Abstract
Primordial germ cells are the progenitor cells that give rise to the gametes. In some animals, the germline is induced by zygotic transcription factors, whereas in others, primordial germ cell specification occurs via inheritance of maternally provided gene products known as germ plasm. Once specified, the primordial germ cells of some animals must acquire motility and migrate to the gonad in order to survive. In all animals examined, perinuclear structures called germ granules form within germ cells. This review focuses on some of the recent studies, conducted by several groups using diverse systems, from invertebrates to vertebrates, which have provided mechanistic insight into the molecular regulation of germ cell specification and migration.
Keywords: Primordial Germ Cell, Primordial Germ Cell Specification, Migration, gametes, germ plasm, Germ Plasm Assemblers, Germ Granules, Germline Identity
Introduction
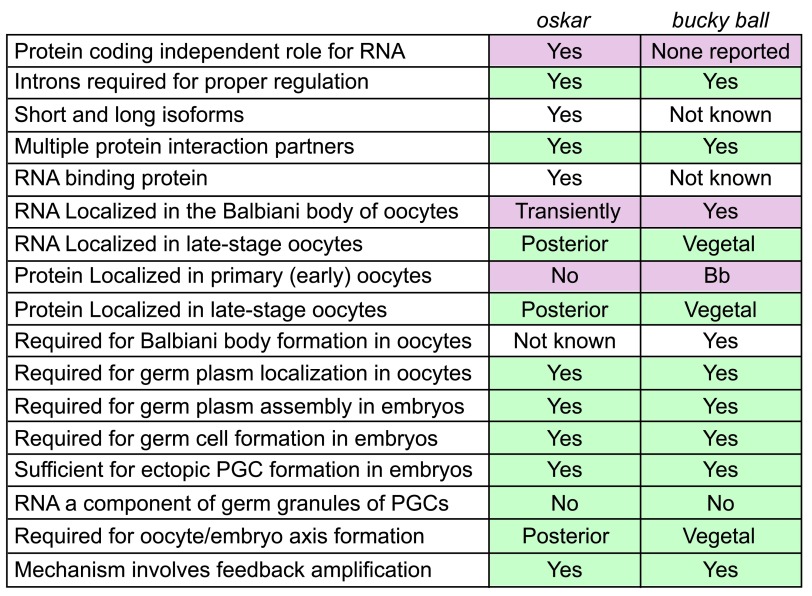
In 1892, August Weismann challenged the notion that the germline (reproductive cells) was derived from the soma (cells of the body). Instead, Weismann proposed that the germ cells possessed a special immortal substance called “ancestral germ plasm” that was inherited from germ cells of one generation to the next in his “theory of the continuity of the germ plasm” 1, 2. Germ plasm is a maternally supplied substance comprised of RNAs, proteins, and organelles that are amassed in oocytes and later is sequestered during the first embryonic cleavages within a few cells that will become the primordial germ cells (PGCs). In the next century, the identification of conserved germline-specific markers and the germ cell accumulation of alkaline phosphatase made it possible to trace the origins of the germline from its earliest emergence through PGC migration to the presumptive gonad where they differentiate as male or female gametes. Such lineage tracing revealed that indeed some animals establish their germline by inheritance of maternal factors and post-transcriptional regulation in the context of a silenced genome but that others do not. In the latter case, these animals lack detectable maternal germ plasm and induce their germ cells by zygotic transcription factors. Genetic and overexpression screens to identify germ cell inducers have uncovered only a few factors with the capacity to generate ectopic PGCs. Oskar 3 and bucky ball 4 are “drivers” of germline fate among animals that use a maternal inheritance mode of PGC specification, but these genes are specific to different subsets of species 5– 7, and it remains to be determined whether their mechanisms of action and specific activities are conserved ( Figure 1). In humans, which use an inductive mode of specification, sox17 is sufficient to specify human primordial germ cell-like cells (hPGCLCs) 8, but in mouse no germ cell inducer has been identified. This suggests that only a few genes possess germ cell-inducing activity or that the coordinated action of multiple genes is required to establish the germline or both. Consistent with this notion, many factors involved in germ cell development are involved in RNA regulation, including Vasa, a universal marker of and regulator of germ cell development 9– 26. Recently, investigators proposed a “last cell standing model”, whereby early PGC determination, as occurs in maternal germ plasm inducers, is not an innovation to protect germline traits. Instead, they proposed that germ plasm provides a means to specify the germline lineage earlier, before gastrulation, and thereby liberate the somatic cells of the embryo to more rapidly evolve 27. According to this model, innovation in animals that do not specify the germline early—prior to or in early gastrulation either by maternal germ plasm or inductive modes—but instead specify PGCs late in gastrulation by zygotic inductive modes are constrained by signaling and morphogenesis requirements associated with germ layer specification and gastrulation 27. Significantly, many of the same or related genes regulate PGC development independently of specification mode. This review focuses on the earliest events of PGC specification in the zygote, the mechanisms that induce early PGC-like cells in culture.
Figure 1. Comparison of two maternal germ plasm assemblers: oskar and bucky ball.
The left column lists activities, localization, or other properties. Those attributed to Drosophila oskar or zebrafish bucky ball or both are indicated in the relevant columns. PGC, primordial germ cell.
Perinuclear accumulations in germ cells
Weismann recognized the germ plasm as a peculiar and complicated structure, but at that time the molecular components were not known. Since then, germ cell-specific accumulations have been detected in germ cells at all stages of the germline cycle. Further adding to the complexity, germ cell substances that have been identified throughout the germline cycle have been referred to by a variety of terms—nuage, germ plasm, and germ granules, or P-granules—at different developmental stages and in different animals. In recent years, molecular identification of the components of these germ cell substances has revealed that the molecular components overlap somewhat and thus nuage, germ plasm, and germ granules have at times been treated as largely equivalent. This assumption has made it challenging to decipher the functions of these germ cell manifestations, particularly when attempting to compare studies of these germ cell substances conducted at different developmental stages and in different organisms. To facilitate comparisons of functional studies between species, definitions of nuage, germ plasm, and germ granules in the context of this review are provided below.
Nuage is the perinuclear granulo-fibrillar electron-dense material that has been identified through histological and ultrastructural examination of oocytes from invertebrates through vertebrates and that is present in various conformations in cells of the male and female gametes 28. Although nuage occupies a distinct subcellular space, it is generally not asymmetric in its distribution. In some animals, a subset of nuage components become asymetrically localized to a specific subcellular location in oocytes where the germ plasm that is transmitted from oocytes to the embryo forms ( Figure 1). This substance is present prior to zygotic genome activation and contains germ cell-inducing activity ( Figure 1). After germ cell specification and zygotic genome activation, germline-specific aggregates of proteins and RNA-binding proteins called germ granules form next to the nucleus ( Figure 1). The functions of each of these germ cell-specific substances and their distinguishing features are active areas of study.
As an initial step toward defining nuage function, the identity of nuage molecules has been sought. The products of conserved germline-specific genes that are necessary for germ cell development, such as Vasa protein, are enriched in perinuclear regions where nuage is found 16, 18, 29, 30. Nuage has been postulated to be involved in the maternal germ plasm pathway; however, nuage is present even in animals that do not use inheritance of maternal germ plasm to specify their germline, indicating that nuage components may have other functions. It is also possible that nuage in animals that use inductive modes also contains germ plasm precursor materials, but that a nucleator or assembly/scaffold factor that can assemble nuage components into active germ plasm is lacking. Notably, evidence suggests that specification by zygotic induction is the ancestral mode of germline determination in insects and vertebrates 6, 7, 29, 31. Maternal specification by germ plasm in some insects has been associated with the presence of the germ plasm inducer oskar, discussed in the following section. Oskar is thought to have been co-opted from an ancestral neural role and to have facilitated the transition from zygotic germ cell induction to maternal specification via germ plasm 6, 7. Based on the conserved presence and perinuclear localization of nuage and the nature of the molecules that localize there, nuage functions in processes other than maternal germ plasm assembly have been proposed.
Among molecules that are enriched in nuage are proteins involved in genesis of piwi-interacting RNAs (piRNAs). piRNAs are components of a gonad-specific RNA silencing pathway that is thought to protect genome integrity by counteracting transposable or selfish genetic elements that promote their own transmission at the expense of other elements (reviewed in 32, 33). piRNAs are small RNAs that are produced from the cleavage of precursor RNAs by endonuclease activity of germline-specific members of the Argonaut family, called Piwi proteins, and secondary amplification 34, 35. Argonaut family members, including Piwi and Aubergine in flies, and human Argonaut homologs have been implicated in transcriptional and post-transcriptional regulation of gene expression 36. Like Vasa protein, Piwi protein homologs reside adjacent to the nucleus within the nuage in some species (reviewed in 37). Based on the conserved localization of piRNA pathway components as well as genetic and other functional data, a conserved nuage role as the site of piRNA amplification has been proposed 38– 56. While this may indeed be the case, not all piRNA components localize to the nuage (reviewed in 37). Moreover, the phenotypes of some piRNA pathway components suggest additional functions in diverse processes, including nuclear functions in chromosome rearrangements, chromosome dynamics, roles in RNA metabolism and storage, stem cell maintenance, regulation of cell divisions at stages before germ plasm would assemble in oocytes, or later roles in PGC maintenance; these roles have generated models whereby nuage and later germ granules serve to extend the nuclear environment and have been reviewed elsewhere 57. Owing to its dynamic nature, its varied composition at different stages, phenotypic differences between genders and species, and multiple stage-specific activities indicated by nuage component mutants, including piRNA pathway molecules, the developmental functions of nuage are not fully understood.
