Abstract
Recently, we marked TRIO for the first time as a candidate gene for intellectual disability (ID). Across diverse vertebrate species, TRIO is a well-conserved Rho GTPase regulator that is highly expressed in the developing brain. However, little is known about the specific events regulated by TRIO during brain development and its clinical impact in humans when mutated. Routine clinical diagnostic testing identified an intragenic de novo deletion of TRIO in a boy with ID. Targeted sequencing of this gene in over 2300 individuals with ID, identified three additional truncating mutations. All index cases had mild to borderline ID combined with behavioral problems consisting of autistic, hyperactive and/or aggressive behavior. Studies in dissociated rat hippocampal neurons demonstrated the enhancement of dendritic formation by suppressing endogenous TRIO, and similarly decreasing endogenous TRIO in organotypic hippocampal brain slices significantly increased synaptic strength by increasing functional synapses. Together, our findings provide new mechanistic insight into how genetic deficits in TRIO can lead to early neuronal network formation by directly affecting both neurite outgrowth and synapse development.
Introduction
Intellectual disability (ID), defined as an IQ of ≤70, has an estimated prevalence of 2–3% in the population (1). The genetic etiology of ID is highly heterogeneous, with to date approximately >700 genes known to be associated with this common disorder (2), the majority of which are very rarely mutated. Novel sequencing approaches, such as massive parallel sequencing, have proven to be successful in identifying novel genes for Mendelian disorders (3). Especially family-based whole exome sequencing (WES) in proband and parents has been instrumental to identify de novo pathogenic variants in sporadic cases with ID, thereby increasing the diagnostic yield in patients with severe ID (IQ <50) to up to 36% (4–10). For mild and borderline ID, however, family-based sequencing is more complex as the distinction between a normal or a mildly affected parent can be difficult to make. This complicates the interpretation of variants from family-based WES as the phenotype can also be the result of inherited variants. To determine the role of mutations in candidate ID genes in individuals with ID, it is necessary to find additional individuals with a mutation in the same gene and a comparable phenotype. Furthermore, interpretation can be supported by functional studies to investigate the mutational effect on protein function. Recently, we marked TRIO (MIM 601893; NM_007118.2) for the first time as one of these candidate genes for ID, based on the identification of two potentially pathogenic de novo missense mutations in this gene in independent individuals with severe ID (6). Both individuals, however, also carried a second de novo mutation in a known ID gene, TCF4 and another candidate ID gene, GFPT2, respectively, complicating a straightforward interpretation of the contribution of the TRIO mutation to the patients' phenotype (6,11). In a routine diagnostic setting using a genomic microarray, we also identified an individual with mild developmental delay carrying an 235 kb intragenic de novo deletion, disrupting TRIO (12) [chr5:14160447–14395478 (hg19)]. Finally, seven de novo mutations were found in TRIO in the context of large scale sequencing projects focused on various neurodevelopmental conditions, including ID, epilepsy and autism (8,13,14). Whereas these seven de novo mutations were not reported as conclusive cause of disease, they do support a possible role for TRIO in the development of ID (Fig. 1B).
Figure 1.
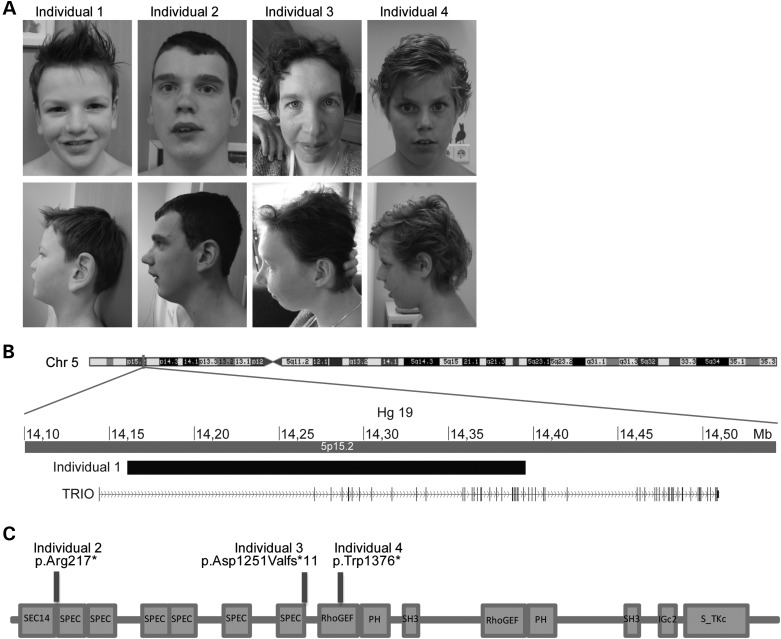
Individuals with LoF TRIO mutations. (A) Frontal and lateral photographs of Individual 1 with the deletion disrupting TRIO and Individuals 2–4 with LoF mutations in TRIO. Only mild facial dysmorphisms were observed. (B) Schematic overview of the 235 kb de novo deletion on chromosome 5, partially disrupting TRIO in Individual 1. (C) Schematic overview of TRIO, including the known domains (N-terminal SEC14 domain, several spectrin repeats, two DH-PH Rho-GEF units, Ig-like domain and C-terminal putative serine/threonine kinase domain). The positions of the three identified mutations in Individuals 2–4 (p.Trp1376*, p.Asp1251Valfs*11 and p.(Arg217*) are depicted.
TRIO is a large protein encoded by 57 coding exons (3097 amino acids) and containing several domains, including an N-terminal SEC14 domain, several spectrin repeats, two Dbl-homology-Pleckstrin-homology (DH-PH) Rho-guanine exchange factor (GEF) units, an Ig-like domain, and a C-terminal serine/threonine kinase domain (Fig. 1C). The serine/threonine kinase domain is presumed to be active but quite selective; both DH-PH Rho-GEF domains are enzymatically active (15,16). The first DH-PH domain has been shown to activate Rac1 and RhoG, whereas the second DH-DP domain activates RhoA (17,18). Rho GTPases regulate changes in cell morphology in response to many factors including stimulation by extracellular ligands. They are activated by GEF catalyzing GDP dissociation and allowing the binding of GTP (19). Interestingly, Rho GTPase signaling pathways have been identified as a major hub-signaling pathway in ID, affecting synaptic structure and function (20,21). In Drosophila, TRIO has been shown to be involved in axon guidance and dendritic arborization (22–24) during neuronal development. Additionally, TRIO is required for axonal growth in vitro and in vivo (25–27) and may contribute to the regulation of cell–cell contact (28,29). Yet, surprisingly little is known about the function of TRIO in early dendrite formation and synaptic function in the mammalian system (20).
