Abstract
The rapid advances in the field of genome editing using targeted endonucleases have called considerable attention to the potential of this technology for human gene therapy. Targeted correction of disease-causing mutations could ensure lifelong, tissue-specific expression of the relevant gene, thereby alleviating or resolving a specific disease phenotype. In this review, we aim to explore the potential of this technology for the therapy of β-thalassemia. This blood disorder is caused by mutations in the gene encoding the β-globin chain of hemoglobin, leading to severe anemia in affected patients. Curative allogeneic bone marrow transplantation is available only to a small subset of patients, leaving the majority of patients dependent on regular blood transfusions and iron chelation therapy. The transfer of gene-corrected autologous hematopoietic stem cells could provide a therapeutic alternative, as recent results from gene therapy trials using a lentiviral gene addition approach have demonstrated. Genome editing has the potential to further advance this approach as it eliminates the need for semi-randomly integrating viral vectors and their associated risk of insertional mutagenesis. In the following pages we will highlight the advantages and risks of genome editing compared to standard therapy for β-thalassemia and elaborate on lessons learned from recent gene therapy trials.
Keywords: thalassemia, genome, gene therapy
β-Thalassemia
β-Thalassemia is a common congenital blood disorder caused by mutations in the β-globin gene. Reduced or absent β-globin expression leads to an imbalance of the α-globin and β-globin subunits that form the hemoglobin tetramer. The toxic accumulation of excess α-globin chains in developing erythrocytes results in severe anemia due to ineffective erythropoiesis 1. In its most serious form, β-thalassemia major, the condition is fatal if left untreated 2. Currently, allogenic bone marrow transplantation (BMT) is the only curative therapeutic option. However, due to the rarity of suitable donors, this treatment is available only to a small subset of patients and the procedure itself entails a risk of potentially life-threatening immunological complications and graft failure, especially for patients over 3 years of age 3, 4.
The majority of β-thalassemia patients depend on regular blood transfusions combined with iron chelation therapy for their survival 5. Even under optimal care, this treatment regimen provides a suboptimal quality of life and leaves patients at an increased risk of death from cardiomyopathies and infection 6, 7. New therapeutic strategies are therefore needed to better manage β-thalassemia.
Gene therapy for β-Thalassemia
In the past 25 years, the field of gene therapy has made considerable progress. Gene therapy aims at the functional cure of disorders through modification of a patient’s genome. Depending on the nature of the causative mutation, this could be achieved through the introduction of a therapeutic gene, correction of the disease-causing mutation, or the elimination of deleterious gene products (reviewed by Kay et al., 2011) 8.
The major obstacle all gene therapy approaches face is safe and efficient gene delivery to the affected tissue or cell type. In vivo delivery is particularly difficult due to poor tissue accessibility, vector immunogenicity, and limited target cell specificity 9. Monogenic blood disorders such as severe combined immune deficiency (SCID), sickle-cell anemia, and β-thalassemia are remarkably attractive targets for gene therapy due to the unique accessibility of hematopoietic progenitor cells, which can be isolated from patient bone marrow 10. The ex vivo correction and re-introduction of autologous hematopoietic stem cells (HSCs) has no associated risk of graft-versus-host disease, the major adverse effect of allogenic BMT. Eliminating the necessity of a matched donor potentially makes this approach applicable to all patients. Gene therapy could therefore provide a safer and more generally available curative treatment for blood disorders than allogenic BMT.
Past and ongoing gene therapy trials are mostly focused on the delivery of a therapeutic gene using integrating viral vectors. This gene addition approach has been successfully applied in severe combined immunodeficiencies 11– 13, retinal disorders 14– 16, and hemophilia 17. The first successful gene therapy trial for β-thalassemia was reported in 2010 18. The trial employed a lentiviral vector for ex vivo delivery of a β-globin transgene into patient HSCs, which were subsequently returned to the patient. The treatment was successful in one patient who remained transfusion-independent for up to 7 years 19, 20. A second trial was subsequently initiated using a modified vector. Although long-term results are yet to be released, promising preliminary data describe two patients remaining transfusion-independent for 14 and 16 months, respectively 21. These trials demonstrate that gene therapy has the potential to provide effective long-term therapy following a single treatment.
The greatest caveat in the use of integrating lentiviral and retroviral vectors lies in the inability to control for target site selection, which can result in considerable genotoxicity from the transactivation of nearby proto-oncogenes 22, 23. This was tragically confirmed when four out of nine children treated in the first gene therapy trial for SCID-X1 developed leukemia as a result of gamma-retrovirus vector integration, causing the death of one patient 24. Following this setback, vector design was improved by the development of self-inactivating lentiviruses, insulator elements, and tissue-specific promoters 25– 27. Nonetheless, insertional mutagenesis still remains the major concern with retroviral and lentiviral gene therapy approaches 28. The importance of understanding and managing this risk was again demonstrated by the appearance of a dominant clone with a transactivating insertion event near the HMGA2 gene in the HSCs of the first successfully treated β-thalassemia gene therapy patient 18. This event, though only transient, has again emphasized the necessity for careful monitoring of patients following treatment with integrating vectors.
