Abstract
Adult-onset inherited myopathies with similar pathological features, including hereditary inclusion body myopathy (hIBM) and limb-girdle muscular dystrophy (LGMD), are a genetically heterogeneous group of muscle diseases. It is unclear whether these inherited myopathies initiated by mutations in distinct classes of genes are etiologically related. Here, we exploit a genetic model system to establish a mechanistic link between diseases caused by mutations in two distinct genes, hnRNPA2B1 and DNAJB6. Hrb98DE and mrj are the Drosophila melanogaster homologs of human hnRNPA2B1 and DNAJB6, respectively. We introduced disease-homologous mutations to Hrb98DE, thus capturing mutation-dependent phenotypes in a genetically tractable model system. Ectopic expression of the disease-associated mutant form of hnRNPA2B1 or Hrb98DE in fly muscle resulted in progressive, age-dependent cytoplasmic inclusion pathology, as observed in humans with hnRNPA2B1-related myopathy. Cytoplasmic inclusions consisted of hnRNPA2B1 or Hrb98DE protein in association with the stress granule marker ROX8 and additional endogenous RNA-binding proteins (RBPs), suggesting that these pathological inclusions are related to stress granules. Notably, TDP-43 was also recruited to these cytoplasmic inclusions. Remarkably, overexpression of MRJ rescued this phenotype and suppressed the formation of cytoplasmic inclusions, whereas reduction of endogenous MRJ by a classical loss of function allele enhanced it. Moreover, wild-type, but not disease-associated, mutant forms of MRJ interacted with RBPs after heat shock and prevented their accumulation in aggregates. These results indicate both genetic and physical interactions between disease-linked RBPs and DNAJB6/mrj, suggesting etiologic overlap between the pathogenesis of hIBM and LGMD initiated by mutations in hnRNPA2B1 and DNAJB6.
Introduction
The inherited myopathies and muscular dystrophies are a diverse group of muscle diseases. Depending on the phenotype, distribution of weakness, age of onset and pathological features, these inherited myopathies are grouped into different categories. Hereditary inclusion body myopathy (hIBM) and limb-girdle muscular dystrophy (LGMD) are a genetically heterogeneous group of inherited myopathies. However, patients with hIBM or LGMD develop both adult-onset proximal and distal muscle weakness that accompanies proteinopathy in affected tissues, resulting in clinical and pathological resemblance to one another (1).
A variety of distinct classes of genes have been associated with inherited myopathies (2). Among these, genes encoding RNA-binding proteins (RBPs), including hnRNPA2B1, hnRNPA1, hnRNPDL and TIA-1, are particularly interesting because these gene products are all paralogous RBPs, which are found in pathological inclusions, and their mutations have been implicated in a broad spectrum of diseases (3–5). Many disease-associated RBPs contain a prion-like domain (PrLD) that has a very strong tendency to self-associate (6). PrLDs have been characterized as ‘prion-like’ not only because they contain high amino acid homology with fungal prions, but also because human PrLDs can functionally substitute for bona fide prion domains in yeast (3). This observation indicates that regulated higher order assemblies of RBPs mediated by PrLDs likely have some native function in human cells. Indeed, RBPs containing PrLDs, such as hnRNPA1 and TDP-43, have been shown to mediate the assembly of RNA granules through liquid–liquid phase separation (7–9). Importantly, maintenance of RNA granules in a highly concentrated condensed liquid state for extended periods of time was shown to promote formation of RBP-containing amyloid-like fibrils, analogous to the inclusions observed in hIBM and other degenerative diseases (7–9). Disease-causing mutations in RBPs generally impact the PrLD, resulting in enhanced prionogenicity and increased propensity for self-association and aggregation. Therefore, mutations in the PrLD of RBPs may promote the accumulation of amyloid-like fibrils, leading to pathogenesis. In addition, mutations in regulatory factors that clear/disassemble the higher order complexes present in RNA granules [e.g. VCP (10)] may further accelerate toxicity through loss of quality control mechanisms.
