Abstract
In mammals, sperm–oocyte fusion initiates Ca2+ oscillations leading to a series of events called oocyte activation, which is the first stage of embryo development. Ca2+ signaling is elicited by the delivery of an oocyte-activating factor by the sperm. A sperm-specific phospholipase C (PLCZ1) has emerged as the likely candidate to induce oocyte activation. Recently, PAWP, a sperm-born tryptophan domain-binding protein coded by WBP2NL, was proposed to serve the same purpose. Here, we studied two infertile brothers exhibiting normal sperm morphology but complete fertilization failure after intracytoplasmic sperm injection. Whole exomic sequencing evidenced a missense homozygous mutation in PLCZ1, c.1465A>T; p.Ile489Phe, converting Ile 489 into Phe. We showed the mutation is deleterious, leading to the absence of the protein in sperm, mislocalization of the protein when injected in mouse GV and MII oocytes, highly abnormal Ca2+ transients and early embryonic arrest. Altogether these alterations are consistent with our patients' sperm inability to induce oocyte activation and initiate embryo development. In contrast, no deleterious variants were identified in WBP2NL and PAWP presented normal expression and localization. Overall we demonstrate in humans, the absence of PLCZ1 alone is sufficient to prevent oocyte activation irrespective of the presence of PAWP. Additionally, it is the first mutation located in the C2 domain of PLCZ1, a domain involved in targeting proteins to cell membranes. This opens the door to structure–function studies to identify the conserved amino acids of the C2 domain that regulate the targeting of PLCZ1 and its selectivity for its lipid substrate(s).
Introduction
Increases in the intracellular concentration of free calcium (Ca2+) was demonstrated to be a sufficient and necessary stimulus to trigger oocyte activation and embryo development in all species studied to date (1). This finding along with subsequent confirmatory studies (2) stimulated interest to elucidate the signaling cascade responsible for Ca2+ release at fertilization. In 1990, studies showed that a soluble component of mammalian sperm extracts, aptly named the sperm factor, was sufficient to induce oocyte activation and replicate the periodical Ca2+ responses, also known as Ca2+ oscillations, which are a hallmark of mammalian fertilization (3,4). PLCZ1 was later identified as the candidate molecule to be the active factor in sperm responsible for the oscillations (5,6). Research from several laboratories confirmed the unique properties of PLCZ1 to induce Ca2+ oscillations in oocytes as well as the association of its absence with infertility (7–12). However, the inability thus far to obtain a PLCZ1 KO animal model capable of producing mature sperm (13) has prevented assigning to this molecule the exclusive role for oocyte activation, while leaving open the possibility that other sperm factors may be required (14). Toward that end, PAWP has been proposed as an alternative or complementary pathway for oocyte activation based on findings that injection of recombinant PAWP induced oscillations comparable to those of fertilization (15,16). The relationship between the proposed function of PAWP and its structure is not yet understood. PAWP displays sequence homology to WW domain-binding protein 2 (WBP2) in its N terminal end and a variable number of PPXY motifs (one in human, six in mouse) in its C-terminal end, a motif known to interact with WW domain. Moreover, the C terminal end contains an unidentified repeated motif (YGXPPXG) (17). It is presently unknown, however, how these motifs may engage the oocyte's signaling machinery to induce Ca2+ oscillations. In this vein, attempts to replicate those studies in mouse oocytes failed (18,19), which raised concerns regarding the importance of PAWP in fertilization. However, its action in human oocyte activation was not formally ruled out, especially as numerous studies have shown that testis and reproductive tissues evolve faster than other tissues leading to noticeable differences between species (20).
Here, we studied two infertile brothers showing complete fertilization failure after intracytoplasmic sperm injection (ICSI). Whole-exome sequencing enabled us to find a missense homozygous mutation in PLCZ1, c.1465A>T; p.Ile489Phe, converting Ile 489 into Phe. Structure–function models revealed that the mutation causes a conformational change that might affect the enzyme's ability to bind to its substrate(s). Using western blotting (WB), immunofluorescence (IF), live fluorescence and Ca2+ imaging, we show that the mutation is deleterious, leading to mislocalization of the protein, lower Ca2+ signaling and lower rates of oocyte activation and embryo development. In contrast, no mutations were identified in WBP2NL and PAWP showed normal expression and localization. Overall we demonstrate the absence of PLCZ1 alone in humans is sufficient to prevent oocyte activation irrespective of PAWP. Moreover, it is the first mutation located in the C2 domain of the enzyme, an important domain targeting proteins to lipidic membranes, opening the door for better structure–function analysis of PLCZ1 and of other proteins carrying this domain.
Results
Patients' description: medical records and spermatocytogram
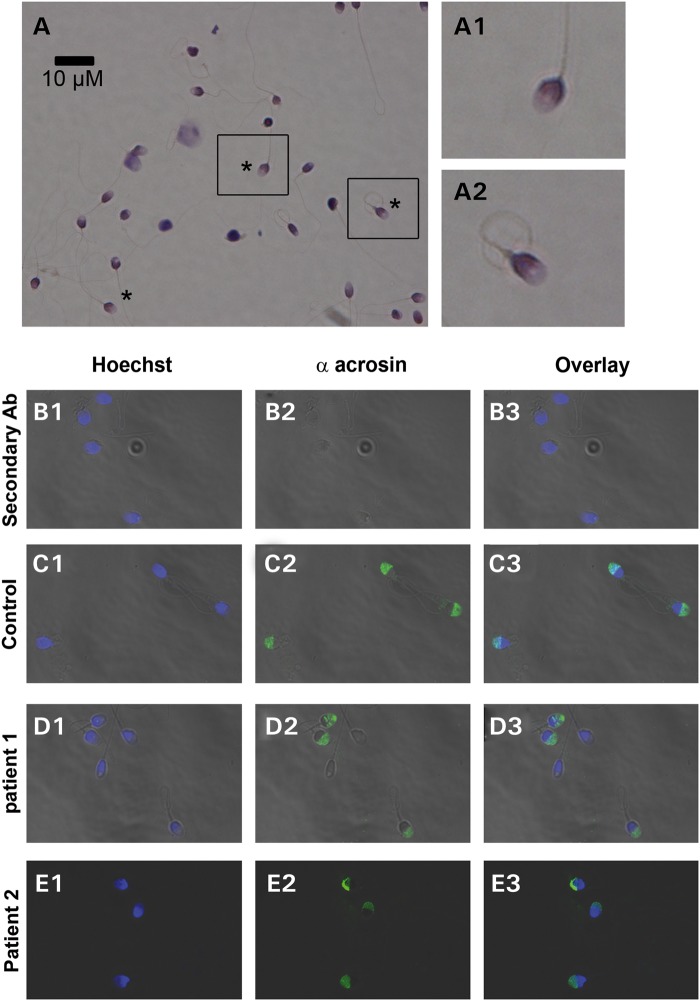
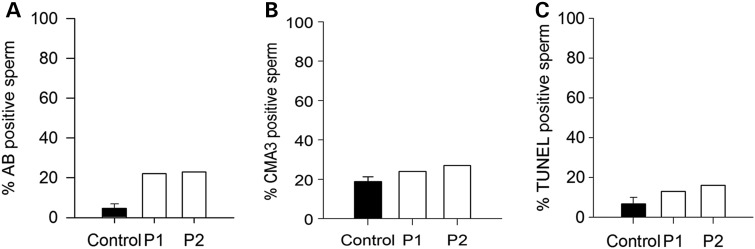
Two Tunisians brothers and their respective wives sought medical advice from infertility clinic in Tunis between 2011 and 2014 after unsuccessful attempts for a full year to conceive a pregnancy. The brothers were born from first cousin parents and have one fertile brother with children conceived spontaneously and two fertile sisters. The morphology of patients' sperm was assessed with Shorr staining (Fig. 1A) and sperm parameters were well above the low reference limits set by the WHO guidelines (21), although Patient 1 (P1) had higher than average number of sperm with acrosome defects (Table 1). To investigate this in more detail, we examined the presence and morphology of the acrosome by IF using an anti-acrosin antibody. We found that the acrosome presented a normal shape in ∼50% of the brothers' sperm (Fig. 1D and E). While not examined, we estimate that the absence of staining on the other half is due to a premature acrosome reaction rather than to an abnormal acrosome biogenesis, as the shape of the sperm heads was normal and not globozoospermic (Fig. 1). We also assessed DNA quality using three different methods: chromomycin A3, aniline blue (AB) and terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate-nick-end labeling (TUNEL), which allowed evaluation of DNA protamination, histone content and DNA fragmentation, respectively. The percentages of positive sperm were higher in patients than in fertile controls, but all values were ∼20% (Fig. 2) and remained below abnormal thresholds defined by several studies (22–24). Remarkably, although these sperm parameters were compatible with spontaneous conception, for both brothers the clinical outcomes following assisted reproduction technologies procedures was oocyte activation failure (OAF); P1 had two unsuccessful attempts of artificial insemination and then sperm from both patients were used for ICSI. In total, 3 ICSI cycles were carried out and 20 MII oocytes were injected, but none showed signs of oocyte activation (Table 2).
Figure 1.