Germ plasm assemblers
In primary oocytes of some animals, maternal germ plasm first assembles within an ancient perinuclear oocyte structure known as the Balbiani body 28, 58, 59 and later is found at the oocyte cortex, the posterior pole in some insects or the vegetal pole of some vertebrates 28, 58, 59 ( Figure 1). Expression-based screens have identified germ plasm components and candidate regulators on the basis of their localization to sites of germ plasm assembly in oocytes, such as the Balbiani body of early oocytes or the cortex of late-stage oocytes (reviewed in 15, 59, 60) . However, localization to the germ plasm is only suggestive of potential function as not all molecules that localize to germ plasm are essential for its assembly or activity 16– 64. Functional assays to define the component(s) of germ plasm that can impart germ cell identity, the germ plasm nucleators or assemblers, have included isolation and transplantation of cytoplasm from oocytes to embryos to identify the substance with PGC-inducing activity in model systems such as Drosophila 65, zebrafish 66, and Xenopus 67. To date, only a limited number of factors that can induce germ cells have been identified, and how the germ plasm assembles remains a key question in the field.
The molecular constituents of the germ plasm are best understood in Drosophila because of the powerful genetic screens which led to the identification of key factors of germline assembly and germ cell development 68– 70, including the germ plasm assembler, oskar 71. Alternative translation generates two forms of Oskar with distinct activities 72. The short form of Oskar is required for germ plasm assembly and function, whereas long Oskar lacks the assembly activity and instead is required to anchor germ plasm 72– 74. Until recently, Oskar (Osk) was viewed as a scaffolding protein that gathered germ plasm via interactions with a myriad of partners 25, 74– 94. The regions of Oskar mediating its association with partners such as Vasa defined Oskar functional domains 73. The recently elucidated Oskar crystal structure revealed that both Oskar dimerization and interaction with Vasa are mediated via the Osk N-terminal LOTUS domain, named after Limkain, Oskar and Tudor domain-containing proteins 5 and 7 83. LOTUS is a globular domain present in several germ plasm/granule components 95, including the conserved Tudor family, first discovered for Tudor’s function in germ plasm assembly in Drosophila 68. This finding is in contrast to previous work that mapped the Vasa interaction to Oskar’s C-terminus 73. In the new study, Vasa-Oskar interactions were tested without RNAs to exclude RNA-mediated association and this may explain the different binding sites. These findings further support interaction between Vasa and Oskar and raise new questions and models to explain how these different Oskar complexes promote germ plasm formation and activity.
Another exciting aspect of the recent study is the evidence that short, but not long, Oskar associates directly with RNA. Surprisingly, this binding interaction is not via a canonical RNA-binding motif, but instead through a domain that resembles an enzymatically inactive SGNH hydrolase domain 83. SGNH hydrolases are a large enzyme family with thousands of members that are found in all life forms. An interesting property among some bacterial SGNH domain-containing proteins that may be of relevance to germ plasm assembly is their propensity to oligomerize to form amorphous aggregates or amyloid-like fibrils 96, 97. In addition, proteins encoded by some LINE (long interspersed nuclear elements) mobile genetic elements contain SGNH-like domains and mediate RNP assembly 98, 99. For example, the zebrafish LINE protein ZfL2-1 ORF1p forms multimers, binds nucleotides, and like Oskar possesses an SGNH-like domain that lacks overt RNA-binding domain structure 98. In addition, ORF1ps, including ZfL2-1, have been shown to function as chaperones. In this context, ORF1ps interact with the LINE RNA and are postulated to mediate rearrangement of the RNA into a stable conformation that protects the RNA from degradation, but later can be reversed to facilitate reverse transcription 98. Based on in vitro structure function studies, ZfL2-1 is postulated to mediate RNP assembly via interactions with positively charged peptides that bind RNA structural elements 99. It remains to be determined whether Oskar has similar chaperone functions, but it is easy to imagine how such an activity could apply to germ plasm RNAs, which must be translationally silent and protected from degradation during transport but later are translated in a specific 96 subcellular location. The unique functional domains and distinct interaction properties of Oskar isoforms discussed above provide new models to test the mechanism by which Oskar could promote RNP and germ plasm assembly either directly via its RNA-binding domain or directly or indirectly via its disordered LOTUS and interaction with Vasa.
Vertebrates, even those that use maternal inheritance to specify their germline, lack oskar and instead have a vertebrate-specific gene called bucky ball ( buc) or vegetally localized 1 ( velo1) 4, 100. Buc shares features with Oskar in that both are localized in oocytes as RNAs and proteins 4, 81, 101– 103, both are required for and can organize germ plasm 3, 4, both have been viewed as unstructured proteins, both display complex post-transcriptional regulation at the level of splicing 81, 104 and RNA localization 72, 81, 105, and both have properties of self-assembling aggregates and interact with RNA-binding proteins and other factors at the RNA and protein levels 73, 75– 90, 92, 94. Whether or not Buc protein, like Osk, can directly bind RNAs is unknown. Based on homology searches, Buc lacks identifiable functional domains and thus the mechanisms by which it promotes Balbiani body formation, germ plasm assembly, and oocyte axis specification are not clear and have relied on identifying Buc interaction partners and mapping their interaction sites on Buc 78, 81, 106. However, Bucky ball and Oskar germ plasm factors also have some differences ( Figure 1). Specifically, Buc protein is present in early-stage oocytes 81, 101, whereas Osk protein is detected in late-stage oocytes 72, and the few reported Buc interaction partners are vertebrate-specific 78. Finally, osk RNA has functions independent of its protein-coding role 84, 107, whereas no evidence of protein-independent RNA functions of buc have been reported. These differences support convergent evolution or co-option of these genes as germ plasm assemblers; however, further analysis, including cross-species comparisons and rescue experiments, are required to determine the extent to which the activities of these germ plasm assemblers overlap.
Much less is understood about the molecular regulators of maternal germ plasm specification in vertebrates; however, recently, endogenous Buc protein was shown to localize to the cleavage furrows of early embryos 101, 106 by a maternal Kinesin 1 (Mkif5Ba)-dependent mechanism 106. Moreover, the ability of Buc to induce ectopic PGCs requires Mkif5Ba and Buc protein localization to the cleavage furrows 106. Similarly, formation of germ plasm aggregates in Xenopus requires the Kinesin-like protein Xklp2 108, indicating a conserved role for microtubule motor-dependent assembly of germ plasm in these vertebrates. Because germ plasm is present in zebrafish M kif5Ba mutant embryos but germ cells are absent, this mutant provides evidence that inheritance of maternal germ plasm from oocytes alone is not sufficient to specify PGCs. Moreover, germline establishment requires proper spatiotemporal localization of PGC determinants.
Germ plasm, germ granules, and germline identity
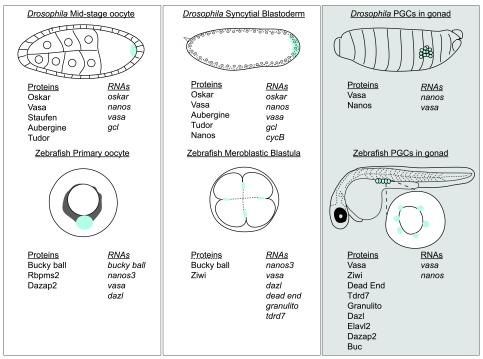
The capacity of germ plasm to specify PGC fate has clear support from studies in Drosophila 65, zebrafish 66, and Xenopus 67, in which transplantation of the germ plasm can induce PGCs. In contrast, when germ granules are not properly segregated in Caenorhabditis elegans mutants, excess PGCs do not form nor do all cells adopt germline fates; instead, the cells take somatic fates 109. This example suggests that germ granules are not sufficient for PGC fate. These differences in germ cell induction capacity between granules and plasm raise the question of whether germ plasm and granules are based on their shared enrichment of RNA-binding proteins (RNAbps), RNAs, and other overlapping component 4, 73, 78, 101, 106, 110 manifestations of the same mechanism/structure operating at different stages or instead are distinct entities with unique properties. Alternatively, not all granules are equivalent. The maternal germ plasm associated with the maternal inheritance specification mode is continuous from oocyte to embryo, whereas perinuclear germ granules assemble only in PGCs, even in zygotically induced/discontinuous modes of germline specification. Thus, maybe these germline entities are not functionally equivalent. If this is the case, germ plasm from oocytes should contain germ cell-inducing factors, which germ granules of PGCs lack. Consistent with this notion, the perinuclear nuage in Drosophila oocytes is distinct from the polar granules nucleated by Oskar 111 during specification of pole cells/PGCs 65. Moreover, recent studies in Drosophila, C. elegans, and zebrafish provide evidence that germ granules are heterogeneous. In Drosophila, single RNA fluorescent in situ hybridization (FISH) analyses of granules revealed their organized architecture of “core” germ plasm proteins (e.g., Vasa and Oskar) distributed throughout granules and specific RNAs spatially arranged within granules 112. Significantly, although Oskar protein is present in granules, oskar RNA is not 112, 113 ( Figure 2). When chimeric osk with a nanos 3’ untranslated region (UTR) is directed to granules, germ cell numbers and Vasa expression are reduced 113, indicating that osk segregation from granules is needed for normal PGC development. Taken together, these findings are consistent with plasm and granules being distinct entities in Drosophila. Similarly, endogenous Buc protein localizes to germ plasm of oocytes and early embryos but is not detected in germ granules of PGCs 106. Moreover, buc RNA is a component of germ plasm of oocytes but not embryos, and even when overexpressed, Buc protein, but not buc RNA, localizes to germ granules of zebrafish PGCs 4, 101, 106 ( Figure 2). In addition, in zebrafish, vasa RNA but not Vasa protein is a component of the Balbiani body, where germ plasm localizes in oocytes 18. In contrast, Vasa protein is a PGC granule component in zebrafish 11, 18, 114 and other animals examined (reviewed in 15, 115, 116). In zebrafish, as in Drosophila, analysis of MS2-tagged and endogenous vasa and nanos RNAs indicates that granules are not equivalent 117, 118; therefore, it is likely that some but not all have the capacity to induce the germ cell fate or that these granules have another function.