DNAJ/HSP40s are crucial co-factors for HSP70-mediated molecular chaperones. DNAJ/HSP40s are homodimeric proteins characterized by the presence of the highly conserved approximately 70 amino acid J-domain, facilitating their interaction with HSP70 family members via binding to the HSP70 N-terminal ATPase domain (11–14). This interaction regulates and stimulates not only the intrinsic ATPase activity of HSP70s, but also the recruitment of client proteins, conferring client specificity and functional diversity of the HSP-mediated chaperone machinery. It has recently been suggested in several reports that DNAJ/HSP40s might specifically function in the regulation of misfolded protein-associated neurodegenerative diseases. DNAJA1, a member of the DNAJ/HSP40 family, was found in aggregates in affected neurons from spinocerebellar ataxia type-1 (SCA-1) patients (15). Furthermore, it was shown that overexpression of wild-type (WT) DNAJA1 in a cellular model of SCA-1 prevented ataxin-1 aggregation, whereas the J domain mutants were unable to suppress aggregation of ataxin-1 (16). Besides DNAJA1, many other family members of DNAJ/HSP40, DNAJB and DNAJC proteins have also shown to play a role in polyglutamine and other protein aggregation diseases (17). In addition, a yeast homolog of DNAJB1, Sis1, was found to regulate the disassembly of fungal prions that are strikingly similar to the PrLDs of human RBPs (18). The direct link of DNAJ/HSP40 families to protein aggregation-associated degenerative diseases came from recent findings that mutations in five different DNAJ proteins are the cause of some neuropathies and myopathies (17). Among these, mutations in the glycine–phenylalanine-rich domain (G/F domain) of DNAJB6 were found to cause autosomal-dominant LGMD type 1E (LGMD1E), which shares histopathological features with disease initiated by mutations in hnRNPA1, hnRNPA2B1 and hnRNPDL, including deposition of fibrillar pathology of hnRNPA1, hnRNPA2B1 and TDP-43 (19–21). Also, DNAJB6 was co-purified in complex with the PrLD-containing RBP, TDP-43 (22). These findings not only demonstrate the functional disruption of DNAJ/HSP40s during the development of diseases with protein aggregation, but also suggest the possibility of exploiting DNAJ/HSP40s as a therapeutic target to prevent aggregation process and thus to reduce the toxicity of the aggregation-prone proteins such as RBPs with PrLD. Yet, the genetic interaction of these two functionally unrelated groups of proteins, RBPs and molecular chaperones, in the context of disease remains unclear.
Here, we exploit a genetic model system in order to establish a mechanistic link between diseases caused by mutations in two distinct genes, hnRNPA2B1 and DNAJB6. We first identified Hrb98DE as the Drosophila melanogaster homolog of human hnRNPA2B1 and introduced disease-homologous mutations to Hrb98DE, thus capturing mutation-dependent phenotypes in a genetically tractable model system. This system was then used to interrogate the functional relationship of Hrb98DE and mrj, a Drosophila homolog of human DNAJB6, as well as to establish the epistasis between disease-specific mutations. We show that ectopic expression of the disease-causing mutant form of either hnRNPA2B1 or Hrb98DE leads to progressive, age-dependent cytoplasmic inclusion pathology. The mutant proteins co-localize with the stress granule marker, ROX8, and sequester other RBPs, including TDP-43, in vivo. As expected, WT MRJ, but not disease-associated MRJ mutants, rescues mutant Hrb98DE and hnRNPA2B1-associated cytoplasmic mislocalization and abnormal wing phenotype and interacts with Hrb98DE after heat shock. Thus, our study provides the first evidence of the genetic interaction between two adult-onset inherited myopathy-related genes in D. melanogaster.
Results
Hrb98DE is the ortholog of human hnRNPA2B1 and hnRNPA1
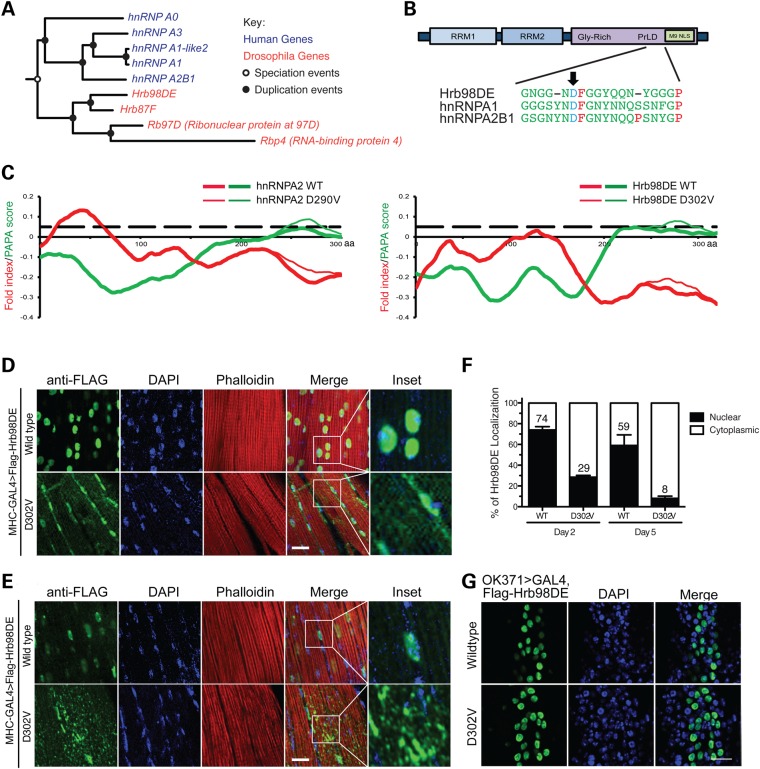
To identify the Drosophila orthologs of human hnRNPA2B1 and hnRNPA1, we performed phylogenetic analysis to resolve the orthologous and paralogous relationships between the hnRNP genes in D. melanogaster and those in Homo sapiens. The branch of genes related to hnRNPA2B1 and hnRNPA1 is shown in Figure 1A. As suggested by the constrained tree, the distinct hnRNP family members diverged after the split of insects from the lineage leading to vertebrates. This pattern suggests that many of these genes have formed recently via gene duplications that occurred separately in either of these two lineages. As a result, Drosophila genes, including Hrb98DE, Hrb87F, ribonuclear protein at 97D (Rb97D) and RBP 4 (Rbp4), showed homology to hnRNPA2B1 and hnRNPA1 and were not able to be further resolved by sequence homology.
Figure 1.
Ectopic expression of Hrb98DE D302V in Drosophila muscle induces age-related accumulation of cytoplasmic aggregates. (A) Phylogenetic tree of the orthologic/paralogic relationships among genes of human and Drosophila HNRNPA2B1 family members. (B) Schematics show conserved domains in Hrb98DE and its amino acid sequence alignment with hnRNPA1 and hnRNPA2B1. Arrow shows the disease-related mutations, which are D290V in hnRNPA2 (dominant isoform of hnRNPA2B1) and D262V in hnRNPA1, and their corresponding mutation D302V in Hrb98DE. RRM, RNA recognition motif; Gly-rich, glycine-rich domain; PrLD, prion-like domain; M9 NLS, M9 nuclear localization signal. (C) The mutation significantly enhances the predicted prion propensity of hnRNPA2 and Hrb98DE. PAPA scores (green) and fold indexes (red) were calculated by using the PAPA algorithm, and those of WT and D302V mutant Hrb98DE were compared. Black dashed line indicates the prion propensity cutoff of the PAPA algorithm, which is 0.05. (D) Cellular distribution of Flag-Hrb98DE WT and D302V in day 2 adult Drosophila indirect flight muscle. Both WT and D302V Flag-Hrb98DE (green) co-localize with DAPI-stained nuclei (blue). (E) Cellular distribution of Flag-Hrb98DE WT and D302V in day 5 adult Drosophila indirect flight muscle. (F) Quantitative analysis of cellular distribution of Flag-Hrb98DE WT and D302V in day 2 and 5 Drosophila. The signal of either WT or D302V Flag-Hrb98DE was referred to DAPI to determine its nuclear localization. The percentages of WT and D302V Flag-Hrb98DE in nuclei (filled bar) or cytoplasm (opened bar) are shown in the bar graph. Scale bar represents 20 µm. (G) Immunofluorescence shows nuclear distribution of WT or D302V Flag-Hrb98DE in motor neurons under the control of the OK371>GAL4 promoter.