Morphology of the patients' sperm. (A) Sperm samples were spread over a slide and dried at RT, fixed in ether/ethanol 1:1 for Harris–Schorr staining. Sperm were then scored according to WHO's laboratory manual for examination and processing of human semen (5th edition). Asterisk indicates fully normal spermatozoa. A1–A2 correspond to enlargement of the black squares in (A), in order to show the morphology of normal sperm. (B–E) Morphology of acrosome stained with anti-acrosin antibody (B) Negative control experiment with the addition of secondary antibody without primary antibody. (C–E) Acrosin staining (green staining, C2–E2) of sperm from subjects with normal fertility (C) and from OAF patients (D and E) and counterstained with Hoechst (blue, B1–E1) to evidence the nucleus/acrosome ratio. The third column corresponds to overlay (B3–E3).
Table 1.
Semen parameters and spermatocytogram of P1 and P2
| Semen parameters | P1 | P2 |
|---|---|---|
| Semen volume (ml) | 3 | 5.8 |
| Sperm concentration (106/ml) | 150 | 101 |
| Total motility 1 h (%) | 50 | 40 |
| Vitality (%) | 63 | 70 |
| Normal spermatozoa (%) | 20 | 7 |
| Anomalies of the flagella (%) | 10 | 20 |
| Abnormal acrosome (%) | 50 | 78 |
| Other anomalies of the head (%) | 10 | 20 |
| Multiple anomalies index (%) | 1.3 | 1.4 |
Values are the average of two separate analyses.
Figure 2.
Assessment of nucleus compaction and DNA breaks of sperm from OAF patients. (A) Histones content of sperm was assessed using the AB test. The histogram shows the percentage of stained sperm in samples from control (n = 5 ± SD) and patients (n = 1). (B) Protamination of sperm was evaluated using the chromomycin A3 test, and results are displayed in the histogram, which shows the percentage of stained sperm in control (n = 5 ± SD) and patients' (n = 1) samples. (C) The histogram of DNA fragmentation analysis with TUNEL assay showing the level of TUNEL-positive sperm in control (n = 5 ± SD) and patients' (n = 1) samples.
Table 2.
ICSI outcomes following stimulation cycles with sperm from P1 and P2
| Patient and procedure | Years | No. of follicles (n) | No. of abnormal oocytes (GV, M1, atretic) (n) | No. of mature oocytes injected (n) | No. of 2PN oocytes (n) |
|---|---|---|---|---|---|
| P1 ICSI—Jasmin clinic | 2011 | 9 | 4 | 5 | 0 |
| P1 ICSI—Jasmin clinic | 2012 | 9 | 2 | 7 | 0 |
| P2 ICSI—Jasmin clinic | 2012 | 14 | 6 | 8 | 0 |
Number (n), P1 and P2 are brothers.
Whole-exome sequencing identified a homozygous missense mutation in PLCZ1
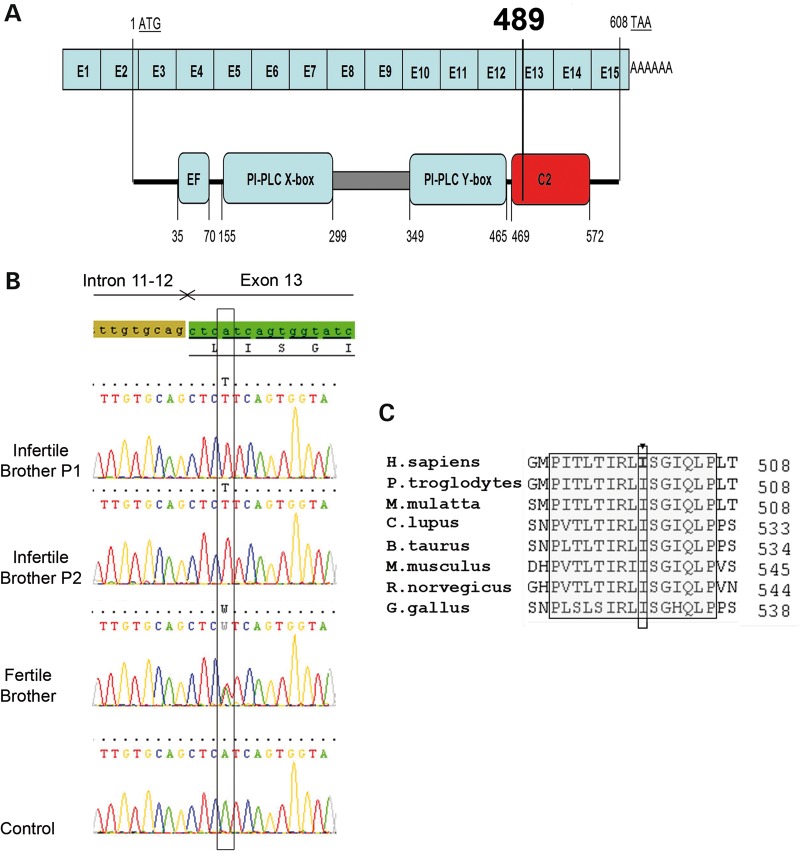
Since both brothers were married to unrelated women, we excluded the possibility of a female factor and focused our research on the brothers. Given the family history of consanguinity, we postulated that this infertility was caused by a homozygous mutation. We, therefore, proceeded to whole-exome sequencing to identify a possible genetic defect(s) that could explain the observed OAF. After exclusion of frequent variants, only four homozygous variants were identified in both brothers (Table 3). Three variants, located in EPS8 (also known as DFNB102), RP11-1021N1.1 and LKAAEAR1 (also known as C20orf201), had no expected deleterious effect. The fourth variant was a missense mutation on PLCZ1, c.1465A>T located in exon 13 (NM_033123.3), changing an Ile at position 489 into a Phe (Ile489Phe) (Fig. 3A). No other variants were identified in the PLCZ1-coding sequence, UTR regions or close intronic regions. Given that PLCZ1 has been suggested to be necessary for oocyte activation in mammals, this mutation could underlie the patients' phenotype. Sanger sequencing confirmed the homozygous mutation for both infertile brothers and showed that the third fertile brother was heterozygous (Fig. 3B). The c.1465A>T variant was absent from over 60 000 individuals described in the ExAC database (exac.broadinstitute.org), which confirms it is not a polymorphism and that missense variations occurring at this localization would likely cause negative selection throughout evolution. Moreover, we found Ile489 to be well conserved throughout evolution (Fig. 3C), suggesting that this mutation could be deleterious.
Table 3.
List of common homozygous variants present in P1 and P2
| Gene | Variant coordinates | Transcript | cDNA variation | Amino acid variation | Prediction |
|---|---|---|---|---|---|
| PLCZ1 | chr12:18841149, T>A | NM_033123.3 | c.1465A>T | p.Ile489Phe | Damaging |
| EPS8 | chr12:15776164, A>C | NM_004447.5 | c.2283A>C | p.Asp761Glu | Benign |
| RP11-1021N1.1 | chr16:15528278, T>C | ENST00000568222a | c.286T>C | p.Tyr96His | Benign |
| LKAAEAR1 | chr20:62715318, T>G | NM_001007125.1 | c.255T>G | p.Glu85Asp | Benign |
Coordinates of all variations are based on the UCSC GRCh37/hg19 assembly.
aNo RefSeq transcript accession number currently available.
Figure 3.
The Ile489Phe mutation is located in the C2 domain of PLCZ1. (A) Schematic representation of the exomic structure of human PLCZ1 cDNA sequence and corresponding functional domains of PLCZ1 (http://www.uniprot.org/uniprot/Q86YW0). The first coding exon is exon 2 (exon sizes are not to scale). The mutation c.1465A>T; p.Ile489Phe (NM_033123.3) is located in exon 13 and changes Isoleucine 489 located in the C2 domain into a phenylalanine. (B) The presence of the identified variation c.1465A>T; p.Ile489Phe (NM_033123.3) was verified by Sanger sequencing of PLCZ1 exon 13. Electropherogram of PLCZ1 exon 13 showing the mutated sequence and sequence obtained from a control individual. The two infertile brothers carried a homozygous missense mutation (p.Ile489Phe) in PLCZ1 exon 13 whereas the fertile brother harbors the mutation in a heterozygous state. (C) The mutation is located in a cluster of 15 highly conserved amino acids.