Figure 2. Comparison of maternal germ plasm and zygotic germ granule components between two organisms, Drosophila and zebrafish, that use maternal inheritance to specify their primordial germ cells (PGCs).
The first column depicts the localization of germ plasm components (listed beneath the cartoon) at the posterior pole of mid-stage oocytes of Drosophila (top) and in the Balbiani body of early-stage oocytes of zebrafish. After fertilization, maternal germ plasm components (listed beneath the schematic) localize to specific membranes within the syncitial blastoderm of Drosophila and meroblastic cleavage (cells are incompletely separated and connected to the yolk) stage zebrafish embryos. The cells that receive these membranes develop as the PGCs. White rectangles depict maternal germ plasm components. The last column (grey rectangle) depicts PGCs of Drosophila and zebrafish after their migration to the gonad and indicates components of embryonic PGCs and germ granules. Dazap2, deleted in azoospermia-associated protein 2; Dazl, deleted in azoospermia-like; Elav, HuC; gcl, germ cell-less; Rbpms2, ribonuclear-binding protein with multiple splice isoforms 2.
In addition to their varied composition discussed above, other evidence indicates that germ plasm and germ granules may not be functionally equivalent. For example, the germ plasm and germ granules occupy distinct subcellar locations. In zebrafish and Xenopus, germ plasm associates with the endoplasmic reticulum in oocytes 28. In zebrafish embryos, germ plasm first accumulates at furrows of cleavage-stage embryos by a mechanism that involves RNA recruitment and clearance 106, 119, whereas germ granules are perinuclear and form after genome activation 120. In addition, the period when cells are competent to develop as PGCs, regardless of specification mode, precedes granule assembly and is developmentally restricted and brief, indicating that germ cell-inducing activity is tightly regulated. In Drosophila, rescue of osk mutants and the number of excess PGCs in overexpression contexts depend on Osk levels 3, 71. In zebrafish, expression of exogenous Buc after fertilization produces only a few additional PGCs, indicating that levels or timing may be limiting 4, 106. Similarly, in mice, high levels of the zinc finger transcriptional protein Blimp/Prdm1 are required to drive the epigenetic and cellular features of germ cells in vivo 121, and BLIMP1 specifies hPGCLCs in a similar dosage-dependent manner 122. These observations suggest that a limiting threshold or additional non-mutually exclusive factors (molecular, spatial, or temporal), or both, are essential for PGC identity. Accordingly, splitting determinants among sister cells, as occurs in C. elegans maternal-effect sterile ( mes-1) mutants 109, 123, would produce two cells lacking sufficient factors for germline fate. Notably, granules are also heterogeneous and dynamic structures in C. elegans 124, 125. Finally, other evidence from C. elegans shows that specific germ granule factors, including PGL-1 and PGL-3, induce aggregate/granule formation in non-germline cells without converting those cells to PGCs, further indicating that granules alone are not sufficient for germ cell identity/specification 126, 127.
Germ granules are enriched for RNA-binding proteins and proteins with other roles in post-transcriptional regulation; therefore, if germ granules do impart germline identity, it is possible that post-transcriptional regulation of other factors that may or may not be granule components is involved. If so, granules could impart germline fate only to cells that already express that factor; for example, Osk recruits components that regulate localization and translation of two germ granule components nanos and pgc, which promote patterning and pole cell development in flies 110, 112, 128– 130, and Buc must be localized to induce PGCs in zebrafish 106. Consistent with an essential role for granules in germ cells, depletion of one or more germ granule components in C. elegans causes sterility 131– 133. Recently, maternal dazap2 was shown to maintain germ granules of zebrafish PGCs by acting epistatic to Tudor-7 and antagonistic to Dynein activity 78. Because PGCs are specified, and granules form but later are not maintained in PGCs lacking maternal Dazap2, M dazap2 mutant germ cells provide an opportunity to explore potential roles of granules in maintenance of vertebrate PGCs. The historical view of nuage, germ plasm, and germ granules was that each of these entities would promote the germ cell fate beginning with their specification to maintenance of germline identity. Efforts to gain a deeper understanding of the components of germ granules and functional assessment of these conserved elements of PGCs have provided strong evidence for mechanisms to preserve or protect germline identity; therefore, it is worth considering the possibility that the germ granules of specified embryonic PGCs contribute to a mechanism that preserves germline identity rather than specification of PGC fate. Understanding the contribution of granules to germ cell development and fertility remains an active area of investigation.
Germline specification in mammals
In mammals, the germ cells are not specified by inheritance of maternal cytoplasm (germ plasm), but instead are specified later by inductive signals. In the mouse embryo just before gastrulation, signals from extra-embryonic ectoderm and visceral endoderm are necessary to specify germ cells within the posterior epiblast that is adjacent to the forming primitive streak ( Figure 3). Unlike in flies and fish, in mice, no single factor that is necessary and sufficient to specify germ cells has been discovered. The earliest markers of mouse PGCs, including Blimp1 (Prdm1) and Prdm14, two critical factors that suppress somatic gene expression (thus promoting germ cell-specific gene programs) 133– 137 and developmental pluripotency-associated 3 (DPP3/Stella), are required to maintain rather than specify the PGCs 138. Strikingly, nearly all cells of the mouse pregastrula epiblast can express Prdm1 or Prdm14 when induced by ubiquitous expression of the bone morphogenetic protein ligands Bmp4 139 and Bmp8b 140. Importantly, induced cells in culture can reconstitute functional sperm to germ cell-depleted neonates 141. However, in vivo, only a few cells within the posterior epiblast that are positioned proximal to the extra-embryonic ectoderm become PGCs 142. A Wnt3 signal from the epiblast primes these cells to respond to BMP produced by the extra-embryonic ectoderm 141. The mechanisms that limit germ cell induction in anterior regions are not fully understood. However, PGC formation involves anterior endoderm factors and additional BMP family members that repress Blimp1 and thus promote acquisition of pluripotency and PGC development specifically in the posterior epiblast 141. Other factors, such as microRNAs and their antagonists, contribute to PGC development (for example, by regulating Prdm1 expression) 143. However, the functions of these molecules are not confined to PGCs and so they do not represent germline determinants. Significant unresolved questions are how and what factors limit selection of just a few cells within this region to become PGCs. Moreover, the identity of the factor or factors that specify them remains unknown.
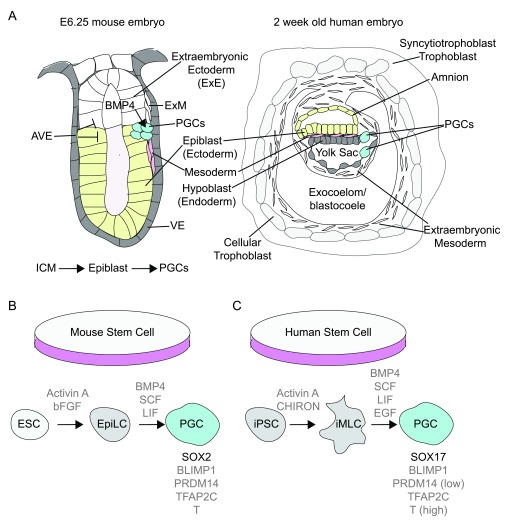
Figure 3. Primordial germ cell (PGC) induction in mammals.