This issue has driven the search for safer gene therapy approaches. One possible solution is the targeted integration of a therapeutic gene into a genomic “safe harbor” site that supports long-term transgene expression without affecting transcriptional activity at endogenous loci. The natural preference of adeno-associated viruses (AAVs) for integration at the AAVS1 site on chromosome 19 could potentially provide an alternative to the semi-random integration profile of lentiviral and retroviral vectors 29. However, their small transgene capacity limits the usefulness of AAVs as gene therapy vectors 30– 32. Hybrid strategies combining the site-selective recombinase activity of the AAV rep protein with larger vectors have the potential to overcome this limitation. Based on this principle, we have previously achieved targeted integration of a bacterial artificial chromosome carrying the whole human β-globin locus into the AAVS1 site in K562 cells 33. Another approach, gene repair through homologous recombination, has been proposed already in the 1980s 34, 35. In 1985, Smithies et al. demonstrated the introduction of heterologous DNA sequences into the β-globin locus of human cell lines using homologous recombination 36. These results led to the first speculation that targeted genome modification via homologous recombination in HSCs could provide a cure for β-hemoglobinopathies. However, before the emergence of targeted endonucleases, this approach remained limited by low efficiency.
Genome editing
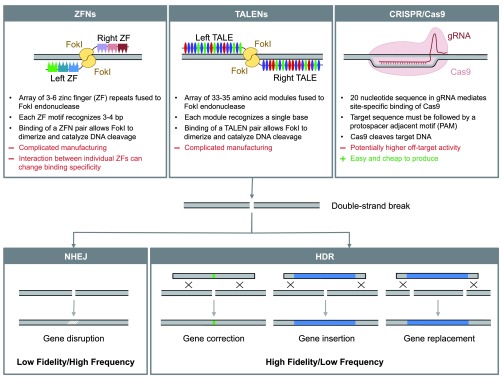
The discovery and development of targetable endonucleases has kindled a new enthusiasm for the previously niche area of genome modification through homologous repair. These enzymes can be engineered to introduce a site-specific double-strand break (DSB) into a target genome, which can subsequently be repaired by endogenous DNA repair mechanisms ( Figure 1). Mammalian cells possess two major DSB repair pathways: non-homologous end-joining (NHEJ) and homology-directed repair (HDR) 37, 38. NHEJ is error prone and leads to the creation of small insertions or deletions at the DSB site. This has been used very efficiently for targeted gene knockout in a variety of cell types and for the generation of knockout animal models 39– 44. HDR uses a homologous DNA template to repair the broken strand with high fidelity. Fusion of a reporter to a gene of interest and gene insertion, as well as targeted gene correction, have been demonstrated using this approach 45– 47.
Figure 1. Genome editing technologies.
ZFNs, TALENs, and CRISPR/Cas9 are used to introduce site-specific DSBs into a target genome. Subsequently, cellular repair mechanisms can be harnessed to introduce precise genetic modifications. Small insertions and deletions generated by NHEJ can be used for gene knockout. In the presence of a homologous repair template, new sequences can be incorporated via HDR, allowing for gene repair, transgene insertion, and gene replacement.
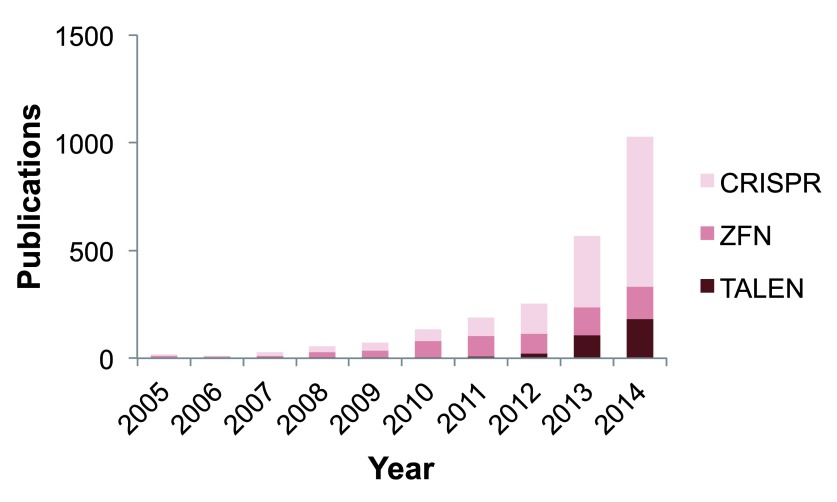
There are three different types of programmable endonucleases. Zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) are generated by the fusion of repetitive arrays of specific DNA-binding amino acid motifs to a FokI endonuclease domain ( Figure 1) 48– 50. Binding of a pair of ZFNs or TALENs on opposite DNA strands allows their FokI domains to dimerize and become catalytically active, introducing a DSB with 5’ overhangs into the target site. ZFNs and TALENs both require complicated cloning approaches to achieve the arrangement of repetitive motifs, imposing penalties of time and expense upon their development. The recent adaptation of clustered-interspaced short palindromic repeats (CRISPRs) and CRISPR-associated protein 9 (Cas9) for use in mammalian cells has greatly facilitated genome editing applications. The Cas9 endonuclease is targeted to a specific DNA sequence by a complementary 20-nucleotide sequence in a guide RNA (gRNA) bound to the Cas9 protein. New gRNAs can be generated quickly and at low cost using standard cloning techniques 51, 52. Due to the ease of use of the Cas9 system, its use has rapidly surpassed that of ZFNs and TALENs in the past years ( Figure 2).
Figure 2. Publications on genome editing between 2005 and 2014.
Data obtained from Medline trend using the search terms “CRISPR Cas9”, “Zinc-finger nuclease”, and “TALEN” show an increase in the use of programmable endonucleases during this period 99.
Similar to lentiviral gene therapy, genome editing could be used to correct patient HSCs ex vivo for the gene therapy of β-thalassemia. Ideally, scarless correction of the β-globin gene in HSCs could be achieved through HDR, resulting in the production of healthy erythrocytes. Several studies have shown that the human β-globin locus is amenable to genome editing ( Table 1) 53– 61. However, technical limitations and safety concerns need to be overcome for this novel approach to become clinically applicable.
Table 1. Recent studies employing novel strategies for therapeutic genome editing at the human β-globin locus.
(iPSCs: induced pluripotent stem cells).
| Strategy | Cell type | Platform | Reference |
|---|---|---|---|
| Correction of β-thalassemia mutations | Patient iPSCs | CRISPR/Cas9 | Xie et al., 2014 55 |
| Patient iPSCs | TALENS | Ma et al., 2013 57 | |
| Correction of sickle-cell mutation | Patient iPSCs | TALENs | Sun et al., 2014 62 |
| Patient iPSCs | TALENs | Ramalingam et al., 2014 58 | |
| Patient iPSCs | CRISPR/Cas9 | Huang et al., 2015 54 | |
| HSCs | ZFNs | Hoban et al., 2015 60 | |
| Gene insertion of β-globin cDNA | K562 | TALENs | Voit et al., 2014 53 |
| γ-globin reactivation | MEL | TALENs | Wienert et al., 2015 59 |
In contrast to viral gene addition approaches, genome editing does not require the use of integrating vectors, as transient expression of a targeted endonuclease is sufficient to achieve the necessary DNA cleavage. This eliminates the issue of insertional mutagenesis. However, off-target cleavage at sites other than that intended is a major concern with genome editing approaches 63– 65. For the CRISPR/Cas9 system, strategies have been developed to reduce the relatively high off-target cleavage associated with wild-type Cas9. A mutated Cas9 protein that introduces a single-stranded nick rather than a DSB can be used to increase cleavage specificity. Consequently, two gRNAs designed to mediate nicking on opposite strands at the target site are required to form a DSB 63. However, a single gRNA is still sufficient to introduce a DNA nick at off-target sites, which may have adverse effects in the target cell. Alternatively, an inactive Cas9 protein can be fused to FokI, which only becomes enzymatically active upon dimerization. With this approach, two Cas9/FokI hybrid units need to be brought together by specific gRNAs to allow cleavage at the target site 66, 67. The target specificity of Cas9 can be further increased through the use of a truncated guide sequence of 17 instead of 20 nucleotides 68. However, off-target activity of any nuclease type still varies between different genomic targets and cell types 69, 70. Therefore, as with all gene therapy strategies, careful vector design and thorough evaluation of risks is necessary.
Off-target site prediction tools that rank potential unintended cleavage sites based on similarity scores were developed to facilitate the evaluation of cleavage stringency for different nuclease platforms. It remains to be determined if the targeted analysis of selected putative off-target sites is sufficient for the determination of nuclease-associated risks. Further validation of the reliability of these prediction tools via unbiased genome-wide detection of off-target cleavage is therefore required. Approaches taking advantage of the occasional capture of foreign sequences in genomic DSBs show promise to close this information gap 71– 73. Many studies report minimal to no detectable off-target activity across a variety of nuclease platforms and target sites 60, 66, 67, 74– 77. A 2015 publication using ZFNs to correct the sickle-cell mutation in primary patient HSCs indicates that therapeutic genome editing of the β-globin gene can be achieved without producing deleterious unintended mutations. The only off-target events detected in a genome-wide analysis were located in the highly homologous δ-globin gene, which is non-essential 60. Also, the first clinical phase I human genome editing trial using ZFNs to disrupt the CCR5 co-receptor for HIV entry in autologous CD4 T cells has not produced any adverse events that could be attributed to the use of ZFNs 78. While further confirmation is still required, these findings suggest that off-target effects will not restrict genome editing from clinical applications.