Deleterious effects of the identified mutation
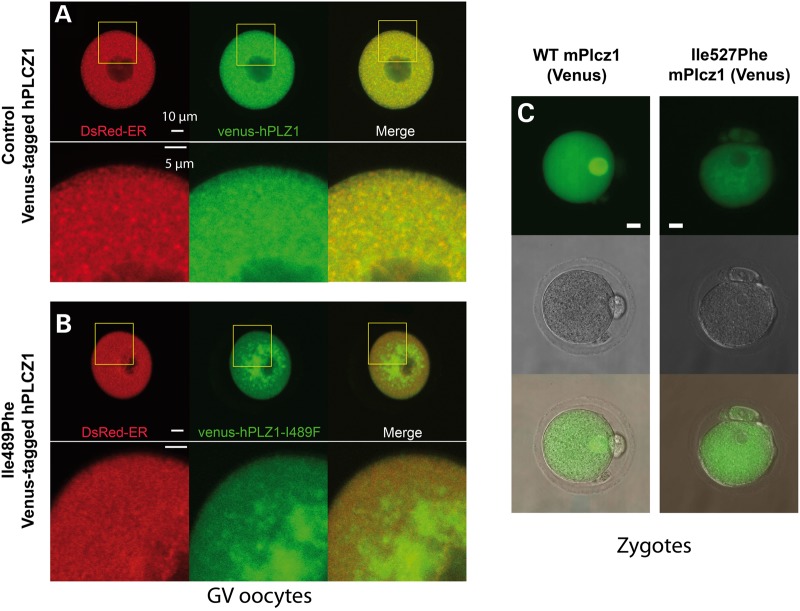
In order to assess the impact of the mutation, PLCZ1 expression and localization were first studied on sperm from both patients (Fig. 4A). We used an antibody targeting human PLCZ1 (anti-hPLCZ1). This antibody has been published and used in three previous publications and the respective western blots showed remarkably specificity (7,11,25). Whereas the sperm of fertile patients showed a band of strong staining on the post-acrosomal area and more diffuse staining over the acrosome (Fig. 4A1, inset), there was no staining detectable in the patients' sperm, except for a few of them showing a faint punctuate staining over the acrosome (Fig. 4A2, inset), suggesting that the mutant PLCZ1 was absent or present at very low concentrations. The absence of PLCZ1 in the patients' sperm was confirmed by WB, where a single band at the expected MW of PLCZ1 was noticeable in the control lane, whereas there was no reactivity in the patients' lanes (Fig. 4B), although similar sperm number were loaded per lane (Fig. 4C). These results suggest that during spermatogenesis Ile489Phe PLCZ1 displays defects in stability, trafficking and/or anchoring, which prevent its presence in mature sperm. To examine these possibilities, cRNAs of human WT and Ile489Phe Venus-tagged PLCZ1 were injected into mouse GV oocytes, which were maintained arrested at the GV stage with IBMX; this was done to ascertain the distribution of the enzyme independently of the stage of the cell cycle. It is noteworthy that is presently unknown how PLCZ1 distribution is regulated in oocytes, although unlike other PLCs (26), it does not appear to localize to the plasma membrane (27), where most of phosphatidylinositol 4,5-bisphosphate (PIP2), the enzyme's substrate, is found. In agreement with this remark, WT PLCZ1 exhibited a homogeneous distribution in the ooplasm of GV oocytes that partly overlapped with distribution of the endoplasmic reticulum (ER), which was marked by ER-DsRed (Fig. 5A). In contrast, Ile489Phe PLCZ1 displayed an uneven distribution characterized by large patches near the nucleus and decreased peripheral localization (Fig. 5B). Furthermore, Ile489Phe PLCZ1 distribution did not overlap with the ER. These results suggest that the mutation does not affect the stability of PLCZ1, although its trafficking and/or anchoring properties were clearly altered. To further test the latter, we took advantage of the fact that following fertilization, Plcz1 is translocated into the pronuclei (PN) (28,29). The sequestration of Plcz1 into the PNs is thought to contribute to the cessation of Ca2+ oscillations, which in the mouse closely corresponds with their formation. WT and Ile527Phe mPlcz1 cRNAs [the equivalent mutation of Ile489Phe human PLCZ1 (hPLCZ1)] were injected into MII oocytes, which were activated by the oscillations initiated by the translated proteins. Following PN formation, as expected, WT mPlcz1 accumulated into the PN. In contrast, following injection of Ile527Phe mPlcz1 cRNA, PN formation was delayed by ∼3 h, presumably due to the lower frequency of oscillations, and the mutant protein failed to localize to the PN (Fig. 5C). These results confirm that the mutation modifies PLCZ1's anchoring and/or trafficking properties in oocytes and zygotes.
Figure 4.
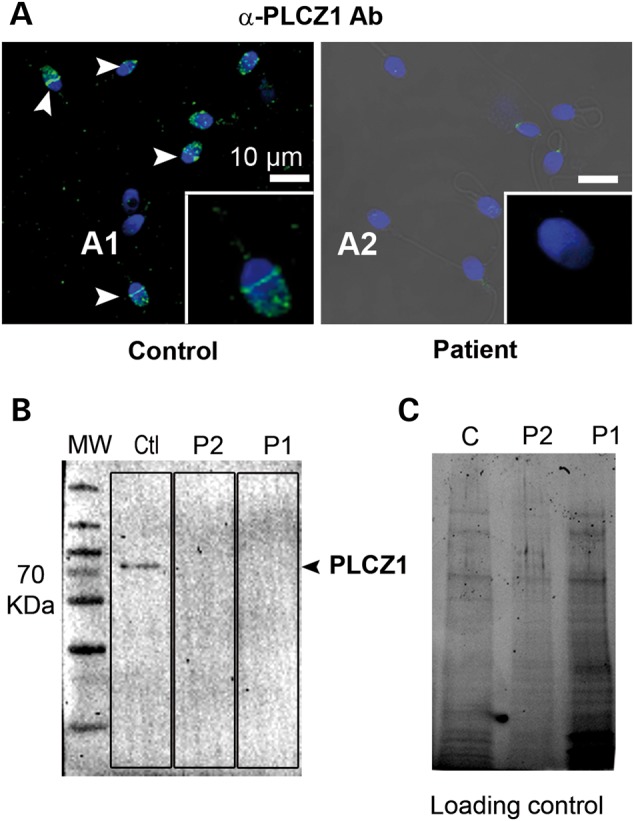
PLCZ1 expression is undetectable by IF and WB in sperm of patients exhibiting OAF after ICSI. (A1) Overlay of Hoechst staining (blue) and PLCZ1 staining (green) of sperm from a fertile control patient showing strong immunoreactivity in the post-acrosomal area (white arrow heads and inset) and to a lesser extent in the acrosome. (A2) Representative staining of sperm from patients with OAF showing the absence of PLCZ1 staining in the post-acrosomal area and very faint staining over the acrosome. More than 100 sperm were observed per patient. (B) WB using protein extracts of sperm from a fertile control (Ctl), or Patient 1 (P1) and Patient 2 (P2) and an anti-PLCZ1 antibody that fails to detect reactivity in the patients' lanes. (C) Protein gel representing loading control for Figure 1B showing that all lanes were similarly loaded. Protein loads were controlled with TGX stain free™ precast gels.
Figure 5.
Mislocalization of mutant PLCZ1 following expression in GV and MII oocytes. (A) Venus-tagged human PLCZ1 cRNA (green) was co-injected in mouse GV stage oocytes with ER-DsRed cRNA, as a marker of the ER (red); the GV arrest was maintained by incubation in 100 μm IBMX. Both PLCZ1 and ER-DsRed exhibited a widespread punctiform staining throughout the cytoplasm and the overlay shows mostly overlapping distribution of the signals. (B) Co-injection of Ile489Phe PLCZ1 and ER-DsRed resulted in different distribution, with most of the PLCZ1 staining (green) concentrated around the nucleus (GV), whereas ER-DsRed remained uniformly distributed in the cytoplasm; there was minimal overlap of the signals at the oocyte periphery. (C) cRNAs encoding for WT mPlcz1 and the equivalent mutant in mice, Ile527Phe, were injected in MII oocytes and their ability to translocate into the PN compared. Whereas WT Plcz1 (left images) concentrated on the PN, Ile527Phe Plcz1 did not (right images).