( A) The early stages of PGC emergence in mouse and humans. In mouse, cues from extra-embryonic ectoderm, including bone morphogentic proteins (BMPs), induce transcriptional regulators that promote PGC identity in the cells of the adjacent posterior epiblast. In humans, PGCs are first detected around the onset of gastrulation within the endodermal yolk sac wall. The signals that induce human PGCs in vivo are not known. ( B, C) The molecular players involved in PGC specification from stem cells in culture. ( B) For mouse cells, the program to induce primordial germ cell-like cells (PGCLCs) from embryonic stem cells (ESCs) in culture requires transforming growth factor-beta (TGF-β) family members (Activin) and basic fibroblast growth factors (bFGFs) to promote an epiblast-like cell (EpiLC) state. Exposure of the EpiLCs to BMPs (BMP4) and stem cell factor (SCF) and leukemia inhibitory factor (LIF) converts the EpiLCs to cells with mouse PGC-specific gene expression profiles. ( C) For human cells, the program to induce PGCLCs from human-induced pluripotency cells (iPSCs) in culture like mouse requires TGF-β family members (Activin) and CHIRON to inhibit glycogen synthase kinase 3, an inhibitor of Wnt activity. This combination of factors promotes an induced mesoderm-like cell (iMLC) fate, which upon exposure to BMP4, LIF, SCF, plus epidermal growth factor (EGF) generates cells with human PGC-specific gene expression profiles. Notably, the transcription factor Sox2 is required in mouse cells, whereas Sox17 is necessary for human cells. In addition, PRDM14 is highly expressed in mouse PGCs and has been reported to be low or not expressed in human PGCs. AVE, anterior visceral endoderm; ExM, extra-embryonic mesoderm; ICM, inner cell mass; VE, visceral endoderm.
PGCs in humans, as in mice, are specified in extragonadal regions around the time of gastrulation onset. However, there are important differences in the timing and development of extra-embryonic tissues and likely PGC specification between human and mouse. During the second week of development, the human embryo is composed of epiblast and primitive endoderm, which gives rise to the yolk sac ( Figure 3). Owing to technical and ethical boundaries, lineage tracing to capture the first emergence of human PGCs prior to gastrulation has not been feasible. Nonetheless, human PGCs were identified in extragonadal regions more than 100 years ago and have been detected in human embryos in the Carnegie collection as early as stage 6 (around 2-week-old embryos) in the yolk sac endoderm in the vicinity of the developing allantois, an amniote structure involved in nutrition and waste removal ( Figure 3) 144. Functional studies of the earliest stages of PGC specification in humans would require manipulation and analysis of embryos within the first month following fertilization and this is not feasible. As an alternative to in vivo mammalian contexts, more tractable in vitro systems to study specification of mouse and human PGCs have been sought. In mice, key advances in PGC reprogramming paradigms have been facilitated by the development of germline reporters to identify and select for the lineage, including a mouse Vasa homolog (MVH) transgenic embryonic stem (ES) cell line, wherein the MVH promoter drives GFP reporter expression 145 and a germ cell-specific gcOct4-GFP 146. These tools enabled identification of germ cell-like cells in culture within just a few days. With markers in hand and using the germline-promoting factors defined from developmental studies, it was not long before mouse sperm 147 and egg 146 cells were derived from long-term ES cells. As germline stem cells (GSCs), PGCs are expected to generate cells that ultimately develop as functional gametes, sperm, or eggs that can produce a normal healthy fertile animal. The first animals generated from PGC-like cells (PGCLCs) were abnormal 148, a limitation to using such cells to study normal germline development or infertility. However, in 2011, healthy offspring were produced from male-derived PGCLCs building on the observation that nearly all of the pregastrula cells in the mouse could express key PGC factors Blimp1 (Prdm1) and Prdm14 in response to BMP4 149. In that study, the authors recapitulated gametogenesis in vitro by first reprogramming cells to a pregastrula epiblast state before exposure to germ cell-differentiation cues ( Figure 3A,B). Later, the same group generated female PGCLCs with meiotic potential in reconstituted ovaries that produced fertile progeny after in vitro fertilization 150, indicating that the in vitro produced cells could develop as functional female gametes in the correct environment.
Encouraged by the success in establishing in vitro models of mouse PGC generation, several groups pursued in vitro models of human PGC development. With the mouse methods based on developmental paradigms in hand, the opportunity to discover the molecular programs responsible for producing the human germline seemed imminent. However, initial attempts to derive human PGCs quickly revealed that the mouse programming strategies were not effective, suggesting that despite conserved germ cell factors there must be differences in their mechanisms to generate germline cells ( Figure 3A). Thus, several groups sought and recently reported conditions to efficiently generate hPGCLCs 8, 122, 151 ( Figure 3C). Similar transcriptional programs associated with successful generation of hPGCLCs were discovered in two independent studies; one study used transcription activator-like effector nucleases to engineer human cell lines that express fluorescent reporters for BLIMP1 and TFAP2C to select for germ cells 122, and the second 8 imparted pluripotency by using a combination of four inhibitors and selected for germline cells by using a reporter for the conserved PGC-specific protein Nanos 62, 63, 152– 154. Interestingly, these studies indicate that PGC specification is somewhat more direct in human cells compared with mouse cells. In human cells, Sox17 induces and is necessary to specify hPGCLCs, with Blimp1 acting downstream to promote PGC gene expression and repress expression of mesendodermal, neuronal, and epigenetic reprogramming genes as the cells differentiate into hPGCLCs 8, 122. In mice, ES cells first transition to an epiblast state, and Blimp1, but not Sox17 155, 156, represses somatic programs in nascent mPGCLCs 149 ( Figure 3). In both, epigenetic reprogramming is associated with PGCLC differentiation. However, the observed overlap in transcriptional programs between the human and mouse PGCLCs was surprisingly limited 122, indicating distinct PGC specification programs between mouse and human despite their reliance on shared signaling molecules. Notably, the hPGCLCs generated so far resemble early hPGCs and thus provide an unprecedented opportunity to study the earliest events in hPGC specification. However, late-stage hPGCLCs have not been obtained on the basis of the absence of markers that are normally expressed after PGCs migrate to the gonad, indicating that further differentiation or refinement of the protocols is required to obtain and study development of these later stages. Recapitulating development from stem cell to mature germ cell remains a challenge and an important step to realize the full potential of hPGCLCs to provide insight into human diseases, including infertility with a genetic basis or environmental/toxicological basis, as well as developmental disorders.
Once specified, PGCs in flies, fish, and mammals, but not C. elegans, must travel from their extragonadal site of specification to the presumptive gonad where the PGCs will differentiate into sperm in males or oocytes in females (reviewed in 157, 158). Successful migration to the gonad anlagen is essential for further PGC development and survival. With PGC markers and molecular and genetic approaches, key steps in PGC migration and the underlying cell behaviors have been defined and have been comprehensively reviewed elsewhere (for detailed reviews, see 157, 158). The RNA-binding protein Nanos regulates PGC migration and survival in Drosophila 63, 159, C. elegans 160, and zebrafish 62, 161, and nanos orthologs have a conserved role from invertebrates to humans in GSC maintenance 62– 64, 152, 159, 160, 162– 166. Despite conserved requirements for nanos orthologs, the relevant RNA targets of nanos genes and downstream mechanisms promoting germ cell survival are not fully understood. In non-mammalian animals, GSCs persist into adulthood and continue to produce new gametes throughout the reproductive life of the animal. In mammals, GSCs maintain spermatogenesis throughout the lifetime of adult males, but whether or not female mammals have the capacity to generate new oocytes after birth has been highly controversial and an area of active study (reviewed in 167). Coming back to August Weismann’s words, “The importance of such a theory lies primarily in its suggestiveness, by which alone it becomes a step towards the ideal at which we aim, namely, the formulation of the true and complete theory” 1, 2. Clearly, the existence of GSCs, either naturally occurring in adult mammalian females or in vitro produced, has important therapeutic implications for the field of reproductive medicine; however, further development of tools and studies of the mechanisms to specify and maintain stem cell niches and ovarian reserve in diverse systems are essential for an improved understanding of germline development and reproductive health.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Bruce Draper, epartment of Molecular and Cellular Biology, University of California Davis, Davis, CA, USA
Prasanth Rangan, Department of Biological Sciences, University at Albany, Albany, NY, USA
Ruth Lehmann, Department of Cell Biology, New York University School of Medicine, New York, NY, USA
Funding Statement
The author(s) declared that no grants were involved in supporting this work.