Here, we report the identification of four cases with intragenic truncating mutations in TRIO, presenting with mild to borderline ID combined with behavioral problems consisting of autistic, hyperactive and/or aggressive behavior. Additionally, we show that during early neurodevelopment TRIO negatively regulates neurite outgrowth and synaptic strength by interfering with α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPAr) endocytosis.
Results
Identification of individuals with mutations in TRIO
Based on the identification of multiple de novo mutations affecting TRIO, we decided to perform targeted sequencing for TRIO in >2300 patients with ID using molecular inversion probes (MIPs) (in the ‘Materials and Methods’ section) (30). Filtering for putative loss-of-function (LoF) mutations (initiator codon, nonsense, frameshift and canonical splice sites), we identified three additional TRIO truncating mutations (Fig. 1C), including one de novo nonsense mutation, c.4128G>A, predicted to result in p.(Trp1376*), in a 10-year-old boy; a de novo frameshift mutation, c.3752del, predicted to result in p.(Asp1251Valfs*11) in a 35-year-old woman and a nonsense mutation, c.649A>T leading to p.(Arg217*) in a 20-year-old male. The latter mutation was inherited from a similarly affected father (Supplementary Material, Fig. S1). This family also has a second child with learning difficulties and behavioral problems, who did not show the mutation upon segregation analysis, suggesting that his phenotype may have another (genetic or environmental) origin. A third, unaffected brother, did not show the variant either. DNA of a fourth brother, the grandparents and/or the 13 siblings from the father, was unavailable for testing. All these individuals, however, appeared to be normal.
As also de novo missense mutations have been implicated in ID (6), we subsequently selected for rare, possible pathogenic (combined annotation dependent depletion score ≥ 20), missense mutations in TRIO detected in our cohort. This analysis yielded seven missense variants (Supplementary Material, Table S1), for which three could be tested for segregation in parental samples. Two of them appeared inherited from an unaffected parent, whereas the other was also identified in a father with borderline ID and behavioral problems, similar to his child. For the remaining four missense variants parental samples were not available. As from these data the impact of (de novo) missense mutations cannot unambiguously be assessed, we decided to further focus on the LoF aspects of TRIO.
De novo truncating mutations have not been observed in over 2000 healthy controls (14), but 13 different LoF variants are reported in ExAC, a large databases collecting next generation sequencing variants as proxy for the general population (Supplementary Material, Table S2) [URL: http://exac.broadinstitute.org (November 2015)]. Notably, only 2 of 13 have been observed more than once (highest minor allele frequency 3.11E-04). These two variants represent a single-base deletion and -duplication at the same genomic location, in a homopolymer stretch, potentially suggesting a sequencing and/or mapping artifact rather than the true nature of the reported variant. Comparison of the observed de novo mutation frequency of TRIO in ID to the gene specific de novo mutation rate (31), however, indicates that we observed more de novo mutations in our ID cohort than expected based on chance alone (exact Poisson test, P < 0.005).
In total, we identified five individuals from four families with a truncating mutation of TRIO, suggestive for loss of the TRIO function. The clinical details for these four families are given in Table 1. To avoid reporting inherited familial traits, the father of Individual 2, who also had learning difficulties and behavior problems, was not included in this table. In summary, all of the index cases (age range, 7–35 years) showed mild to borderline ID (IQ range, 68–81) with autistic, hyperactive and aggressive behavioral problems. All individuals attended special education and needed special therapy to improve their language and motor development. Other observed features were recurrent infections (3/4 individuals), micrognathia (3/4), minor hand abnormalities (4/4), microcephaly (2/4) or low normal head circumference (2/4). No congenital abnormalities were observed and only mild facial dysmorphisms were noted (Fig. 1A).
Table 1.
Molecular and clinical details of individuals with TRIO mutations
| Individual 1 | Individual 2 | Individual 3 | Individual 4 | |
|---|---|---|---|---|
| Gender | Male | Male | Female | Male |
| Age at last visit | 7 | 20 | 35 | 10 |
| Mutations | ||||
| Deletion breakpoints | Chr5:14160447-14395478 | − | − | − |
| Deleted genes | TRIO | − | − | − |
| cDNA change | − | c.649A>T | c.3752del | c.4128G>A |
| Amino acid change | − | p.Arg217* | p.Asp1251Valfs*11 | p.Trp1376* |
| Chromosome position | − | 14290933 | 14387728 | 14390409 |
| Growth | ||||
| Birth weight | 0 SD | +0.25 SD | −0.75 SD | 0 SD |
| Height | 0 SD | −0.75 SD | −1.5 SD | +1.25 SD |
| Weight | NR | NR | +1 SD | +3 SD |
| Head circumference | −1 SD | −2.5 SD | −2.5 SD | −1.5 SD |
| Development | ||||
| ID | Borderline (IQ81) | Borderline (IQ78) | Mild (IQ62) | Mild (IQ68) |
| Speech delay | + 7 years: 5–6 words |
NR NR |
+ 3 years: first words |
+ NR |
| Motor delay | + Fine motor delay |
+ 3 years: first steps |
+ 2 years: first steps |
+ Fine motor delay |
| Neurological | ||||
| Behavioral problems | + PDD-NOS |
+ ADHD |
+ Autistic-like traits |
+ ADHD |
| High reflexes | − | + | + | − |
| Hyperacusis | + | − | + | − |
| Swallow difficulties | − | NR | + | + |
| Facial | ||||
| Facial asymmetry | − | + | + | − |
| Periorbital fullness | + | − | + | − |
| Full lips | − | + | + | − |
| High palate | − | + | NR | + |
| Dental crowding | − | + | + | + |
| Micrognathia | − | + | + | + |
| Other facial dysmorphism | High forehead, large protruding ears, short philtrum | − | High forehead, Full eyebrows, Downslanted palperbral fissures, full nasal tip |
High forehead, Long philtrum |
| Extremities | ||||
| Minor hand abnormalities | + Short, broad with Simian creases and volaire pads |
+ Broad proximal interphalangeal joints, camptodactyly dig V |
+ Brachydactyly dig I, tapering fingers |
+ Tapering fingers |
| Other | ||||
| Recurrent infections | + | NR | + | + |
| Kyphosis | − | − | + | + |
| Fatigue | + | − | + | − |
| Neonatal feeding difficulties | + | − | NR | + |
| Other clinical features | Anal fistula, scapula alatae, fourth palmar crease, clinodactyly digitus 5 and sandal gap | Torsio testis, pectus excavatum and tremor | Constipation, hypercalcemia, hypotonia, pes planus and varices | Automutilation |
NR, not reported.