Gene therapy trials for SCID are simplified due to the selective advantage of gene corrected cells over unmodified HSCs 11. In the case of the β-hemoglobinopathies, β-globin expression does not convey an advantage for HSCs. Consequently, a substantial fraction of HSCs needs to be modified to achieve a therapeutic effect. Lentiviral or retroviral delivery and nucleofection of DNA or mRNA can achieve transfection rates greater than 80% in primary human HSCs 79– 81. These methods are also suitable for the delivery of genome editing tools. A high transduction efficiency, leading to a high frequency of target cleavage, is essential for efficient genome editing. However, low HDR frequency in naïve HSCs, accompanied by a background of disruptive NHEJ, currently impedes the generation of therapeutic levels of edited cells 60, 82. Although NHEJ is unlikely to produce adverse effects in an already non-functional gene, it will be crucial to increase the fraction of cells that undergo HDR genome editing to be successful in the clinic. Several groups have developed screening methods that permit simultaneous quantification of NHEJ and HDR 83– 87. These can be used for the identification of conditions that favor HDR. Most notably, inhibition of DNA ligase 4, which is required for the NHEJ pathway, has been shown to not only decrease NHEJ but also increase HDR frequencies in cell lines and mouse embryos 88, 89. As the repair pathway choice in a cell is largely dependent on the cell cycle stage, cell synchronization and timed nuclease delivery could also bias cells towards HDR 90. Increasing the frequency of gene correction in HSCs will be crucial in determining the feasibility of therapeutic genome editing in the hematopoietic system.
A future in the clinic
Although low HDR efficiency and safety concerns regarding off-target effects are currently obstructing the therapeutic application of genome editing, strategies to resolve these limitations are rapidly progressing. As with all novel therapeutics, every custom genome editing vector will be subject to careful clinical trials. It is therefore crucial to design therapeutic genome editing strategies to be as inclusive as possible, i.e. to minimize the number of different vectors required to treat the maximum number of patients. While over 200 mutations are known to cause β-thalassemia, a relatively small number of mutations account for the majority of cases 91. Therefore, a small number of Cas9/gRNA vectors could be sufficient to address the majority of patients. Alternatively, the introduction of two DSBs at either side of the β-globin gene could allow for gene replacement without the need for allele-specific vectors, thus placing a therapeutic β-globin under the control of endogenous regulatory elements at the β-globin locus 53, 92. Like lentiviral gene therapy, genome editing can also be applied to gene addition. A single genome editing vector targeting a safe harbor site could be combined with a separate HDR template containing a therapeutic β-globin gene. This approach has the potential to provide a universally applicable strategy, as a single genome editing vector could be used for a large range of monogenic disorders by simply exchanging the HDR template. Genome editing also has the potential to introduce mutations that modify the severity of β-thalassemia. It is known from individuals with hereditary persistence of fetal hemoglobin that elevated expression of γ-globin, a developmentally silenced β-globin-like gene, can be protective of the pathologic effects associated with the absence of β-globin expression 93, 94. Replication of this phenotype through genome editing could therefore alleviate the symptoms in β-thalassemic patients. A recent study employed TALENs to introduce a single point mutation within the β-globin locus to increase the expression of γ-globin 59. Interference with the expression of BCL11A, a major regulator of β-globin gene expression, has also been shown to promote the expression of γ-globin 95, 96. An erythroid-specific enhancer for BCL11A expression was recently identified by Bauer et al. 97. Targeted elimination of this enhancer in patient-derived HSCs could allow the induction of γ-globin expression in erythroid cells without affecting BCL11A-dependent processes in other lineages 98. This could be achieved through an NHEJ approach, unimpeded by the low frequency that currently limits strategies depending on HDR. However, in the future, the latter could be applied to the correction of patient-derived iPSCs, thus circumventing the issue of HDR efficiency, since a large number of cells can be generated from a few corrected clones. With this range of possibilities, genome editing is diversifying gene therapy research with the potential to greatly relieve the global health burden of the β-hemoglobinopathies.
Abbreviations
AAV, adeno-associated virus; BMT, bone marrow transplantation; Cas9, CRISPR-associated protein 9; CRISPRs, clustered interspaced palindromic repeats; DSB, double strand break; gRNA, guide RNA; HDR, homology-directed repair; HSCs, hematopoietic stem cells; iPSCs induced pluripotent stem cells; NHEJ, non-homologous end-joining; TALENs, transcription activator-like effector nucleases; SCID, severe combined immune deficiency; ZFNs, zinc-finger nucleases
Acknowledgments
The National Health and Medical Research Council, the Murdoch Childrens Research Institute, the Victorian Government’s Operational Infrastructure Support Program, and Thalassaemia Australia.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Donald Kohn, Department of Microbiology, Immunology and Molecular Genetics and Department of Pediatrics, University of California, Los Angeles, Los Angeles, CA, USA
Alan Schechter, Molecular Medicine Branch, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, Maryland, 20892, USA
Funding Statement
The author(s) declared that no grants were involved in supporting this work.