[version 1; referees: 3 approved]
References
- 1. Weismann A: Aufsätze über Vererbung und verwandte biologische Fragen. (Jena,: G. Fisher),1892. Reference Source [Google Scholar]
- 2. Weismann A, Parker WN, Rönnfeldt H: The germ-plasm: a theory of heredity. (New York: C. Scribner's sons),1893. 10.5962/bhl.title.29345 [DOI] [Google Scholar]
- 3. Ephrussi A, Lehmann R: Induction of germ cell formation by oskar. Nature. 1992;358(6385):387–92. 10.1038/358387a0 [DOI] [PubMed] [Google Scholar]
- 4. Bontems F, Stein A, Marlow F, et al. : Bucky ball organizes germ plasm assembly in zebrafish. Curr Biol. 2009;19(5):414–22. 10.1016/j.cub.2009.01.038 [DOI] [PubMed] [Google Scholar]
- 5. Ahuja A, Extavour CG: Patterns of molecular evolution of the germ line specification gene oskar suggest that a novel domain may contribute to functional divergence in Drosophila. Dev Genes Evol. 2014;224(2):65–77. 10.1007/s00427-013-0463-7 [DOI] [PubMed] [Google Scholar]
- 6. Ewen-Campen B, Srouji JR, Schwager EE, et al. : Oskar predates the evolution of germ plasm in insects. Curr Biol. 2012;22(23):2278–83. 10.1016/j.cub.2012.10.019 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 7. Lynch JA, Ozüak O, Khila A, et al. : The phylogenetic origin of oskar coincided with the origin of maternally provisioned germ plasm and pole cells at the base of the Holometabola. PLoS Genet. 2011;7(4):e1002029. 10.1371/journal.pgen.1002029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Irie N, Weinberger L, Tang WW, et al. : SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160(1–2):253–68. 10.1016/j.cell.2014.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 9. Abe K, Noce T: A DEAD-family protein gene, Ddx4, encoding a murine homolog of Drosophila vasa maps to the distal end of mouse chromosome 13. Mamm Genome. 1997;8(8):622–3. 10.1007/s003359900521 [DOI] [PubMed] [Google Scholar]
- 10. Anderson RA, Fulton N, Cowan G, et al. : Conserved and divergent patterns of expression of DAZL, VASA and OCT4 in the germ cells of the human fetal ovary and testis. BMC Dev Biol. 2007;7:136. 10.1186/1471-213X-7-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Braat AK, Zandbergen T, van de Water S, et al. : Characterization of zebrafish primordial germ cells: morphology and early distribution of vasa RNA. Dev Dyn. 1999;216(2):153–67. [DOI] [PubMed] [Google Scholar]
- 12. Cao M, Yang Y, Xu H, et al. : Germ cell specific expression of Vasa in rare minnow, Gobiocypris rarus. Comp Biochem Physiol A Mol Integr Physiol. 2012;162(3):163–70. 10.1016/j.cbpa.2012.02.007 [DOI] [PubMed] [Google Scholar]
- 13. Cui S, Nanayakkara K, Selwood L: A marsupial, Trichosurus vulpecula, DDX4/VASAGene (Tv DDX4) of the DEAD box protein family: molecular conservation and germline expression. Cytogenet Genome Res. 2009;126(4):348–58. 10.1159/000266170 [DOI] [PubMed] [Google Scholar]
- 14. Gustafson EA, Yajima M, Juliano CE, et al. : Post-translational regulation by gustavus contributes to selective Vasa protein accumulation in multipotent cells during embryogenesis. Dev Biol. 2011;349(2):440–50. 10.1016/j.ydbio.2010.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hartung O, Forbes MM, Marlow FL: Zebrafish vasa is required for germ-cell differentiation and maintenance. Mol Reprod Dev. 2014;81(10):946–61. 10.1002/mrd.22414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hickford DE, Frankenberg S, Pask AJ, et al. : DDX4 (VASA) is conserved in germ cell development in marsupials and monotremes. Biol Reprod. 2011;85(4):733–43. 10.1095/biolreprod.111.091629 [DOI] [PubMed] [Google Scholar]
- 17. Ikenishi K, Tanaka TS: Involvement of the protein of Xenopus vasa homolog ( Xenopus vasa-like gene 1, XVLG1) in the differentiation of primordial germ cells. Dev Growth Differ. 1997;39(5):625–33. 10.1046/j.1440-169X.1997.t01-4-00010.x [DOI] [PubMed] [Google Scholar]
- 18. Knaut H, Pelegri F, Bohmann K, et al. : Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J Cell Biol. 2000;149(4):875–88. 10.1083/jcb.149.4.875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Komiya T, Itoh K, Ikenishi K, et al. : Isolation and characterization of a novel gene of the DEAD box protein family which is specifically expressed in germ cells of Xenopus laevis. Dev Biol. 1994;162(2):354–63. 10.1006/dbio.1994.1093 [DOI] [PubMed] [Google Scholar]
- 20. Komiya T, Tanigawa Y: Cloning of a gene of the DEAD box protein family which is specifically expressed in germ cells in rats. Biochem Biophys Res Commun. 1995;207(1):405–10. 10.1006/bbrc.1995.1202 [DOI] [PubMed] [Google Scholar]
- 21. Kuznicki KA, Smith PA, Leung-Chiu WM, et al. : Combinatorial RNA interference indicates GLH-4 can compensate for GLH-1; these two P granule components are critical for fertility in C. elegans. Development. 2000;127(13):2907–16. [DOI] [PubMed] [Google Scholar]
- 22. Lasko PF, Ashburner M: The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature. 1988;335(6191):611–7. 10.1038/335611a0 [DOI] [PubMed] [Google Scholar]
- 23. Roussell DL, Bennett KL: glh-1, a germ-line putative RNA helicase from Caenorhabditis, has four zinc fingers. Proc Natl Acad Sci U S A. 1993;90(20):9300–4. 10.1073/pnas.90.20.9300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith P, Leung-Chiu WM, Montgomery R, et al. : The GLH proteins, Caenorhabditis elegans P granule components, associate with CSN-5 and KGB-1, proteins necessary for fertility, and with ZYX-1, a predicted cytoskeletal protein. Dev Biol. 2002;251(2):333–47. 10.1006/dbio.2002.0832 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 25. Tsunekawa N, Naito M, Sakai Y, et al. : Isolation of chicken vasa homolog gene and tracing the origin of primordial germ cells. Development. 2000;127(12):2741–50. [DOI] [PubMed] [Google Scholar]
- 26. Yoon C, Kawakami K, Hopkins N: Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development. 1997;124(16):3157–65. [DOI] [PubMed] [Google Scholar]
- 27. Johnson AD, Alberio R: Primordial germ cells: the first cell lineage or the last cells standing? Development. 2015;142(16):2730–9. 10.1242/dev.113993 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 28. Kloc M, Jedrzejowska I, Tworzydlo W, et al. : Balbiani body, nuage and sponge bodies--term plasm pathway players. Arthropod Struct Dev. 2014;43(4):341–8. 10.1016/j.asd.2013.12.003 [DOI] [PubMed] [Google Scholar]
- 29. Ewen-Campen B, Donoughe S, Clarke DN, et al. : Germ cell specification requires zygotic mechanisms rather than germ plasm in a basally branching insect. Curr Biol. 2013;23(10):835–42. 10.1016/j.cub.2013.03.063 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 30. Liang L, Diehl-Jones W, Lasko P: Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development. 1994;120(5):1201–11. [DOI] [PubMed] [Google Scholar]
- 31. Evans T, Wade CM, Chapman FA, et al. : Acquisition of germ plasm accelerates vertebrate evolution. Science. 2014;344(6180):200–3. 10.1126/science.1249325 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 32. Banisch TU, Goudarzi M, Raz E: Small RNAs in germ cell development. Curr Top Dev Biol. 2012;99:79–113. 10.1016/B978-0-12-387038-4.00004-5 [DOI] [PubMed] [Google Scholar]
- 33. Hartung O, Marlow F: Get it together: How RNA-Binding Proteins Assemble and Regulate Germ Plasm in the Oocyte and Embryo. (New York: Nova Science Publishers, Inc).2014. Reference Source [Google Scholar]
- 34. Pek JW, Patil VS, Kai T: piRNA pathway and the potential processing site, the nuage, in the Drosophila germline. Dev Growth Differ. 2012;54(1):66–77. 10.1111/j.1440-169X.2011.01316.x [DOI] [PubMed] [Google Scholar]
- 35. Saxe JP, Lin H: Small noncoding RNAs in the germline. Cold Spring Harb Perspect Biol. 2011;3(9):a002717. 10.1101/cshperspect.a002717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Höck J, Meister G: The Argonaute protein family. Genome Biol. 2008;9(2):210. 10.1186/gb-2008-9-2-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Voronina E, Seydoux G, Sassone-Corsi P, et al. : RNA granules in germ cells. Cold Spring Harb Perspect Biol. 2011;3(12): pii: a002774. 10.1101/cshperspect.a002774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Aravin A, Gaidatzis D, Pfeffer S, et al. : A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442(7099):203–7. 10.1038/nature04916 [DOI] [PubMed] [Google Scholar]
- 39. Cox DN, Chao A, Baker J, et al. : A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12(23):3715–27. 10.1101/gad.12.23.3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cox DN, Chao A, Lin H: piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127(3):503–14. [DOI] [PubMed] [Google Scholar]
- 41. Grivna ST, Pyhtila B, Lin H: MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci U S A. 2006;103(36):13415–20. 10.1073/pnas.0605506103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Houwing S, Berezikov E, Ketting RF: Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 2008;27(20):2702–11. 10.1038/emboj.2008.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Houwing S, Kamminga LM, Berezikov E, et al. : A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129(1):69–82. 10.1016/j.cell.2007.03.026 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 44. Huang HY, Houwing S, Kaaij LJ, et al. : Tdrd1 acts as a molecular scaffold for Piwi proteins and piRNA targets in zebrafish. EMBO J. 2011;30(16):3298–308. 10.1038/emboj.2011.228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kuramochi-Miyagawa S, Kimura T, Yomogida K, et al. : Two mouse piwi-related genes: miwi and mili. Mech Dev. 2001;108(1–2):121–33. 10.1016/S0925-4773(01)00499-3 [DOI] [PubMed] [Google Scholar]