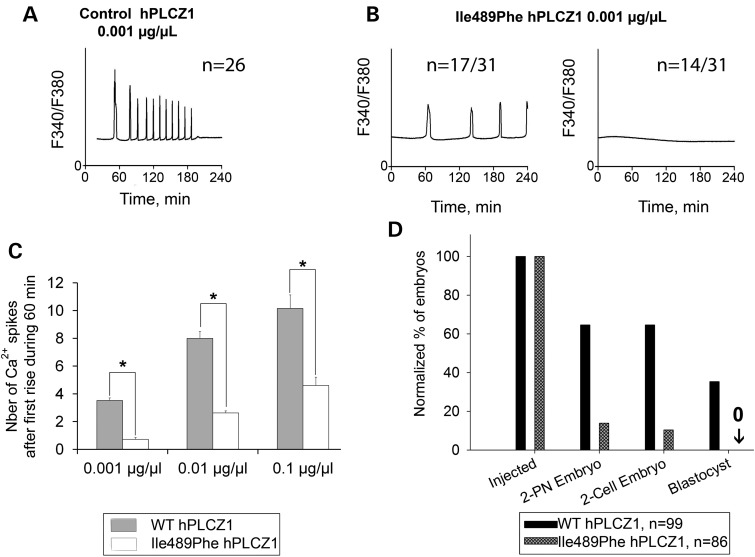
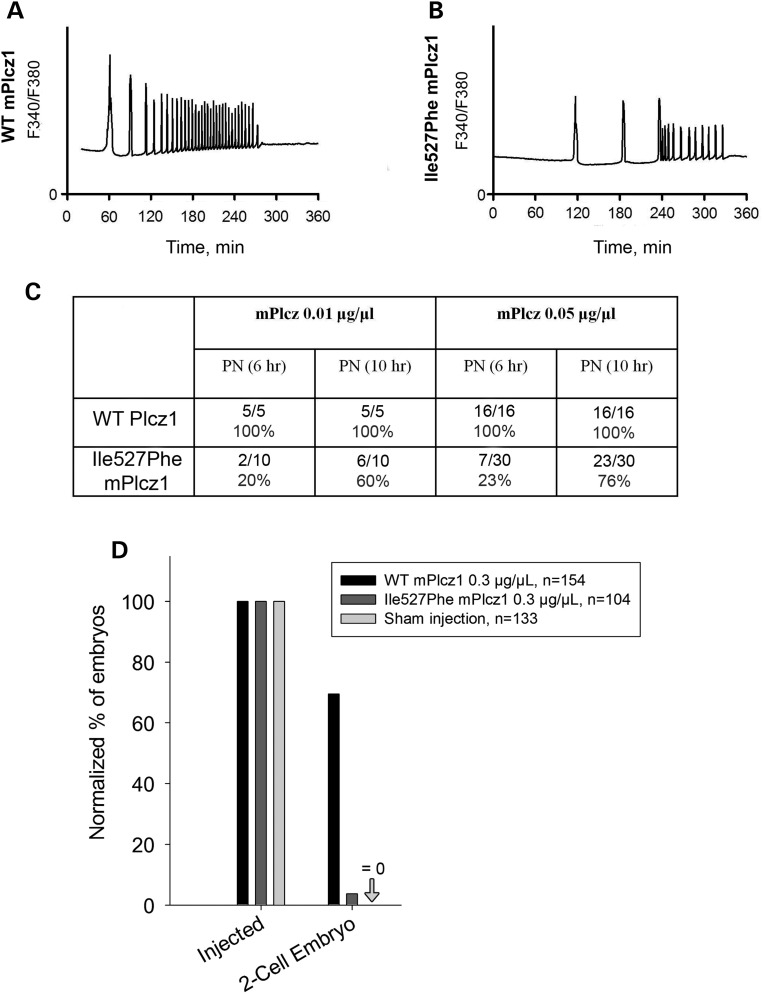
We next assessed the enzymatic activity of the mutated human (h) and mouse (m) PLCZ1s, by examining Ca2+ responses elicited by the injection of their respective cRNAs into mouse MII oocytes. WT hPLCZ1 cRNA (0.001 μg/μl) initiated high-frequency oscillations in all injected oocytes (Fig. 6A) whereas the mutant hPLCZ1 cRNA failed to initiate oscillations in 14/31 oocytes (46%) or induced responses with a low frequency in 17/31 oocytes (54%) (Fig. 6B). Furthermore, the enzymatic activity of the mutant versus WT hPLCZ1 was reduced irrespective of the concentrations of cRNA injected (Fig. 6C). These results suggest that possible trace amounts of hPLCZ1 are likely not sufficient to activate oocytes. To confirm this hypothesis, both WT and mutant cRNAs were injected into mouse oocytes, and the development of parthenote embryos was followed by evaluating the rates of PN formation, cleavage to the two-cell stage and blastocysts. Following injection of WT hPLCZ1 64.6% of the oocytes showed signs of activation (2PN) and 35% reached the blastocyst stage. In contrast, Ile489Phe hPLCZ1 showed a greatly reduced ability to induce oocyte activation and allowed the fertilization of only 13.9% of the oocytes and none developed to the blastocyst stage (Fig. 6D). Similar results were obtained with the mutant mPlcz1, as Ca2+ responses triggered by the mIle527Phe mutant were weaker than those observed with WT Plcz1, showing longer lag time and reduced frequency (Fig. 7A, B). Consistent with this, the timing of PN formation was delayed and the percentages of zygotes reaching the 2PN stage at 6 and 10 h post-insemination was decreased (Fig. 7C) when Ile527Phe mPlcz1 cRNA was injected compared with WT Plcz1 cRNA injection; the altered Ca2+ signaling also prevented most zygotes from cleaving past the two-cell stage (Fig. 7D).
Figure 6.
Ile489Phe PLCZ1 induces fewer Ca2+ oscillations compromising embryo development. (A) Injection of hPLCZ1 cRNA (0.001 μg/μl) in MII mouse oocytes triggered high-frequency Ca2+ oscillations, and between 6 and 10 rises per hour that end by 120–180 min after injection. (B) In contrast, injection of cRNA of Ile489Phe hPLCZ1 (0.001 μg/μl) triggered oscillations that were either delayed and with a low frequency (17/31) or failed to initiate them (14/31). (C) Histogram showing means + SD of the frequency of Ca2+ rises in function of control or Ile489Phe cRNA PLCZ1 concentrations injected into oocytes; asterisks indicate significant differences determined by t-test, P < 0.01. (D) Rates of 2PN, two-cell embryos and blastocysts obtained after injection of WT or Ile489Phe cRNA PLCZ1 (0.01 μg/μl) in mouse MII oocytes; n corresponds to the number of injected oocytes.
Figure 7.
Ca2+ responses induced by Ile527Phe mouse Plcz1 are altered and embryo development is compromised. (A) Profile of Ca2+ responses induced by injection of WT mPlcz1 cRNA (0.01 µg/µl) into mouse oocytes. (B) Ca2+ profiles recorded after injection of Ile527Phe mPlcz1 cRNA (the mouse equivalent of Ile489Phe hPLCZ1) show delayed and reduced response. (C) 2PN formation rates are reduced and delayed when both Ile527Phe mPlcz1 0.01 and 0.05 µg cRNA/µl concentrations are used. (D) Injection of Ile527Phe mPlcz1 cRNAs does not support embryo development, as the rate of two-cell embryos formation was strongly reduced or abolished; n corresponds to the number of injected MII oocytes.
The Ile489Phe mutation alters the conformation of the C2 domain
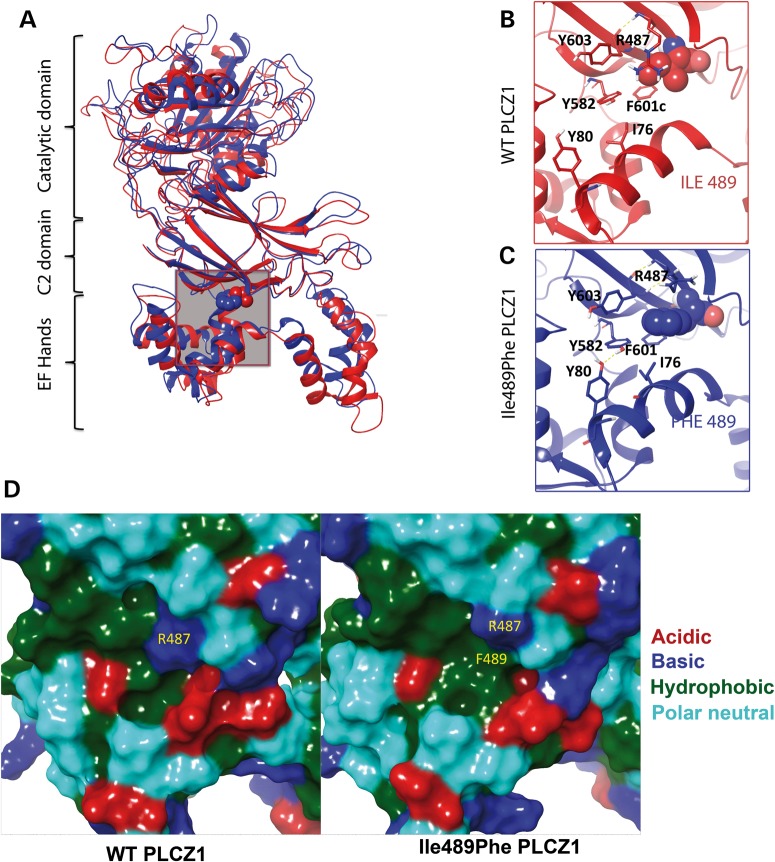
To assess the possible impact of the mutation on the structure of hPLCZ1, a 3D structure of hPLCZ1 was modeled from the crystallographic structure of rPLCδ1 (42% identity and 59% homology). The model shows that Ile489 is at the interface of the EF hand and C2 domains, and it appears as being too far from the catalytic domain to directly affect its function (Fig. 8A). Using molecular dynamics simulations of the hPLCZ1 and hPLCZ1-Ile489Phe models, we observed that the larger size of Phe over Ile (203 versus 169 Å3), which requires more accommodating space (Supplementary Material, Fig. S1), results first in a displacement of its intra-domain neighbors (Y582, F601, Y603 and R487) and second in establishing a unique inter-domain hydrophobic contact with I76 from the proximal EF-hand 2 helix (Fig. 8B and C). The first notable outcome of these perturbations is that causes a significant shift of the EF2 domain toward the C2 domain (12.2 versus 10.7 Å for CαI76–CαI489 and CαI76–CαF489 distances, respectively), which is reinforced by the newly formed H-bond between the newly displaced Y582 and Y80 of EF2 (Fig. 8C). Significantly, this tighter inter-domain arrangement returns back to the original looser status when Phe489 is mutated back to Ile489 in the course of a simulation (Supplementary Material, Fig. S2). The second notable outcome is the formation in the C2 domain of an aromatic-rich concave-like sub-site capable of associating with lipophilic molecules or protein surfaces (Fig. 8D). Because EF hands have been shown to be important for both PLCZ1 anchoring on phospholipids and its nuclear translocation (30,31), this unique new inter-domains interaction in the mutant enzyme could support the observed deleterious effects of the mutation.
Figure 8.
Molecular dynamic simulation of the effect of the Ile489Phe mutation on the conformation of PLCZ1. (A) The site of the Ile489Phe mutation is located at the interface of C2 and EF-hand domains of PLCZ1 (gray box) (B and C) Enlargement of the red/blue boxes for WT and mutant PLCZ1. As suggested by molecule dynamics simulation, the mutation results in a displacement of the surrounding C2 residues (Y582, F601, Y603 and R487) by the larger side chain of Phe. This reorganization, in turn, leads to novel contacts between C2 and EF2 hand via persistent hydrophobic (Phe489–Ile76) and H-bonding (Tyr582–Tyr80) side-chain interactions, which shifts the entire EF-hand domain toward C2 by an ∼1.5 Å by the end of the 1.2-ns simulation. (D) The molecular surfaces of WT and mutant PLCZ1 in the vicinity of the 489 residue, demonstrating the formation of a novel hydrophobic sub-site at the C2/EF-hand junction. The basic, acidic hydrophobic and polar neutral residues shown in blue, red, green and cyan, respectively.