[version 1; referees: 2 approved]
References
- 1. Ribeil JA, Arlet JB, Dussiot M, et al. : Ineffective erythropoiesis in β -thalassemia. ScientificWorldJournal. 2013;2013:394295. 10.1155/2013/394295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lanzkowsky P: Manual of pediatric hematology and oncology.2011;5 Reference Source [Google Scholar]
- 3. King A, Shenoy S: Evidence-based focused review of the status of hematopoietic stem cell transplantation as treatment of sickle cell disease and thalassemia. Blood. 2014;123(20):3089–94; quiz 3210. 10.1182/blood-2013-01-435776 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 4. Gennery AR, Slatter MA, Grandin L, et al. : Transplantation of hematopoietic stem cells and long-term survival for primary immunodeficiencies in Europe: entering a new century, do we do better? J Allergy Clin Immunol. 2010;126(3):602–10.e1-11. 10.1016/j.jaci.2010.06.015 [DOI] [PubMed] [Google Scholar]
- 5. Galanello R, Origa R: Beta-thalassemia. Orphanet J Rare Dis. 2010;5:11. 10.1186/1750-1172-5-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kremastinos DT, Farmakis D, Aessopos A, et al. : Beta-thalassemia cardiomyopathy: history, present considerations, and future perspectives. Circ Heart Fail. 2010;3(3):451–8. 10.1161/CIRCHEARTFAILURE.109.913863 [DOI] [PubMed] [Google Scholar]
- 7. Cappellini MD, Cohen A, Eleftheriou A, et al. : Guidelines for the Clinical Management of Thalassaemia [Internet].2008. [PubMed] [Google Scholar]
- 8. Kay MA: State-of-the-art gene-based therapies: the road ahead. Nat Rev Genet. 2011;12(5):316–28. 10.1038/nrg2971 [DOI] [PubMed] [Google Scholar]
- 9. Mali S: Delivery systems for gene therapy. Indian J Hum Genet. 2013;19(1):3–8. 10.4103/0971-6866.112870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Herzog RW, Hagstrom JN: Gene therapy for hereditary hematological disorders. Am J Pharmacogenomics. 2001;1(2):137–44. [DOI] [PubMed] [Google Scholar]
- 11. Fischer A, Hacein-Bey Abina S, Touzot F, et al. : Gene therapy for primary immunodeficiencies. Clin Genet. 2015;88(6):507–15. 10.1111/cge.12576 [DOI] [PubMed] [Google Scholar]
- 12. Hacein-Bey-Abina S, Hauer J, Lim A, et al. : Efficacy of gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2010;363(4):355–64. 10.1056/NEJMoa1000164 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 13. Gaspar HB, Cooray S, Gilmour KC, et al. : Hematopoietic stem cell gene therapy for adenosine deaminase-deficient severe combined immunodeficiency leads to long-term immunological recovery and metabolic correction. Sci Transl Med. 2011;3(97):97ra80. [DOI] [PubMed] [Google Scholar]
- 14. Bainbridge JW, Smith AJ, Barker SS, et al. : Effect of gene therapy on visual function in Leber's congenital amaurosis. N Engl J Med. 2008;358(21):2231–9. 10.1056/NEJMoa0802268 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 15. Hauswirth WW, Aleman TS, Kaushal S, et al. : Treatment of leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: short-term results of a phase I trial. Hum Gene Ther. 2008;19(10):979–90. 10.1089/hum.2008.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maguire AM, Simonelli F, Pierce EA, et al. : Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med. 2008;358(21):2240–8. 10.1056/NEJMoa0802315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nathwani AC, Tuddenham EG, Rangarajan S, et al. : Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med. 2011;365(25):2357–65. 10.1056/NEJMoa1108046 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 18. Cavazzana-Calvo M, Payen E, Negre O, et al. : Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467(7313):318–22. 10.1038/nature09328 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 19. Press release. First two patients in the HGB-205 Study achieved transfusion independence within two weeks of an autologous transplant with bluebird’s lentiviral gene therapy.2014. Reference Source [Google Scholar]
- 20. Finotti A, Breda L, Lederer CW, et al. : Recent trends in the gene therapy of β-thalassemia. J Blood Med. 2015;6:69–85. 10.2147/JBM.S46256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Press Release. Bluebird Bio Reports New Beta-thalassemia Major and Severe Sickle Cell Disease Data from HGB-205 Study at EHA.2015. Reference Source [Google Scholar]
- 22. Trobridge GD: Genotoxicity of retroviral hematopoietic stem cell gene therapy. Expert Opin Biol Ther. 2011;11(5):581–93. 10.1517/14712598.2011.562496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hacein-Bey-Abina S, Von Kalle C, Schmidt M, et al. : LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302(5644):415–9. 10.1126/science.1088547 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 24. Hacein-Bey-Abina S, Garrigue A, Wang GP, et al. : Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118(9):3132–42. 10.1172/JCI35700 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 25. Emery DW: The use of chromatin insulators to improve the expression and safety of integrating gene transfer vectors. Hum Gene Ther. 2011;22(6):761–74. 10.1089/hum.2010.233 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 26. Yi Y, Hahm SH, Lee KH: Retroviral gene therapy: safety issues and possible solutions. Curr Gene Ther. 2005;5(1):25–35. 10.2174/1566523052997514 [DOI] [PubMed] [Google Scholar]
- 27. Zhou S, Mody D, DeRavin SS, et al. : A self-inactivating lentiviral vector for SCID-X1 gene therapy that does not activate LMO2 expression in human T cells. Blood. 2010;116(6):900–8. 10.1182/blood-2009-10-250209 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 28. Wu C, Dunbar CE: Stem cell gene therapy: the risks of insertional mutagenesis and approaches to minimize genotoxicity. Front Med. 2011;5(4):356–71. 10.1007/s11684-011-0159-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Samulski RJ, Zhu X, Xiao X, et al. : Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10(12):3941–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dong JY, Fan PD, Frizzell RA: Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum Gene Ther. 1996;7(17):2101–12. 10.1089/hum.1996.7.17-2101 [DOI] [PubMed] [Google Scholar]
- 31. Grieger JC, Samulski RJ: Packaging capacity of adeno-associated virus serotypes: impact of larger genomes on infectivity and postentry steps. J Virol. 2005;79(15):9933–44. 10.1128/JVI.79.15.9933-9944.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hermonat PL, Quirk JG, Bishop BM, et al. : The packaging capacity of adeno-associated virus (AAV) and the potential for wild-type-plus AAV gene therapy vectors. FEBS Lett. 1997;407(1):78–84. 10.1016/S0014-5793(97)00311-6 [DOI] [PubMed] [Google Scholar]
- 33. Howden SE, Voullaire L, Wardan H, et al. : Site-specific, Rep-mediated integration of the intact beta-globin locus in the human erythroleukaemic cell line K562. Gene Ther. 2008;15(20):1372–83. 10.1038/gt.2008.84 [DOI] [PubMed] [Google Scholar]
- 34. Vega MA: Prospects for homologous recombination in human gene therapy. Hum Genet. 1991;87(3):245–53. 10.1007/BF00200899 [DOI] [PubMed] [Google Scholar]
- 35. Capecchi MR: Altering the genome by homologous recombination. Science. 1989;244(4910):1288–92. 10.1126/science.2660260 [DOI] [PubMed] [Google Scholar]
- 36. Smithies O, Gregg RG, Boggs SS, et al. : Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985;317(6034):230–4. 10.1038/317230a0 [DOI] [PubMed] [Google Scholar]
- 37. Chapman JR, Taylor MR, Boulton SJ, et al. : Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47(4):497–510. 10.1016/j.molcel.2012.07.029 [DOI] [PubMed] [Google Scholar]
- 38. Zhang Y, Vanoli F, LaRocque JR, et al. : Biallelic targeting of expressed genes in mouse embryonic stem cells using the Cas9 system. Methods. 2014;69(2):171–8. 10.1016/j.ymeth.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bauer DE, Canver MC, Orkin SH: Generation of genomic deletions in mammalian cell lines via CRISPR/Cas9. J Vis Exp. 2015; (95). 10.3791/52118 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 40. Brandl C, Ortiz O, Röttig B, et al. : Creation of targeted genomic deletions using TALEN or CRISPR/Cas nuclease pairs in one-cell mouse embryos. FEBS Open Bio. 2015;5:26–35. 10.1016/j.fob.2014.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 41. Peng D, Kurup SP, Yao PY, et al. : CRISPR-Cas9-mediated single-gene and gene family disruption in Trypanosoma cruzi. MBio. 2015;6(1):e02097–14. 10.1128/mBio.02097-14 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 42. Gratz SJ, Wildonger J, Harrison MM, et al. : CRISPR/Cas9-mediated genome engineering and the promise of designer flies on demand. Fly (Austin). 2013;7(4):249–55. 10.4161/fly.26566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mandal PK, Ferreira LM, Collins R, et al. : Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell. 2014;15(5):643–52. 10.1016/j.stem.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 44. Wang W, Ye C, Liu J, et al. : CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS One. 2014;9(12):e115987. 10.1371/journal.pone.0115987 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 45. Wang H, Yang H, Shivalila CS, et al. : One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–8. 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 46. Yin H, Xue W, Chen S, et al. : Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32(6):551–3. 10.1038/nbt.2884 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 47. Li HL, Nakano T, Hotta A: Genetic correction using engineered nucleases for gene therapy applications. Dev Growth Differ. 2014;56(1):63–77. 10.1111/dgd.12107 [DOI] [PubMed] [Google Scholar]
- 48. Kim YG, Cha J, Chandrasegaran S: Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A. 1996;93(3):1156–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Boch J, Scholze H, Schornack S, et al. : Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326(5959):1509–12. 10.1126/science.1178811 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 50. Christian M, Cermak T, Doyle EL, et al. : Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186(2):757–61. 10.1534/genetics.110.120717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mali P, Yang L, Esvelt KM, et al. : RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–6. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 52. Cong L, Ran FA, Cox D, et al. : Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 53. Voit RA, Hendel A, Pruett-Miller SM, et al. : Nuclease-mediated gene editing by homologous recombination of the human globin locus. Nucleic Acids Res. 2014;42(2):1365–78. 10.1093/nar/gkt947 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 54. Huang X, Wang Y, Yan W, et al. : Production of Gene-Corrected Adult Beta Globin Protein in Human Erythrocytes Differentiated from Patient iPSCs After Genome Editing of the Sickle Point Mutation. Stem Cells. 2015;33(5):1470–9. 10.1002/stem.1969 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 55. Xie F, Ye L, Chang JC, et al. : Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 2014;24(9):1526–33. 10.1101/gr.173427.114 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 56. Sun N, Zhao H: Seamless correction of the sickle cell disease mutation of the HBB gene in human induced pluripotent stem cells using TALENs. Biotechnol Bioeng. 2014;111(5):1048–53. 10.1002/bit.25018 [DOI] [PubMed] [Google Scholar]
- 57. Ma N, Liao B, Zhang H, et al. : Transcription activator-like effector nuclease (TALEN)-mediated gene correction in integration-free β-thalassemia induced pluripotent stem cells. J Biol Chem. 2013;288(48):34671–9. 10.1074/jbc.M113.496174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ramalingam S, Annaluru N, Kandavelou K, et al. : TALEN-mediated generation and genetic correction of disease-specific human induced pluripotent stem cells. Curr Gene Ther. 2014;14(6):461–72. 10.2174/1566523214666140918101725 [DOI] [PubMed] [Google Scholar]
- 59. Wienert B, Funnell AP, Norton LJ, et al. : Editing the genome to introduce a beneficial naturally occurring mutation associated with increased fetal globin. Nat Commun. 2015;6:7085. 10.1038/ncomms8085 [DOI] [PubMed] [Google Scholar]
- 60. Hoban MD, Cost GJ, Mendel MC, et al. : Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood. 2015;125(17):2597–604. 10.1182/blood-2014-12-615948 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 61. Liang P, Xu Y, Zhang X, et al. : CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6(5):363–72. 10.1007/s13238-015-0153-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sun N, Liang J, Abil Z, et al. : Optimized TAL effector nucleases (TALENs) for use in treatment of sickle cell disease. Mol Biosyst. 2012;8(4):1255–63. 10.1039/c2mb05461b [DOI] [PubMed] [Google Scholar]
- 63. Lin Y, Cradick TJ, Brown MT, et al. : CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42(11):7473–85. 10.1093/nar/gku402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fu Y, Foden JA, Khayter C, et al. : High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–6. 10.1038/nbt.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cradick TJ, Fine EJ, Antico CJ, et al. : CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41(20):9584–92. 10.1093/nar/gkt714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ran FA, Hsu PD, Lin CY, et al. : Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–9. 10.1016/j.cell.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 67. Tsai SQ, Wyvekens N, Khayter C, et al. : Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32(6):569–76. 10.1038/nbt.2908 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 68. Fu Y, Sander JD, Reyon D, et al. : Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32(3):279–84. 10.1038/nbt.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 69. Smith C, Gore A, Yan W, et al. : Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell. 2014;15(1):12–3. 10.1016/j.stem.2014.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Veres A, Gosis BS, Ding Q, et al. : Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15(1):27–30. 10.1016/j.stem.2014.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang X, Wang Y, Wu X, et al. : Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat Biotechnol. 2015;33(2):175–8. 10.1038/nbt.3127 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 72. O'Geen H, Henry IM, Bhakta MS, et al. : A genome-wide analysis of Cas9 binding specificity using ChIP-seq and targeted sequence capture. Nucleic Acids Res. 2015;43(6):3389–404. 10.1093/nar/gkv137 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 73. Corrigan-Curay J, O'Reilly M, Kohn DB, et al. : Genome editing technologies: defining a path to clinic. Mol Ther. 2015;23(5):796–806. 10.1038/mt.2015.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Li L, Krymskaya L, Wang J, et al. : Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Mol Ther. 2013;21(6):1259–69. 10.1038/mt.2013.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mussolino C, Alzubi J, Fine EJ, et al. : TALENs facilitate targeted genome editing in human cells with high specificity and low cytotoxicity. Nucleic Acids Res. 2014;42(10):6762–73. 10.1093/nar/gku305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Li HL, Fujimoto N, Sasakawa N, et al. : Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports. 2015;4(1):143–54. 10.1016/j.stemcr.2014.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tan EP, Li Y, Velasco-Herrera Mdel C, et al. : Off-target assessment of CRISPR-Cas9 guiding RNAs in human iPS and mouse ES cells. Genesis. 2015;53(2):225–36. 10.1002/dvg.22835 [DOI] [PubMed] [Google Scholar]