- 46. Kuramochi-Miyagawa S, Watanabe T, Gotoh K, et al. : DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22(7):908–17. 10.1101/gad.1640708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lim AK, Lorthongpanich C, Chew TG, et al. : The nuage mediates retrotransposon silencing in mouse primordial ovarian follicles. Development. 2013;140(18):3819–25. 10.1242/dev.099184 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 48. Lim RS, Anand A, Nishimiya-Fujisawa C, et al. : Analysis of Hydra PIWI proteins and piRNAs uncover early evolutionary origins of the piRNA pathway. Dev Biol. 2014;386(1):237–51. 10.1016/j.ydbio.2013.12.007 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 49. Megosh HB, Cox DN, Campbell C, et al. : The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol. 2006;16(19):1884–94. 10.1016/j.cub.2006.08.051 [DOI] [PubMed] [Google Scholar]
- 50. Patil VS, Anand A, Chakrabarti A, et al. : The Tudor domain protein Tapas, a homolog of the vertebrate Tdrd7, functions in the piRNA pathway to regulate retrotransposons in germline of Drosophila melanogaster. BMC Biol. 2014;12:61. 10.1186/s12915-014-0061-9 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 51. Patil VS, Kai T: Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr Biol. 2010;20(8):724–30. 10.1016/j.cub.2010.02.046 [DOI] [PubMed] [Google Scholar]
- 52. Reddien PW, Oviedo NJ, Jennings JR, et al. : SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310(5752):1327–30. 10.1126/science.1116110 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 53. Rossi L, Salvetti A, Lena A, et al. : DjPiwi-1, a member of the PAZ-Piwi gene family, defines a subpopulation of planarian stem cells. Dev Genes Evol. 2006;216(6):335–46. 10.1007/s00427-006-0060-0 [DOI] [PubMed] [Google Scholar]
- 54. Rouhana L, Weiss JA, King RS, et al. : PIWI homologs mediate histone H4 mRNA localization to planarian chromatoid bodies. Development. 2014;141(13):2592–601. 10.1242/dev.101618 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 55. Takebe M, Onohara Y, Yokota S: Expression of MAEL in nuage and non-nuage compartments of rat spermatogenic cells and colocalization with DDX4, DDX25 and MIWI. Histochem Cell Biol. 2013;140(2):169–81. 10.1007/s00418-012-1067-4 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 56. Tan CH, Lee TC, Weeraratne SD, et al. : Ziwi, the zebrafish homologue of the Drosophila piwi: co-localization with vasa at the embryonic genital ridge and gonad-specific expression in the adults. Gene Expr Patterns. 2002;2(3–4):257–60. 10.1016/S1567-133X(02)00052-2 [DOI] [PubMed] [Google Scholar]
- 57. Seto AG, Kingston RE, Lau NC: The coming of age for Piwi proteins. Mol Cell. 2007;26(5):603–9. 10.1016/j.molcel.2007.05.021 [DOI] [PubMed] [Google Scholar]
- 58. Kloc M, Bilinski S, Etkin LD: The Balbiani body and germ cell determinants: 150 years later. Curr Top Dev Biol. 2004;59:1–36. 10.1016/S0070-2153(04)59001-4 [DOI] [PubMed] [Google Scholar]
- 59. Marlow FL: In Maternal Control of Development in Vertebrates: My Mother Made Me Do It!(San Rafael (CA)).2010. 10.4199/C00023ED1V01Y201012DEB005 [DOI] [PubMed] [Google Scholar]
- 60. Kloc M, Bilinski S, Chan AP, et al. : RNA localization and germ cell determination in Xenopus. Int Rev Cytol. 2001;203:63–91. 10.1016/S0074-7696(01)03004-2 [DOI] [PubMed] [Google Scholar]
- 61. Bhat KM: The posterior determinant gene nanos is required for the maintenance of the adult germline stem cells during Drosophila oogenesis. Genetics. 1999;151(4):1479–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Draper BW, McCallum CM, Moens CB: nanos1 is required to maintain oocyte production in adult zebrafish. Dev Biol. 2007;305(2):589–98. 10.1016/j.ydbio.2007.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Forbes A, Lehmann R: Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 1998;125(4):679–90. [DOI] [PubMed] [Google Scholar]
- 64. Tsuda M, Sasaoka Y, Kiso M, et al. : Conserved role of nanos proteins in germ cell development. Science. 2003;301(5637):1239–41. 10.1126/science.1085222 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 65. Illmensee K, Mahowald AP: Transplantation of posterior polar plasm in Drosophila. Induction of germ cells at the anterior pole of the egg. Proc Natl Acad Sci U S A. 1974;71(4):1016–20. 10.1073/pnas.71.4.1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hashimoto Y, Maegawa S, Nagai T, et al. : Localized maternal factors are required for zebrafish germ cell formation. Dev Biol. 2004;268(1):152–61. 10.1016/j.ydbio.2003.12.013 [DOI] [PubMed] [Google Scholar]
- 67. Wylie CC, Holwill S, O'Driscoll M, et al. : Germ plasm and germ cell determination in Xenopus laevis as studied by cell transplantation analysis. Cold Spring Harb Symp Quant Biol. 1985;50:37–43. 10.1101/SQB.1985.050.01.007 [DOI] [PubMed] [Google Scholar]
- 68. Boswell RE, Mahowald AP: tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell. 1985;43(1):97–104. 10.1016/0092-8674(85)90015-7 [DOI] [PubMed] [Google Scholar]
- 69. Nüsslein-Volhard C, Wieschaus E: Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287(5785):795–801. 10.1038/287795a0 [DOI] [PubMed] [Google Scholar]
- 70. Schupbach T, Wieschaus E: Germline autonomy of maternal-effect mutations altering the embryonic body pattern of Drosophila. Dev Biol. 1986;113(2):443–8. 10.1016/0012-1606(86)90179-X [DOI] [PubMed] [Google Scholar]
- 71. Lehmann R, Nüsslein-Volhard C: Abdominal segmentation, pole cell formation, and embryonic polarity require the localized activity of oskar, a maternal gene in Drosophila. Cell. 1986;47(1):141–52. 10.1016/0092-8674(86)90375-2 [DOI] [PubMed] [Google Scholar]
- 72. Markussen FH, Michon AM, Breitwieser W, et al. : Translational control of oskar generates short OSK, the isoform that induces pole plasma assembly. Development. 1995;121(11):3723–32. [DOI] [PubMed] [Google Scholar]
- 73. Breitwieser W, Markussen FH, Horstmann H, et al. : Oskar protein interaction with Vasa represents an essential step in polar granule assembly. Genes Dev. 1996;10(17):2179–88. 10.1101/gad.10.17.2179 [DOI] [PubMed] [Google Scholar]
- 74. Vanzo NF, Ephrussi A: Oskar anchoring restricts pole plasm formation to the posterior of the Drosophila oocyte. Development. 2002;129(15):3705–14. [DOI] [PubMed] [Google Scholar]
- 75. Besse F, López de Quinto S, Marchand V, et al. : Drosophila PTB promotes formation of high-order RNP particles and represses oskar translation. Genes Dev. 2009;23(2):195–207. 10.1101/gad.505709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chang JS, Tan L, Schedl P: The Drosophila CPEB homolog, orb, is required for oskar protein expression in oocytes. Dev Biol. 1999;215(1):91–106. 10.1006/dbio.1999.9444 [DOI] [PubMed] [Google Scholar]
- 77. Chekulaeva M, Hentze MW, Ephrussi A: Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124(3):521–33. 10.1016/j.cell.2006.01.031 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 78. Forbes MM, Rothhämel S, Jenny A, et al. : Maternal dazap2 Regulates Germ Granules by Counteracting Dynein in Zebrafish Primordial Germ Cells. Cell Rep. 2015;12(1):49–57. 10.1016/j.celrep.2015.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 79. Gonsalvez GB, Rajendra TK, Wen Y, et al. : Sm proteins specify germ cell fate by facilitating oskar mRNA localization. Development. 2010;137(14):2341–51. 10.1242/dev.042721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gunkel N, Yano T, Markussen FH, et al. : Localization-dependent translation requires a functional interaction between the 5' and 3' ends of oskar mRNA. Genes Dev. 1998;12(11):1652–64. 10.1101/gad.12.11.1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Heim AE, Hartung O, Rothhämel S, et al. : Oocyte polarity requires a Bucky ball-dependent feedback amplification loop. Development. 2014;141(4):842–54. 10.1242/dev.090449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Jambor H, Brunel C, Ephrussi A: Dimerization of oskar 3' UTRs promotes hitchhiking for RNA localization in the Drosophila oocyte. RNA. 2011;17(12):2049–57. 10.1261/rna.2686411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jeske M, Bordi M, Glatt S, et al. : The Crystal Structure of the Drosophila Germline Inducer Oskar Identifies Two Domains with Distinct Vasa Helicase- and RNA-Binding Activities. Cell Rep. 2015;12(4):587–98. 10.1016/j.celrep.2015.06.055 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 84. Kanke M, Jambor H, Reich J, et al. : oskar RNA plays multiple noncoding roles to support oogenesis and maintain integrity of the germline/soma distinction. RNA. 2015;21(6):1096–109. 10.1261/rna.048298.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kim-Ha J, Kerr K, Macdonald PM: Translational regulation of oskar mRNA by Bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81(3):403–12. 10.1016/0092-8674(95)90393-3 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 86. Micklem DR, Adams J, Grünert S, et al. : Distinct roles of two conserved Staufen domains in oskar mRNA localization and translation. EMBO J. 2000;19(6):1366–77. 10.1093/emboj/19.6.1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Morais-de-Sá E, Vega-Rioja A, Trovisco V, et al. : Oskar is targeted for degradation by the sequential action of Par-1, GSK-3, and the SCF -Slimb ubiquitin ligase. Dev Cell. 2013;26(3):303–14. 10.1016/j.devcel.2013.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 88. Munro TP, Kwon S, Schnapp BJ, et al. : A repeated IMP-binding motif controls oskar mRNA translation and anchoring independently of Drosophila melanogaster IMP. J Cell Biol. 2006;172(4):577–88. 10.1083/jcb.200510044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sanghavi P, Laxani S, Li X, et al. : Dynein associates with oskar mRNPs and is required for their efficient net plus-end localization in Drosophila oocytes. PLoS One. 2013;8(11):e80605. 10.1371/journal.pone.0080605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Snee M, Benz D, Jen J, et al. : Two distinct domains of Bruno bind specifically to the oskar mRNA. RNA Biol. 2008;5(1):1–9. 10.4161/rna.5.1.5735 [DOI] [PubMed] [Google Scholar]