Normal PAWP expression in patients with Ile489Phe PLCZ1 mutation
We next examined the sequence of WBP2NL, which encodes for PAWP. Analysis of exome data from P1 and P2 did not reveal any sequence variation in the WBP2NL-coding sequence, UTR regions or close intronic regions indicating that P1 and P2 should produce a fully functional PAWP protein. Because exome sequencing only allows covering 80–90% of all targeted sequences, WBP2NL coverage was verified: all exons and exon borders for both patients was at least 40×, which unambiguously confirmed the absence of deleterious variants in WBP2NL in the patients (Supplementary Material, Fig. S3). Consistent with these results, we confirmed by WB normal expression of PAWP (Fig. 9A) using extracts of sperm from patients and a fertile control prepared with similar number of sperm (Fig. 9B). We also examined PAWP localization by IF and in agreement with previous reports, PAWP reactivity appeared as a compact band around the equatorial/post-acrosomal area in the sperm of a control fertile human (Fig. 9C1) as well as in the sperm of both patients (Fig. 9C2). Altogether, these results show that PAWP is unable to support activation of human oocytes when PLCZ1 is absent and/or non-functional.
Figure 9.
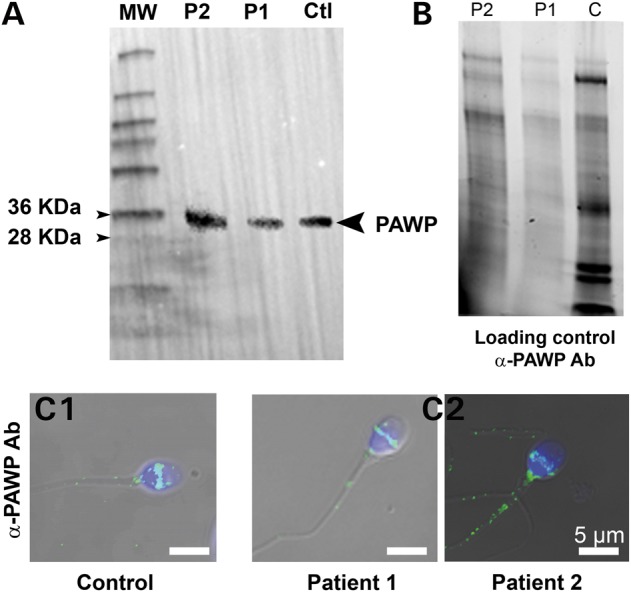
Normal expression and localization of PAWP in patients with OAF. (A) WB using protein extracts from sperm of a fertile control (Ctl), P1 and P2 and an anti-PAWP antibody. Immunoreactivity is observed in the lanes of control and patients' sperm extracts. (B) Protein gel representing loading control for (A) showing that all lanes were similarly loaded. Protein loads were controlled with TGX stain free™ precast gels. (C1) Sperm from a fertile control were stained with anti-PAWP antibody and counterstained with Hoechst. Overlay shows that the antibody stains strongly the post-acrosomal area. (C2) Similar staining of sperm from P1 and P2, showing the same reactivity to that of the control fertile sperm.
To extend these results, we examined the sperm from Dpy19l2 KO males. Dpy19l2 homozygous KO male mice are infertile, they have round-headed acrosomeless spermatozoa, which fail to induce oocyte activation following ICSI due to loss of Plcz1 (11,32). Here, we show by WB that PAWP expression is normal in these round-headed sperm (Supplementary Material, Fig. S4), demonstrating that its presence is not sufficient to trigger oocyte activation when ICSI is performed. Therefore, it appears the presence of PAWP is incapable of rescuing the lack of activation caused by the absence of PLCZ1 expression in human or mouse sperm.
Discussion
Infertility and Ile489Phe PLCZ1
Herein, we have used whole-exome sequencing to identify a homozygous missense mutation in PLCZ1 in two infertile brothers presenting OAF. The mutation led to an almost complete disappearance of the protein in the patients' sperm and based on the targeting and/or anchoring defects observed following injection of cRNAs into oocytes, we estimate it hampers the retention of PLCZ1 in mature sperm. During spermatogenesis, as in all cells, proteins are synthesized in the reticulum and targeted to their final location according to their function. However, the endoplasmic reticulum (ER) and all components necessary for proteins synthesis are eventually discarded at the end of the spermatozoon differentiation, leading to the release of a giant anucleate vesicle known as the residual body (33). We have previously shown that PLCZ1 is specifically located in the perinuclear theca in the vicinity of the inner acrosomal membrane of mature human sperm (11). The absence of PLCZ1 in the patients' sperm suggests that the mutation may prevent PLCZ1 from reaching and attaching to the perinuclear theca/inner acrosomal membrane, which renders it susceptible to disposal through the residual body. It is worth noting that PLCZ1 is also absent in globozoospermic sperm, which display defects in the perinuclear theca region and lack the acrosome vesicle (11). Moreover, we showed that the mutant protein had a strongly impaired ability to produce inositol 1,4,5-trisphosphate (IP3), as witnessed by the failure to induce Ca2+ oscillations and to sustain normal embryonic development, contrary to the WT protein. The OAF of these patients is thus due to the dramatic decrease of both PLCZ1 concentration and IP3 production.
Ile489Phe PLCZ1 and function of the C2 domain of PLCZ1
PLCs belong to a large family of enzymes able to bind to lipids in membranes where they hydrolyze-specific phospholipids, mostly PIP2. Several classical molecular domains are found in PLCs, including two catalytic domains, called X and Y, and working in tandem, lipid-binding domains such as PH and C2 and EF-hand domains, which are Ca2+-dependent domains (34). PLCZ1 is the shortest PLC and contains only four domains: four EF hands, the XY catalytic tandem and a C2 domain (Fig. 3) and the lack of a PH domain favor its more widespread distribution in the ooplasm. The C2 domain, which is the site of the homozygous mutation in our patients, is a lipid-binding domain shared by more than 100 human proteins. Originally, C2 domains were shown to bind membranes in a Ca2+-dependent manner (35,36). Crystallographic studies have shown that C2 domains have a common fold of conserved eight-stranded antiparallel β-sandwich (37,38), which, according to our modeling studies, are also present in PLCZ1 (Fig. 8). There are two areas important for lipid binding in this domain, which are the highly variable loops between β-strands and a cationic patch in the concave face of the β-sandwich that corresponds to β3- and β4-strands (39). However, the sequence conservation among C2 domains is low and a significant number of C2 domains with little to no Ca2+ affinity have been identified, which is the case for the C2 domain of PLCZ1 (39). Moreover, some C2 domains have low-membrane affinity and are involved in protein–protein interaction. Although it has been shown that PLCZ1 deleted of the full C2 domain does not produce Ca2+ oscillations, demonstrating thus that the C2 domain of PLCZ1 is necessary for the overall enzymatic activity (30,40), its molecular function remains uncharacterized so far. The Ile489Phe mutation, which is located in the β1-strand of the C2 domain, opens the door for structure–function studies to define how the conserved amino acids on the C2 domain are involved in protein targeting and selectivity for lipid substrate(s).
Our results, following injection of PLCZ1 cRNAs into oocytes, have showed that the WT protein was distributed evenly in the ooplasm and its area of distribution largely overlapped that of the ER, whereas the Ile489Phe PLCZ1 was unevenly distributed and accumulated around the nucleus. It is worth noting that the subcellular localization of C2-domain containing proteins is controlled by the lipid selectivity of the C2 domain and it is also known that lipid selectivity is highly variable among C2 domains (39,41). For instance, PLCδ is located in the phosphatidylserine (PS) rich PM because its C2 domain selectively binds PS (26), whereas cPLA2α translocates to the perinuclear ER, which is rich in phosphatidylcholine, the target of its C2 domain (41). We can, therefore, speculate that the C2 domain of lle489Phe PLCZ1 displays changed lipid affinity, which alters the intracellular distribution of the enzyme, and this may conspire to its retention in mature sperm and its reduced catalytic activity in oocytes. Altogether, our results suggest for the first time that the C2 domain of PLCZ1 participates in the targeting of the enzyme to lipid-containing membranes and our results demonstrate a pivotal role of a stretch of highly conserved residues in the C2 domain surrounding Ile489. It is also the first description of a functional role for residues located in the β1-strand of a C2 domain (42).
Our molecular modeling studies of WT and mutant PLCZ1 offers insights into the atomic-level perturbations caused by the Ile489Phe mutation. Dynamic simulations showed the Ile489Phe mutation is likely to induce a shift of the EF2-hand helix 1 toward C2, and given that EF2 hand is involved in binding to lipid membrane (30,31), this shift may reduce enzymatic activity. Further, the opening of a new aromatic-rich surface patch in the mutant protein capable of associating with additional hydrophobic counterparts may also alter the distribution and/or activity of the enzyme. Finally, the nuclear translocation of the mIle527Phe Plcz1 is hampered, suggesting that binding to nuclear import proteins necessary for PN translocation is defective (43), which may suggest a role of C2 domain in protein–protein interactions. Therefore, by changing the structural properties of the C2 domain and its interaction with the EF-hand domains, Ile489Phe-mutation is likely to modify the interactions of PLCZ1 with lipid membranes and possibly proteins, directly affecting its targeting and/or anchoring properties and enzymatic activity, all of which undermine the fertility of patients bearing this mutation.
PLCZ1 and oocyte activation
The function of PLCZ1 as the sperm factor has recently been challenged, and another protein, PAWP, has been proposed as an alternative candidate (15,16). Controversial results, however, have been published (18,19) and it is presently unclear how this protein is involved in human fertilization. The study of genetic diseases often provides the opportunity to better understand protein functions and our understanding of reproduction, including gametogenesis and fertilization, has benefited from the genetic characterization of several phenotypes of male and female infertilities (44). In human, a link between OAF and PLCZ1 was first reported in patients displaying abnormal expression and localization of PLCZ1 in sperm (7). However, these patients exhibited severe teratozoospermia, which raised concerns about miss-expression and/or malfunction of several proteins. Heytens et al. (8) provided the first genetic evidence linking PLCZ1 to infertility, as they found a heterozygous mutation at position 398 of the Y catalytic domain of PLCZ1 that reduced its ability to induce Ca2+ oscillations. Nevertheless, it was unclear how this heterozygous mutation could cause infertility, although, subsequently, another heterozygous mutation was found in the same patient at position 233 of the X catalytic domain, resulting in compound heterozygosity, which reinforced the link between PLCZ1, OAF and infertility (45). Here, we show the first identified homozygous mutation of PLCZ1 leading to OAF and infertility. Importantly, it is the first exomic analysis of patients presenting OAF. We identified only four homozygous variants shared by both brothers: three of them were not expected to have any deleterious effects, which is in contrast to the missense mutation found in PLCZ1. Importantly, this mutation was not associated with teratozoospermia and the numbers of normal sperm were well above the lower accepted reference values (21). Moreover, the sperm of both brothers showed only slightly reduced DNA quality, ruling this out as the cause of OAF (46,47). Altogether, these results indicate that PLCZ1 plays a direct and primary role in the activation of mammalian oocytes. It is worth noting that a Plcz1 KO animal model does not presently exist, due to an early spermatogenesis arrest in this model (13), and this absence has been used to raise doubts about the role of PLCZ1 in oocyte activation. Here, we present the first functional knock-down of PLCZ1 in human sperm without effects on spermatogenesis and the results demonstrate a required role for oocyte activation and fertility in this species. Finally, unlike PLCZ1, the expression and localization of PAWP were unchanged, dismissing its contribution to the phenotype of our patients. This conclusion was reinforced by our results showing that sperm from the Dpy19l2 KO mouse model despite exhibiting normal PAWP expression cannot induce oocyte activation after ICSI, as they lack Plcz1 (11,32). These results are also consistent with the report showing that PAWP null mice are fertile, their sperm show no morphological defects and that they can trigger oocyte activation (48). Our results thus do not support the notion that PAWP triggers Ca2+ release and oocyte activation (15), and instead confirm the importance of PLCZ1 for this function in human.
In summary, whole-exome sequencing of two infertile brothers identified for the first time a homozygous mutation in the C2 domain of PLCZ1 responsible for ICSI failure and infertility. We also show PAWP expression was unaffected in these patients indicating it is unable to induce Ca2+ responses or oocyte activation. Therefore, given the required role of PLCZ1 for fertility and our findings demonstrating the significance of specific residues in the C2 domain for the enzyme's function, future studies should establish the molecular target(s) in the ooplasm and the host organelle that guide the enzyme to sites of accessible and abundant substrate, which is required to support the long-lasting oscillations that underlie egg activation in mammals.
Materials and Methods
Biological samples
Sperm were obtained following informed consent from patients consulting with the Department of fertility at Grenoble (France) or with the Clinique des Jasmins (Tunis, Tunisia) after approval by the Ethics committee of the university. In addition, all patients gave informed consent for preservation of unused sperm in the Germetheque biobank and subsequent use for studies on human fertility in accordance with the Helsinki Declaration on human experimentation. Dpy19l2 KO mice were obtained from the Mutant Mouse Regional Resource Center, University of California, Davis, CA, USA.
Exome sequencing and bioinformatics analysis
Genomic DNA was isolated from saliva using Oragen DNA extraction kit (DNAgenotech®, Ottawa, Canada). Coding regions and intron/exon boundaries were enriched using the ‘all Exon V5 kit’ (Agilent Technologies, Wokingham, UK). DNA sequencing was undertaken at the Genoscope, Evry, France, on the HiSeq 2000 from Illumina®. Sequence reads were aligned to the reference genome (hg19) using MAGIC (49). Duplicate reads and reads that mapped to multiple locations in the exome were excluded from further analysis. Positions whose sequence coverage was <10 on either the forward or reverse strand were excluded. Single-nucleotide variations and small insertions/deletions (indels) were identified and quality-filtered using in-house scripts. The most promising candidate variants were identified using an in-house bioinformatics pipeline. Variants with a minor allele frequency >5% in the NHLBI ESP6500 or in 1000 Genomes Project phase 1 data sets, or >1% in ExAC, were discarded. We also compared these variants with an in-house database of 56 control exomes obtained from subjects from the same geographic origin as our two patients (North Africa). All variants present in homozygous state in this database were excluded. We used variant effect predictor to predict the impact of the selected variants. We only retained variants impacting splice donor/acceptor or causing frameshift, inframe insertions/deletions, stop gain, stop loss or missense variants except those scored as ‘tolerated’ by SIFT (sift.jcvi.org) and as ‘benign’ by Polyphen-2 (genetics.bwh.harvard.edu/pph2).
Sanger sequencing
The presence of the identified variation was verified by Sanger sequencing of PLCZ1 exon 13. Primers were as followed: PLCZ1_14F: TCAATGTTTGTGGGAGCTGA and PLCZ1_14R: GGACATAATGGAAAACCCTTG. Thirty-five cycles of polymerase chain reaction amplification were carried out with an hybridization temperature of 60°C. Sequencing reactions were carried out with BigDye Terminator v3.1 (Applied Biosystems). Sequence analysis were carried out on ABI 3130XL (Applied Biosystems).
Primary antibodies
Anti-human acrosin and PAWP antibodies were from Sigma-Aldrich and Proteintech, respectively; anti-PLCZ1 antibodies were raised against a 15-mer peptide sequence (305KETHERKGSDKRGDN319) of the human PLCZ1 (7), affinity purified and stored at a concentration of 1 µg/µl and used at 1/100 for IF and 1/1000 for WB.
Western blotting
WB was carried out as described by our laboratory (11). Briefly, sperm were first washed in phosphate-buffered saline (PBS) and resuspended in Laemmli sample buffer and boiled. Protein extracts equivalent to 1–2 × 106 sperm were loaded per lane into 4–20% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and resolved proteins were transferred onto PVDF membranes. The membranes were blocked and incubated overnight with anti-PLCZ1 Ab (1/1000) and next with horseradish peroxidase labeled secondary Ab (1 h). Immunoreactivity was detected using chemiluminescence detection kit reagents and a Chimidoc™ Station (Bio-Rad).
Immunofluorescence
IF was carried out as described by our laboratory (11). Sperm were fixed with 4% paraformaldehyde, washed in PBS, and spotted onto poly-L-lysine precoated slides. Sperm were then permeabilized with 0.1% (v/v) Triton X-100. Slides were then blocked in 5% normal goat serum–Dulbecco's PBS (DPBS) (GIBCO, Invitrogen) and incubated overnight at 4°C with primary antibodies. Washes were performed with 0.1% (v/v) Tween 20–DPBS, followed by 1 h incubation with secondary Ab (1/400). Samples were counterstained with Hoechst 33342 and mounted with DAKO mounting media (Life Technology). Fluorescence images were captured with a confocal microscope (Zeiss LSM 710).
Generation of constructs and preparation of cRNA
Human and mouse PLCZ1 constructs were kind gifts from Dr K. Fukami (Tokyo University of Pharmacy and Life Science, Japan) and Dr K. Jones (University of Southampton, UK), respectively. WT h and mPLCZ1-venus sequences were subcloned into a pcDNA6/Myc-His B (Invitrogen) between EcoRI and XbaI restrictions sites. hPLCZ1-Ile489Phe and mPlcz1 Ile527Phe were generated by substituting Ile to Phe using the Gibson Assembly Cloning Kit (New England Bio Labs), as previously reported (50). All constructs were finally sequenced. pDsRed2-ER was kindly provided by Dr M Trebak (Pen State Hershey College of Medicine, PA, USA). The ER-targeting sequence of calreticulin, DsRed2 and the KDEL ER retention sequence were ligated to pcDNA6/Myc-His B. Plasmids were linearized outside of the coding region with PmeI and in vitro transcribed using mMESSAGE/mMACHINE T7 Kit (Ambion). Poly-A tail was added to the mRNAs using a Tailing Kit (Ambion).
Confocal microscopy of florescent-PLCZ1
Live-cell images of oocytes and zygotes expressing fluorescently tagged proteins were captured with a confocal laser-scanning microscope (LSM 510 META, Carl Zeiss) using a 63 × 1.4 numerical aperture oil-immersion objective lens. Images were reconstructed using the LSM software (Carl Zeiss). Oocytes were maintained in HCZB medium and those expressing Venus-hPLCZ1 and DsRed-ER proteins were imaged at the GV stage, whereas expression of Venus-mPlcz1 and DsRed-ER proteins was imaged in PN stage zygotes.
Ca2+ monitoring
Ca2+ monitoring was carried out as described by our laboratory (7). Briefly, mouse oocytes were loaded with fura-2-AM (molecular probes) prior to injecting the cRNAs, after which they were transferred into a monitoring dish containing 50 µl drops of TL–HEPES medium under mineral oil. Excitation wavelengths of 340/380 nm were alternated using a filter wheel (Ludl Electronic Products Ltd). Fluorescence ratios were taken every 20 s. After passing through a 510 nm barrier filter, the emitted light was collected by a cooled Photometrics SenSys CCD camera (Roper Scientific). The SimplePCI imaging software was used to run all the hardware and capture images (Hamamatsu). [Ca2+]i values are reported as the ratio of 340/380 nm fluorescence in the whole oocyte.
Parthenogenetic oocyte activation
Oocytes were activated by injection of PLCZ1 cRNA into the ooplasm (concentration as indicated). After cRNA microinjection, oocytes were cultured in KSOM/EAA, supplemented with 5 µg/ml cytochalasin B to diploidize the parthenotes. The injection volume was ∼5–10 pl, which is ∼1–3% of the total oocyte volume. PN formation was checked 6 h after injection and development was followed up to the blastocyst stage.
Homology modeling
A homology model for hPLCZ1 was generated with the Prime homology modeling workflow (version 3.8, Schrödinger, LLC) using the structure of rPLCδ1, in complex with inositol-1,4,5-trisphosphate and associated Ca2+ (PDB code: 1DJX) as the template. Briefly, all sequences (1DJX, rPLCD1 and hPLCZ1) were initially aligned by the BLAST homology search. First, a contiguous template structure was generated by replacing the missing 445–486 loop in the rPLCD1 structure with a peptide bond between the proximal (per 1DJX) G444 and K487 and minimization of the resulting loop using the OPLS2005 force field within Prime. Next, a similar operation resulted in the exclusion of the basic loop from the catalytic domain of hPLCZ1 (304–344) by linking residues 303 and 345 to recreate a contiguous mode. A single-chain, liganded model, comprising the residues 64–303 and 345–608, was then built by the energy-based algorithm in Prime to construct and refine non-identical residues and loops with deletions and insertions.
Molecular dynamics simulations
Both the resulting complex and pre-minimized I489F mutant were subjected to an unconstrained 1.2 ns, molecular dynamics procedure within Desmond (version 4.0, D. E. Shaw Research & Schrödinger, LLC). An NPT ensemble was built at 300 K and 1 atm with the neutralized (by 10 Cl– ions) system and further Na+/Cl– ions to simulate 150 mm concentration in an explicit SPC solvent model. The interactions that both I489 and the corresponding phenylalanine were participating throughout the simulations were identified using the simulated interaction diagram routine within Desmond. Topological changes were detected using simulated event analysis program by monitoring the CαI76–CαI489 and CαI76–CαF489 distances and root mean square fluctuations as a function of simulation time. The mutant structure from the final frame was modified back to contain I489, and a simulation under the identical conditions was conducted to ascertain the persistence of the mutant's conformation.
Supplementary Material
Authors’ Contributions
R.Z., L.H. and C.T. identified patients, J.E., S.Y., H.C.L. and G.M. performed research; C.C., T.K., Z.K., N.T.-M. contributed to genetic analyses; S.N.S. performed PLCZ1 structure analyses; R.F., P.F.R. and C.A. designed research, analyzed data and wrote the paper.
Funding
This work was supported by following grants: ANR Genopat 2009, project ICG2I to P.R. and C.A. and NIH R01 HD051872 to R.A.F.
Supplementary Material
Acknowledgements
We thank S. Crouzy (CEA Grenoble) for valuable discussions.
Conflict of Interest statement. None declared.
References
- 1.Steinhardt R.A., Epel D., Carroll E.J. Jr., Yanagimachi R. (1974) Is calcium ionophore a universal activator for unfertilised eggs? Nature, 252, 41–43. [DOI] [PubMed] [Google Scholar]
- 2.Ridgway E.B., Gilkey J.C., Jaffe L.F. (1977) Free calcium increases explosively in activating medaka eggs. Proc. Natl Acad. Sci. USA, 74, 623–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swann K. (1990) A cytosolic sperm factor stimulates repetitive calcium increases and mimics fertilization in hamster eggs. Development, 110, 1295–1302. [DOI] [PubMed] [Google Scholar]
- 4.Stice S.L., Robl J.M. (1990) Activation of mammalian oocytes by a factor obtained from rabbit sperm. Mol. Reprod. Dev., 25, 272–280. [DOI] [PubMed] [Google Scholar]
- 5.Parrington J., Jones K.T., Lai A., Swann K. (1999) The soluble sperm factor that causes Ca2+ release from sea-urchin (Lytechinus pictus) egg homogenates also triggers Ca2+ oscillations after injection into mouse eggs. Biochem. J., 341, 1–4. [PMC free article] [PubMed] [Google Scholar]
- 6.Jones K.T., Cruttwell C., Parrington J., Swann K. (1998) A mammalian sperm cytosolic phospholipase C activity generates inositol trisphosphate and causes Ca2+ release in sea urchin egg homogenates. FEBS Lett., 437, 297–300. [DOI] [PubMed] [Google Scholar]
- 7.Yoon S.Y., Jellerette T., Salicioni A.M., Lee H.C., Yoo M.S., Coward K., Parrington J., Grow D., Cibelli J.B., Visconti P.E. et al. (2008) Human sperm devoid of PLC, zeta 1 fail to induce Ca(2+) release and are unable to initiate the first step of embryo development. J. Clin. Invest., 118, 3671–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heytens E., Parrington J., Coward K., Young C., Lambrecht S., Yoon S.Y., Fissore R.A., Hamer R., Deane C.M., Ruas M. et al. (2009) Reduced amounts and abnormal forms of phospholipase C zeta (PLCzeta) in spermatozoa from infertile men. Hum. Reprod., 24, 2417–2428. [DOI] [PubMed] [Google Scholar]
- 9.Taylor S.L., Yoon S.Y., Morshedi M.S., Lacey D.R., Jellerette T., Fissore R.A., Oehninger S. (2010) Complete globozoospermia associated with PLCzeta deficiency treated with calcium ionophore and ICSI results in pregnancy. Reprod. Biomed. Online, 20, 559–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amdani S.N., Jones C., Coward K. (2013) Phospholipase C zeta (PLCzeta): oocyte activation and clinical links to male factor infertility. Adv. Biol. Regul., 53, 292–308. [DOI] [PubMed] [Google Scholar]
- 11.Escoffier J., Yassine S., Lee H.C., Martinez G., Delaroche J., Coutton C., Karaouzene T., Zouari R., Metzler- Guillemain C., Pernet- Gallay K. et al. (2015) Subcellular localization of phospholipase Czeta in human sperm and its absence in DPY19L2-deficient sperm are consistent with its role in oocyte activation. Mol. Hum. Reprod., 21, 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nomikos M., Kashir J., Swann K., Lai F.A. (2013) Sperm PLCzeta: from structure to Ca2+ oscillations, egg activation and therapeutic potential. FEBS Lett., 587, 3609–3616. [DOI] [PubMed] [Google Scholar]
- 13.Ito M., Nagaoka K., Kuroda K., Kawano N., Yoshida K., Harada Y., Shikano T., Miyado M., Oda S., Toshimori K. et al. (2010) Arrest of spermatogenesis at round spermatids in PLCz1 deficient mice. Abstracts of the 11th International Symposium on Spermatology; June 26– 29, 2010; Okinawa, Japan. [Google Scholar]
- 14.Aarabi M., Yu Y., Xu W., Tse M.Y., Pang S.C., Yi Y.J., Sutovsky P., Oko R. (2012) The testicular and epididymal expression profile of PLCzeta in mouse and human does not support its role as a sperm-borne oocyte activating factor. PLoS One, 7, e33496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aarabi M., Balakier H., Bashar S., Moskovtsev S.I., Sutovsky P., Librach C.L., Oko R. (2014) Sperm-derived WW domain-binding protein, PAWP, elicits calcium oscillations and oocyte activation in humans and mice. FASEB J., 28, 4434–4440. [DOI] [PubMed] [Google Scholar]
- 16.Aarabi M., Balakier H., Bashar S., Moskovtsev S.I., Sutovsky P., Librach C.L., Oko R. (2014) Sperm content of postacrosomal WW binding protein is related to fertilization outcomes in patients undergoing assisted reproductive technology. Fertil. Steril., 102, 440–447. [DOI] [PubMed] [Google Scholar]
- 17.Wu A.T., Sutovsky P., Manandhar G., Xu W., Katayama M., Day B.N., Park K.W., Yi Y.J., Xi Y.W., Prather R.S., Oko R. (2007) PAWP, a sperm-specific WW domain-binding protein, promotes meiotic resumption and pronuclear development during fertilization. J. Biol. Chem., 282, 12164–12175. [DOI] [PubMed] [Google Scholar]
- 18.Nomikos M., Sanders J.R., Theodoridou M., Kashir J., Matthews E., Nounesis G., Lai F.A., Swann K. (2014) Sperm-specific post-acrosomal WW-domain binding protein (PAWP) does not cause Ca2+ release in mouse oocytes. Mol. Hum. Reprod., 20, 938–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nomikos M., Sanders J.R., Kashir J., Sanusi R., Buntwal L., Love D., Ashley P., Sanders D., Knaggs P., Bunkheila A. et al. (2015) Functional disparity between human PAWP and PLCzeta in the generation of Ca2+ oscillations for oocyte activation. Mol. Hum. Reprod., 21, 702–710. [DOI] [PubMed] [Google Scholar]
- 20.Brawand D., Soumillon M., Necsulea A., Julien P., Csardi G., Harrigan P., Weier M., Liechti A., Aximu-Petri A., Kircher M. et al. (2011) The evolution of gene expression levels in mammalian organs. Nature, 478, 343–348. [DOI] [PubMed] [Google Scholar]
- 21.World Health Organization (2010) WHO laboratory manual for the Examination and processing of human semen. 5th edn WHO Press, Geneva, Switzerland. [Google Scholar]
- 22.Sharma R.K., Sabanegh E., Mahfouz R., Gupta S., Thiyagarajan A., Agarwal A. (2010) TUNEL as a test for sperm DNA damage in the evaluation of male infertility. Urology, 76, 1380–1386. [DOI] [PubMed] [Google Scholar]
- 23.Ribas-Maynou J., Garcia-Peiro A., Fernandez-Encinas A., Abad C., Amengual M.J., Prada E., Navarro J., Benet J. (2013) Comprehensive analysis of sperm DNA fragmentation by five different assays: TUNEL assay, SCSA, SCD test and alkaline and neutral Comet assay. Andrology, 1, 715–722. [DOI] [PubMed] [Google Scholar]
- 24.Erenpreiss J., Bars J., Lipatnikova V., Erenpreisa J., Zalkalns J. (2001) Comparative study of cytochemical tests for sperm chromatin integrity. J. Androl., 22, 45–53. [PubMed] [Google Scholar]
- 25.Lee H.C., Arny M., Grow D., Dumesic D., Fissore R.A., Jellerette-Nolan T. (2014) Protein phospholipase C Zeta1 expression in patients with failed ICSI but with normal sperm parameters. J. Assist. Reprod. Genet., 31, 749–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ananthanarayanan B., Das S., Rhee S.G., Murray D., Cho W. (2002) Membrane targeting of C2 domains of phospholipase C-delta isoforms. J. Biol. Chem., 277, 3568–3575. [DOI] [PubMed] [Google Scholar]
- 27.Yu Y., Nomikos M., Theodoridou M., Nounesis G., Lai F.A., Swann K. (2012) PLCzeta causes Ca(2+) oscillations in mouse eggs by targeting intracellular and not plasma membrane PI(4,5)P(2). Mol. Biol. Cell., 23, 371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoda A., Oda S., Shikano T., Kouchi Z., Awaji T., Shirakawa H., Kinoshita K., Miyazaki S. (2004) Ca2+ oscillation-inducing phospholipase C zeta expressed in mouse eggs is accumulated to the pronucleus during egg activation. Dev. Biol., 268, 245–257. [DOI] [PubMed] [Google Scholar]
- 29.Larman M.G., Saunders C.M., Carroll J., Lai F.A., Swann K. (2004) Cell cycle-dependent Ca2+ oscillations in mouse embryos are regulated by nuclear targeting of PLCzeta. J. Cell Sci., 117, 2513–2521. [DOI] [PubMed] [Google Scholar]
- 30.Kuroda K., Ito M., Shikano T., Awaji T., Yoda A., Takeuchi H., Kinoshita K., Miyazaki S. (2006) The role of X/Y linker region and N-terminal EF-hand domain in nuclear translocation and Ca2+ oscillation-inducing activities of phospholipase Czeta, a mammalian egg-activating factor. J. Biol. Chem., 281, 27794–27805. [DOI] [PubMed] [Google Scholar]
- 31.Nomikos M., Sanders J.R., Parthimos D., Buntwal L., Calver B.L., Stamatiadis P., Smith A., Clue M., Sideratou Z., Swann K., Lai F.A. (2015) Essential role of the EF-hand domain in targeting sperm phospholipase Czeta to membrane PIP2. J. Biol. Chem., 290, 29519–29530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yassine S., Escoffier J., Martinez G., Coutton C., Karaouzene T., Zouari R., Ravanat J.L., Metzler-Guillemain C., Fissore R., Hennebicq S. et al. (2015) Dpy19l2-deficient globozoospermic sperm display altered genome packaging and DNA damage that compromises the initiation of embryo development. Mol. Hum. Reprod., 21, 169–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breucker H., Schafer E., Holstein A.F. (1985) Morphogenesis and fate of the residual body in human spermiogenesis. Cell Tissue Res., 240, 303–309. [DOI] [PubMed] [Google Scholar]
- 34.Saunders C.M., Larman M.G., Parrington J., Cox L.J., Royse J., Blayney L.M., Swann K., Lai F.A. (2002) PLC zeta: a sperm-specific trigger of Ca(2+) oscillations in eggs and embryo development. Development, 129, 3533–3544. [DOI] [PubMed] [Google Scholar]
- 35.Bommert K., Charlton M.P., DeBello W.M., Chin G.J., Betz H., Augustine G.J. (1993) Inhibition of neurotransmitter release by C2-domain peptides implicates synaptotagmin in exocytosis. Nature, 363, 163–165. [DOI] [PubMed] [Google Scholar]
- 36.Clark J.D., Lin L.L., Kriz R.W., Ramesha C.S., Sultzman L.A., Lin A.Y., Milona N., Knopf J.L. (1991) A novel arachidonic acid-selective cytosolic PLA2 contains a Ca(2+)-dependent translocation domain with homology to PKC and GAP. Cell, 65, 1043–1051. [DOI] [PubMed] [Google Scholar]
- 37.Sutton R.B., Davletov B.A., Berghuis A.M., Sudhof T.C., Sprang S.R. (1995) Structure of the first C2 domain of synaptotagmin I: a novel Ca2+/phospholipid-binding fold. Cell, 80, 929–938. [DOI] [PubMed] [Google Scholar]
- 38.Essen L.O., Perisic O., Cheung R., Katan M., Williams R.L. (1996) Crystal structure of a mammalian phosphoinositide-specific phospholipase C delta. Nature, 380, 595–602. [DOI] [PubMed] [Google Scholar]
- 39.Cho W., Stahelin R.V. (2006) Membrane binding and subcellular targeting of C2 domains. Biochim. Biophys. Acta., 1761, 838–849. [DOI] [PubMed] [Google Scholar]
- 40.Kouchi Z., Shikano T., Nakamura Y., Shirakawa H., Fukami K., Miyazaki S. (2005) The role of EF-hand domains and C2 domain in regulation of enzymatic activity of phospholipase Czeta. J. Biol. Chem., 280, 21015–21021. [DOI] [PubMed] [Google Scholar]
- 41.Stahelin R.V., Rafter J.D., Das S., Cho W. (2003) The molecular basis of differential subcellular localization of C2 domains of protein kinase C-alpha and group IVa cytosolic phospholipase A2. J. Biol. Chem., 278, 12452–12460. [DOI] [PubMed] [Google Scholar]
- 42.Corbalan-Garcia S., Gomez-Fernandez J.C. (2014) Signaling through C2 domains: more than one lipid target. Biochim. Biophys. Acta., 1838, 1536–1547. [DOI] [PubMed] [Google Scholar]
- 43.Piazzi M., Blalock W.L., Bavelloni A., Faenza I., D'Angelo A., Maraldi N.M., Cocco L. (2013) Phosphoinositide-specific phospholipase C beta 1b (PI-PLCbeta1b) interactome: affinity purification-mass spectrometry analysis of PI-PLCbeta1b with nuclear protein. Mol. Cell Proteomics, 12, 2220–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coutton C., Escoffier J., Martinez G., Arnoult C., Ray P.F. (2015) Teratozoospermia: spotlight on the main genetic actors in the human. Hum. Reprod. Update, 21, 455–485. [DOI] [PubMed] [Google Scholar]
- 45.Kashir J., Konstantinidis M., Jones C., Lemmon B., Chang L.H., Hamer R., Heindryckx B., Deane C.M., De Sutter P., Fissore R.A. et al. (2012) A maternally inherited autosomal point mutation in human phospholipase C zeta (PLCzeta) leads to male infertility. Hum. Reprod., 27, 222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martianov I., Brancorsini S., Catena R., Gansmuller A., Kotaja N., Parvinen M., Sassone-Corsi P., Davidson I. (2005) Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc. Natl Acad. Sci. USA, 102, 2808–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao M., Shirley C.R., Hayashi S., Marcon L., Mohapatra B., Suganuma R., Behringer R.R., Boissonneault G., Yanagimachi R., Meistrich M.L. (2004) Transition nuclear proteins are required for normal chromatin condensation and functional sperm development. Genesis, 38, 200–213. [DOI] [PubMed] [Google Scholar]
- 48.Satouh Y., Nozawa K., Ikawa M. (2015) Sperm postacrosomal WW domain-binding protein is not required for mouse egg activation. Biol. Reprod., 93, 94–100. [DOI] [PubMed] [Google Scholar]
- 49.SEQC/MAQC-III Consortium (2014) A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat. Biotechnol., 32, 903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A. III, Smith H.O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods, 6, 343–345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.