Abstract
With a combined carrier frequency of 1:200, heteroplasmic mitochondrial DNA (mtDNA) mutations cause human disease in ∼1:5000 of the population. Rapid shifts in the level of heteroplasmy seen within a single generation contribute to the wide range in the severity of clinical phenotypes seen in families transmitting mtDNA disease, consistent with a genetic bottleneck during transmission. Although preliminary evidence from human pedigrees points towards a random drift process underlying the shifting heteroplasmy, some reports describe differences in segregation pattern between different mtDNA mutations. However, based on limited observations and with no direct comparisons, it is not clear whether these observations simply reflect pedigree ascertainment and publication bias. To address this issue, we studied 577 mother–child pairs transmitting the m.11778G>A, m.3460G>A, m.8344A>G, m.8993T>G/C and m.3243A>G mtDNA mutations. Our analysis controlled for inter-assay differences, inter-laboratory variation and ascertainment bias. We found no evidence of selection during transmission but show that different mtDNA mutations segregate at different rates in human pedigrees. m.8993T>G/C segregated significantly faster than m.11778G>A, m.8344A>G and m.3243A>G, consistent with a tighter mtDNA genetic bottleneck in m.8993T>G/C pedigrees. Our observations support the existence of different genetic bottlenecks primarily determined by the underlying mtDNA mutation, explaining the different inheritance patterns observed in human pedigrees transmitting pathogenic mtDNA mutations.
Introduction
First described in 1988, point mutations of mitochondrial DNA (mtDNA) have emerged as a major cause of maternally inherited human disease (1,2). Pathogenic mtDNA mutations causing a severe multisystem phenotype are usually heteroplasmic, with a mixture of mutated and wild-type mtDNA present in the same individual. The percentage level of mutated mtDNA largely determines whether a biochemical defect is expressed on a cellular level, and the inherited level of heteroplasmy correlates more or less with the severity of the clinical phenotype (3).
Rapid intergenerational shifts in the level of mtDNA heteroplasmy levels were first observed in Holstein cows (4,5) and subsequently documented in human pedigrees transmitting pathogenic mtDNA mutations. A restriction in the number of mitochondrial genomes repopulating the female germ line (the mtDNA bottleneck) is thought to explain this phenomenon, supported by observations in mice (6,7). Although the initial analysis of human pedigrees implied differences in the mtDNA bottleneck between different families (8), the analysis of aggregate data did not support earlier reports (9). Limited data analysis pointed towards a random genetic drift mechanism acting on all heteroplasmic mtDNA mutations (9), but in some instances, there appeared to be evidence of selection in favour of mutated genomes (10), even after minimising the effects of ascertainment bias. These conflicting data lead to two fundamental unanswered questions: do all mtDNA mutations behave the same during inheritance, and is there selection for or against different levels of heteroplasmy? Clarifying the underlying trends is key to providing reliable recurrence risks for patients with mtDNA diseases, and understanding the underlying mechanisms involved may open new avenues for preventative treatment in the future (11).
To address this issue, we have performed the largest analysis of inherited mtDNA heteroplasmic mutations in humans to date. Our findings show that differences in the behaviour of the mtDNA bottleneck between specific pathogenic mtDNA mutations, explaining the variability in clinical inheritance pattern observed in human pedigrees transmitting different mtDNA mutations.
Results
Determining the potential impact of ascertainment bias
Given previous concerns about ascertainment bias when studying the inheritance of heteroplasmy in human pedigrees (9), first we performed a simulation experiment to determine the possible consequences of identifying pedigrees through a clinically affected child. We then determined whether the standard approach of omitting the affected proband minimizes any bias to an acceptable level.
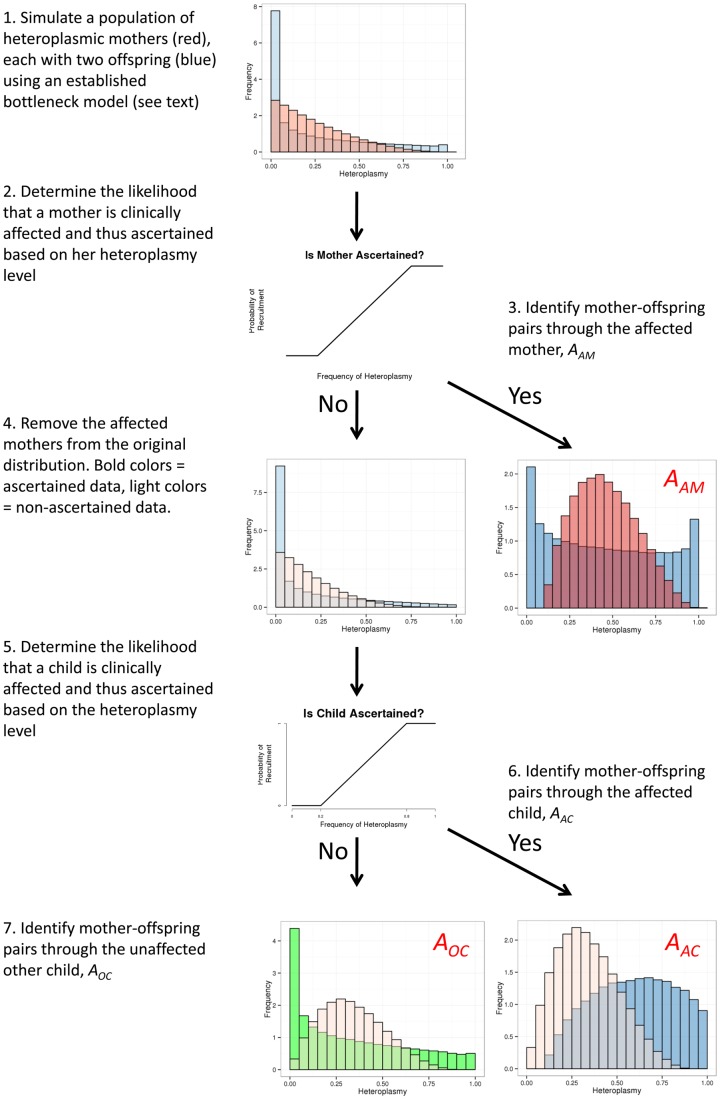
The simulations were based on an established model for the mtDNA genetic bottleneck using measurements of heteroplasmy made in human oocytes for neutral alleles (i.e. with no selection) (12,13). We studied the difference in heteroplasmy level between a mother and child (ΔM-O) in simulated pedigrees in silico, with three possible strengths of the mtDNA genetic bottleneck (where bottleneck parameter, b= 0.9, is a weak bottleneck; b= 0.7 is an intermediate strength bottleneck; and b= 0.2 is a strong bottleneck). We then modelled the effects of ascertaining the whole pedigrees (Fig. 1) using three forms of ascertainment: (1) through a mother (AAM), (2) through an affected child (AAC) or (3) through the other unaffected child, but disregarding data contributed by the affected child (AOC)—the latter mimicking the conventional approach used to minimize ascertainment bias by omitting to analyse data from an affected proband. For a full description, see the Materials and Methods and Figure 1.
Figure 1.
Overview of the inheritance model. Simulations used to determine the potential effects of ascertainment bias through a mother, and affected child, or an unaffected child. Mothers= red; offspring= blue; unaffected offspring= green. Darker distributions represent ascertained pedigrees; lighter distributions represent non-ascertained pedigrees. The probability of a family being recruited depends on an individual developing the disease, which in turn is correlated with the heteroplasmy level. The probability that an individual is recruited, pr, based on a distribution that is zero below a level of heteroplasmy d1, and one above heteroplasmy d2, with a linear increase between these points. The probability of being recruited with a heteroplasmy of (d1 + d2)/2 is 0.5, reflecting the increased likelihood of developing symptoms and thus presenting clinically. AAM= families ascertained through an affected mother; AAC families ascertained through an affected child; AOC families ascertained through an affected child, only but including the transmission from the mother to the other child, thus mimicking the effect of deleting probands from the ascertainment of real pedigrees.