- 91. Styhler S, Nakamura A, Swan A, et al. : vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development. 1998;125(9):1569–78. [DOI] [PubMed] [Google Scholar]
- 92. Suyama R, Jenny A, Curado S, et al. : The actin-binding protein Lasp promotes Oskar accumulation at the posterior pole of the Drosophila embryo. Development. 2009;136(1):95–105. 10.1242/dev.027698 [DOI] [PubMed] [Google Scholar]
- 93. Voronina E, Lopez M, Juliano CE, et al. : Vasa protein expression is restricted to the small micromeres of the sea urchin, but is inducible in other lineages early in development. Dev Biol. 2008;314(2):276–86. 10.1016/j.ydbio.2007.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zimyanin V, Lowe N, St Johnston D: An oskar-dependent positive feedback loop maintains the polarity of the Drosophila oocyte. Curr Biol. 2007;17(4):353–9. 10.1016/j.cub.2006.12.044 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 95. Callebaut I, Mornon JP: LOTUS, a new domain associated with small RNA pathways in the germline. Bioinformatics. 2010;26(9):1140–4. 10.1093/bioinformatics/btq122 [DOI] [PubMed] [Google Scholar]
- 96. Hwang H, Kim S, Yoon S, et al. : Characterization of a novel oligomeric SGNH-arylesterase from Sinorhizobium meliloti 1021. Int J Biol Macromol. 2010;46(2):145–52. 10.1016/j.ijbiomac.2009.12.010 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 97. Kim S, Bae SY, Kim SJ, et al. : Characterization, amyloid formation, and immobilization of a novel SGNH hydrolase from Listeria innocua 11262. Int J Biol Macromol. 2012;50(1):103–11. 10.1016/j.ijbiomac.2011.10.003 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 98. Nakamura M, Okada N, Kajikawa M: Self-interaction, nucleic acid binding, and nucleic acid chaperone activities are unexpectedly retained in the unique ORF1p of zebrafish LINE. Mol Cell Biol. 2012;32(2):458–69. 10.1128/MCB.06162-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Schneider AM, Schmidt S, Jonas S, et al. : Structure and properties of the esterase from non-LTR retrotransposons suggest a role for lipids in retrotransposition. Nucleic Acids Res. 2013;41(22):10563–72. 10.1093/nar/gkt786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Claussen M, Pieler T: Xvelo1 uses a novel 75-nucleotide signal sequence that drives vegetal localization along the late pathway in Xenopus oocytes. Dev Biol. 2004;266(2):270–84. 10.1016/j.ydbio.2003.09.043 [DOI] [PubMed] [Google Scholar]
- 101. Riemer S, Bontems F, Krishnakumar P, et al. : A functional Bucky ball-GFP transgene visualizes germ plasm in living zebrafish. Gene Expr Patterns. 2015;18(1–2):44–52. 10.1016/j.gep.2015.05.003 [DOI] [PubMed] [Google Scholar]
- 102. Snee MJ, Harrison D, Yan N, et al. : A late phase of Oskar accumulation is crucial for posterior patterning of the Drosophila embryo, and is blocked by ectopic expression of Bruno. Differentiation. 2007;75(3):246–55. 10.1111/j.1432-0436.2006.00136.x [DOI] [PubMed] [Google Scholar]
- 103. Zimyanin VL, Belaya K, Pecreaux J, et al. : In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell. 2008;134(5):843–53. 10.1016/j.cell.2008.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 104. Hachet O, Ephrussi A: Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428(6986):959–63. 10.1038/nature02521 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 105. Rongo C, Gavis ER, Lehmann R: Localization of oskar RNA regulates oskar translation and requires Oskar protein. Development. 1995;121(9):2737–46. [DOI] [PubMed] [Google Scholar]
- 106. Campbell PD, Heim AE, Smith MZ, et al. : Kinesin-1 interacts with Bucky ball to form germ cells and is required to pattern the zebrafish body axis. Development. 2015b;142(17):2996–3008. 10.1242/dev.124586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Jenny A, Hachet O, Závorszky P, et al. : A translation-independent role of oskar RNA in early Drosophila oogenesis. Development. 2006;133(15):2827–33. 10.1242/dev.02456 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 108. Robb DL, Heasman J, Raats J, et al. : A kinesin-like protein is required for germ plasm aggregation in Xenopus. Cell. 1996;87(5):823–31. 10.1016/S0092-8674(00)81990-X [DOI] [PubMed] [Google Scholar]
- 109. Strome S, Martin P, Schierenberg E, et al. : Transformation of the germ line into muscle in mes-1 mutant embryos of C. elegans. Development. 1995;121(9):2961–72. [DOI] [PubMed] [Google Scholar]
- 110. Ephrussi A, Dickinson LK, Lehmann R: Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66(1):37–50. 10.1016/0092-8674(91)90137-N [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 111. Snee MJ, Macdonald PM: Live imaging of nuage and polar granules: evidence against a precursor-product relationship and a novel role for Oskar in stabilization of polar granule components. J Cell Sci. 2004;117(Pt 10):2109–20. 10.1242/jcs.01059 [DOI] [PubMed] [Google Scholar]
- 112. Trcek T, Grosch M, York A, et al. : Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat Commun. 2015;6:7962. 10.1038/ncomms8962 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 113. Little SC, Sinsimer KS, Lee JJ, et al. : Independent and coordinate trafficking of single Drosophila germ plasm mRNAs. Nat Cell Biol. 2015;17(5):558–68. 10.1038/ncb3143 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 114. Knaut H, Steinbeisser H, Schwarz H, et al. : An evolutionary conserved region in the vasa 3'UTR targets RNA translation to the germ cells in the zebrafish. Curr Biol. 2002;12(6):454–66. 10.1016/S0960-9822(02)00723-6 [DOI] [PubMed] [Google Scholar]
- 115. Raz E: The function and regulation of vasa-like genes in germ-cell development. Genome Biol. 2000;1(3):REVIEWS1017. 10.1186/gb-2000-1-3-reviews1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Yajima M, Wessel GM: The multiple hats of Vasa: its functions in the germline and in cell cycle progression. Mol Reprod Dev. 2011;78(10–11):861–7. 10.1002/mrd.21363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Campbell PD, Chao JA, Singer RH, et al. : Dynamic visualization of transcription and RNA subcellular localization in zebrafish. Development. 2015a;142(7):1368–74. 10.1242/dev.118968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Gross-Thebing T, Paksa A, Raz E: Simultaneous high-resolution detection of multiple transcripts combined with localization of proteins in whole-mount embryos. BMC Biol. 2014;12:55. 10.1186/s12915-014-0055-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Eno C, Pelegri F: Gradual recruitment and selective clearing generate germ plasm aggregates in the zebrafish embryo. Bioarchitecture. 2013;3(4):125–32. 10.4161/bioa.26538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Strasser MJ, Mackenzie NC, Dumstrei K, et al. : Control over the morphology and segregation of Zebrafish germ cell granules during embryonic development. BMC Dev Biol. 2008;8:58. 10.1186/1471-213X-8-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Kurimoto K, Yamaji M, Seki Y, et al. : Specification of the germ cell lineage in mice: a process orchestrated by the PR-domain proteins, Blimp1 and Prdm14. Cell Cycle. 2008b;7(22):3514–8. 10.4161/cc.7.22.6979 [DOI] [PubMed] [Google Scholar]