Developmental expression of TRIO and its role in neurite outgrowth
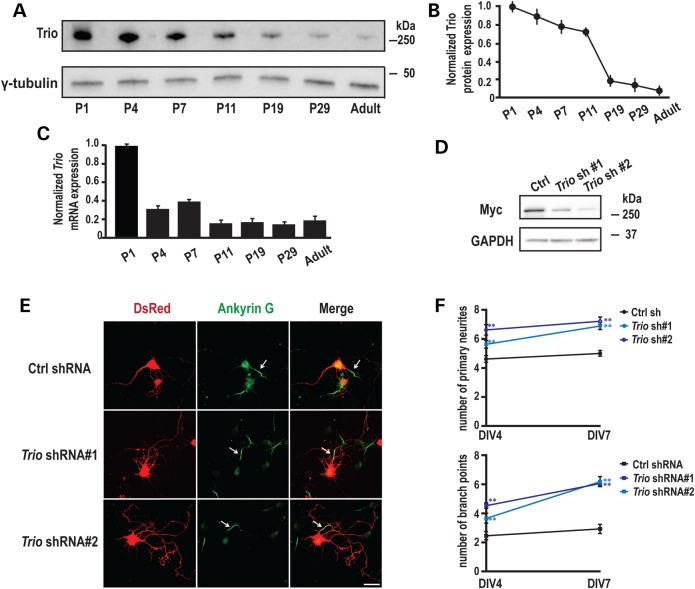
To find supportive evidence for TRIO as underlying cause in the phenotypes observed in our individuals, we investigated the neuronal function of TRIO in mammalian cells by examining the expression of TRIO during rat hippocampal development. Western blot analysis of protein extracts revealed that full-length TRIO is expressed during the early postnatal (P) period, but rapidly decreases after postnatal (P) day 11 (Fig. 2A and B). The rapid reduction in TRIO during postnatal development was further confirmed by measuring Trio mRNA expression (Fig. 2C). This decreased expression of TRIO is consistent with previous results (32) and suggests a role for TRIO in early neuronal development.
Figure 2.
Developmental expression of TRIO and its role in neurite outgrowth. (A) Rat hippocampi were collected at the indicated ages; equal amounts of protein (50 μg) were subjected to western blot analysis and representative blots are shown. (B) Quantification of Trio protein levels at different postnatal ages, normalized to the levels at P1 (n = 3). (C) Quantification of Trio mRNA levels using Q-PCR at different postnatal ages (n = 3). (D) 293 cells co-transfected with vectors encoding MYC-TRIO and Trio shRNA 1 and 2 were harvested after 24 h. Western blot analysis using a MYC antibody demonstrated that both Trio shRNA 1 and 2 efficiently reduced Trio expression. (E) Rat hippocampal neurons nucleofected at the time of plating with a control (scrambled) shRNA or Trio shRNA 1 or 2 were fixed and analyzed at the indicated times. shRNA transfected neurons were identified by DsRed co-expression; axon initial segments were identified by staining for Ankyrin G; representative images at DIV7 are shown. Scale bar, 50 µm. (F) Quantification of primary neurites and branch points at DIV4 and DIV7. Neurons expressing Trio shRNAs exhibited more primary neurites and more branch points than neurons expressing the control shRNA. Five separate experiments were performed; 20–30 neurons per group per experiment were counted. **P < 0.01 with respect to the control (scramble) shRNA vector.
To study the function of endogenous TRIO during early neuronal development, we designed short hairpin RNAs (shRNAs) targeting the spectrin repeat region of Trio, a region that is present in most of the alternately spliced Trio transcripts (33). The efficiency of Trio shRNA 1 and 2 was verified by co-expression with a vector encoding myelocytomatosis viral oncogene homolog tagged TRIO (MYC-TRIO) (full-length) in HEK293 cells (Fig. 2D). Dissociated rat hippocampal neurons were nucleofected at days in vitro (DIV) 0 with either a Trio shRNA or a scrambled shRNA control; expression of DsRed, which is encoded by the same vector, allowed identification of transfected cells and assessment of neuronal morphology. We first evaluated whether TRIO contributes to axon specification in early development by immunostaining neurons with a monoclonal antibody against Ankyrin G, which localizes to the axon initial segment (34). A single-Ankyrin-G-positive process was identified in the control group and in the Trio knock-down groups at DIV 4 and DIV 7 (Fig. 2E), indicating that axon polarity is not affected by the absence of endogenous TRIO. Next, we examined dendritic arbor formation. Compared with control neurons, expression of Trio shRNAs significantly increased both the number of primary dendrites and branch points observed at DIV 4 and at DIV 7 (Fig. 2E and F). Similar results were obtained at DIV 2 (data not shown). These results demonstrate that endogenous TRIO limits dendrite formation, without affecting the establishment of axon polarity. Kalirin, a TRIO orthologue with the 64% identical protein structure (33), is emerging as a key regulator of structural and functional plasticity. Interestingly, Kalirin has been reported to exhibit the opposite effect, stimulating both dendritic outgrowth and branching at this stage of neuronal development (35).
Post-synaptic TRIO regulates synaptic transmission
Previous proteomics studies of adult tissue have shown that TRIO is present at both the human and mouse post-synaptic density (36), suggesting that TRIO functions at this site. In addition, the major isoform of Kalirin found in the adult nervous system has been shown to play a critical role in synaptic transmission, specifically through the regulation of ionotropic AMPAr (37) and N-methyl-D-aspartate receptor (NMDAr) (38) trafficking. Modifications in the number and/or function of glutamate receptors are a core mechanism of regulating synaptic efficiency (39). We, therefore, assessed whether TRIO plays a role in synaptic transmission at the hippocampal CA3–CA1 pathway.
