] [Google Scholar]
- 40.Turner RM, Bird SM, Higgins JP. The impact of study size on meta-analyses: examination of underpowered studies in Cochrane reviews. PLoS One 2013; 8:e59202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brok J, Thorlund K, Wetterslev J, et al. Apparently conclusive meta-analyses may be inconclusive–Trial sequential analysis adjustment of random error risk due to repetitive testing of accumulating data in apparently conclusive neonatal meta-analyses. Int J Epidemiol 2009; 38:287–298. [DOI] [PubMed] [Google Scholar]
- 42.Wetterslev J, Thorlund K, Brok J, et al. Estimating required information size by quantifying diversity in random-effects model meta-analyses. BMC Med Res Methodol 2009; 9:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holst LB, Petersen MW, Haase N, et al. Restrictive versus liberal transfusion strategy for red blood cell transfusion: systematic review of randomised trials with meta-analysis and trial sequential analysis. BMJ 2015; 350:h1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elsaid A, Abdel-Aziz AF, Elmougy R, et al. Association of polymorphisms G(-174)C in IL-6 gene and G(-1082)A in IL-10 gene with traditional cardiovascular risk factors in patients with coronary artery disease. Indian J Biochem Biophys 2014; 51:282–292. [PubMed] [Google Scholar]
- 45.Ianni M, Callegari S, Rizzo A, et al. Pro-inflammatory genetic profile and familiarity of acute myocardial infarction. Immun Ageing 2012; 9:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lorenzova A, Stanek V, Gebauerova M, et al. High-sensitivity C-reactive protein concentration in patients with myocardial infarction-environmental factors, and polymorphisms in interleukin-10 and CD14 genes. Clin Chem Lab Med 2007; 45:855–861. [DOI] [PubMed] [Google Scholar]
- 47.O’Halloran AM, Stanton A, O’Brien E, et al. The impact on coronary artery disease of common polymorphisms known to modulate responses to pathogens. Ann Hum Genet 2006; 70:934–945. [DOI] [PubMed] [Google Scholar]
- 48.Sultana S, Kolla VK, Jeedigunta Y, et al. Tumour necrosis factor alpha and interleukin 10 gene polymorphisms and the risk of ischemic stroke in south Indian population. J Genet 2011; 90:361–364. [DOI] [PubMed] [Google Scholar]
- 49.Chao L, Lei H, Fei J. A meta-analysis of interleukin-10-1082 promoter genetic polymorphism associated with atherosclerotic risk. Neurol India 2014; 62:130–136. [DOI] [PubMed] [Google Scholar]
- 50.Jin J, Li W, Peng L, et al. Relationship between interleukin-10 -1082A/G polymorphism and risk of ischemic stroke: a meta-analysis. PLoS One 2014; 9:e94631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yadav S, Hasan N, Marjot T, et al. Detailed analysis of gene polymorphisms associated with ischemic stroke in South Asians. PLoS One 2013; 8:e57305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin GT, Ma YT, Zheng YY, et al. Polymorphisms of interleukin-10 genes on the risk of ischemic stroke in a meta-analysis. Int J Clin Exp Med 2015; 8:1888–1895. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