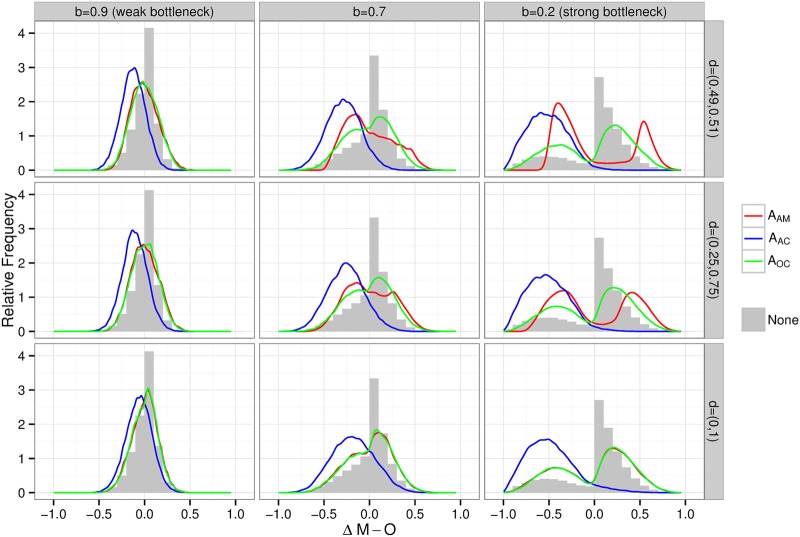
The simulations confirmed previous assumptions (9) that ascertainment through an affected child (AAC) leads to a skewed distribution of ΔM-O, creating the false impression that there is selection bias in favour of the mtDNA mutation (Fig. 2, blue line compared with the grey histogram). This was most prominent when the bottleneck was narrow (i.e. b was low, a strong bottleneck). However, when families were ascertained through the mother (AAM), or the data from the affected child (AOC) were not included, the ΔM-O distribution resembled the entire data set before sampling (Fig. 2, red and green lines compared with the grey histogram), showing that omission of the proband (AAC) minimized the ascertainment bias.
Figure 2.
Modelling the effects of ascertainment bias on simulated pedigrees transmitting mtDNA heteroplasmy. Frequency distribution histograms for the difference between maternal and offspring heteroplasmy levels (ΔM-O) for three different values of the bottleneck strength (bottleneck parameter, b= 0.9 a weak bottleneck; b= 0.7 an intermediate bottleneck; and b= 0.2 a strong bottleneck), and for three different methods of ascertainment: sampling the families where there was an affected mother—(ascertainment − affected mother, AAM, red); sampling the families where there was an affected child—(ascertainment − affected child, AAC, blue); and sampling the families through an affected child, only but including the transmission from the mother to the other child—(ascertainment − other child, AOC, green). The grey histogram gives the simulated distribution before any ascertainment, thus corresponding to data without any ascertainment bias. Each row reflects different thresholds heteroplasmy values required to cause disease. The parameter d models the threshold for disease resulting from the pathogenic mutation. The range given for d is the range of heteroplasmies where the probability of disease increases linearly from 0 (lower level) to 1 (upper level) (see legend to Fig. 1 for details of the simulation model).
Analysis of human pedigree data
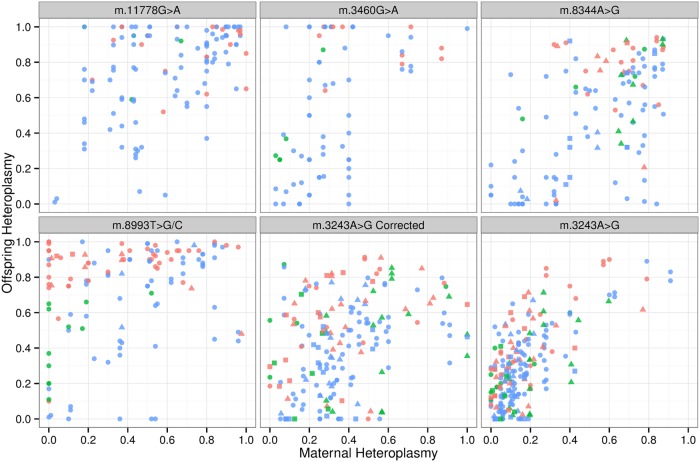
To generate the largest data set possible, we studied pedigrees derived from a meta-analysis of published data [n= 532 mother–child pairs transmitting five common heteroplasmic mtDNA mutations: m.11778G>A (n= 117), m.3460G>A (n= 74), m.8344A>G (n= 96), m.8993T>G/C (n= 117) and m.3423A>G (n= 128), citations shown in the Material and Methods]; and 45 new unpublished mother–child pairs [m.8344A>G (n= 9), m.8993T>G/C (n= 2) and m.3243A>G (n= 34)] measured in two centres. Given that there was no obvious difference between the two data sets (Fig. 3), we merged all of the data and minimized ascertainment bias by analysing separately data with and without the clinically affected probands. The final data set included 467 mother–child pairs for the uncorrected data. m.3243A>G was analysed before and after correcting for the known decrease in leucocyte heteroplasmy levels for this specific mutation using the published approach (14). For this correction, only individuals with a heteroplasmy level of <95% were included to avoid pairs where the mother or offspring values corrected to >100%. This reduced the sample size of m.3243A>G from 137 to 99 pairs. The entire data set was used to determine the likely size of the bottleneck parameter, b, using the model described previously (12,13), incorporating the laboratory assay and laboratory site as covariates. Bayesian statistical analyses were performed using JAGS (15).
Figure 3.
Relationship between the level of mtDNA heteroplasmy in mothers and offspring for five pathogenic mtDNA mutations. Maternal and offspring heteroplasmy levels are displayed as a proportion. Red symbols= affected proband, green symbols= mother of the proband, blue= other relatives. Circles= meta-analysis data, triangles= new pedigree data measured at Centre 1, squares= new pedigree data measured at Centre 2. m.3243A>G corrected refers to heteroplasmy values after a correction for the known decline in heteroplasmy levels with age (14).
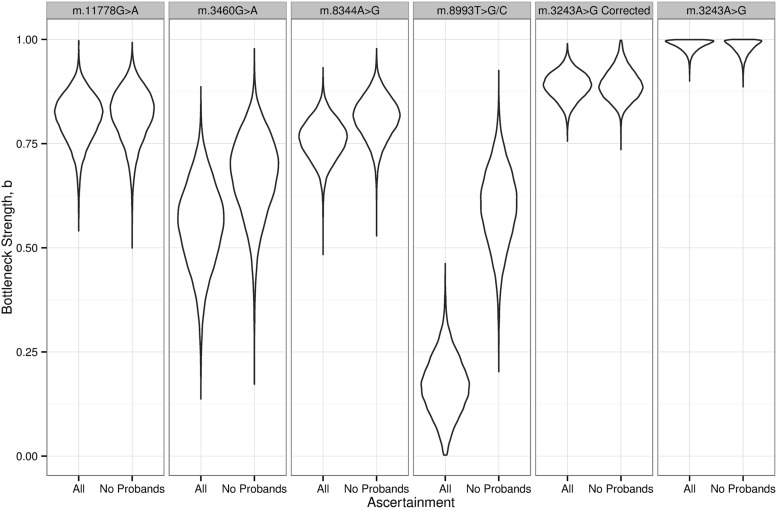
For each mutation, the average change in heteroplasmy was not significantly different from zero, consistent with no selection for or against the mtDNA mutations during transmission. The posterior differences in bottleneck strength, b, are shown in Figure 4. As predicted from the simulations, the bottleneck strength was overestimated (i.e. b is low, a narrower bottleneck) when we included pairs ascertained through a clinically affected proband. However, after the exclusion of affected probands, we observed a difference in the strength of the bottleneck parameter, b, estimated for different mtDNA mutations. m.8993T>G/C showed the largest difference, closely followed by m.3460A>G. A two tailed Bayesian hypothesis test was used to determine whether the bottleneck parameter, b, was significantly stronger for m.8993T>G/C than for m.11778G>A, m.8344A>G and m.3460G>A. This test gave estimated posterior probabilities of 0.031, 0.044 and 0.54. The bottleneck parameter distribution for the uncorrected m.3243A>G data was distorted, likely due to the known confounding effect of changing heteroplasmy levels with age (Fig. 4, far right hand panel) (14). We therefore used age-corrected data in the Bayesian hypothesis test comparing the bottleneck parameter, b, for m.8993T>G/C to m.3243A>G, which revealed an estimated posterior probability of 0.001. No other tests reached statistical significance at the 5% level.
Figure 4.
Differences in the size of the mitochondrial DNA genetic bottleneck of five pathogenic mtDNA mutations. Violin plots show the probability density for a given bottleneck strength parameter, b, calculated from actual heteroplasmy measurements in human pedigrees shown in Figure 3. All= all of the mother–child pairs. No probands= mother–child pairs remaining after the exclusion of an affected proband, thus minimising ascertainment bias. ‘m.3243A>G corrected’ refers to heteroplasmy values after a correction for the known decline in heteroplasmy levels with age (14).
Discussion
Our analysis accounted for several sources of potential variability, including different analytical techniques to measure heteroplasmy levels, different laboratories, differences between individual pedigrees themselves and differences in maternal age (16). Having minimised the likely effects of ascertainment bias, we observed different rates of heteroplasmy segregation during maternal transmission between mtDNA mutations. Patients harbouring the m.8993T>G/C mutation showed more rapid segregation of heteroplasmy levels than any of the other mutations. Although this trend has been suspected for some time (17), the analysis we present here provides the first direct comparison and a formal demonstration that m.8993T>G/C behaves differently to other pathogenic mtDNA mutations. Given that the size of the mitochondrial genetic bottleneck is the primary factor determining the rate of segregation during transmission, our findings indicate differences in the behaviour of the mtDNA bottleneck based on the underlying mtDNA genotype.
The more rapid segregation observed in m.8993T>G/C pedigrees is consistent with earlier reports of major shifts in heteroplasmy observed in pedigrees transmitting this mutation (17,18). This explains why severely affected children are often the only affected individuals in families transmitting this mutation, and why their mothers are usually asymptomatic. Conversely the less dramatic rates of segregation seen for other mtDNA mutations explain why a wide range of heteroplasmy values are seen in different family members, with oligo-symptomatic individuals transmitting the mutation in larger pedigrees, with multiple moderate and severely affected family members (19,20). Our findings also provide a potential explanation for the relative frequency of different pathogenic mutations in epidemiological studies (for example, m.3243A>G being much more common than m.8993T>G/C), despite the same background frequency of healthy carriers in the population with low heteroplasmy levels (21,22). It should be noted, however, that this discussion relates to the observed statistical trends, and although uncommon, rapidly segregating families with m.3243A>G (23) and slowly segregating families with m.8993T>G/C have been described (24,25) and are also consistent with our findings.
Studies in mice have shown a dramatic reduction in the amount of mtDNA within single cells at an early stage in mammalian germ cell development (6,7). This reduction is sufficient to explain the segregation of mtDNA heteroplasmy in mice (6). Mathematical models predict that subtle differences in the size of the mtDNA genetic bottleneck at this critical period will have a dramatic impact on the rate of segregation of mtDNA heteroplasmy (26). Given emerging evidence that the amount of mtDNA within cells can be influenced by the mtDNA sequence (27,28), it is plausible that the different mtDNA mutations we have studied cause differences in the amount of mtDNA within the developing germ line (29), either through a replication advantage or as a compensation for the lower rates of oxidative phosphorylation and higher levels of reactive oxygen species production (28,30). The differences in mtDNA level would lead to differences in the rate of segregation through the genetic bottleneck. Alternatively, selection either for or against a particular mutation would lead to differences in the rate of segregation (31). First described by population genetic theory (13,32), the effective bottleneck size, Ne, can be defined like the effective population size during genetic drift. Studies in isolated reduced populations have shown that selective pressures can influence Ne, without directly influencing the true population size. The apparent difference in bottleneck size between the different mtDNA mutations could thus reflect the selection pressure and need not necessarily be caused by a difference in the actual amount of mtDNA during germ cell development. Although it will be fascinating to determine the underlying molecular mechanisms, this will not alter our conclusions, or the relevance of our findings for women transmitting the mtDNA mutations we have studied.
These findings have important implications for our understanding for the recurrence risks of heteroplasmic mtDNA diseases, for the prevention of mtDNA disease using conventional techniques such as prenatal and pre-implantation diagnosis, and also suggest that therapies aimed at manipulating a germ cell mtDNA content could influence the underlying rate of segregation of pathogenic mtDNA mutations in humans. On the one hand, increasing the mtDNA content in germ cells could slow down segregation and thereby prevent the expression of severe disease in subsequent generations. On the other hand, factors known to reduce mtDNA content could lead to more rapid segregation of heteroplasmic alleles increasing the probability of generating germ cells with very low heteroplasmy levels (as well as very high heteroplasmy levels, which may not be viable at an early stage in pregnancy). Finally, environmental factors leading to selection pressure could also alter the behaviour of the genetic bottleneck, as described earlier. Given the recent observation of widespread low-level mtDNA heteroplasmy (33), these reductions in mtDNA content could lead to the emergence of pre-existing pathogenic variants, previously present at a very low level in the female germline. With recent evidence that common genetic polymorphisms of mtDNA can influence mtDNA levels (28), it is conceivable that geographic, ethnic and even inter-familial differences in segregation occur, influenced by non-synonymous single base-pair substitutions on the background mtDNA haplotype. This could explain differences in the rates of segregation of the same pathogenic mtDNA mutation, which have been described in different families, and also influence the segregation of low-level heteroplasmy, providing a potential mechanism for the association of mtDNA haplogroups with common late-onset human diseases (34).
Materials and Methods
Heteroplasmy levels in mothers and offspring
We studied the transmission of mtDNA heteroplasmy in both published and new unpublished pedigrees.
Ascertainment of published data
Published pedigrees transmitting m.11778G>A, m.3460G>A, m.8344A>G, m.8993T>G/C and m.3423A>G were identified through a systematic review of the literature. We included all papers where the age, clinical status and laboratory methods were clearly recorded, and there was at least one heteroplasmic individual. These parameters, along with the laboratory location, were incorporated in subsequent analyses. We identified 77 publications, containing blood DNA heteroplasmy levels from 532 mother–child pairs transmitting 5 common heteroplasmic mitochondrial DNA mutations: m.11778G>A (n= 117 mother–child pairs) (35–45), m.3460G>A (n= 74) (35,37,43,46–51), m.8344A>G (n= 96) (52–62), m.8993T>G/C (n= 117) (24,62–84) and m.3423A>G (n= 128) (20,54,85,86,87,88,89,90,91,92,93,94,95–105).
Unpublished data
Unpublished pedigrees transmitting the m.8344A>G (n= 9 mother–child pairs), m.8993T>G/C (n= 2) and m.3423A>G (n= 34) mutations were identified from the following accredited diagnostic laboratories: Newcastle, UK; Maastricht, NL; Milan and Bologna, Italy; Bergen, Norway and Munich, Germany. Genomics DNA was analysed in two laboratories: Newcastle, UK; and Maastricht, NL.
Quantification of heteroplasmy: Newcastle
Heteroplasmy was measured by quantitative pyrosequencing. Pyromark Assay Design Software v.2.0 (Qiagen) was used to design primers for template generation, one of which contains a biotinylated tag (*BIO), and the pyrosequencing reaction (Seq). For m.3243A>G: F-*BIO-TAAGGCCTACTTCACAAAGCG, R-GCGATTAGAATGGGTACAATGAG, Seq-ATGCGATTACCGGGC; for m.8344A>G: F-*BIO-CATGCCCATCGTCCTAGAAT, R-TTTTTATGGGCTTTGGTGAGG, Seq-TAAGTTAAAGATTAAGAGA; and for m.8993T>G F-AGGCACACCTACACCCCTTA, R-*TGTGAAAACGTAGGCTTGGAT, Seq-CATTCAACCAATAGCCC. Product templates were generated with a GoTaq® DNA Polymerase (Promega) reaction according to manufacturer's protocol. Pyrosequencing was performed using the Pyromark Q24 platform according to the manufacturer's protocol, using the designed pyrosequencing primers for each mutation. Pyromark Q24 software was used to quantify the heteroplasmy levels of each mutation through comparison of the relevant peak heights of both wild-type and mutant mtDNA. The accuracy of the pyrosequencing assay was determined by generating wild-type and mutant clones, which when mixed at the correct proportions mimicked a range of heteroplasmy levels between 0 and 100% for each mutation. Each mixed sample was assessed for mutation load using pyrosequencing and used to generate a standard curve (Supplementary Material, Fig. S1).
Quantification of heteroplasmy: Maastricht
Heteroplasmy was measured by semiquantitative restriction fragment length polymorphism (RFLP) analysis using mutation-specific restriction enzymes. Primers used for m.3243A>G were as follows: CAACTTAGTATTATACCCACACCAACTTAGTATTATACCCACAC (forward) and TTTCGTTCGGTAAGCATTAG (reverse); for m.8344A>G: TCGTCCTAGAATTAATTCCC (forward) and GTAGTATTTAGTTGGGGCATTTCACTGTAAAGCCGTGTTG (reverse); and for m.8993T>C/G: CACACCTACACCCCTTATCCC (forward) and TCATTATGTGTTGTCGTGCAG (reverse). For each forward primer, an unlabelled and FAM-labelled primer was available, polymerase chain reaction (PCR) was performed on the GeneAmp PCR System 9700 (Perkin–Elmer Applied Biosystems) in a total volume of 50 µl, containing 1× PCR buffer (Invitrogen) and 1 U of Taq DNA polymerase (Invitrogen), 2 mm MgCl2 (Invitrogen) and 0.1 mm dNTP (Pharmacia). Unlabelled forward (15 pmol) and reverse primer (3pmal) sets were used in the first round. First round PCR started with 5-min denaturation at 94°C followed by 32 cycles of 1-min denaturation at 92°C, 45-s annealing at 53°C and 45-s elongation at 72°C, followed by a final elongation step of 7 min at 72°C. 15 ml of first round amplification product was adjusted to 50 ml with second round PCR mix, containing 1× PCR buffer (Invitrogen) 1 U of DNA polymerase, 2 mm MgCl2 (Invitrogen) and a fluorescently labelled forward primer (15 pmol). One final amplification cycle was performed of 5-min denaturation at 94°C, 1-min annealing at 53°C and 7-min elongation at 72°C. The labelled second round PCR product (15 ml) was digested in the appropriate digestion buffer in a total volume of 50 ml containing 10 U HaeIII (10 U/µl; Roche) for the m.3243A>G mutation, 10 U BglI (10 U/µl) for the m.8344A>G mutation and 10 U HpaII (10 U/ µl; Roche) for the m.8993T>G mutation. The m.3243A>G-amplicon contains an additional HaeIII restriction site as an internal control for restriction enzyme digestion completion. For each mutation, a sample with known mutation load was included as reference. After digestion, samples were purified using a QIAquick PCR purification kit (Qiagen). Samples were analysed by capillary electrophoresis on an ABI Prism 3730 Genetic Analyser followed by GeneScan analysis. To calculate the mutation load, the area of the mutation peak was divided by the sum of the peak area of the wild-type and mutation peak. Each forward primer contains a biotinylated tag. PCR was performed using Taq DNA Polymerase (Invitrogen) according to manufacturer's protocol. The PCR fragments were digested by mutation-specific restriction enzymes: Hae III (10 U/µl) Roche for m.3243A>G; Bgl I (10 U/µl) Biolabs for m.8344A>G and Hpa II (10 U/µl) Roche for m.8993T>C/G. Digestion products were purified (Qiagen Qiaquick PCR Purification). Fragments were separated and analysed by capillary electrophoresis (automatic DNA Sequencer 3730, Applied Biosystems) to determine mutation load.
Simulation of mtDNA heteroplasmy inheritance
An overview of the simulations is shown in Figure 1. First, we generated a population of simulated mothers with different heteroplasmy levels. We then modelled the inheritance of heteroplasmy from each of these mothers to two offspring (see below). Next we determined the likelihood of the individuals being clinically affected. We then sampled the simulated families based on the following ascertainment criteria, in the following order:
Sampling the families where there was an affected mother—(AAM). Here, we included the transmission from the mother to both offspring.
Sampling the families where there was an affected child—(AAC), but the mother was not affected. Here, we also included the transmission from the mother to both offspring.
Sampling the families through an affected child, only but including the transmission from the mother to the other child—(AOC).
This sampling algorithm mimicked the different ways that mother–child pairs could be identified in a study of real human pedigrees, and the resulting difference in distributions for the three different pairs (Supplementary Material, Fig. S2) resembles those from the real data from the human pedigrees shown in Figure 3.
We then compared the sampled mother–child pairs with the original simulated pedigrees to determine whether the ascertainment method influenced the observed distribution of heteroplasmy transmissions.
Simulating the transmission of mtDNA heteroplasmy (the mtDNA genetic bottleneck)
We modelled the mtDNA genetic bottleneck using established model derived from measurements of heteroplasmy made in human oocytes (12,13). Here, the bottleneck is modelled as an infinite population of mtDNA molecules that was instantaneously reduced to the minimum bottleneck size N for g generations, before expanding back to a large size. We simulated the transmission of mtDNA heteroplasmy of a neutral allele from the mother to two offspring through the bottleneck by approximating with a beta distribution with parameters α= pM b/(1−b) and β= (1−pM)b/(1−b), where b is the drift parameter b= exp(−g/N). A large b indicates a weak (or wide) genetic bottleneck. When g is large compared with N, b approaches 0, corresponding to a very strong (or narrow) bottleneck. Using this model, we generated a series of simulated heteroplasmy transmissions from mothers starting with a range of heteroplasmy values.
Statistical methods
Changes in heteroplasmy between mother and child were modelled using the same beta distribution as for the simulation, using a hierarchical model with a different bottleneck parameter b for each mtDNA mutation. Measurements of heteroplasmy were assumed to be normally distributed about the true values, with censored at 0 and 1, and with an error rate σi for publication i, to allow for different measurement technologies. Writing mi for the observed maternal heteroplasmy for mother i, and oij for the heteroplasmy for child j of mother i, where j= 1, … , ni, and ni is the number of children for Mother I,
where mi gives the mutation for Mother i and ri gives the publication for mother i. To test for systematic increases or decreases in the level of heteroplasmy (i.e. selection), an additional model was built. This allowed for changes in the expected heteroplasmy in the offspring, by multiplying the expected child heteroplasmy, by a random factor, which models selection by increasing or decreasing the average heteroplasmy value. The JAGS statistical package (15) was used to make inferences about the strength of the bottleneck using the statistical model above. A Bayesian framework was used with uniform priors between 0 and 1 for the bk, and broad priors for the error parameters The model and all JAGS and R scripts are available through the following URL: github.com/ijwilson/mtBottleneck.
Bayesian comparison of bottleneck sizes
Maternal heteroplasmy values were corrected for age as described (14). Bayesian hypothesis tests of bj versus bk were calculated using the empirical joint posterior distributions of bj and bk from JAGS (15) and using The JAGS script is available through the following URL: github.com/ijwilson/mtBottleneck.
Supplementary Material
Funding
Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Supplementary Material
Acknowledgements
P.F.C. is a Wellcome Trust Senior Fellow in Clinical Science (101876/Z/13/Z), and a UK NIHR Senior Investigator, who receives support from the Medical Research Council Mitochondrial Biology Unit (MC_UP_1501/2), the Wellcome Trust Centre for Mitochondrial Research (096919Z/11/Z), the Medical Research Council (UK) Centre for Translational Muscle Disease research (G0601943), EU FP7 TIRCON, and the National Institute for Health Research (NIHR) Biomedical Research Centre based at Cambridge University Hospitals NHS Foundation Trust and the University of Cambridge. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. R.H. is supported by the European Research Council (309548). P.F.C. and R.H. are supported by the Mitochondrial European Educational Training, ITN Marie Curie People (317433). R.W.T. also receives support from The Lily Foundation and the UK NHS Specialist Commissioners, which funds the ‘Rare Mitochondrial Disorders of Adults and Children’ Diagnostic Service in Newcastle upon Tyne. C.L.A. is the recipient of an NIHR doctoral fellowship (NIHR-HCS-D12-03-04). We acknowledge the ‘Cell lines and DNA Bank of Paediatric Movement Disorders and Neurodegenerative Diseases’ of the Telethon Network of Genetic Biobanks (grant GTB12001J) and the EurobiobanK Network. This work was supported by the Medical Research Council, the Pierfranco and Luisa Mariani Foundation, Telethon Grant GGP11011 and ERC Advanced Grant FP7-322424. The publication is the work of the authors, and Patrick Chinnery will serve as guarantor for the contents of this paper.
Conflict of Interest statement. None declared.
References
- 1.Vafai S.B., Mootha V.K. (2013) Medicine. A common pathway for a rare disease? Science, 342, 1453–1454. [DOI] [PubMed] [Google Scholar]
- 2.Gorman G.S., Schaefer A.M., Ng Y. et al. (2015) Prevalence of nuclear and mtDNA mutations related to adult mitochondrial disease. Ann. Neurol., 77, 753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiMauro S., Schon E.A., Carelli V., Hirano M. (2013) The clinical maze of mitochondrial neurology. Nat. Rev. Neurol., 9, 429–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Upholt W.B., Dawid I.B. (1977) Mapping of mitochondrial DNA of individual sheep and goats: rapid evolution in the D loop region. Cell, 11, 571–583. [DOI] [PubMed] [Google Scholar]
- 5.Olivo P.D., Van de Walle M.J., Laipis P.J., Hauswirth W.W. (1983) Nucleotide sequence evidence for rapid genotypic shifts in the bovine mitochondrial DNA D-loop. Nature, 306, 400–402. [DOI] [PubMed] [Google Scholar]
- 6.Cree L.M., Samuels D.C., de Sousa Lopes S.C. et al. (2008) A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat. Genet., 40, 249–254. [DOI] [PubMed] [Google Scholar]
- 7.Wai T., Teoli D., Shoubridge E.A. (2008) The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat. Genet., 40, 1484–1488. [DOI] [PubMed] [Google Scholar]
- 8.Howell N., Halvorson S., Kubacka I., McCullough D.A., Bindoff L.A., Turnbull D.M. (1992) Mitochondrial gene segregation in mammals: is the bottleneck always narrow? Hum. Genet., 90, 117–120. [DOI] [PubMed] [Google Scholar]
- 9.Chinnery P.F., Thorburn D.R., Samuels D.C. et al. (2000) The inheritance of mtDNA heteroplasmy: random drift, selection or both? Trends Genet., 16, 500–505. [DOI] [PubMed] [Google Scholar]
- 10.Uusimaa J., Moilanen J.S., Vainionpaa L. et al. (2007) Prevalence, segregation, and phenotype of the mitochondrial DNA 3243A>G mutation in children. Ann. Neurol., 62, 278–287. [DOI] [PubMed] [Google Scholar]
- 11.Hellebrekers D.M., Wolfe R., Hendrickx A.T. et al. (2012) PGD and heteroplasmic mitochondrial DNA point mutations: a systematic review estimating the chance of healthy offspring. Hum. Reprod., update 18, 341–349. [DOI] [PubMed] [Google Scholar]
- 12.Brown D.T., Samuels D.C., Michael E.M., Turnbull D.M., Chinnery P.F. (2001) Random genetic drift determines the level of mutant mitochondrial DNA in human primary oocytes. Am. J. Hum. Genet., 68, 553–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright S. (1969) Evolution and the Genetics of Populations. University of Chicago Press, Chicago. [Google Scholar]
- 14.Rajasimha H.K., Chinnery P.F., Samuels D.C. (2008) Selection against pathogenic mtDNA mutations in a stem cell population leads to the loss of the 3243A-->G mutation in blood. Am. J. Hum. Genet., 82, 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plummer M. (2003) JAGS: a program for analysis of Bayesian graphical models using Gibbs sampling. In Proceedings of the 3rd International Workshop on Distributed Statistical Computing Technische Universit at Wien, Vol. 124, p. 125. [Google Scholar]
- 16.Rebolledo-Jaramillo B., Su M.S., Stoler N. et al. (2014) Maternal age effect and severe germ-line bottleneck in the inheritance of human mitochondrial DNA. Proc. Natl Acad. Sci. USA, 111, 15474–15479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blok R.B., Gook D.A., Thorburn D.R., Dahl H.H. (1997) Skewed segregation of the mtDNA nt 8993 (T to G) mutation in human oocytes. Am. J. Hum. Genet., 60, 1495–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White S.L., Collins V.A., Woolfe R. et al. (1999) Genetic counseling and prenatal diagnosis for the mitochondrial DNA mutations at nucleotide 8993. Am. J. Hum. Genet., 65, 474–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsson N.G., Tulinius M.H., Holme E. et al. (1992) Segregation and manifestations of the mtDNA tRNA(Lys) A-->G(8344) mutation of myoclonus epilepsy and ragged-red fibers (MERRF) syndrome. Am. J. Hum. Genet., 51, 1201–1212. [PMC free article] [PubMed] [Google Scholar]
- 20.Ciafaloni E., Ricci E., Shanske S. et al. (1992) MELAS: clinical features, biochemistry, and molecular genetics. Ann. Neurol., 31, 391–398. [DOI] [PubMed] [Google Scholar]
- 21.Elliott H.R., Samuels D.C., Eden J.A., Relton C.L., Chinnery P.F. (2008) Pathogenic mitochondrial DNA mutations are common in the general population. Am. J. Hum. Genet., 83, 254–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chinnery P.F., Johnson M.A., Wardell T.M. et al. (2000) Epidemiology of pathogenic mitochondrial DNA mutations. Ann. Neurol., 48, 188–193. [PubMed] [Google Scholar]
- 23.Hammans S.R., Sweeney M.G., Brockington M. et al. (1993) The mitochondrial DNA transfer RNA(Lys)A-->G(8344) mutation and the syndrome of myoclonic epilepsy with ragged red fibres (MERRF). Relationship of clinical phenotype to proportion of mutant mitochondrial DNA. Brain, 116, 617–632. [DOI] [PubMed] [Google Scholar]
- 24.Uziel G., Moroni I., Lamantea E. et al. (1997) Mitochondrial disease associated with the T8993G mutation of the mitochondrial ATPase 6 gene: a clinical, biochemical, and molecular study in six families. J. Neurol. Neurosurg. Psychiatry, 63, 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfeffer G., Blakely E.L., Alston C.L. et al. (2012) Adult-onset spinocerebellar ataxia syndromes due to MTATP6 mutations. J. Neurol. Neurosurg. Psychiatry, 83, 883–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wonnapinij P., Chinnery P.F., Samuels D.C. (2010) Previous estimates of mitochondrial DNA mutation level variance did not account for sampling error: comparing the mtDNA genetic bottleneck in mice and humans. Am. J. Hum. Genet., 86, 540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreno-Loshuertos R., Acin-Perez R., Fernandez-Silva P. et al. (2006) Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat. Genet., 38, 1261–1268. [DOI] [PubMed] [Google Scholar]
- 28.Gomez-Duran A., Pacheu-Grau D., Lopez-Gallardo E. et al. (2010) Unmasking the causes of multifactorial disorders: OXPHOS differences between mitochondrial haplogroups. Hum. Mol. Genet., 19, 3343–3353. [DOI] [PubMed] [Google Scholar]
- 29.Monnot S., Samuels D.C., Hesters L. et al. (2013) Mutation dependance of the mitochondrial DNA copy number in the first stages of human embryogenesis. Hum. Mol. Genet., 22, 1867–1872. [DOI] [PubMed] [Google Scholar]
- 30.Pello R., Martin M.A., Carelli V. et al. (2008) Mitochondrial DNA background modulates the assembly kinetics of OXPHOS complexes in a cellular model of mitochondrial disease. Hum. Mol. Genet., 17, 4001–4011. [DOI] [PubMed] [Google Scholar]
- 31.Stewart J.B., Larsson N.G. (2014) Keeping mtDNA in shape between generations. PLoS Genet., 10, e1004670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solignac M., Monnerot M., Mounolou J.C. (1983) Mitochondrial DNA heteroplasmy in Drosophila mauritiana. Proc. Natl Acad. Sci. USA, 80, 6942–6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Payne B.A., Wilson I.J., Yu-Wai-Man P. et al. (2013) Universal heteroplasmy of human mitochondrial DNA. Hum. Mol. Genet., 22, 384–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hudson G., Gomez-Duran A., Wilson I.J., Chinnery P.F. (2014) Recent mitochondrial DNA mutations increase the risk of developing common late-onset human diseases. PLoS Genet., 10, e1004369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carelli V., Ghelli A., Ratta M. et al. (1997) Leber's hereditary optic neuropathy: biochemical effect of 11778/ND4 and 3460/ND1 mutations and correlation with the mitochondrial genotype. Neurology, 48, 1623–1632. [DOI] [PubMed] [Google Scholar]
- 36.Chuenkongkaew W.L., Suphavilai R., Vaeusorn L., Phasukkijwatana N., Lertrit P., Suktitipat B. (2005) Proportion of 11778 mutant mitochondrial DNA and clinical expression in a thai population with leber hereditary optic neuropathy. J. Neuro-Ophthalmol., 25, 173–175. [DOI] [PubMed] [Google Scholar]
- 37.Harding A.E., Sweeney M.G., Govan G.G., Riordan-Eva P. (1995) Pedigree analysis in Leber hereditary optic neuropathy families with a pathogenic mtDNA mutation. Am. J. Hum. Genet., 57, 77–86. [PMC free article] [PubMed] [Google Scholar]
- 38.Holt I.J., Miller D.H., Harding A.E. (1989) Genetic heterogeneity and mitochondrial DNA heteroplasmy in Leber's hereditary optic neuropathy. J. Med. Genet., 26, 739–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howell N., Xu M., Halvorson S., Bodis-Wollner I., Sherman J. (1994) A heteroplasmic LHON family: tissue distribution and transmission of the 11778 mutation. Am. J. Hum. Genet., 55, 203–206. [PMC free article] [PubMed] [Google Scholar]
- 40.Juvonen V., Nikoskelainen E., Lamminen T., Penttinen M., Aula P., Savontaus M.L. (1997) Tissue distribution of the ND4/11778 mutation in heteroplasmic lineages with Leber hereditary optic neuropathy. Hum. Mutat., 9, 412–417. [DOI] [PubMed] [Google Scholar]
- 41.Phasukkijwatana N., Chuenkongkaew W.L., Suphavilai R., Luangtrakool K., Kunhapan B., Lertrit P. (2006) Transmission of heteroplasmic G11778A in extensive pedigrees of Thai Leber hereditary optic neuropathy. J. Hum. Genet., 51, 1110–1117. [DOI] [PubMed] [Google Scholar]
- 42.Simon D.K., Pulst S.M., Sutton J.P., Browne S.E., Beal M.F., Johns D.R. (1999) Familial multisystem degeneration with parkinsonism associated with the 11778 mitochondrial DNA mutation. Neurology, 53, 1787–1793. [DOI] [PubMed] [Google Scholar]
- 43.Sweeney M.G., Davis M.B., Lashwood A., Brockington M., Toscano A., Harding A.E. (1992) Evidence against an X-linked locus close to DXS7 determining visual loss susceptibility in British and Italian families with Leber hereditary optic neuropathy. Am. J. Hum. Genet., 51, 741–748. [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka A., Kiyosawa M., Mashima Y., Tokoro T. (1998) A family with Leber's hereditary optic neuropathy with mitochondrial DNA heteroplasmy related to disease expression. J Neuro-Ophthalmol., 18, 81–83. [PubMed] [Google Scholar]
- 45.Zhu D.P., Economou E.P., Antonarakis S.E., Maumenee I.H. (1992) Mitochondrial DNA mutation and heteroplasmy in type I Leber hereditary optic neuropathy. Am. J. Med. Genet., 42, 17317–9. [DOI] [PubMed] [Google Scholar]
- 46.Black G.C., Morten K., Laborde A., Poulton J. (1996) Leber's hereditary optic neuropathy: heteroplasmy is likely to be significant in the expression of LHON in families with the 3460 ND1 mutation. Brit. J. Ophthalmol., 80, 915–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghosh S.S., Fahy E., Bodis-Wollner I., Sherman J., Howell N. (1996) Longitudinal study of a heteroplasmic 3460 Leber hereditary optic neuropathy family by multiplexed primer-extension analysis and nucleotide sequencing. Am. J. Hum. Genet., 58, 325–334. [PMC free article] [PubMed] [Google Scholar]
- 48.Howell N., Bindoff L.A., McCullough D.A. et al. (1991) Leber hereditary optic neuropathy: identification of the same mitochondrial ND1 mutation in six pedigrees. Am. J. Hum. Genet., 49, 939–950. [PMC free article] [PubMed] [Google Scholar]
- 49.Howell N., McCullough D., Bodis-Wollner I. (1992) Molecular genetic analysis of a sporadic case of Leber hereditary optic neuropathy. Am. J. Hum. Genet., 50, 443–446. [PMC free article] [PubMed] [Google Scholar]
- 50.Kaplanova V., Zeman J., Hansikova H. et al. (2004) Segregation pattern and biochemical effect of the G3460A mtDNA mutation in 27 members of LHON family. J. Neurol. Sci., 223, 149–155. [DOI] [PubMed] [Google Scholar]
- 51.Volod'ko N.V., L'Vova M.A., Starikovskaia E.B. et al. (2006) Spectrum of pathogenic mtDNA mutations in Leber hereditary optic neuropathy families from Siberia. Genetika, 42, 89–97. [PubMed] [Google Scholar]
- 52.Campos Y., Esteban J., Cabello A., Arenas J. (1994) Genetic analysis of one family with myoclonic epilepsy and ragged-red fibers (MERRF). Muscle Nerve, 17, 1229–1231. [PubMed] [Google Scholar]
- 53.Gamez J., Playan A., Andreu A.L. et al. (1998) Familial multiple symmetric lipomatosis associated with the A8344G mutation of mitochondrial DNA. Neurology, 51, 258–260. [DOI] [PubMed] [Google Scholar]
- 54.Hammans S.R., Sweeney M.G., Hanna M.G., Brockington M., Morgan-Hughes J.A., Harding A.E. (1995) The mitochondrial DNA transfer RNALeu(UUR) A-->G(3243) mutation. A clinical and genetic study. Brain, 118(Pt 3), 721–734. [DOI] [PubMed] [Google Scholar]
- 55.Howell N., Kubacka I., Smith R., Frerman F., Parks J.K., Parker W.D. Jr (1996) Association of the mitochondrial 8344 MERRF mutation with maternally inherited spinocerebellar degeneration and Leigh disease. Neurology, 46, 219–222. [DOI] [PubMed] [Google Scholar]
- 56.Munoz-Malaga A., Bautista J., Salazar J.A. et al. (2000) Lipomatosis, proximal myopathy, and the mitochondrial 8344 mutation. A lipid storage myopathy? Muscle Nerve, 23, 538–542. [DOI] [PubMed] [Google Scholar]
- 57.Piccolo G., Focher F., Verri A. et al. (1993) Myoclonus epilepsy and ragged-red fibers: blood mitochondrial DNA heteroplasmy in affected and asymptomatic members of a family. Acta. Neurol. Scand., 88, 406–409. [DOI] [PubMed] [Google Scholar]
- 58.Seibel P., Flierl A., Kottlors M., Reichmann H. (1994) A rapid and sensitive PCR screening method for point mutations associated with mitochondrial encephalomyopathies. Biochem. Biophys. Res. Commun., 200, 938–942. [DOI] [PubMed] [Google Scholar]
- 59.Silvestri G., Ciafaloni E., Santorelli F.M. et al. (1993) Clinical features associated with the A-->G transition at nucleotide 8344 of mtDNA (‘MERRF mutation’). Neurology, 43, 1200–1206. [DOI] [PubMed] [Google Scholar]
- 60.Traff J., Holme E., Ekbom K., Nilsson B.Y. (1995) Ekbom's syndrome of photomyoclonus, cerebellar ataxia and cervical lipoma is associated with the tRNA(Lys) A8344G mutation in mitochondrial DNA. Acta Neurol. Scand., 92, 394–397. [DOI] [PubMed] [Google Scholar]
- 61.van de Glind G., de Vries M., Rodenburg R., Hol F., Smeitink J., Morava E. (2007) Resting muscle pain as the first clinical symptom in children carrying the MTTK A8344G mutation. Eur. J. Paed. Neurol., 11, 243–246. [DOI] [PubMed] [Google Scholar]
- 62.Wong L.J., Wong H., Liu A. (2002) Intergenerational transmission of pathogenic heteroplasmic mitochondrial DNA. Genet. Med., 4, 78–83. [DOI] [PubMed] [Google Scholar]
- 63.Bartley J., Senadheera D., Park P., Brar H., D A., Wong L. (1996) Prenatal diagnosis of T8993G mitochondrial DNA point mutation in amniocytes by heteroplasmy detection. Am. J. Hum. Genet., 59. [Google Scholar]
- 64.Ciafaloni E., Santorelli F.M., Shanske S. et al. (1993) Maternally inherited Leigh syndrome. J. Pediatr., 122, 419–422. [DOI] [PubMed] [Google Scholar]
- 65.Degoul F., Diry M., Rodriguez D. et al. (1995) Clinical, biochemical, and molecular analysis of a maternally inherited case of Leigh syndrome (MILS) associated with the mtDNA T8993G point mutation. J. Inher. Met. Dis., 18, 682–688. [DOI] [PubMed] [Google Scholar]
- 66.Enns G.M., Bai R.K., Beck A.E., Wong L.J. (2006) Molecular-clinical correlations in a family with variable tissue mitochondrial DNA T8993G mutant load. Mol. Genet. Metab., 88, 364–371. [DOI] [PubMed] [Google Scholar]
- 67.Ferlin T., Landrieu P., Rambaud C. et al. (1997) Segregation of the G8993 mutant mitochondrial DNA through generations and embryonic tissues in a family at risk of Leigh syndrome. J. Pediatr., 131, 447–449. [DOI] [PubMed] [Google Scholar]
- 68.Fryer A., Appleton R., Sweeney M.G., Rosenbloom L., Harding A.E. (1994) Mitochondrial DNA 8993 (NARP) mutation presenting with a heterogeneous phenotype including ‘cerebral palsy’. Arch. Dis. Childhood, 71, 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Holt I.J., Harding A.E., Petty R.K., Morgan-Hughes J.A. (1990) A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am. J. Hum. Genet., 46, 428–433. [PMC free article] [PubMed] [Google Scholar]
- 70.Houstek J., Klement P., Hermanska J. et al. (1995) Altered properties of mitochondrial ATP-synthase in patients with a T-->G mutation in the ATPase 6 (subunit a) gene at position 8993 of mtDNA. Biochim. Biophys. Acta, 1271, 349–357. [DOI] [PubMed] [Google Scholar]
- 71.Lodi R., Montagna P., Iotti S. et al. (1994) Brain and muscle energy metabolism studied in vivo by 31P-magnetic resonance spectroscopy in NARP syndrome. J. Neurol. Neurosurg. Psychiatry, 57, 1492–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mak S.C., Chi C.S., Liu C.Y., Pang C.Y., Wei Y.H. (1996) Leigh syndrome associated with mitochondrial DNA 8993 T-->G mutation and ragged-red fibers. Pediatr. Neurol., 15, 72–75. [DOI] [PubMed] [Google Scholar]
- 73.Makela-Bengs P., Suomalainen A., Majander A. et al. (1995) Correlation between the clinical symptoms and the proportion of mitochondrial DNA carrying the 8993 point mutation in the NARP syndrome. Pediatric Res., 37, 634–639. [DOI] [PubMed] [Google Scholar]
- 74.Mkaouar-Rebai E., Chaari W., Younes S., Bousoffara R., Sfar M.T., Fakhfakh F. (2009) Maternally inherited Leigh syndrome: T8993G mutation in a Tunisian family. Pediatr. Neurol., 40, 437–442. [DOI] [PubMed] [Google Scholar]
- 75.Pastores G.M., Santorelli F.M., Shanske S. et al. (1994) Leigh syndrome and hypertrophic cardiomyopathy in an infant with a mitochondrial DNA point mutation (T8993G). Am. J. Med. Genet., 50, 265–271. [DOI] [PubMed] [Google Scholar]
- 76.Porto F.B., Mack G., Sterboul M.J. et al. (2001) Isolated late-onset cone-rod dystrophy revealing a familial neurogenic muscle weakness, ataxia, and retinitis pigmentosa syndrome with the T8993G mitochondrial mutation. Am. J. Ophthalmol., 132, 935–937. [DOI] [PubMed] [Google Scholar]
- 77.Puddu P., Barboni P., Mantovani V. et al. (1993) Retinitis pigmentosa, ataxia, and mental retardation associated with mitochondrial DNA mutation in an Italian family. Brit. J. Ophthalmol., 77, 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sakuta R., Goto Y., Horai S. et al. (1992) Mitochondrial DNA mutation and Leigh's syndrome. Ann. Neurol., 32, 597–598. [DOI] [PubMed] [Google Scholar]
- 79.Santorelli F.M., Shanske S., Macaya A., DeVivo D.C., DiMauro S. (1993) The mutation at nt 8993 of mitochondrial DNA is a common cause of Leigh's syndrome. Ann. Neurol., 34, 827–834. [DOI] [PubMed] [Google Scholar]
- 80.Shoffner J.M., Fernhoff P.M., Krawiecki N.S. et al. (1992) Subacute necrotizing encephalopathy: oxidative phosphorylation defects and the ATPase 6 point mutation. Neurology, 42, 2168–2174. [DOI] [PubMed] [Google Scholar]
- 81.Steffann J., Gigarel N., Corcos J. et al. (2007) Stability of the m.8993T->G mtDNA mutation load during human embryofetal development has implications for the feasibility of prenatal diagnosis in NARP syndrome. J. Med. Genet., 44, 664–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tatuch Y., Christodoulou J., Feigenbaum A. et al. (1992) Heteroplasmic mtDNA mutation (T----G) at 8993 can cause Leigh disease when the percentage of abnormal mtDNA is high. Am. J. Hum. Genet., 50, 852–858. [PMC free article] [PubMed] [Google Scholar]
- 83.Tulinius M.H., Houshmand M., Larsson N.G. et al. (1995) De novo mutation in the mitochondrial ATP synthase subunit 6 gene (T8993G) with rapid segregation resulting in Leigh syndrome in the offspring. Hum. Genet., 96, 290–294. [DOI] [PubMed] [Google Scholar]
- 84.White S.L., Shanske S., McGill J.J. et al. (1999) Mitochondrial DNA mutations at nucleotide 8993 show a lack of tissue- or age-related variation. J. Inher. Metab. Dis., 22, 899–914. [DOI] [PubMed] [Google Scholar]
- 85.Cervin C., Liljestrom B., Tuomi T. et al. (2004) Cosegregation of MIDD and MODY in a pedigree: functional and clinical consequences. Diabetes, 53, 1894–1899. [DOI] [PubMed] [Google Scholar]
- 86.Chinnery P.F., Zwijnenburg P.J., Walker M. et al. (1999) Nonrandom tissue distribution of mutant mtDNA. Am. J. Med. Genet., 85, 498–501. [PubMed] [Google Scholar]
- 87.Chou Y.J., Ou C.Y., Hsu T.Y. et al. (2004) Prenatal diagnosis of a fetus harboring an intermediate load of the A3243G mtDNA mutation in a maternal carrier diagnosed with MELAS syndrome. Prenatal Diagn., 24, 367–370. [DOI] [PubMed] [Google Scholar]
- 88.Dubeau F., De Stefano N., Zifkin B.G., Arnold D.L., Shoubridge E.A. (2000) Oxidative phosphorylation defect in the brains of carriers of the tRNAleu(UUR) A3243G mutation in a MELAS pedigree. Ann. Neurol., 47, 179–185. [PubMed] [Google Scholar]
- 89.Fabrizi G.M., Cardaioli E., Grieco G.S. et al. (1996) The A to G transition at nt 3243 of the mitochondrial tRNALeu(UUR) may cause an MERRF syndrome. J. Neurol. Neurosurg. Psychiatry, 61, 47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hotta O., Inoue C.N., Miyabayashi S., Furuta T., Takeuchi A., Taguma Y. (2001) Clinical and pathologic features of focal segmental glomerulosclerosis with mitochondrial tRNALeu(UUR) gene mutation. Kidney Int., 59, 1236–1243. [DOI] [PubMed] [Google Scholar]
- 91.Huang C.C., Chen R.S., Chen C.M. et al. (1994) MELAS syndrome with mitochondrial tRNA(Leu(UUR)) gene mutation in a Chinese family. J. Neurol. Neurosurg. Psychiatry, 57, 586–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang C.C., Chen R.S., Chu N.S., Pang C.Y., Wei Y.H. (1996) Random mitotic segregation of mitochondrial DNA in MELAS syndrome. Acta Neurol. Scand., 93, 198–202. [DOI] [PubMed] [Google Scholar]
- 93.Iwanishi M., Obata T., Yamada S. et al. (1995) Clinical and laboratory characteristics in the families with diabetes and a mitochondrial tRNA(LEU(UUR)) gene mutation. Diabetes Res. Clin. Pract., 29, 75–82. [DOI] [PubMed] [Google Scholar]
- 94.Jansen J.J., Maassen J.A., van der Woude F.J. et al. (1997) Mutation in mitochondrial tRNA(Leu(UUR)) gene associated with progressive kidney disease. J. Am. Soc. Nephrol. JASN, 8, 1118–1124. [DOI] [PubMed] [Google Scholar]
- 95.Ko C.H., Lam C.W., Tse P.W., Kong C.K., Chan A.K., Wong L.J. (2001) De novo mutation in the mitochondrial tRNALeu(UUR) gene (A3243G) with rapid segregation resulting in MELAS in the offspring. J. Paedr. Child Health, 37, 87–90. [DOI] [PubMed] [Google Scholar]
- 96.Lien L.M., Lee H.C., Wang K.L., Chiu J.C., Chiu H.C., Wei Y.H. (2001) Involvement of nervous system in maternally inherited diabetes and deafness (MIDD) with the A3243G mutation of mitochondrial DNA. Acta Neurol. Scand., 103, 159–165. [DOI] [PubMed] [Google Scholar]
- 97.Liou C.W., Huang C.C., Chee E.C. et al. (1994) MELAS syndrome: correlation between clinical features and molecular genetic analysis. Acta Neurol. Scand., 90, 354–359. [DOI] [PubMed] [Google Scholar]
- 98.Lu C.Y., Tso D.J., Yang T., Jong Y.J., Wei Y.H. (2002) Detection of DNA mutations associated with mitochondrial diseases by Agilent 2100 bioanalyzer. Clin. Chim. Acta, 318, 97–105. [DOI] [PubMed] [Google Scholar]
- 99.Martinuzzi A., Bartolomei L., Carrozzo R. et al. (1992) Correlation between clinical and molecular features in two MELAS families. J. Neurol. Sci., 113, 222–229. [DOI] [PubMed] [Google Scholar]
- 100.Morovvati S., Nakagawa M., Sato Y., Hamada K., Higuchi I., Osame M. (2002) Phenotypes and mitochondrial DNA substitutions in families with A3243G mutation. Acta Neurol. Scand., 106, 104–108. [DOI] [PubMed] [Google Scholar]
- 101.Olsson C., Zethelius B., Lagerstrom-Fermer M., Asplund J., Berne C., Landegren U. (1998) Level of heteroplasmy for the mitochondrial mutation A3243G correlates with age at onset of diabetes and deafness. Hum. Mutat., 12, 52–58. [DOI] [PubMed] [Google Scholar]
- 102.Onishi H., Hanihara T., Sugiyama N. et al. (1998) Pancreatic exocrine dysfunction associated with mitochondrial tRNA(Leu)(UUR) mutation. J Med Genet, 35, 255–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rusanen H., Majamaa K., Tolonen U., Remes A.M., Myllyla R., Hassinen I.E. (1995) Demyelinating polyneuropathy in a patient with the tRNA(Leu)(UUR) mutation at base pair 3243 of the mitochondrial DNA. Neurology, 45, 1188–1192. [DOI] [PubMed] [Google Scholar]
- 104.Vilarinho L., Santorelli F.M., Rosas M.J., Tavares C., Melo-Pires M., DiMauro S. (1997) The mitochondrial A3243G mutation presenting as severe cardiomyopathy. J. Med. Genet., 34, 607–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wilichowski E., Korenke G.C., Ruitenbeek W. et al. (1998) Pyruvate dehydrogenase complex deficiency and altered respiratory chain function in a patient with Kearns-Sayre/MELAS overlap syndrome and A3243G mtDNA mutation. J. Neurol. Sci., 157, 206–213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.