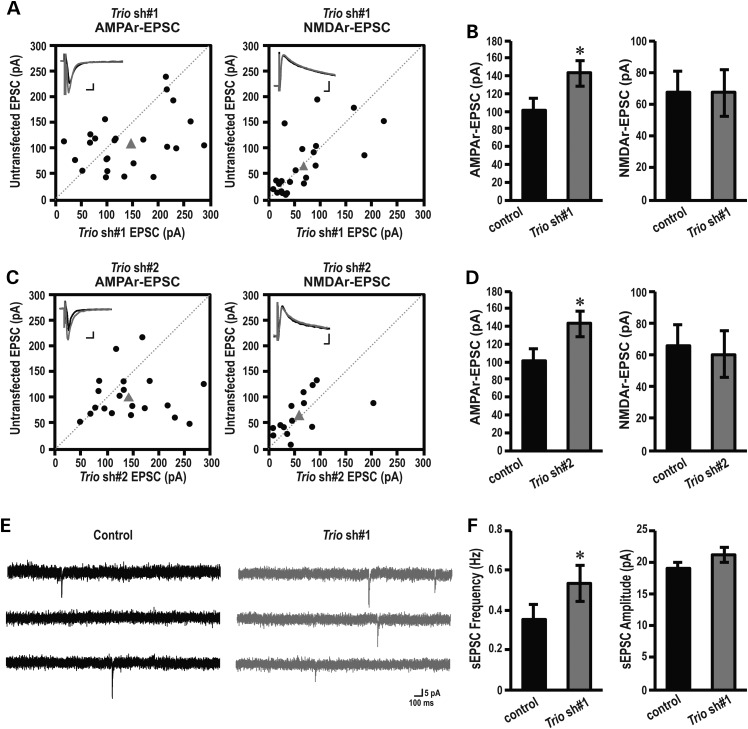
Organotypic hippocampal brain slices were prepared from P6 pups, a time period at which dendrites have already formed, allowing us to bypass the effect of TRIO on dendritic arborization (Fig. 2). Hippocampal brain slices were biolistically transfected with Trio shRNAs. Four days post-transfection, simultaneous recordings of evoked excitatory postsynaptic currents (eEPSCs) from a DsRed-labeled, shRNA expressing CA1 pyramidal neuron and an adjacent non-transfected neuron were performed (Fig. 3A). We stimulated the Schaffer collaterals with two independent bipolar electrodes and measured the evoked response while holding CA1 neurons at either −60 mV to measure AMPAr-mediated responses or at +40 mV to measure NMDAr-mediated responses. Neurons transfected with either Trio shRNA 1 or Trio shRNA 2 showed significantly increased AMPAr-mediated, but not NMDAr-mediated, transmission compared with control (Fig. 3A–D). These data indicate that TRIO, at least at excitatory CA1 synapses, is specific for AMPAr’s versus NMDARs. Importantly, we previously showed that expressing GFP or a scrambled shRNA by means of biolistics does not affect AMPAr-mediated synaptic transmission (40,41).
Figure 3.
Post-synaptic TRIO regulates synaptic transmission. (A, B) Amplitudes of AMPAr (left panel) and NMDAr (right panel) eEPSCs in control, non-transfected neurons are plotted against simultaneously recorded neighboring neurons expressing (A) Trio shRNA 1 (AMPAr, n = 25; NMDAr, n = 22), (B) Trio shRNA 2 (AMPAr, n = 20; NMDAr, n = 19). Black symbols represent single pairs of recordings; grey symbols show mean amplitudes. Inserts in each panel show sample averaged traces; grey traces, transfected neurons; black traces, non-transfected neighboring neurons. Scale bars represent 10 ms and 25 pA. (C and D) Summary (mean ± SEM) of effects of expressing Trio shRNA 1, on AMPAr (left) and NMDAr (right) eEPSCs calculated as the averaged ratios obtained from pairs of infected and uninfected neighboring neurons. Data are shown as mean ± SEM. *P < 0.05, paired Student's t-test, Trio shRNA 1 (AMPAr, n = 25; NMDAr, n = 22), (B) Trio shRNA 2 (AMPAr, n = 20; NMDAr, n = 19). (E) Representative traces of sEPSC recorded at −60 mV in an individual neuron from untransfected (control) or Trio shRNA 1 transfected group. Scale bars represent 100 ms and 5 pA. (F) Quantification of sEPSC frequency (left) and amplitude (right) for control and Trio shRNA 1-expressing neurons. Data are shown as mean ± SEM. *P < 0.05, paired Student's t-test, n = 8 pairs.
The changes in AMPAr-mediated transmission could be a result of either a change in synaptic AMPAr’s at individual synapses, or a change in the number of functional synapses or both. In order to determine the precise mechanism of action of the above possibilities we measured spontaneous sEPSCs. We observed that the frequency, but not the amplitude, of the sEPSCs was significantly increased when neurons were transfected with Trio shRNA 1 (Fig. 3E and F). Increases in sEPSC frequency could be due to either increased intrinsic excitability of the pre-synaptic CA3 neurons, or increased active synapse between CA3 and CA1 neurons. Since biolistic transfection resulted in low transfection efficiency (a few neurons/slice), the major contribution of knocking down Trio therefore comes from the recorded transfected postsynaptic cells (CA1) of the CA3–CA1 synapse. Our data, thus, suggest that depleting endogenous TRIO in CA1 neurons is sufficient to increase synaptic strength by increasing the amount of functional synapses.
Down-regulation of Trio reduces AMPAr endocytosis
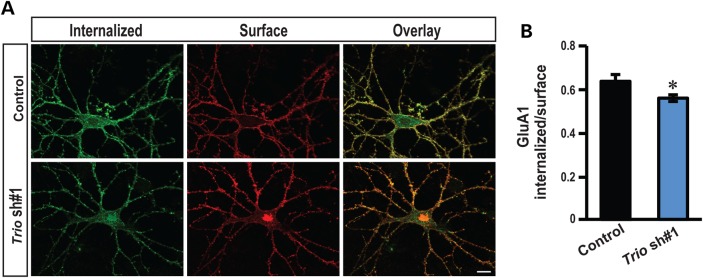
To further understand the mechanism by which TRIO affects hippocampal glutamatergic synapses, we tested whether down-regulation of Trio correlated with persistent changes in AMPAr internalization rate. Previously, an isoform of Trio has been shown to colocalize with Rab5 in endosomes (42) and Rab5 is required for AMPAr endocytosis at excitatory CA1 synapses (43). We, therefore, assessed directly the function of Trio in AMPAr endocytosis. Primary hippocampal neurons were transfected with Trio shRNA 1 at DIV 3. At DIV 14 surface AMPAr GluA1 subunits were lively labeled with an antibody recognizing the extracellular epitopes of GluA1 and allowed AMPAr’s to internalize at 37°C for 2 h. Neurons transfected with Trio shRNA 1 showed a marked reduction in endocystosis as assessed by the ratio between internalized and extracellular GluA1, suggesting that down-regulation of Trio decreases AMPAr endocytosis rate (Fig. 4A and B). Collectively, our results indicate that TRIO-mediated signaling controls synaptic function by regulating AMPAr endocytosis at CA1 excitatory synapses.
Figure 4.
Down-regulation of Trio reduces AMPAr endocytosis. (A) Representative double-label images of internalized (green) and surface (red) AMPAr GluA1 subunit in low-density hippocampal neurons. (B) Ratiometric analysis of the intensity of internalized GluA1 to surface GluA1 in indicated conditions. Control shRNA: n = 15; Trio shRNA 1: n = 14; scale bars, 10 µm; *P < 0.05, t-test; error bars represent SEM.
Discussion
Here, we show that LoF mutations in TRIO are responsible for a clinical phenotype involving mild to borderline ID with behavioral problems, and that TRIO has an important function during neurite outgrowth and basal synaptic transmission. Specifically, we demonstrated for the first time that endogenous Trio negatively regulates hippocampal synaptic strength by specifically affecting AMPAr internalization at CA1 excitatory synapses.
Large scale reference data sets such as present in ExAC have now catalogued human genetic diversity at the unprecedented level (44). Whereas the LoF mutations identified in this study are not present in ExAC, 13 other truncating variants in TRIO (NM_007118.2) are. Four of these are located in the last exon, thereby escaping nonsense mediated mRNA decay. This leaves nine different potential LoF variants, reported to occur in 33 of >60 000 individuals represented in ExAC. Remarkably, ExAC recently reported that only 3230 human genes (∼15%) are near-completely depleted from truncating variants. This implicates that for the vast majority of genes such truncating variants are observed, and therefore likely, also include well-known disease genes causing disease by haploinsufficiency. For instance, analysis of ARID1B revealed 16 different LoF variants, identified in 29 of >60 000 individuals in ExAC (data not shown). Interestingly, however, LoF mutations in ARID1B cause autosomal dominant mental retardation type 12 (MIM 614562). Whereas such individuals could go undiagnosed in the general population for which ExAC is a proxy, the frequency in which these ARID1B LoF mutations are observed relatively high to remain undiagnosed in the general population. In contrast, the aspecific phenotype and mild-to-borderline ID observed in patients with truncating variants in TRIO could remain undiagnosed in the population. Hence, LoF of TRIO is not unexpected to be present in ExAC.
The molecular mechanisms that promote excitatory synapse development have been extensively studied. However, the molecular events that prevent premature excitatory synapse development so that synapses form at the correct time and place are less well-understood. Here, we find that knockdown of Trio, during early development prevents both neurite outgrowth and synapse formation. Together with our observation that TRIO is highly expressed at early stages of development and that its expression decreases at the time when synapses are being formed, our data suggest that TRIO serves as an endogenous break for synaptic development. Indeed, down-regulation of TRIO in hippocampal brain slices, using two independent shRNAs, increased AMPAr-mediated, but not NMDAr-mediated transmission. This Increased synaptic AMPAr-mediated response, was accompanied by an increase in the frequency of spontaneous excitatory currents as well as by a reduction of AMPAr internalization. Interestingly, an isoform of Trio (Trio8) has been shown to modulate early-endosome dynamics in COS-7 cells and to colocalize with Rab5 in neurons, whereas Rab5 activation drives the specific internalization of synaptic AMPAr’s in a clathrin-dependent manner (42,43). It is, therefore, plausible that TRIO controls AMPAr endocytosis via a functional interaction with Rab5 in hippocampal neurons.
It is known that the redistribution of postsynaptic AMPAR’s plays a key role in controlling excitatory synaptic efficacy during early development. Early in development a majority of the synapses are silent, referring to the fact that they contain NMDArs but no functional AMPAr’s (45). The proportion of these silent synapses rapidly decreases during the first two weeks of post-natal development in the hippocampus, coinciding with the maturation process of the synapses (46) and occurs as a result of AMPAr incorporation. The loss of TRIO could thus accelerate this unsilencing process. Of interest, premature maturation of excitatory synapses has been observed in several models for ASD. For example, accelerated maturation of excitatory synapses in an early period of hippocampal development contributes to the learning deficits in a mouse model of human SYNGAP1 haploinsuffciency (47). Similarly, accelerated maturation of glutamatergic synapses has been seen upon the loss of the ID and Schizophrenia-associated microRNA, miR-137 (48).
Kalirin and TRIO belong to the dbl family of RhoGEFs and are unique within this family in that they display two GEF domains of distinct specificity. Whereas Kalirin has been extensively studied for its synaptic function, very little is known regarding the role of TRIO at the synapse. To our surprise we found that TRIO plays an opposing role compared with Kalirin both, at the level of neurite outgrowth and synaptic strength. Whereas Kalirin stimulates neurite outgrowth (35) we found that knockdown of Trio enhanced neurite outgrowth. Similarly, whereas Kalirin is required for activity-dependent spine enlargement and enhancement of AMPA-mediated synaptic transmission we found that TRIO negatively regulated AMPAr-mediated, but did not affect NMDAr-mediated transmission (37). The distinct effects of TRIO and Kalirin in mammals indicate that these genes contribute to different aspects of neuronal development. Their opposing roles could result in the fine balance needed to form complex dendritic arbors during development, which are critical for proper synaptogenesis and precise cognitive function. In addition, several Trio splicing variants have been identified (33), and the expression of each isoform is regulated in a temporal and spatial-specific manner. These isoforms display either one or both GEF domains, suggesting that they could differentially regulate Rac1 and RhoA. The loss of TRIO in different brain structures could therefore account for different phenotypes. Functions of these isoforms are however largely unknown. Future studies will be needed to explore the function of the multiple TRIO domains in dendritic arborization and synapse formation and how the actions of the different domains are coordinated to regulate neuronal development.
In summary, our current data identified TRIO in patients with mild to borderline ID combined with behavioral problems and provide novel insight into how genetic deficits in TRIO can lead to glutamatergic dysfunction. Over the coming years, we expect many more mutations in TRIO to be identified. Therefore, we established a website (http://www.triogene.com) to collect detailed phenotypic data of individuals harboring TRIO mutations not only to gain insight into the clinical spectrum that these mutations might cause but also to obtain fundamental understanding of the pathogenic mechanisms underlying TRIO-related ID.
Materials and Methods
Identification of additional individuals with TRIO mutations
We performed targeted sequencing of the coding sequence of candidate ID genes, including TRIO, in a cohort of 2326 individuals with unexplained ID, using MIPs as described previously (30). The individuals in this cohort were selected from the in-house collection of the Department of Human Genetics of Radboud University Medical Center (Nijmegen), containing more than 5000 samples from individuals with unexplained ID. DNA was extracted from peripheral blood. After confirmation with Sanger sequencing, de novo occurrence of the mutation was determined by using parental DNA. This study was approved by the institutional review board Commissie Mensgebonden Onderzoek Regio Arnhem-Nijmegen NL36191.091.11. Written informed consent was obtained from all individuals.
Cell culture
Hippocampal neuron cultures were prepared from embryonic day 18 rats (Sprague-Dawley, Charles River) and nucleofected with various Ds-Red vectors as described previously (35). Briefly, hippocampal tissue was separated and meninges were removed carefully with forceps under the dissecting microscope. Tissues were digested with 0.25% trypsin (Corning) for 15 min before dissociation. Culture media contained 1 µg/ml fungizone (amphotericin B, Gibco) and 1 µm Ara-C (Sigma) until harvest. Cultures were examined at DIV 2, 4 and 7, enumerating the numbers of primary neurites and branch points in the transfected cells, using fluorescence to identify transfected neurons (35).
Organotypic hippocampal slice cultures were prepared from postnatal day 6 rat pups as described (49). Slices were biolistically transfected at four DIV using a Helios Gene Gun (Bio-Rad) (41) and recorded 4 days post-transfection.
Knockdown experiments
To reduce expression of endogenous Trio in the cultures, shRNAs targeted the spectrin repeat region, since most of the alternately spliced Trio transcripts contain the spectrin repeats2; the effects of shRNAs targeting Trio were compared with a control shRNA (scrambled shRNA), which matches nothing in rodent genomes or transcripts. Oligonucleotides were annealed and ligated into the RNAi-Ready-pSiren-DNR-DsRed-Express vector (Clontech, Mountain View, CA, USA). This vector utilizes the human U6 promoter to control shRNA expression and the CMV promoter to control DsRed expression3. The sequences targeted were: scramble shRNA, AATGCACGCTCAGCACAAGC; Trio shRNA 1, TTCTGGCAGAAACAGAGGA; Trio shRNA 2, TTCAGACGCAGCACAATCA. All constructs were verified by sequencing. Transfection with 2 μg DNA was performed using a Nucleofector™ 2b device (Lonza) at the time of plating and 2.5 × 106 neurons for each transfection (35). Neurons were fixed at DIV 2, 4 or 7 using 4% paraformaldehyde in phosphate buffered saline.
Immunocytochemistry and image acquisition
Fixed neurons were permeabilized with 0.3% Triton and stained with antisera to Ankyrin G (1: 1000; mouse, Neuromab). Images were taken using a Zeiss LSM 510 Meta confocal microscope and a 40× objective. For each neuron, numbers of primary neurites and branch points were measured from the same image. Primary neurites that are longer than one soma diameter were counted at all days in vitro indicated.
Western blot analysis
For time-course studies, hippocampal tissues collected from rats at different ages were sonicated into the sodium dodecyl sulphate lysis buffer, heated for 10 min at 95°C and insoluble debris was removed by centrifugation. The supernatant protein concentration was determined using the bicinchoninic acid assay (Pierce, Rockford, IL, USA) and bovine serum albumin (BSA) as the standard; 50 µg of total protein was loaded onto each lane for analysis of Trio expression. Westerns were performed with Trio antibody (UltraCruz) at 1:10002. Similar expression patterns were observed in a separate set of animals.
RNA isolation, reverse transcription and real-Time polymerase chain reaction
Total RNA from the tissue was isolated using Nucleospin RNA kit (Macherey-Nagel). cDNA was synthesized from 0.5 to 1 mg RNA using iScript™ cDNA Synthesis Kit. For detecting Trio expression, real-time polymerase chain reaction (PCR) was performed in 7900HT Fast Real-Time PCR System using GoTaq qPCR Master mix (Promega). The following primers were used: forward 1, GTCACGGAACATGTTGAAGG, reverse 1, CCTGTATCACCTCACGGATG; forward 2, TCGAGGAAGTTGCACAGAAC, reverse 2, ATCTCTTCGGGGTCACATTC; forward 3, AGTTGGAGAACGGGTACAGG; reverse 3, ACCTCGCTCAATGGGATAAC. Relative Trio expression compared with the control was calculated by 2−ΔΔCt.
Electrophysiology
Whole-cell recordings were obtained with Multiclamp 700B amplifiers (Axon Instruments). To monitor TRIO's effects on synaptic transmission, slices were biolistically transfected at DIV 4. Two days later, whole-cell recordings were obtained simultaneously from a transfected and an adjacent non-transfected neuron in the CA1 region under visual guidance using epifluorescence (red fluorescence) and transmitted light illumination. The recording chamber was perfused with artificial cerebrospinal fluid (ACSF) containing 119 mm NaCl, 2.5 mm KCl, 4 mm CaCl2, 4 mm MgCl2, 26 mm NaHCO3, 1 mm NaH2PO4, 11 mm glucose, 0.1 mm picrotoxin and 4 μm 2-chloroadenosine (pH 7.4), and gassed with 5% CO2/95% O2. Recordings were made at 30°C. Patch recording pipettes (3–5 MΩ) were filled with intracellular solution containing 115 mm cesium methanesulfonate, 20 mm CsCl, 10 mm 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 2.5 mm MgCl2, 4 mm Na2ATP, 0.4 mm Na3GTP, 10 mm sodium phosphocreatine, and 0.6 mM ethylene glycol tetraacetic acid (pH 7.25). Evoked responses were induced using bipolar electrodes placed on Schaffer collateral pathway. Responses were recorded at both −60 mV (for AMPAr-mediated responses) and +40 mV (for NMDAr-mediated responses). NMDAr-mediated responses were quantified as the mean between 60 and 65 ms after stimulation. All recordings were done by stimulating two independent synaptic inputs; results from each pathway were averaged and counted as n = 1.
Spontaneous responses were recorded at −60 mV (sEPSC) in ACSF containing 2.5 mM CaCl2 and 1.2 mm MgCl2 at 30°C. sEPSCs were recorded in the presence of 0.1 mm picrotoxin. Five to 10 min of recordings were analyzed from each cell. Data were acquired at 5 kHz, filtered at 2 kHz, and analyzed using the mini analysis program (Synaptosoft). All data are reported as mean ± SEM. A statistical significance was determined by the paired Student's t-test. Significance was set to P < 0.05.
AMPAr endocytosis assay
Endocytosis assay was performed as previously described (50,51) with modifications. Briefly, living primary hippocampal neurons transfected with indicated constructs were pretreated for 30 min with 100 µg/ml leupeptin and then labeled with anti-GluA1 antibody (Millipore). Internalization of the labeled GluA1 subunits was then allowed for 2 h at 37°C. After fixation with 4% paraformaldehyde and blocking with 5% BSA, surface-labeled GluA1 subunits were exposed to Alexa-488 secondary antibody (Invitrogen). After washing, the remaining surface GluA1 subunits were incubated with an unlabeled secondary antibody at a concentration of 0.13 mg/ml overnight to saturate surface anti-GluA1 antibodies. Neurons were then permeabilized and incubated with Alexa-647 secondary antibody to label internalized GluA1 subunits. GluA1 endocytosis was calculated by dividing the intensity of internalized GluA1 by the intensity of surface GluA1 using imageJ (I488/I647).
Supplementary Material
Funding
This work was supported by the Netherlands Organization for Health Research and Development grant 912-12-109 (to J.A.V. and B.B.A.d.V.) and 916-14-043 (to C.G.), the European research council (ERC Starting grant DENOVO 281964 to J.A.V.). The research of NNK supported by grants from the ‘Donders Center for Neuroscience fellowship award of the Radboudumc’ (to N.N.K.); the ‘FP7-Marie Curie International Reintegration Grant’ (N.N.K. grant number: 277091); the Jerome Lejeune Foundation (to N.N.K). NIH grant funding for this study was provided in part by the National Institutes for Mental Health (no. 1R01MH101221 to E.E.E.). E.E.E. is an investigator of the Howard Hughes Medical Institute.
Supplementary Material
Acknowledgements
We are thankful to the individuals involved and their parents for their participation.
Conflict of Interest statement. None declared.
References
- 1.Leonard H., Wen X. (2002) The epidemiology of mental retardation: challenges and opportunities in the new millennium. Ment. Retard. Dev. Disabil. Res. Rev., 8, 117–134. [DOI] [PubMed] [Google Scholar]
- 2.Vissers L.E.L.M., Gilissen C., Veltman J.A. (2015) Genetic studies in intellectual disability and related disorders. Nat. Rev. Genet. 10.1038/nrg3999. [DOI] [PubMed] [Google Scholar]
- 3.Bamshad M.J., Ng S.B., Bigham A.W., Tabor H.K., Emond M.J., Nickerson D.A., Shendure J. (2011) Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet., 12, 745–755. [DOI] [PubMed] [Google Scholar]
- 4.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z. et al. (2013) Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N. Engl. J. Med., 369, 1502–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vissers L.E.L.M., de Ligt J., Gilissen C., Janssen I., Steehouwer M., de Vries P., van Lier B., Arts P., Wieskamp N., del Rosario M. et al. (2010) A de novo paradigm for mental retardation. Nat. Genet., 42, 1109–1112. [DOI] [PubMed] [Google Scholar]
- 6.de Ligt J., Willemsen M.H., van Bon B.W.M., Kleefstra T., Yntema H.G., Kroes T., Vulto-van Silfhout A.T., Koolen D.A., de Vries P., Gilissen C. et al. (2012) Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med., 367, 1921–1929. [DOI] [PubMed] [Google Scholar]
- 7.Rauch A., Wieczorek D., Graf E., Wieland T., Endele S., Schwarzmayr T., Albrecht B., Bartholdi D., Beygo J., Di Donato N. et al. (2012) Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet, 380, 1674–1682. [DOI] [PubMed] [Google Scholar]
- 8.The Deciphering Developmental Disorders Study (2014) Large-scale discovery of novel genetic causes of developmental disorders. Nature, 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright C.F., Fitzgerald T.W., Jones W.D., Clayton S., McRae J.F., van Kogelenberg M., King D.A., Ambridge K., Barrett D.M., Bayzetinova T. et al. (2014) Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet, 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y., Muzny D.M., Xia F., Niu Z., Person R., Ding Y., Ward P., Braxton A., Wang M., Buhay C. et al. (2014) Molecular findings among patients referred for clinical whole-exome sequencing. JAMA, 312, 1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilissen C., Hehir-Kwa J.Y., Thung D.T., van de Vorst M., van Bon B.W.M., Willemsen M.H., Kwint M., Janssen I.M., Hoischen A., Schenck A. et al. (2014) Genome sequencing identifies major causes of severe intellectual disability. Nature, 511, 344–347. [DOI] [PubMed] [Google Scholar]
- 12.Vulto-van Silfhout A.T., Hehir-Kwa J.Y., van Bon B.W.M., Schuurs-Hoeijmakers J.H.M., Meader S., Hellebrekers C.J.M., Thoonen I.J.M., de Brouwer A.P.M., Brunner H.G., Webber C. et al. (2013) Clinical significance of de novo and inherited copy-number variation. Hum. Mutat., 34, 1679–1687. [DOI] [PubMed] [Google Scholar]
- 13.EuroEPINOMICS-RES Consortium, Epilepsy Phenome/Genome ProjectEpi4 K Consortium (2014) De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am. J. Hum. Genet., 95, 360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iossifov I., O'Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E. et al. (2014) The contribution of de novo coding mutations to autism spectrum disorder. Nature, 515, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Debant A., Serra-Pagès C., Seipel K., O'Brien S., Tang M., Park S.H., Streuli M. (1996) The multidomain protein Trio binds the LAR transmembrane tyrosine phosphatase, contains a protein kinase domain, and has separate rac-specific and rho-specific guanine nucleotide exchange factor domains. Proc. Natl Acad. Sci. USA, 93, 5466–5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medley Q.G., Serra-Pagès C., Iannotti E., Seipel K., Tang M., O'Brien S.P., Streuli M. (2000) The trio guanine nucleotide exchange factor is a RhoA target. Binding of RhoA to the trio immunoglobulin-like domain. J. Biol. Chem., 275, 36116–36123. [DOI] [PubMed] [Google Scholar]
- 17.Jaiswal M., Dvorsky R., Ahmadian M.R. (2013) Deciphering the molecular and functional basis of Dbl family proteins: a novel systematic approach toward classification of selective activation of the Rho family proteins. J. Biol. Chem., 288, 4486–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blangy A., Vignal E., Schmidt S., Debant A., Gauthier-Rouvière C., Fort P. (2000) TrioGEF1 controls Rac- and Cdc42-dependent cell structures through the direct activation of rhoG. J. Cell Sci., 113, 729–739. [DOI] [PubMed] [Google Scholar]
- 19.Rossman K.L., Der C.J., Sondek J. (2005) GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol., 6, 167–180. [DOI] [PubMed] [Google Scholar]
- 20.Choi B.J., Imlach W.L., Jiao W., Wolfram V., Wu Y., Grbic M., Cela C., Baines R.A., Nitabach M.N., McCabe B.D. (2014) Miniature neurotransmission regulates Drosophila synaptic structural maturation. Neuron, 82, 618–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ba W., van der Raadt J., Nadif Kasri N. (2013) Rho GTPase signaling at the synapse: implications for intellectual disability. Exp. Cell Res., 319, 2368–2374. [DOI] [PubMed] [Google Scholar]
- 22.Newsome T.P., Schmidt S., Dietzl G., Keleman K., Asling B., Debant A., Dickson B.J. (2000) Trio combines with dock to regulate Pak activity during photoreceptor axon pathfinding in Drosophila. Cell, 101, 283–294. [DOI] [PubMed] [Google Scholar]
- 23.Iyer S.C., Wang D., Iyer E.P.R., Trunnell S.A., Meduri R., Shinwari R., Sulkowski M.J., Cox D.N. (2012) The RhoGEF trio functions in sculpting class specific dendrite morphogenesis in Drosophila sensory neurons. PLoS One, 7, e33634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shivalkar M., Giniger E. (2012) Control of dendritic morphogenesis by Trio in Drosophila melanogaster. PLoS One, 7, e33737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Haren J., Boudeau J., Schmidt S., Basu S., Liu Z., Lammers D., Demmers J., Benhari J., Grosveld F., Debant A. et al. (2014) Dynamic microtubules catalyze formation of navigator-TRIO complexes to regulate neurite extension. Curr. Biol., 24, 1778–1785. [DOI] [PubMed] [Google Scholar]
- 26.DeGeer J., Boudeau J., Schmidt S., Bedford F., Lamarche-Vane N., Debant A. (2013) Tyrosine phosphorylation of the Rho guanine nucleotide exchange factor Trio regulates netrin-1/DCC-mediated cortical axon outgrowth. Mol. Cell Biol., 33, 739–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Briançon-Marjollet A., Ghogha A., Nawabi H., Triki I., Auziol C., Fromont S., Piché C., Enslen H., Chebli K., Cloutier J.-F. et al. (2008) Trio mediates netrin-1-induced Rac1 activation in axon outgrowth and guidance. Mol. Cell Biol., 28, 2314–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charrasse S., Comunale F., Fortier M., Portales-Casamar E., Debant A., Gauthier-Rouvière C. (2007) M-cadherin activates Rac1 GTPase through the Rho-GEF trio during myoblast fusion. Mol. Biol. Cell, 18, 1734–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt S., Debant A. (2014) Function and regulation of the Rho guanine nucleotide exchange factor Trio. Small GTPases, 5, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Roak B.J., Vives L., Fu W., Egertson J.D., Stanaway I.B., Phelps I.G., Carvill G., Kumar A., Lee C., Ankenman K. et al. (2012) Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science, 338, 1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. et al. (2014) A framework for the interpretation of de novo mutation in human disease. Nat. Genet., 46, 944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma X.-M., Huang J.-P., Eipper B.A., Mains R.E. (2005) Expression of Trio, a member of the Dbl family of Rho GEFs in the developing rat brain. J. Comp. Neurol., 482, 333–348. [DOI] [PubMed] [Google Scholar]
- 33.McPherson C.E., Eipper B.A., Mains R.E. (2005) Multiple novel isoforms of Trio are expressed in the developing rat brain. Gene, 347, 125–135. [DOI] [PubMed] [Google Scholar]
- 34.Zhou D., Lambert S., Malen P.L., Carpenter S., Boland L.M., Bennett V. (1998) AnkyrinG is required for clustering of voltage-gated Na channels at axon initial segments and for normal action potential firing. J. Cell Biol., 143, 1295–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan Y., Eipper B.A., Mains R.E. (2015) Kalirin-9 and Kalirin-12 play essential roles in dendritic outgrowth and branching. Cereb. Cortex 10, 3487–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bayés A., Collins M.O., Croning M.D.R., van de Lagemaat L.N., Choudhary J.S., Grant S.G.N. (2012) Comparative study of human and mouse postsynaptic proteomes finds high compositional conservation and abundance differences for key synaptic proteins. PLoS One, 7, e46683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie Z., Srivastava D.P., Photowala H., Kai L., Cahill M.E., Woolfrey K.M., Shum C.Y., Surmeier D.J., Penzes P. (2007) Kalirin-7 controls activity-dependent structural and functional plasticity of dendritic spines. Neuron, 56, 640–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiraly D.D., Lemtiri-Chlieh F., Levine E.S., Mains R.E., Eipper B.A. (2011) Kalirin binds the NR2B subunit of the NMDA receptor, altering its synaptic localization and function. J. Neurosci., 31, 12554–12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kessels H.W., Malinow R. (2009) Synaptic AMPA receptor plasticity and behavior. Neuron, 61, 340–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nadif Kasri N., Nakano-Kobayashi A., van Aelst L. (2011) Rapid synthesis of the X-linked mental retardation protein OPHN1 mediates mGluR-dependent LTD through interaction with the endocytic machinery. Neuron, 72, 300–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nadif Kasri N., Nakano-Kobayashi A., Malinow R., Li B., van Aelst L. (2009) The Rho-linked mental retardation protein oligophrenin-1 controls synapse maturation and plasticity by stabilizing AMPA receptors. Genes Dev., 23, 1289–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun Y.-J., Nishikawa K., Yuda H., Wang Y.-L., Osaka H., Fukazawa N., Naito A., Kudo Y., Wada K., Aoki S. (2006) Solo/Trio8, a membrane-associated short isoform of Trio, modulates endosome dynamics and neurite elongation. Mol. Cell Biol., 26, 6923–6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown T.C., Tran I.C., Backos D.S., Esteban J.A. (2005) NMDA receptor-dependent activation of the small GTPase Rab5 drives the removal of synaptic AMPA receptors during hippocampal LTD. Neuron, 45, 81–94. [DOI] [PubMed] [Google Scholar]
- 44.Lek, M. Exome Aggregation Consortium (2015) Analysis of Protein-Coding Genetic Variation in 60,706 Humans 10.1101/030338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanse E., Seth H., Riebe I. (2013) AMPA-silent synapses in brain development and pathology. Nat. Rev. Neurosci. 14, 839–850. [DOI] [PubMed] [Google Scholar]
- 46.Liao D., Zhang X., O'Brien R., Ehlers M.D., Huganir R.L. (1999) Regulation of morphological postsynaptic silent synapses in developing hippocampal neurons. Nat. Neurosci., 2, 37–43. [DOI] [PubMed] [Google Scholar]
- 47.Clement J.P., Aceti M., Creson T.K., Ozkan E.D., Shi Y., Reish N.J., Almonte A.G., Miller B.H., Wiltgen B.J., Miller C.A. et al. (2012) Pathogenic SYNGAP1 mutations impair cognitive development by disrupting maturation of dendritic spine synapses. Cell, 151, 709–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olde Loohuis N.F.M., Ba W., Stoerchel P.H., Kos A., Jager A., Schratt G., Martens G.J.M., van Bokhoven H., Nadif Kasri N., Aschrafi A. (2015) MicroRNA-137 controls AMPA-receptor-mediated transmission and mGluR-dependent LTD. Cell Rep., 11, 1876–1884. [DOI] [PubMed] [Google Scholar]
- 49.Kasri N.N., Govek E.-E., Aelst L.V. (2008) Characterization of oligophrenin-1, a RhoGAP lost in patients affected with mental retardation: lentiviral injection in organotypic brain slice cultures. Methods Enzymol., 439, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carrodus N.L., Teng K.S.-L., Munro K.M., Kennedy M.J., Gunnersen J.M. (2014) Differential labeling of cell-surface and internalized proteins after antibody feeding of live cultured neurons. J.Vis. Exp.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raynaud F., Janossy A., Dahl J., Bertaso F., Perroy J., Varrault A., Vidal M., Worley P.F., Boeckers T.M., Bockaert J. et al. (2013) Shank3-Rich2 interaction regulates AMPA receptor recycling and synaptic long-term potentiation. J. Neurosci., 33, 9699–9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.