- 78. Tebas P, Stein D, Tang WW, et al. : Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370(10):901–10. 10.1056/NEJMoa1300662 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 79. von Levetzow G, Spanholtz J, Beckmann J, et al. : Nucleofection, an efficient nonviral method to transfer genes into human hematopoietic stem and progenitor cells. Stem Cells Dev. 2006;15(2):278–85. 10.1089/scd.2006.15.278 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 80. Wiehe JM, Ponsaerts P, Rojewski MT, et al. : mRNA-mediated gene delivery into human progenitor cells promotes highly efficient protein expression. J Cell Mol Med. 2007;11(3):521–30. 10.1111/j.1582-4934.2007.00038.x [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 81. Leurs C, Jansen M, Pollok KE, et al. : Comparison of three retroviral vector systems for transduction of nonobese diabetic/severe combined immunodeficiency mice repopulating human CD34 + cord blood cells. Hum Gene Ther. 2003;14(6):509–19. 10.1089/104303403764539305 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 82. Genovese P, Schiroli G, Escobar G, et al. : Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510(7504):235–40. 10.1038/nature13420 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 83. Yu C, Liu Y, Ma T, et al. : Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16(2):142–7. 10.1016/j.stem.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 84. Certo MT, Ryu BY, Annis JE, et al. : Tracking genome engineering outcome at individual DNA breakpoints. Nat Methods. 2011;8(8):671–6. 10.1038/nmeth.1648 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 85. Shahar OD, Kalousi A, Eini L, et al. : A high-throughput chemical screen with FDA approved drugs reveals that the antihypertensive drug Spironolactone impairs cancer cell survival by inhibiting homology directed repair. Nucleic Acids Res. 2014;42(9):5689–701. 10.1093/nar/gku217 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 86. Hendel A, Kildebeck EJ, Fine EJ, et al. : Quantifying genome-editing outcomes at endogenous loci with SMRT sequencing. Cell Rep. 2014;7(1):293–305. 10.1016/j.celrep.2014.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 87. Kuhar R, Gwiazda KS, Humbert O, et al. : Novel fluorescent genome editing reporters for monitoring DNA repair pathway utilization at endonuclease-induced breaks. Nucleic Acids Res. 2014;42(1):e4. 10.1093/nar/gkt872 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 88. Chu VT, Weber T, Wefers B, et al. : Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33(5):543–8. 10.1038/nbt.3198 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 89. Maruyama T, Dougan SK, Truttmann MC, et al. : Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33(5):538–42. 10.1038/nbt.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 90. Lin S, Staahl BT, Alla RK, et al. : Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014;3:e04766. 10.7554/eLife.04766 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 91. Weatherall DJ, Clegg JB: The Thalassaemia Syndromes. Blackwell Science Ltd.,2001;4 10.1002/9780470696705 [DOI] [Google Scholar]
- 92. Byrne SM, Ortiz L, Mali P, et al. : Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells. Nucleic Acids Res. 2015;43(3):e21. 10.1093/nar/gku1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Thein SL: Genetic modifiers of beta-thalassemia. Haematologica. 2005;90(5):649–60. [PubMed] [Google Scholar]
- 94. Galanello R, Cao A: Relationship between genotype and phenotype. Thalassemia intermedia. Ann N Y Acad Sci. 1998;850:325–33. 10.1111/j.1749-6632.1998.tb10489.x [DOI] [PubMed] [Google Scholar]
- 95. Sankaran VG, Menne TF, Xu J, et al. : Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322(5909):1839–42. 10.1126/science.1165409 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 96. Sankaran VG, Xu J, Ragoczy T, et al. : Developmental and species-divergent globin switching are driven by BCL11A. Nature. 2009;460(7259):1093–7. 10.1038/nature08243 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 97. Bauer DE, Kamran SC, Lessard S, et al. : An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342(6155):253–7. 10.1126/science.1242088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Canver MC, Smith EC, Sher F, et al. : BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527(7577):192–7. 10.1038/nature15521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Corlan AD: Medline trend: automated yearly statistics of PubMed results for any query.2004. Reference Source [Google Scholar]