- 122. Sasaki K, Yokobayashi S, Nakamura T, et al. : Robust In Vitro Induction of Human Germ Cell Fate from Pluripotent Stem Cells. Cell Stem Cell. 2015;17(2):178–94. 10.1016/j.stem.2015.06.014 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 123. Berkowitz LA, Strome S: MES-1, a protein required for unequal divisions of the germline in early C. elegans embryos, resembles receptor tyrosine kinases and is localized to the boundary between the germline and gut cells. Development. 2000;127(20):4419–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Brangwynne CP, Eckmann CR, Courson DS, et al. : Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324(5935):1729–32. 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 125. Wang JT, Smith J, Chen BC, et al. : Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. eLife. 2014;3:e04591. 10.7554/eLife.04591 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 126. Hanazawa M, Yonetani M, Sugimoto A: PGL proteins self associate and bind RNPs to mediate germ granule assembly in C. elegans. J Cell Biol. 2011;192(6):929–37. 10.1083/jcb.201010106 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 127. Updike DL, Hachey SJ, Kreher J, et al. : P granules extend the nuclear pore complex environment in the C. elegans germ line. J Cell Biol. 2011;192(6):939–48. 10.1083/jcb.201010104 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 128. Rangan P, DeGennaro M, Jaime-Bustamante K, et al. : Temporal and spatial control of germ-plasm RNAs. Curr Biol. 2009;19(1):72–7. 10.1016/j.cub.2008.11.066 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 129. Smith JL, Wilson JE, Macdonald PM: Overexpression of oskar directs ectopic activation of nanos and presumptive pole cell formation in Drosophila embryos. Cell. 1992;70(5):849–59. 10.1016/0092-8674(92)90318-7 [DOI] [PubMed] [Google Scholar]
- 130. Zaessinger S, Busseau I, Simonelig M: Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development. 2006;133(22):4573–83. 10.1242/dev.02649 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 131. Kawasaki I, Amiri A, Fan Y, et al. : The PGL family proteins associate with germ granules and function redundantly in Caenorhabditis elegans germline development. Genetics. 2004;167(2):645–61. 10.1534/genetics.103.023093 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 132. Kawasaki I, Shim YH, Kirchner J, et al. : PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell. 1998;94(5):635–45. 10.1016/S0092-8674(00)81605-0 [DOI] [PubMed] [Google Scholar]
- 133. Updike DL, Knutson AK, Egelhofer TA, et al. : Germ-granule components prevent somatic development in the C. elegans germline. Curr Biol. 2014;24(9):970–5. 10.1016/j.cub.2014.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 134. Kurimoto K, Yabuta Y, Ohinata Y, et al. : Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev. 2008a;22(12):1617–35. 10.1101/gad.1649908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Ohinata Y, Payer B, O'Carroll D, et al. : Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436(7048):207–13. 10.1038/nature03813 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 136. Vincent SD, Dunn NR, Sciammas R, et al. : The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development. 2005;132(6):1315–25. 10.1242/dev.01711 [DOI] [PubMed] [Google Scholar]
- 137. Yamaji M, Seki Y, Kurimoto K, et al. : Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet. 2008;40(8):1016–22. 10.1038/ng.186 [DOI] [PubMed] [Google Scholar]
- 138. Bortvin A, Goodheart M, Liao M, et al. : Dppa3 / Pgc7 / stella is a maternal factor and is not required for germ cell specification in mice. BMC Dev Biol. 2004;4:2. 10.1186/1471-213X-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Lawson KA, Dunn NR, Roelen BA, et al. : Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13(4):424–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Ying Y, Liu XM, Marble A, et al. : Requirement of Bmp8b for the generation of primordial germ cells in the mouse. Mol Endocrinol. 2000;14(7):1053–63. 10.1210/mend.14.7.0479 [DOI] [PubMed] [Google Scholar]
- 141. Ohinata Y, Ohta H, Shigeta M, et al. : A signaling principle for the specification of the germ cell lineage in mice. Cell. 2009;137(3):571–84. 10.1016/j.cell.2009.03.014 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 142. Lawson KA, Hage WJ: Clonal analysis of the origin of primordial germ cells in the mouse. Ciba Found Symp. 1994;182:68–84; discussion 84–91. 10.1002/9780470514573.ch5 [DOI] [PubMed] [Google Scholar]
- 143. West JA, Viswanathan SR, Yabuuchi A, et al. : A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 2009;460(7257):909–13. 10.1038/nature08210 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 144. Witschi, E: Migration of germ cells of human embryos from the yolk sac to the primitive gonadal folds. Contr Embryol Carnegie Inst. 1948;209:67–80. [Google Scholar]
- 145. Toyooka Y, Tsunekawa N, Akasu R, et al. : Embryonic stem cells can form germ cells in vitro. Proc Natl Acad Sci U S A. 2003;100(20):11457–62. 10.1073/pnas.1932826100 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 146. Hübner K, Fuhrmann G, Christenson LK, et al. : Derivation of oocytes from mouse embryonic stem cells. Science. 2003;300(5623):1251–6. 10.1126/science.1083452 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 147. Geijsen N, Horoschak M, Kim K, et al. : Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature. 2004;427(6970):148–54. 10.1038/nature02247 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 148. Nayernia K, Nolte J, Michelmann HW, et al. : In vitro-differentiated embryonic stem cells give rise to male gametes that can generate offspring mice. Dev Cell. 2006;11(1):125–32. 10.1016/j.devcel.2006.05.010 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 149. Hayashi K, Ohta H, Kurimoto K, et al. : Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146(4):519–32. 10.1016/j.cell.2011.06.052 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 150. Hayashi K, Ogushi S, Kurimoto K, et al. : Offspring from oocytes derived from in vitro primordial germ cell-like cells in mice. Science. 2012;338(6109):971–5. 10.1126/science.1226889 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 151. Sugawa F, Araúzo-Bravo MJ, Yoon J, et al. : Human primordial germ cell commitment in vitro associates with a unique PRDM14 expression profile. EMBO J. 2015;34(8):1009–24. 10.15252/embj.201488049 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 152. Beer RL, Draper BW: nanos3 maintains germline stem cells and expression of the conserved germline stem cell gene nanos2 in the zebrafish ovary. Dev Biol. 2013;374(2):308–18. 10.1016/j.ydbio.2012.12.003 [DOI] [PubMed] [Google Scholar]
- 153. Gkountela S, Li Z, Vincent JJ, et al. : The ontogeny of cKIT + human primordial germ cells proves to be a resource for human germ line reprogramming, imprint erasure and in vitro differentiation. Nat Cell Biol. 2013;15(1):113–22. 10.1038/ncb2638 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 154. Julaton VT, Reijo Pera RA: NANOS3 function in human germ cell development. Hum Mol Genet. 2011;20(11):2238–50. 10.1093/hmg/ddr114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Hara K, Kanai-Azuma M, Uemura M, et al. : Evidence for crucial role of hindgut expansion in directing proper migration of primordial germ cells in mouse early embryogenesis. Dev Biol. 2009;330(2):427–39. 10.1016/j.ydbio.2009.04.012 [DOI] [PubMed] [Google Scholar]
- 156. Kanai-Azuma M, Kanai Y, Gad JM, et al. : Depletion of definitive gut endoderm in Sox17-null mutant mice. Development. 2002;129(10):2367–79. [DOI] [PubMed] [Google Scholar]
- 157. Paksa A, Raz E: Zebrafish germ cells: motility and guided migration. Curr Opin Cell Biol. 2015;36:80–5. 10.1016/j.ceb.2015.07.007 [DOI] [PubMed] [Google Scholar]
- 158. Richardson BE, Lehmann R: Mechanisms guiding primordial germ cell migration: strategies from different organisms. Nat Rev Mol Cell Biol. 2010;11(1):37–49. 10.1038/nrm2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Kobayashi S, Yamada M, Asaoka M, et al. : Essential role of the posterior morphogen nanos for germline development in Drosophila. Nature. 1996;380(6576):708–11. 10.1038/380708a0 [DOI] [PubMed] [Google Scholar]
- 160. Subramaniam K, Seydoux G: nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development. 1999;126(21):4861–71. [DOI] [PubMed] [Google Scholar]
- 161. Köprunner M, Thisse C, Thisse B, et al. : A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 2001;15(21):2877–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Kusz K, Tomczyk L, Spik A, et al. : NANOS3 gene mutations in men with isolated sterility phenotype. Mol Reprod Dev. 2009;76(9):804. 10.1002/mrd.21070 [DOI] [PubMed] [Google Scholar]
- 163. Qin Y, Zhao H, Kovanci E, et al. : Mutation analysis of NANOS3 in 80 Chinese and 88 Caucasian women with premature ovarian failure. Fertil Steril. 2007;88(5):1465–7. 10.1016/j.fertnstert.2007.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Santos MG, Machado AZ, Martins CN, et al. : Homozygous inactivating mutation in NANOS3 in two sisters with primary ovarian insufficiency. Biomed Res Int. 2014;2014: 787465. 10.1155/2014/787465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Suzuki H, Tsuda M, Kiso M, et al. : Nanos3 maintains the germ cell lineage in the mouse by suppressing both Bax-dependent and -independent apoptotic pathways. Dev Biol. 2008;318(1):133–42. 10.1016/j.ydbio.2008.03.020 [DOI] [PubMed] [Google Scholar]
- 166. Wu X, Wang B, Dong Z, et al. : A NANOS3 mutation linked to protein degradation causes premature ovarian insufficiency. Cell Death Dis. 2013;4:e825. 10.1038/cddis.2013.368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Grieve KM, McLaughlin M, Dunlop CE, et al. : The controversial existence and functional potential of oogonial stem cells. Maturitas. 2015;82(3):278–81. 10.1016/j.maturitas.2015.07.017 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation