Abstract
Mutations in mitochondrial (mt) genes coding for mt-tRNAs are responsible for a range of syndromes, for which no effective treatment is available. We recently showed that the carboxy-terminal domain (Cterm) of human mt-leucyl tRNA synthetase rescues the pathologic phenotype associated either with the m.3243A>G mutation in mt-tRNALeu(UUR) or with mutations in the mt-tRNAIle, both of which are aminoacylated by Class I mt-aminoacyl-tRNA synthetases (mt-aaRSs). Here we show, by using the human transmitochondrial cybrid model, that the Cterm is also able to improve the phenotype caused by the m.8344A>G mutation in mt-tRNALys, aminoacylated by a Class II aaRS. Importantly, we demonstrate that the same rescuing ability is retained by two Cterm-derived short peptides, β30_31 and β32_33, which are effective towards both the m.8344A>G and the m.3243A>G mutations. Furthermore, we provide in vitro evidence that these peptides bind with high affinity wild-type and mutant human mt-tRNALeu(UUR) and mt-tRNALys, and stabilize mutant mt-tRNALeu(UUR). In conclusion, we demonstrate that small Cterm-derived peptides can be effective tools to rescue cellular defects caused by mutations in a wide range of mt-tRNAs.
Introduction
Mitochondrial (mt) diseases are multi-system disorders due to mutations in nuclear or mtDNA genes. Among the latter, >50% are located in transfer RNA (tRNA) genes [(1); URL: http://www.mitomap.org] and are responsible for a wide range of clinical phenotypes, such as the severe Mitochondrial Encephalopathy with Lactic Acidosis and Stroke like Episodes (MELAS) and Myoclonic Epilepsy with Ragged-Red Fibres (MERRF) syndromes, for which no effective treatments are available at present (2).
Evidence provided by our group and others has shown that overexpression of mt-aminoacyl-tRNA synthetases (mt-aaRSs) attenuates the detrimental effects of point mutations in cognate mt-tRNAs both in human cells (3–6) and in a yeast model (7,8). Our group and others extended these results by showing that class Ia human mt-leucyl-tRNA synthetase (mt-LeuRS) and mt-valyl-tRNA synthetase (mt-ValRS) are able to rescue defects due to pathogenic mutations in non-cognate, as well as cognate, mt-tRNAs, in both human transmitochondrial cybrid cells (herein named cybrids) (9,10) and yeast (11).
We further demonstrated that the rescuing activity of human mt-LeuRS resides in a relatively small (<70 amino acids) protein region: the carboxy-terminal domain (Cterm) (9,10). Cterm overexpression in human cybrids rescued both relatively mild defects caused by two different homoplasmic mutations in the non-cognate mt-tRNAIle gene (MTTI), and severe pathological phenotypes caused by the heteroplasmic m.3243A>G mutation in the cognate mt-tRNALeu(UUR) gene (MTTL1) (9). The latter mutation is the most prevalent heteroplasmic mt-tRNA mutation associated with mt disease, leading to a wide spectrum of clinical manifestations. These are partly determined by the level of heteroplasmy and include maternally inherited diabetes and deafness and the severe biochemical and clinical phenotypes ascribed to MELAS (12,13). We showed that the Cterm is both necessary and sufficient for the entire rescuing activity. It is more effective than the full-length enzyme on all mtDNA mutations tested, whereas the activity of a mt-LeuRS variant deleted of the Cterm is negligible or missing (9). We proposed that the Cterm rescuing activity is mediated by a chaperone-like mechanism: the Cterm would directly interact with the mutated mt-tRNAs and stabilize a conformation suitable to establish interactions with cognate aaRS, or other molecular partners required to perform tRNA function. This hypothesis is supported by in vitro experiments demonstrating that the Cterm directly interacts with both the cognate human mt-tRNALeu(UUR), with high affinity and stability, and with the non-cognate human mt-tRNAIle, although with lower affinity (9).
To identify the minimal Cterm regions responsible for the rescue, we designed short peptides comprising the Cterm regions that interact with tRNALeu in the available three-dimensional (3D) structures (14,15). These peptides were named β30_31 (15 aa) and β32_33 (16 aa), since they encompass β-strands 30 and 31, and β-strands 32 and 33, respectively. Both peptides were endowed with even higher rescuing activity than the full Cterm when directed to a large panel of yeast mt-tRNA mutants (16,17).
In the work here presented, we have investigated the rescuing ability of β30_31 and β32_33 peptides in human cybrids bearing either the aforementioned m.3243A>G mutation in MTTL1, or the m.8344A>G MERRF mutation in mt-tRNALys (MTTK). Both peptides are able to ameliorate viability and energetic proficiency of human cells carrying either mutation and to interact in vitro with mutant human mt-tRNALeu(UUR) and mt-tRNALys. In the case of the ‘cognate’ mt-tRNALeu(UUR) m.3243A>G mutant, both peptides also restore wild-type (WT) levels of thermal stability and structure, which are strongly impaired by the mutations.
These results indicate that β30_31 and β32_33 peptides, derived from the Cterm fragment of mt-LeuRS, are new tools to effectively correct defective phenotypes caused by mutations in mt-tRNA genes. Hence, they represent attractive lead molecules for the development of compounds aimed at therapeutic applications against syndromes associated with pathogenic mt-tRNA mutations.
Results
Pathological phenotype of m.8344A>G MTTK mutant cybrids
First, we investigated the Cterm ability to rescue the pathological phenotype of cybrid cells bearing a mutation in mt-tRNALys, which is aminoacylated by class II LysRS. To this end, we manipulated m.8344A>G osteosarcoma-derived cybrids (kind gift from Valeria Tiranti) using ethidium bromide (EtBr) to obtain different cell lines bearing either high or low levels of mutated mt-DNA (see the Materials and Methods section). The assessment of m.8344A>G mutation load was performed by quantitative pyrosequencing. The selected clones showed 34% (L-8344, low-mutant percentage) and 86% (H-8344, high-mutant percentage) m.8344A>G mutation loads, respectively. To investigate the phenotype of mutant cell lines, cybrids were grown in glucose-free medium supplemented with galactose (galactose medium), a condition forcing cells to rely on the mt respiratory chain for ATP synthesis. In this condition, cell growth of H-8344 cybrids was severely impaired whereas L-8344 cybrids were indistinguishable from WT cells (Supplementary Material, Fig. S1A), providing us with an isogenic control for further experiments. Decreased viability of H-8344 cybrids was associated with an increase in apoptosis and impaired respiratory chain activity, as compared with isogenic L-8344 cybrids (Supplementary Material, Fig. S1A–C). To evaluate the effect of the m.8344A>G mutation on mt-tRNALys steady-state levels we performed high-resolution northern blot analysis. We showed that high-mutation levels are associated with a 50% decrease in mt-tRNALys steady-state levels relative to the amount of 5SrRNA (Supplementary Material, Fig. S1D).
The carboxy-terminal domain of human mt-LeuRS is able to rescue the phenotype of m.8344A>G MTTK mutant cybrids
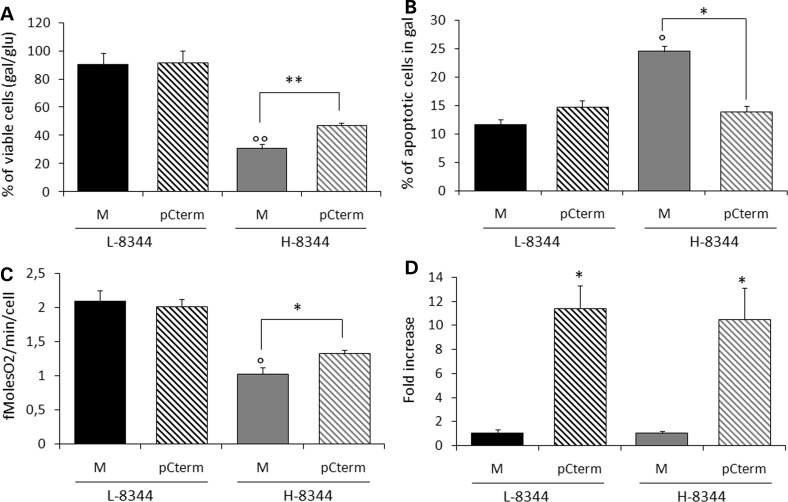
We transiently expressed the Cterm in H-8344 and L-8344 cybrids. This resulted in increased cell viability of H-8344 cybrids after 24 h incubation in galactose medium (up to 1.5-fold as compared to mock transformants) and decreased apoptosis after 6 h (up to 1.6-fold when compared with mock transformants) (Fig. 1A and B). Moreover, Cterm transformants showed a significant increase in oxygen consumption when compared with mock transformants (up to 1.4-fold increase) (Fig. 1C). Conversely, Cterm overexpression did not affect viability of isogenic L-8344 cybrids (Fig. 1A and C). For each experiment, we measured transfection efficiency by quantitative real-time polymerase chain reaction (PCR) (Fig. 1D).
Figure 1.
The carboxy-terminal domain of human mt-LeuRS (Cterm) is able to rescue the phenotype of m.8344A>G MTTK mutant cybrids. (A) Viability of mock (M) and Cterm (pCterm) transformants evaluated after 24 h incubation in galactose medium. The number of viable cells in galactose is normalized to the number of viable cells in glucose at the same time point. (B) Apoptotic cell death of M and pCterm evaluated after 6 h incubation in galactose medium. (C) A respiration rate measured in M and pCterm. (D) Relative expression levels of Cterm in transformant cybrids with respect to 18S gene. Gene expression levels are normalized to the gene expression level of the relative mock. L-8344 and H-8344: low- and high-percentage m.8344A>G mutant cells. Results are the mean ± SEM of at least triplicate transfection experiments. °P < 0.05, °°P < 0.01 for H-8344 mock cells versus L-8344 mock cells. *P < 0.05, **P < 0.01 for transformants versus mock cells (T-test).
Cterm-derived peptides are endowed with rescuing ability towards both m.3243A>G MTTL1 and m.8344A>G MTTK mutant cybrid phenotypes
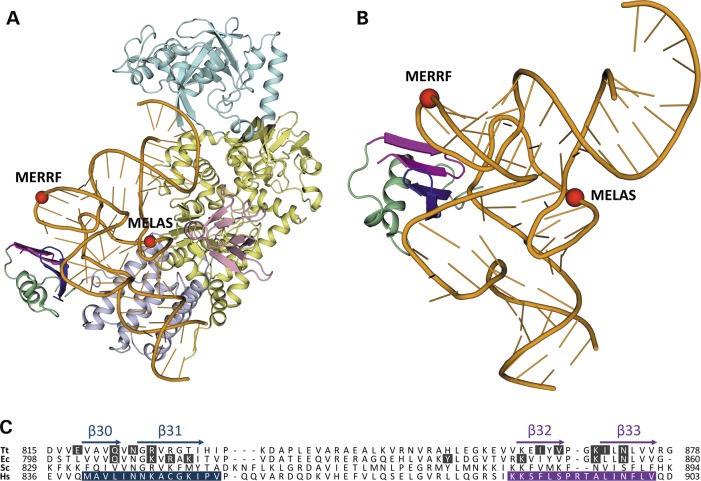
The structural basis of the interaction between the Cterm of human mt-LeuRS and the cognate tRNA can be inferred from the analysis of the experimentally determined 3D structures of LeuRS-tRNALeu complexes from the bacteria Thermus thermophilus (14) and Escherichia coli (15) available from the Protein Data Bank [PDB; (18)]. As shown in Figure 2, two pairs of β-strands in the carboxy-terminal domain of bacterial LeuRS (whose human homologues were named β30_31 and β32_33 (16) comprise most of the residues that are involved in contacts with the elbow region of tRNALeu. Thus, we tested the hypothesis that the short peptides from the human enzyme were responsible for the Cterm ability to interact with, and have rescuing activity towards, mutations in human cognate mt-tRNALeu(UUR) and non-cognate mt-tRNALys.
Figure 2.
Structural basis of the interaction between peptides derived from LeuRS and cognate tRNALeu. (A) A 3D structure of LeuRS-tRNALeu complex from T. thermophilus (14). The structural domains of LeuRS are represented as ribbon and coloured as follows: catalytic, leucine-specific, editing and anticodon-binding domains are yellow, pink, cyan and lilac, respectively; peptides β30_31 and β32_33 are blue and magenta, respectively, and the rest of the carboxy-terminal domain is green. tRNALeu is coloured orange and tRNA bases are shown as sticks. The phosphorous atoms of nucleotides 14 and 55 at the beginning of the D- and T-loop, respectively, corresponding to tRNA regions where pathogenic point mutations occur in MELAS (m.3243A>G) and MERRF (m.8344A>G) diseases, are shown as red spheres. (B) Zoom-in of the interaction between peptides β30_31 and β32_33, within the carboxy-terminal domain of LeuRS, and the ‘elbow’ region of tRNALeu, where the two branches of the ‘L’-shaped tertiary structure join to each other. Colour-coding is as in panel A. (C) Multiple sequence alignment of the carboxy-terminal domains of LeuRS from T. thermophilus (Tt) and E. coli (Ec), and of mt-LeuRS from Saccharomyces cerevisiae (Sc) and Homo sapiens (Hs). The secondary structure elements of Tt-LeuRS are reported above the alignment: α, alpha-helix; β, beta-strand. Residues in contact with tRNALeu in the experimentally determined 3D structures of Tt-LeuRS (14) and Ec-LeuRS (15) have a black background. Residues belonging to peptides β30_31 and β32_33 of human mt-LeuRS have a blue and magenta background, respectively.
The constructs designed for transfection experiments comprise: the mt-targeting sequence (MTS) of human COX8a gene, to ensure mt localization; the nucleotide sequence coding for peptide β30_31, β32_33 or Cterm (as a positive control) and a FLAG domain (for more details see ‘Materials and Methods’ section).
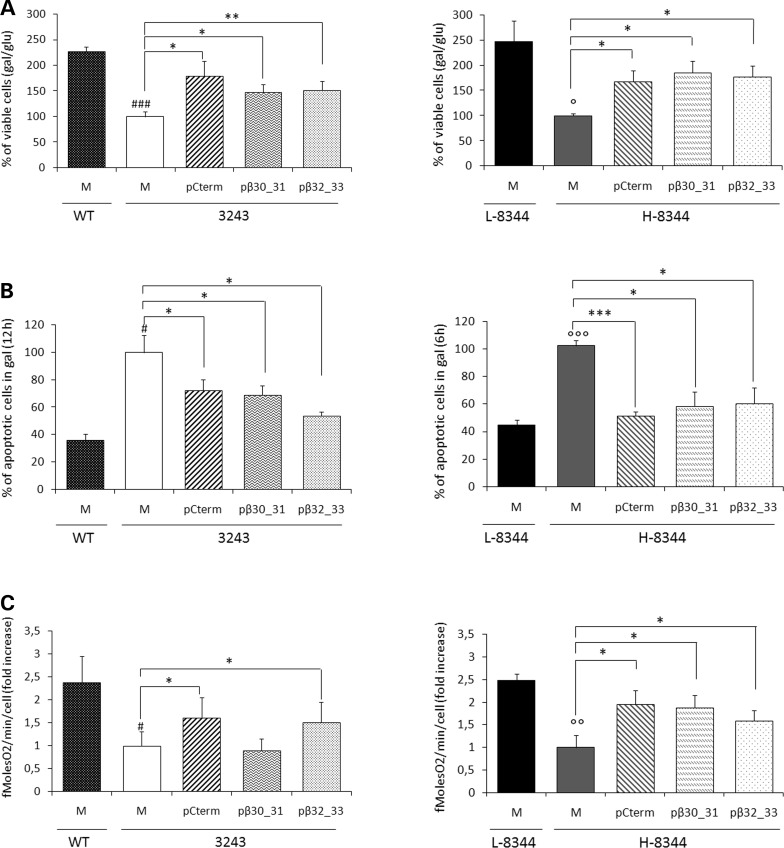
We transiently overexpressed either β30_31 or β32_33 peptide in cybrid lines, which were subsequently stressed by galactose medium incubation. In these conditions, transformant cells showed: (i) increased viability (up to 1.8- and 1.7-fold for m.3243A>G and m.8344A>G cybrids, respectively, when compared with their mock transformants); and (ii) decreased apoptotic cell death (up to 1.6- and 2-fold for m.3243A>G and m.8344A>G cybrids, respectively, when with to their mock transformants) (Fig. 3A and B). Conversely, peptide overexpression had no effect on the viability of WT and L-8344 cybrids (Supplementary Material, Fig. S2). To investigate whether increased cell viability was actually related to improved mt bioenergetics, we measured oxygen consumption rate in transfected cybrids. Peptide β32_33 increased oxygen consumption in both m.3243A>G and m.8344A>G cybrids, when compared with the respective mock transformants; conversely, peptide β30_31 increased oxygen consumption in m.8344A>G cybrids only (Fig. 3C).
Figure 3.
Short peptides derived from the Cterm improve cell viability and energetic proficiency of both m.3243A>G MTTL1 and m.8344A>G MTTK mutant cybrids. (A) Viability of mock, Cterm, β30_31 and β32_33 transformant cybrids (M, pCterm, pβ30_31 and pβ32_33) bearing either the m.3243A>G point mutation (left panel) or high levels of the m.8344A>G point mutation (right panel), evaluated after 24 h incubation in galactose medium. The number of viable cells in galactose medium is normalized to the number of viable cells in glucose at the same time point. (B). Apoptotic cell death of m.3243A>G (left panel) or H-8344 (right panel) M, pCterm, pβ30_31 and pβ32_33 evaluated after 6–12 h incubation in galactose medium. (C) The rate of oxygen consumption of m.3243A>G (left panel) or H-8344 (right panel). WT, wild-type; 3243, m.3243A>G mutant cells; L-8344 and H-8344, low- and high-m.8344A>G mutation load. Data are normalized to the value obtained in mock mutant cells. Results are the mean ± SEM of at least triplicate transfection experiments. #P < 0.05, ###P < 0.001 for 3243 mock mutant cells versus WT mock cells; °P < 0.05, °°P < 0.01, °°°P < 0.001 for H-8344 mock cells versus L-8344 mock cells for H-8344 mock cells; *P < 0.05, **P < 0.01, ***P < 0.001 for transformants versus mock cells (T-test).
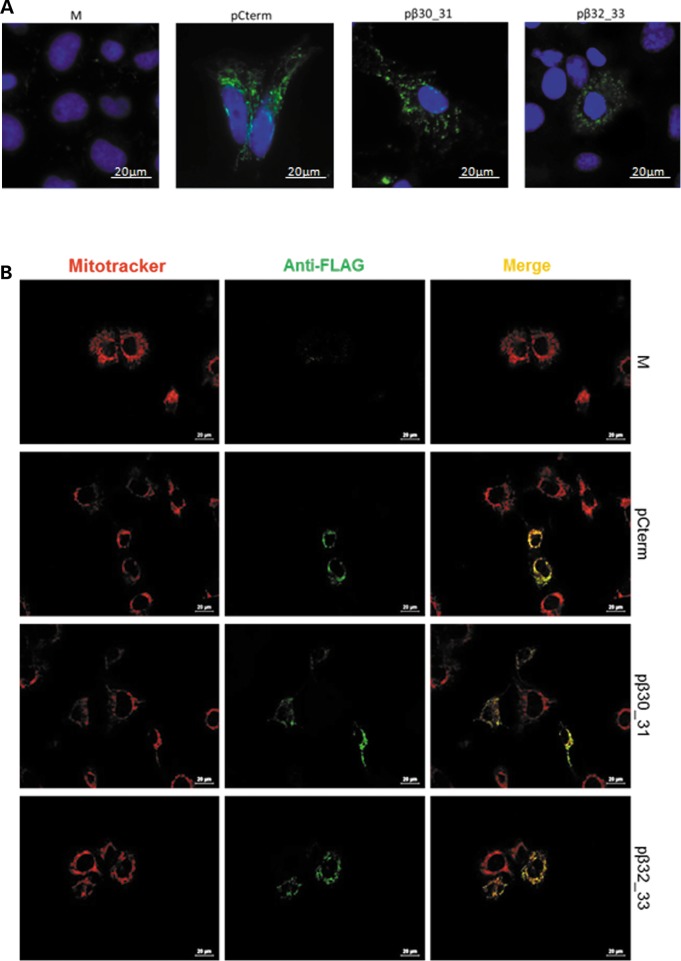
We verified the transient over-expression of both Cterm and Cterm peptides by real-time PCR for all experiments and by immunofluorescence for a subset of experiments (see the ‘Materials and Methods’ section). Upon transfection, both peptide and Cterm transformants showed increased mRNA expression levels (Supplementary Material, Fig. S3) as well as intense cytoplasmic granular stain with specific anti-FLAG antibodies which was evident in about 50% of transfected cells (Fig. 4A). In addition, mt localization following transient transfection was verified by using a specific anti-FLAG antibody in combination with Mitotracker Red CMXRos (see Materials and Methods section). As shown in Figure 4B, the peptide-specific immunofluorescence pattern (green) perfectly superimposes the mt network (red).
Figure 4.
The Cterm- and short-peptide derivatives are over-expressed and colocalize to mitochondria. (A) Representative images showing the over-expression of MTS-Cterm-FLAG, MTS-β30_31-FLAG and MTS-β32_33-FLAG constructs in transformant cybrids. Immunofluorescence shows M, pCterm, pβ30_31 and pβ32_33 stained with a specific anti FLAG antibody. Nuclei are stained with 4′,6-diamidino-2-phenylindole (scale bar: 20 µm). (B) Stain of mock, Cterm, β30_31 and β32_33 transformants (M, pCterm, pβ30_31 and pβ32_33) showing colocalization of the antibody against the FLAG and the Mitotracker Red CMXRos. Overlap is shown in yellow. WT cells were examined and imaged with an Olympus IX50 fluorescence microscope (scale bar: 20 µm).
We also investigated whether transfection with either peptide affected the steady-state levels of mt-tRNA mutants. High-resolution northern blot analysis was performed in WT and m.3243A>G MTTL1 or m.8344A>G MTTK mutant cybrids transiently transfected with peptide β30_31 or β32_33. Values were normalized for 5S rRNA. Consistent with our previous report on m.3243A>G MTTL1 mutant cybrids transfected with the Cterm (9), transient overexpression of either peptide did not result in a significant increase of the mutated mt-tRNALeu(UUR) or mt-tRNALys steady-state levels (Supplementary Material, Fig. S4). We performed additional experiments on m.3243A>G MTTL1 mutant cybrids stably transfected with the Cterm, as a ‘long term’ overexpression model. These resulted in a significant increase in the steady-state levels of mutated mt-tRNALeu(UUR) (Supplementary Material, Fig. S4), although these changes were at the lower limits of detection for northern experiments.
Cterm-derived peptides directly interact with both human mt-tRNALeu(UUR) m.3243A>G and mt-tRNALys m.8344A>G mutants with high affinity in vitro
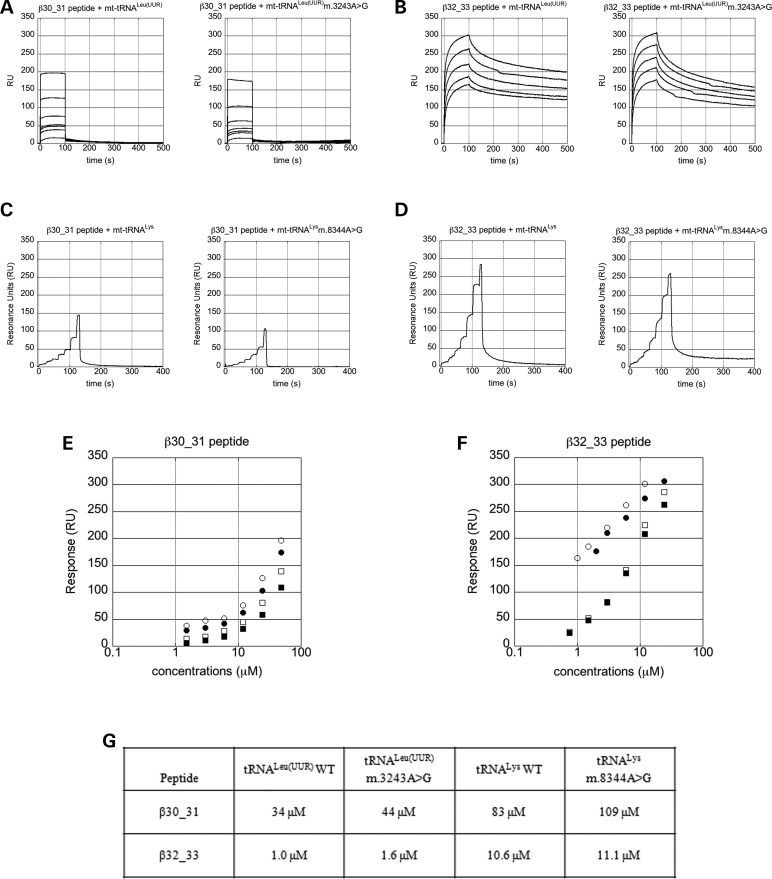
The ability of peptides β30_31 and β32_33 to interact with the WT and mutated tRNAs under investigation was evaluated by surface plasmon resonance (SPR) experiments. These were carried out by immobilizing N-biotinylated peptides onto BioCap sensorchips and adding human mt-tRNALeu(UUR) (WT or m.3243A>G mutant), or human mt-tRNALys (WT or m.8344A>G mutant) as analytes. Figure 5 shows the sensorgrams of each of the eight evaluated interactions (A–D), a logarithmic plot of all experiments with peptides β30_31 (E) and β32_33 (F), and a table with the KD values of each peptide for each tRNA, obtained from Scatchard analysis of all the experiments (G).
Figure 5.
Short peptides derived from the Cterm interact directly and with high affinity with both human mt-tRNALeu(UUR) m.3243A>G and mt-tRNALys m.8344A>G mutants in vitro. (A–D) Sensorgrams of the interaction between N-biotinylated β30_31 (A and C) or β32_33 (B and D) peptide, immobilized on BioCap sensorchips, with human WT (left panels) of mutant (right panels) mt-tRNAs. (A). mt-tRNALeu(UUR) (left) and mt-tRNALeu(UUR) m.3243A>G mutant (right); tRNA concentrations: 1.5; 3.0; 6.0; 12; 24 and 48 µm. (B) mt-tRNALeu(UUR) (left; concentrations = 0.8; 1.5; 3.0; 6.0 and 12 µm) and mt-tRNALeu(UUR) m.3243A>G mutant (right; concentrations: as in (A). (C) mt-tRNALys (left) and mt-tRNALys 8344A>G mutant (right); tRNA concentrations: 0.75; 1.5; 3.0; 6.0; 12; 24 and 48 µm at 0–20; 21–40; 41–60; 61–80; 81–100; 101–120 and 121–130 s, respectively. (D) mt-tRNALys (left) and mt-tRNALys m.8344A>G mutant (right); tRNA concentrations: 0.37 µm; 0.75 µm; 1.5 µm; 3.0 µm; 6.0 µm; 12 µm; 24 µm at the same time points as in (C). (E and F) Logarithmic plot of the interaction of N-biotinylated β30_31 (E) and β32_33 (F) peptide with human WT mt-tRNALeu(UUR) (white circle), mt-tRNALeu(UUR) m.3243A>G mutant (black circle), human WT mt-tRNALys (white square) and mt-tRNALys m.8344A>G mutant (black square). The maximal response is plotted versus the logarithm of tRNA concentration. (G) KD values of the interactions between peptides and tRNAs.
Both peptides interact directly and with high affinity with all four tRNAs, although the strength of the interactions differs substantially. Unsurprisingly, the most efficient interactions are with the cognate human mt-tRNALeu(UUR) forms. Peptide β30_31 shows KD = 34 and 44 µm with WT and m.3243A>G mutant, respectively, while peptide β32_33 interacts with even higher affinity with both forms (KD = 1.0 and 1.6 µm with WT and m.3243A>G mutant). Remarkably, these values are very similar to the affinity of the whole Cterm for mt-tRNALeu(UUR) [i.e. 0.6 µm; (9)], indicating that peptide β32_33 comprises most of the Cterm residues required for cognate mt-tRNA recognition. Interestingly, peptide β32_33 has higher affinity than β30_31 towards non-cognate human mt-tRNALys as well (Fig. 5). These results suggest that peptides β30_31 and β32_33 might be able to recognize general tRNA features and, therefore, bind additional non-cognate tRNA molecules.
Cterm-derived peptides compensate structure and thermal stability impairments of both human mt-tRNA mutants in vitro
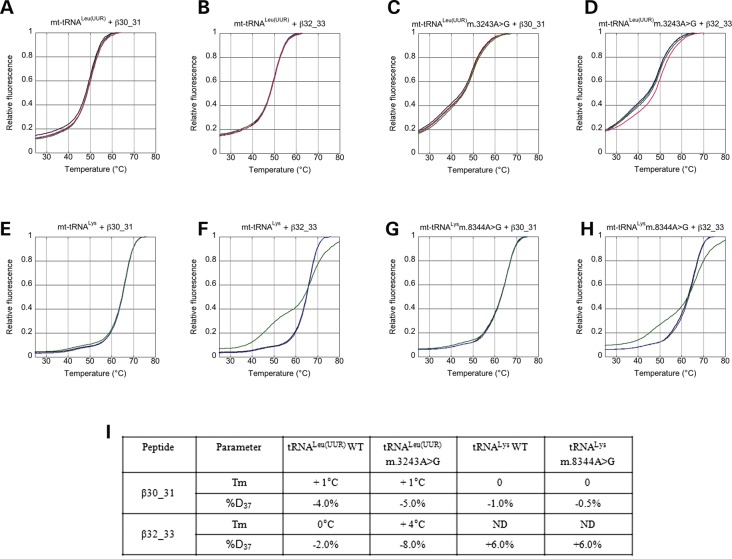
Thermal denaturation (TD) experiments were carried out to elucidate whether, and to what extent, the interaction with peptides β30_31 and β32_33 reverts the structure and thermal stability alterations that occur in human mt-tRNALeu(UUR) and mt-tRNALys as a consequence of the m.3243A>G and m.8344A>G mutations, respectively. To this end, tRNAs were synthesized as molecular beacons (MB-tRNAs) with chemical groups able to emit and quench fluorescence at the 5′ and 3′ ends, respectively.
In all experiments, low fluorescence was observed at low temperature, due to the vicinity of the 6-carboxyfluorescein (FAM) fluorophore and DABCYL quencher (at 5′ and 3′ end, respectively), and high fluorescence upon thermal melting, due to decreased fluorescence quenching caused by tRNA unfolding. Figure 6A–H shows the results of thermal shift assays (average of three experiments) performed on human MB-mt-tRNALeu(UUR) WT and m.3243A>G mutant (A–D), and MB-mt-tRNALys WT and m.8344A>G mutant (E–H), in the presence of different concentrations of peptide β30_31 or β32_33. Figure 6I reports the values of MB-tRNAs melting temperature (Tm) and percentage of folding at 37% (%Folded37) calculated for each series of experiments. The thermal and structural stability of MB-mt-tRNALeu(UUR) m.3243A>G mutant are lower than the WT in the absence of peptides (Tm = 48°C for both forms but %Folded37 = 66% for the mutant and 82% for the WT) and increase significantly upon addition of peptide β30_31 and β32_33 up to 2 µm. Consistent with the results of tRNA-binding affinity, peptide β32_33 has the highest stabilization effect (Tm increases by 4°C versus 1°C and %Folded37 increases by 8 versus 5%). As expected, WT MB-mt-tRNALeu(UUR) undergoes only small changes in thermal and structural stability in the presence of peptide β30_31 up to 3 µm and β32_33 up to 2 µm (see Fig. 6). Intriguingly, the structures of MB-mt-tRNALys (WT and m.8344A>G mutant) are slightly stabilized by peptide β30_31 but not by the addition of peptide β32_33.
Figure 6.
Short peptides derived from the Cterm restore thermal stability and structure of mutated mt-tRNALeu(UUR). (A–H) Thermal shift assays (average of three experiments) performed on human WT mt-tRNALeu(UUR) (A and B), mt-tRNALeu(UUR) m.3243A>G (C and D), WT mt-tRNALys (E and F) and mt-tRNALys m.8344A>G (G and H) in the presence of 1 mm Mg2+, 0.1 mm spermine (Spm) and different concentrations of β30_31 (A, C, E and G) or β32_33 (B, D, F and H) peptide (A–D: black, 0 µm; blue, 0.2 µm, green, 0.5 µm; red, 2 µm; (E–H): black, 0 µm; blue, 0.4 µm; green, 2 µm). (I) Table showing the maximum observed variation in Tm and %Folded37 (see the ‘Materials and Methods’ section) upon treatment with peptides.
All TD experiments were highly reproducible. Melting curves measured in the same experimental conditions from different samples are entirely superimposable, with standard deviations for Tm values below 1°C for each measurement.
Supplementary Material, Figure S5 section shows gel electrophoresis of 1 µm mt-tRNALeu(UUR) m.3243A>G samples incubated in 20 mm HEPES (pH 7.4), 150 mm NaCl, 1 mm MgCl2 and 0.1 mm Spermine (Spm) for 15′ in the presence of β30_31 or β32_33 peptides. The interaction with 10 µm peptides decreases by ∼7–10% the amount of mt-tRNALeu(UUR) m.3243A>G mutant in the dimer form (upper band), first reported by Wittenhagen and Kelley (19), whereas in the same conditions WT mt-tRNALeu(UUR) is fully monomeric.
Discussion
In the present study, we expand recent results previously reported by our group and others about the ability of human mt-LeuRS Cterm to interact with both cognate and non-cognate mt-tRNAs, and rescue pathological phenotypes arising from mt-tRNA mutations in human cells (9,10). While the Cterm had been shown to interact with a number of non-cognate mt-tRNAs aminoacylated by both class I and II aminoacyl-tRNA synthetases (aaRSs) (10), its rescuing ability had been demonstrated only towards mt-tRNAs aminoacylated by aaRSs belonging, like LeuRS, to class Ia (9). Since aaRSs belonging to the same class and subclass have higher structure and sequence similarity to one another than to other aaRSs, whether the therapeutic properties of the Cterm could be extended to mutations in mt-tRNAs aminoacylated by aaRSs other than those in class Ia remained an open question.
Here, we show that, remarkably, the Cterm is able to ameliorate the phenotype of cybrid cells carrying the m.8344A>G mutation in mt-tRNALys, which has the following features: (i) the cognate LysRS belongs to class II and thus is not evolutionarily related to class I enzymes; (ii) it is one of the most prevalent among disease-causing mt-tRNA mutations; (iii) it is responsible for a severe syndrome (MERRF). Together with recent observations in yeast (16,17), these results strongly support the hypothesis that the Cterm may rescue defects due to mutations in a number of additional mt-tRNAs aminoacylated by aaRSs belonging to both classes. This makes the Cterm a promising candidate as ‘universal suppressor’ for mt-tRNA mutation-associated diseases.
Importantly, we demonstrate here that not only LeuRS C-terminal domain but also two short peptides derived from it, β30_31 and β32_33, are endowed with rescuing ability towards two mutations: m.3243A>G in mt-tRNALeu(UUR) and m.8344A>G in mt-tRNALys. The ability of both peptides to rescue pathological phenotypes due to mutations in a number of mt-tRNAs, which are aminoacylated by aaRSs belonging to different classes and subclasses, has been recently suggested in yeast (17). Our results on the activities of peptides β30_31 and β32_33, together with the overall LeuRS sequence conservation from eukaryote mitochondria to bacterial cells indicate that, in spite of the evolutionary distance and different cellular compartment, the modality of tRNA recognition and regions involved in tRNA binding are largely conserved.
The overall rescuing activity of peptides β30_31 and β32_33 is manifested by increased cell viability and decreased apoptotic cell death, as is the case with the Cterm (9). Intriguingly, while peptide β32_33 restores oxygen consumption of both mutant cybrids, upon transfection with peptide β30_31 this effect is observed only in m.8344A>G cybrids, suggesting a different effect of this peptide on the respiratory chain function affected by either mutation.
Despite the significant increase in cell viability, we did not detect an increase of either mutant mt-tRNALeu(UUR) or mt-tRNALys steady-state levels upon transient overexpression of either peptide β30_31 or β32_33. These results are in line with our previous observations on m.3243A>G cybrids transiently transfected with the Cterm (9) and suggest that, in agreement with our hypothesis of a chaperonic mechanism of action, the effect of our molecules is mainly to improve the function of the mutant mt-tRNAs, rather than preventing them from degradation. On the other hand, we observed a slight, albeit significant increase of mutant mt-tRNALeu(UUR) steady-state levels after stable transfection of m.3243A>G cybrids with the Cterm. Thus, on a longer timescale an effect on mutant mt-tRNA steady state levels can be highlighted, which may contribute to the rescuing activity.
The results of our in vitro experiments contribute to elucidate the molecular basis of the rescuing activity of peptides β30_31 and β32_33 and support the hypothesis that they act as molecular chaperones, as previously proposed for the Cterm. In fact, in spite of their small size, the two peptides were able to bind directly and with high affinity to all WT and mutant mt-tRNAs investigated in this work. These results suggest that peptide-tRNA interactions are mediated by structural features shared by tRNA molecules, consistent with the rationale behind peptide design. Accordingly, both peptides bind WT mt-tRNAs with higher affinity than their mutant counterparts, the structure of which is affected by the presence of the mutation. Peptide β32_33 showed a significantly higher binding affinity than peptide β30_31 towards each and all of the investigated tRNA forms, indicating that it is a better candidate for future therapeutic applications.
To investigate the effects of peptide binding on the mt-tRNA structure and stability, we performed thermal melting experiments on tRNAs modified to act as MB-tRNAs. These experiments highlighted that the m.3243A>G mutation induces a thermal and structural destabilization of mt-tRNALeu(UUR), both of which are reversed by association with peptide β32_33 and, to a lesser extent, β30_31. These results support the hypothesis that the two peptides act like molecular chaperones, by stabilizing a mt-tRNALeu(UUR) conformation suitable to establish interactions with partner molecules (e.g. aaRSs, elongation factors, ribosome, mRNAs etc.) that are required to enact tRNA function. In addition, the mutation-dependent dimerization of mt-tRNALeu(UUR) m.3243A>G, which may contribute to impairment of the tRNA function, is substantially decreased by the interaction with peptides β30_31 and β32_33 (Supplementary Material, Fig. S5). Conversely, we could not detect a significant increase in either thermal stability or folding of the mt-tRNALys m.8344A>G mutant, despite the ability of both peptides to directly interact with both WT and mutant tRNALys with high affinity. This result may be ascribed to the absence of post-transcriptional modifications in the in vitro synthesized WT and mutant mt-tRNALys used in our experiment, since editing has been proposed to play an important role in the stabilization of a canonical two-dimensional and 3D mt-tRNALys structure (20,21). Alternatively, in the case of the mt-tRNALys mutant the rescuing effect of the peptides may be mediated by a non-chaperonic mechanism, consisting, for example, in the protection from degradation. Although we cannot exclude that the effect of β32_33 peptide on mt-tRNALys stability depends on peptide binding to regions different from those recognized in ‘cognate’ mt-tRNALeu(UUR), the ability of the β32_33 peptide to rescue defects due to mutations in a large panel of mt-tRNA mutants previously demonstrated in yeast (17) supports the hypothesis that common structural features of tRNA molecules are at least partially involved in the interaction.
In conclusion, we provide evidence that small peptides derived from the mt-LeuRS Cterm can be effective tools to rescue cellular defects caused by mutations in mt-tRNAs. As a consequence, these peptides represent an important advance in the quest for therapeutic agents against diseases caused by mutations in mt-tRNAs. The whole Cterm is amenable to gene therapy-based approaches [see for example (22,23)], whilse small peptides could be more directly administered, possibly aided by suitable carrier molecules and/or mitochondria-targeting agents. Additionally, small peptides may serve as a basis for the development of non-peptide synthetic compounds, particularly if the rescuing activity can be mapped to even smaller peptide regions. Strikingly, the ability of the peptide to rescue defects due to mutations in ‘non-cognate’ in addition to ‘cognate’ mt-tRNAs suggests that they may be used to develop wide-spectrum therapeutics towards mt-tRNA mutation-associated diseases.
Materials and Methods
Cybrid cell lines
For this study, we used previously established osteosarcoma-derived (143B.TK−) cybrid cell lines from patients (24) and controls [(25); generous gift from Dr Valeria Tiranti and Dr Valerio Carelli] (Supplementary Material, Table S1). The m.8344A>G cybrids were established in Valeria Tiranti's laboratory as previously described (26). We then manipulated the ratio of mutant and WT mt-DNA of the m.8344A>G cybrids using EtBr. Cybrid cells were maintained in glucose medium supplemented with 20 ng/ml of EtBr for 10 days as previously described (27) and replated to repopulate mitochondria at low density in glucose medium. Individual cell clones (∼50) were isolated 10–20 days later using glass cylinders and tested for the mutation load. For the present study, we selected two clones, showing high (86%) and low (34%) levels of m.8344A>G mutation, respectively.
Assessment of m.8344 A>G mutation load
Accurate m.8344A>G heteroplasmy levels were obtained by quantitative pyrosequencing using a Pyromark Q24 platform. The PyroMark assay design software v2.0 (Qiagen, Hilden, Germany) and the current mtDNA reference sequence (Genbank accession number: NC_012920.1) were used to design mutation-specific pyrosequencing primer trios [5′biotinylated forward: m. 8240–8264; reverse: m. 8364–8387 and reverse pyrosequencing primer: m.8348–8367 (IDT, Coralville, IA, USA)]. The allele quantification application of the proprietary Q24 software was used to calculate mtDNA heteroplasmy levels using standard parameters.
Cell culture
Cybrid cells were cultured in Dulbecco's modified Eagle's medium (DMEM), supplemented with 4.5 g/l d-glucose, 10% fetal bovine serum (FBS), 2 mm l-glutamine, 50 µg/ml uridine, 100 U/ml penicillin and 100 mg/ml streptomycin (referred to as glucose medium) in a humidified atmosphere of 95% air and 5% CO2 at 37°C. For a subset of experiments cybrids were grown in glucose-free DMEM, supplemented with 5 mm galactose, 110 mg/ml sodium pyruvate and 10% FBS (referred as galactose medium).
Bioinformatics analysis
Structure visualization and analysis were performed using PyMol (http://www.pymol.org/). Cterm residues interacting with tRNALeu in experimentally determined 3D structures (14,15) were calculated using the FACE2FACE server developed by the authors (http://apps.ibpm.cnr.it/f2f/index). The multiple sequence alignment shown in Figure 2C was obtained by merging the alignments of human and yeast mt-LeuRS to LeuRS from T. thermophilus and E. coli obtained using Phyre2 (28).
Plasmid construction
Cybrid cells were transiently transfected with either the C-terminal region of human mt-LeuRS (named pCterm) or the short peptides derived from it (named pβ30_31 and pβ32_33) cloned into pcDNA3.2-TOPO vector (Invitrogen™, Life Technologies Italia, MB, Italy). The cDNA fragments (listed in Supplementary Material, Table S2) were fused at 5′ with the mitochondrial-targeting sequence (MTS) derived from human COX8 gene (29) and at 3′ with the FLAG sequence (5′-GATTACAAGGATGACGACGATAAG-3′). Both the MTS and the FLAG sequences were separated from the peptide sequence by a 5-residue linker (GGSGG).
Transient and stable transfections
Cybrids were plated in 60 mm dishes, incubated in glucose medium for 24 h, and then transfected with 2.5 µg of the full or empty vector (mock) by using LIPOFECTINE 3000 (Life Technologies Italia), according to the manufacturer's protocol. Stable clones were selected and maintained in glucose medium supplemented with G418 (300 µg/ml). Prior to testing the effect of our constructs, we verified that the mutation load of m.3243A>G and m.8344A>G mutant cybrids did not change upon transfection (Supplementary Material, Fig. S6). To assess m.3243A>G mutation load we used the assay previously described by McDonnel et al. (30). Briefly, WT 154 base-pairs (bps) PCR products amplified with forward (5′-TATACCCACACCCACCCAA-3′) and reverse (5′-GCGATTAGAATGGGTACAAT −3′) primers contain a single HaeIII restriction site, which cleaves the amplicons into two fragments of 117 and 37 bps. The m.3243A>G mutation introduces a second HaeIII recognition site that cuts the 117 bps fragment into two smaller fragments of 72 and 45 bps. To assess m.8344A>G mutation load, we used the assay previously described by Larsson et al. (31). Briefly, WT 230 bps PCR products, amplified with forward (5′-CTACGGTCAATGCTCTGAAA-3′) and reverse (5′-ATACGGTAGTATTTAGTTGGGGCATTTCACTGTAAAGCCGTGTTGG-3′) mismatch primers, do not contain any BglI restriction site. The m.8344A>G mutation introduces a BglI recognition site that cuts the 230 bps fragment into two smaller fragments of 184 and 46 bps.
Restriction fragments were separated and identified using the Agilent 2100 bioanalyzer instrument with the 1000 Lab Chip kit (Supplementary Material, Fig. S6).
Evaluation of transient transfection efficiency by real-time PCR
About 24 h after transfection total RNA was isolated, measured and reverse-transcribed to cDNA as previously described (9). To assess the relative expression of MTS-Cterm-FLAGwe used TaqMan probe chemistry by means of custom FAM-labelled TaqMan MGB probe (Applied Biosystems Warrington, UK; Supplementary Material, Table S3), according to the manufacturer's instructions. In all samples, the relative expression of the target gene was evaluated with respect to the average of controls (mock sample) using the comparative threshold cycle (ΔCt) method. All values were normalized for the housekeeper 18S ribosomal RNA (18S) using inventoried FAM-labelled TaqMan MGB probes (Supplementary Material, Table S3). Due to the shortness of Cterm-peptides the primers of the real-time PCR assays were design to anneal, respectively, with the MTS and the FLAG sequence (Supplementary Material, Table S3). These assays prevented the normalization of over-expressed β30_31 and β32_33 relative to the endogenous levels of mt-LeuRS in the mock.
Evaluation of constructs expression levels by immunofluorescence
About 24 h after transfection cells were fixed with 4% formaldehyde freshly prepared from paraformaldehyde in phosphate buffered saline (PBS) (pH 7.4) with 0.1% Triton X-100. Mouse monoclonal anti-FLAG (Sigma, Saint Louis, MO, USA) was used as primary antibody and visualized using secondary fluorescein isothiocyanate-conjugated antibody (Jackson Laboratories, Bar Harbor, ME, USA). Cells were examined and imaged with an Olympus IX50 Fluorescence microscope.
Mt localization of constructs
About 24 h after transfection cells were incubated with 100 nm Mitotracker red CMXRos (Life Technologies Italia) for 20 min at 37°C and then fixed with 4% formaldehyde freshly prepared from paraformaldehyde in PBS (pH 7.4) for 15 min at 37°C. Subsequently cells were incubated with mouse monoclonal anti-FLAG (Sigma) and visualized by mean of secondary Alexa fluor goat anti-mouse 488 (Life Technologies Italia).
Cell proliferation assay
To test growth capability, transformed cells were maintained in glucose medium for 24 h and then harvested and seeded at 30 × 104 in 60 mm dishes in glucose or galactose. Cell viability was measured by the Trypan blue dye exclusion assay. Cells were harvested after 24 h with 0.25% trypsin and 0.2% ethylenediaminetetraacetic acid, washed, suspended in PBS in the presence of Trypan blue solution (Sigma) at 1:1 ratio, and counted using a haemocytometer. The number of viable cells in galactose medium was expressed as a percentage of the number of cells in glucose medium.
Rate of apoptosis by flow cytometer
To test the rate of apoptosis transformed cells were maintained in glucose medium for 24 h and then harvested and seeded at 0.5–1 × 106 in glucose or galactose for 6–24 h. Cells were washed and harvested in binding buffer 1× (BD Biosciences, Franklin Lakes, NJ, USA) by scraper to minimize potentially high annexin V background levels in adherent cells. Transfected cells were stained with APC-conjugated annexin V (BD Biosciences) and analysed on a FACS-Calibur flow cytometer (BD Biosciences) (32).
Respirometry assay
Oxygen consumption rate (OCR) of transfected cybrids was evaluated with Clark type oxygen electrode (Hansatech Instruments, Norfolk UK). After transfection control and mutant cybrids were maintained in glucose medium for 48 h, then OCR was measured in intact cells (3 × 106) in 1 ml DMEM lacking glucose supplemented with 10% sodium pyruvate, as previously described (33).
High-resolution northern
To determine mt-tRNALeu(UUR) and mt-tRNALys steady-state levels cybrids were maintained in glucose medium for 12 h and then collected for RNA extraction. Total RNA was obtained using Trizol reagent (Life Technologies, Paisley, UK). RNA (2 µg) was electrophoresed through 13% denaturing polyacrylamide gel, electroblotted onto Hybond N+ membrane (GE Healthcare Life Sciences, Little Chalfont, UK) in 0.25× TBE and immobilized by ultraviolet cross-linking. Hybridization with radiolabelled probes was performed as previously described (34). Primers for generating the mt-tRNALeu(UUR) and mt-tRNALys probes were L3200 (positions 3200–3219; GenBank accession number: NC_012920.1 for human mtDNA) and H3353 (positions 3353–3334), and L8246 (positions 8246–8266) and H8390 (positions 8390–8371), respectively. Steady-state mt-tRNA levels were normalized to that of 5S rRNA (RN5S17; GenBank accession number: NR_023379.1); primers for generating the 5S probe were F1 (positions 1–23) and R121 (positions 121–101).
SPR experiments
SPR experiments were carried out with a SensiQ Pioneer apparatus. N-biotinylated β30_31 and β32_33 peptides (see Fig. 2) were synthesized by Biomatik (purity > 90%). Streptavidin-coated BioCap sensorchips were chemically activated by a 35 µl injection of 10 mm NaOH at 10 µl/min flow rate. N-biotinylated peptides were immobilized via interaction with the streptavidin surface of the chips. This procedure ensured directional immobilization of the peptides. The amount of immobilized peptides was detected by mass concentration-dependent changes in the refractive index on the sensor chip surface, and corresponded to ∼50 resonance units (RU).
Samples of human WT mt-tRNALeu(UUR), mt-tRNALeu(UUR) m.3243A>G, WT mt-tRNALys and mt-tRNALys m.8344A>G, were synthesized and modified by Trilink Biotechnologies. All tRNA samples were dissolved in sterile HEPES 20 mm pH 7.4, NaCl 150 mm, 1 mm MgCl2, heated to 80°C for 5 min and slowly cooled down to room temperature to allow renaturation.
The tRNA-binding experiments were carried out at 298 K in degassed 20 mm HEPES at pH 7.4, 0.15 M NaCl, 1 mm MgCl2 and 0.005% surfactant P-20 (HBS-MP buffer GE Healthcare Life Sciences, Milan, Italy). Sample injections were performed for experiments on both mt-tRNALeu(UUR) WT and m.3243A>G mutant for 100 s at 30 µl/min. Concentrations of (i) mt-tRNALeu(UUR) WT and m.3243A>G mutant with β30_31 peptide: 1.5; 3.0; 6.0; 12; 24 and 48 µm; (ii) mt-tRNALeu(UUR) WT with β32_33 peptide: 0.8; 1.5; 3.0; 6.0 and 12 µm and (iii) mt-tRNALeu(UUR) m.3243A>G mutant with β32_33 peptide: 1.5; 3.0; 6.0; 12 and 24 µm. For both human mt-tRNALys WT and m.8344A>G mutant, injections were carried out as follows. tRNAs were automatically diluted in HBS-MP and injected by seven serial doubling steps (step contact time = 20 s, nominal flow rate = 100 µl/min). At the following time points: (i) 0–20 s; (ii) 21–40 s; (iii) 41–60 s; (iv) 61–80 s; (v) 81–100 s; (vi) 101–120 s and (vii) 121–130 s, tRNA concentrations were: (i) 0.75; (ii) 1.5; (iii) 3.0; (vi) 6.0; (v) 12 µm; (vi) 24 µm and (vii) 48 µm with peptide β30_31 and (i) 0.37; (ii) 0.75; (iii) 1.5; (iv) 3.0; (v) 6.0; (vi) 12 µm; (vii) 24 µm with peptide β32_33. The increase in RU relative to baseline indicates complex formation between the immobilized N-biotinylated peptide and tRNA. The plateau region represents the steady-state phase of the interaction. The decrease in RU (after 100 s for WT and mutant mt-tRNALeu(UUR) and 130 s for WT and mutant mt-tRNALys) indicates tRNA dissociation from the immobilized peptide after buffer injection. A response change of 1000 RU typically corresponds to 1 ng/mm2 change in analyte concentration on the sensor chip.
As a negative control, sensor chips were treated as described above in the absence of immobilized N-biotinylated peptides. Values of the plateau signal at steady-state (Req) were calculated from kinetic evaluation of the sensorgrams using the Qdat 4.0 programme. The equilibrium constant of ligand–analyte interaction was evaluated by Scatchard analysis of the dependence of Req on analyte concentration.
TD experiments on tRNA-MBs
Samples of human WT mt-tRNALeu(UUR), mt-tRNALeu(UUR) m.3243A>G, WT mt-tRNALys and mt-tRNALys m.8344A>G were synthesized by Trilink Biotechnologies. All tRNAs used for TD experiments were synthesized as MB-tRNAs by using a 3′-DABCYL support for tRNA synthesis, and 5′ FAM modification was added using phosphoramidite chemistry. Dual polyacrylamide gel electrophoresis/high-pressure (or high-performance) liquid chromatography purification was performed for all samples, followed by a desalting step and subsequent lyophilization of the final compound. For all tested samples, purity was >95%. β30_31 and β32_33 peptides were synthesized by Biomatik (purity > 90%).
TD experiments were performed with a Mx3000P Real-Time PCR System (Stratagene, La Jolla, CA, USA) in PCR-96-microplates, using 25 µl sample volume, 0.14 µm MB-tRNAs in 20 mm HEPES pH 7.4 + 150 mm NaCl + 1 mm MgCl2 + 0.1 mm spermine (Spm), and different concentrations of β30_31 or β32_33 peptide. At least two experiments were performed for each condition, and triplicate samples were used within the same experiment. Sample temperature was increased from 25 to 90°C at 0.5°C/min rate. Fluorescence was measured at each temperature step (0.5°C) using FAM filter set (excitation: 492 nm, emission: 516 nm). As negative controls we used sample wells with 20 mm HEPES pH 7.4 + 150 mm NaCl + 1 mm MgCl2 + 0.1 mm spermine alone or in the presence of either MB-tRNA or different concentration of β30_31 or β32_33 peptide.
Normalized fluorescence values of the traces [relative fluorescence (F)] were obtained by dividing the fluorescence intensity of each sample by the maximum fluorescence of the same trace. For each temperature (T), the differential fluorescence (dF) value of each trace was obtained as follows: dF = F(T + 0.5°C)−F(T). For each experimental condition melting temperature values were calculated by determining the temperature of maximal dF, and the percentage of folded sample at 37°C (%Folded37) was calculated as follows: %Folded37 = 100 × [F(37°C) − Fmin]/[1 − Fmin], where Fmin is the minimum value of F for the group of experiments.
Gel electrophoresis
Gel electrophoresis of 1 µm mt-tRNALeu(UUR) m.3243A>G samples, incubated in 20 mm HEPES (pH 7.4), 150 mm NaCl, 1 mm MgCl2 and 0.1 mm Spm for 15′ in the presence of different amounts of β30_31 or β32_33 peptides was carried out in 12% native electrophoresis gel, and stained using SYBR Green. Dimerization percentage in each lane was calculated using the Fiji suite.
Statistical analysis
All data are expressed as mean ± SEM. Data were analysed by standard paired t-test procedures. The significance was considered at P < 0.05. Numerical estimates were obtained with the Graphpad InStat 3 version (Graphpad, Inc., San Diego, CA, USA).
Supplementary Material
Funding
This work is supported by AFM Télethon (to G.d.A.), Istituto Pasteur-Fondazione Cenci Bolognetti, the Associazione Serena Talarico per i giovani nel mondo (to G.d.A. and A.M.) and National Research Council of Italy (CNR) Interomics Flagship Project (to G.C. and V.M.). R.W.T. and H.A.T. were supported by a Wellcome Trust Strategic Award (096919/Z/11/Z); R.W.T. was supported by the MRC Centre for Neuromuscular Diseases (G0601943), the Lily Foundation and the UK NHS Highly Specialised ‘Rare Mitochondrial Disorders of Adults and Children’ Service in Newcastle upon Tyne. The financial support of Telethon – Italy (grant no. GGP13097 to G.d.A.) is gratefully acknowledged. Funding to pay the Open Access publication charges for this article was provided by Telethon-Italy.
Supplementary Material
Acknowledgements
We gratefully acknowledge Laura Frontali and Silvia Francisci (Department of Biology and Biotechnologies ‘Charles Darwin’ Sapienza University of Rome, Rome, I) for helpful discussions, Zofia Chrzanowska-Lightowlers, Robert Lightowlers (The Wellcome Trust Centre for Mitochondrial Research, Institute for Ageing and Health, The Medical School, Newcastle University, Newcastle upon Tyne, UK) and Massimo Zeviani (MRC-Mitochondrial Biology Unit, Cambridge, UK) for carefully reviewing the manuscript.
Conflict of Interest statement. None declared.
References
- 1.Lott M.T., Leipzig J.N., Derbeneva O., Xie H.M., Chalkia D., Sarmady M., Procaccio V., Wallace D.C. (2013) mtDNA variation and analysis using MITOMAP and MITOMASTER. Curr. Protoc. Bioinformatics, 1, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lightowlers R.N., Taylor R.W., Turnbull D.M. (2015) Mutations causing mitochondrial disease: what is new and what challenges remain? Science, 349, 1494–1499. [DOI] [PubMed] [Google Scholar]
- 3.Park H., Davison E., King M. (2008) Overexpressed mitochondrial leucyl-tRNA synthetase suppresses the A3243G mutation in the mitochondrial tRNALeu(UUR) gene. RNA, 14, 2407–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rorbach J., Yusoff A.A., Tuppen H., Abg-Kamaludin D.P., Chrzanowska-Lightowlers Z.M., Taylor R.W., Turnbull D.M., McFarland R., Lightowlers R.N. (2008) Overexpression of human mitochondrial valyl tRNA synthetase can partially restore levels of cognate mt-tRNAVal carrying the pathogenic C25 U mutation. Nucleic Acids Res., 36, 3065–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li R., Guan M.X. (2010) Human mitochondrial leucyl-tRNA synthetase corrects mitochondrial dysfunctions due to the tRNALeu(UUR) A3243G mutation, associated with mitochondrial encephalomyopathy, lactic acidosis, and stroke-like symptoms and diabetes. Mol. Cell. Biol., 30, 2147–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perli E., Giordano C., Tuppen H.A., Montopoli M., Montanari A., Orlandi M., Pisano A., Catanzaro D., Caparrotta L., Musumeci B. et al. (2012) Isoleucyl-tRNA synthetase levels modulate the penetrance of a homoplasmic m.4277T>C mitochondrial tRNAIle mutation causing hypertrophic cardiomyopathy. Hum. Mol. Genet., 21, 85–100. [DOI] [PubMed] [Google Scholar]
- 7.De Luca C., Besagni C., Frontali L., Bolotin-Fukuhara M., Francisci S. (2006) Mutations in yeast mt tRNAs: specific and general suppression by nuclear encoded tRNA interactors. Gene, 377, 169–176. [DOI] [PubMed] [Google Scholar]
- 8.De Luca C., Zhou Y., Montanari A., Morea V., Oliva R., Besagni C., Bolotin-Fukuhara M., Frontali L., Francisci S. (2009) Can yeast be used to study mitochondrial diseases? Biolistic tRNA mutants for the analysis of mechanisms and suppressors. Mitochondrion, 9, 408–417. [DOI] [PubMed] [Google Scholar]
- 9.Perli E., Giordano C., Pisano A., Montanari A., Campese A.F., Reyes A., Ghezzi D., Nasca A., Tuppen H.A., Orlandi M. et al. (2014) The isolated carboxy-terminal domain of human mitochondrial leucyl-tRNA synthetase rescues the pathological phenotype of mitochondrial tRNA mutations in human cells. EMBO Mol. Med., 6, 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hornig-Do H.T., Montanari A., Rozanska A., Tuppen H.A., Almalki A.A., Abg-Kamaludin D.P., Frontali L., Francisci S., Lightowlers R.N., Chrzanowska-Lightowlers Z.M. (2014) Human mitochondrial leucyl tRNA synthetase can suppress non cognate pathogenic mt-tRNA mutations. EMBO Mol. Med., 6, 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montanari A., De Luca C., Frontali L., Francisci S. (2010) Aminoacyl-tRNA synthetases are multivalent suppressors of defects due to human equivalent mutations in yeast mt tRNA genes. Biochim. Biophys. Acta, 1803, 1050–1057. [DOI] [PubMed] [Google Scholar]
- 12.Kaufmann P., Engelstad K., Wei B.S.Y., Kulikova R., Oskoui M., Sproule D.M., Battista V., Koenigsberger D.Y., Pascual J.M., Shanske S. et al. (2011) Natural history of MELAS associated with mitochondrial DNA m.3243A>G genotype. Neurology, 77, 1965–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nesbitt V., Pitceathly R.D., Turnbull D.M., Taylor R.W., Sweeney M.G., Mudanohwo E.E., Rahman S., Hanna M.G., McFarland R. (2013) The UK MRC Mitochondrial Disease Patient Cohort Study: clinical phenotypes associated with the m.3243A>G mutation—implications for diagnosis and management. J. Neurol. Neurosurg. Psychiatry, 84, 936–938. [DOI] [PubMed] [Google Scholar]
- 14.Tukalo M., Yaremchuk A., Fukunaga R., Yokoyama S., Cusack S. (2005) The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer-editing conformation. Nat. Struct. Mol. Biol., 12, 923–930. [DOI] [PubMed] [Google Scholar]
- 15.Palencia A., Crépin T., Vu M.T., Lincecum T.L. Jr., Martinis S.A., Cusack S. (2012) Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase. Nat. Struct. Mol. Biol., 19, 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francisci S., Montanari A., De Luca C., Frontali L. (2011) Peptides from aminoacyl-tRNA synthetases can cure the defects due to mutations in mt tRNA genes. Mitochondrion, 11, 919–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Micco P., Fazzi D'Orsi M., Morea V., Frontali L., Francisci S., Montanari A. (2014) The yeast model suggests the use of short peptides derived from mt LeuRS for the therapy of diseases due to mutations in several mt tRNAs. Biochim. Biophys. Acta, 1843, 3065–3074. [DOI] [PubMed] [Google Scholar]
- 18.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wittenhagen L.M., Kelley S.O. (2002) Dimerization of a pathogenic human mitochondrial tRNA. Nat. Struct. Biol., 9, 586–590. [DOI] [PubMed] [Google Scholar]
- 20.Helm M., Brule H., Degoul F., Cepanec C., Leroux J.P., Giege R., Florenz C. (1998) The presence of modified nucleotides is required for cloverleaf folding of human mitochondrial tRNA. Nucleic Acid. Res., 26, 1636–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobitski A.Y., Hengesbach M., Seidu-Larry S., Dammertz K., Chow C.S., van Aerschot A., Herdewijn P., Nienhaus G.U., Helm M. (2011) Single-molecule FRET reveals a cooperative effect of two methyl group modifications in the folding of human mitochondrial tRNA(Lys). Chem. Biol., 18, 928–936. [DOI] [PubMed] [Google Scholar]
- 22.Nathwani A.C., Tuddenham E.G., Rangarajan S., Rosales C., McIntosh J., Linch D.C., Chowdary P., Riddell A., Pie A.J., Harrington C. et al. (2011) Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med., 365, 2357–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Meo I., Auricchio A., Lamperti C., Burlina A., Viscomi C., Zeviani M. (2012) Effective AAV-mediated gene therapy in a mouse model of ethilmalonic encephalopathy. EMBO Mol. Med., 4, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King M.P., Koga Y., Davidson M., Schon E.A. (1992) Defects in mitochondrial protein synthesis and respiratory chain activity segregate with the tRNA(Leu(UUR)) mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes. Mol. Cell. Biol., 12, 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pello R., Martín M.A., Carelli V., Nijtmans L.G., Achilli A., Pala M., Torroni A., Gómez-Durán A., Ruiz-Pesini E., Martinuzzi A. et al. (2008) Mitochondrial DNA background modulates the assembly kinetics of OXPHOS complexes in a cellular model of mitochondrial disease. Hum. Mol. Genet., 17, 4001–4011. [DOI] [PubMed] [Google Scholar]
- 26.King M.P., Attardi G. (1989) Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science, 246, 500–503. [DOI] [PubMed] [Google Scholar]
- 27.King M.P., Attardi G. (1996) Isolation of human cell lines lacking mitochondrial DNA. Methods Enzymol., 264, 304–313. [DOI] [PubMed] [Google Scholar]
- 28.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc., 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rizzuto R., Simpson A.W., Brini M., Pozzan T. (1992) Rapid changes of mitochondrial Ca2+ revealed by specifically targeted recombinant aequorin. Nature, 358, 325–327. [DOI] [PubMed] [Google Scholar]
- 30.McDonnell M.T., Schaefer A.M., Blakely E.L., McFarland R., Chinnery P.F., Turnbull D.M., Taylor R.W. (2004) Noninvasive diagnosis of the 3243A > G mitochondrial DNA mutation using urinary epithelial cells. Eur. J. Hum. Genet., 12, 778–781. [DOI] [PubMed] [Google Scholar]
- 31.Larsson N.G., Tulinius M.H., Holme E., Oldfors A., Andersen O., Wahlström J., Aasly J. (1992) Segregation and manifestations of the mtDNA tRNA(Lys) A-->G(8344) mutation of myoclonus epilepsy and ragged-red fibers (MERRF) syndrome. Am. J. Hum. Genet., 51, 1201–1212. [PMC free article] [PubMed] [Google Scholar]
- 32.Campese A.F., Grazioli P., Colantoni S., Anastasi E., Mecarozzi M., Checquolo S., De Luca G., Bellavia D., Frati L., Gulino A. et al. (2009) Notch3 and pTalpha/pre-TCR sustain the in vivo function of naturally occurring regulatory T cells. Int. Immunol., 21, 727–743. [DOI] [PubMed] [Google Scholar]
- 33.Carelli V., Vergani L., Bernazzi B., Zampieron C., Bucchi L., Valentino M., Rengo C., Torroni A., Martinuzzi A. (2002) Respiratory function in cybrid cell lines carrying European mtDNA haplogroups: implications for Leber's hereditary optic neuropathy. Biochim. Biophys. Acta, 1588, 7–14. [DOI] [PubMed] [Google Scholar]
- 34.Taylor R.W., Giordano C., Davidson M.M., d'Amati G., Bain H., Hayes C.M., Leonard H., Barron M.J., Casali C., Santorelli F.M. et al. (2003) A homoplasmic mitochondrial transfer ribonucleic acid mutation as a cause of maternally inherited hypertrophic cardiomyopathy. J. Am. Coll. Cardiol., 41, 1786–1796. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.