We previously found that prion-like propensity was attributable to the C-termini of hnRNPA2B1 and hnRNPA1 (3). We, therefore, performed prion propensity prediction analysis on all four Drosophila proteins, using the PAPA algorithm (23). Hrb98DE and Hrb87F have prion propensities similar to those of hnRNPA2B1 and hnRNPA1, with higher scores in their C-termini; however, Rb97D and Rbp4 did not exhibit prion propensity (Supplementary Material, Fig. S1). hnRNPA2B1 is expressed as two alternatively spliced isoforms: A2 and B1. The shorter hnRNPA2, which lacks 12 amino acids in the N-terminal region, is the main isoform, accounting for 90% of the protein in most tissues. The disease-related mutation substitutes a valine residue in the place of an evolutionarily conserved aspartate residue (D290V and D302V in A2 and B1 forms, respectively). The amino acid sequences flanking the aspartate to valine mutation from hnRNPA2B1 and hnRNPA1 were used to align with Hrb98DE and Hrb87F. It appears that only Hrb98DE bares the conserved aspartate at amino acid 302 of its C-terminus, with similar flanking sequence (Fig. 1B). The PAPA algorithm was then used to compare the prion propensity of WT and D302V-mutated Hrb98DE (Fig. 1C). Although the fold indexes predict that the C-terminal halves of both proteins remain unfolded, the D302V mutation enhances the predicted prion propensity (PAPA score = 0.08 at amino acid position 275) beyond that of WT (PAPA score = 0.05 at amino acid position 235). The result showed a pattern similar to that in WT and mutant hnRNPA2 (Fig. 1C). Therefore, we concluded that Hrb98DE might be the Drosophila ortholog of human hnRNPA2B1 and hnRNPA1.
Expression of the Hrb98DE D302V mutant in Drosophila recapitulates MSP muscle pathology
Transgenic Drosophila lines bearing upstream activation sequence (UAS)-controlled WT or D302V mutant form of Hrb98DE were generated by single-copy, site-directed insertion to test the mutation-related phenotype in different tissues. Ectopic expression of Hrb98DE WT or D302V in adult Drosophila eyes or motor neurons did not cause phenotypic differences (Supplementary Material, Fig. S2). Ectopic expression of WT or D302V form of Hrb98DE in adult Drosophila indirect flight muscles under the control of MHC-GAL4 led to muscle dysfunction indicated by abnormal wing posture (Supplementary Material, Fig. S2A and B). However, despite the comparable levels of expression (Supplementary Material, Fig. S2D), only mutant Hrb98DE D302V resulted in the significant accumulation of protein in cytoplasm at day 2 after eclosing from pupae, whereas ectopic expression of Hrb98DE WT was predominantly localized to muscle nuclei (Fig. 1D). The cytoplasmic accumulation increased with age. At day 5, more prominent cytoplasmic aggregates of Hrb98DE D302V were formed, which was accompanied by the disappearance of proteins from the nucleus (Fig. 1E). On day 2, 29% of Hrb98DE D302V was found in the nucleus. On day 5, this percentage was reduced to 8% (P = 0.0001, t-test, Fig. 1F). However, the difference in the percentages of Hrb98DE WT in the nucleus at each time point was not statistically significant (74 and 59% on day 2 and 5, respectively. P = 0.1678, t-test). Intriguingly, the cytoplasmic aggregation of Hrb98DE D302V was cell-type-specific. The Hrb98DE D302V-associated cytoplasmic aggregation was not found in motor neuron cell bodies, in which ectopic expression of Hrb98DE did not cause an apparent phenotype (Fig. 1G), implying that additional cell-type-related modulators might be involved to confer tissue specificity of the disease.
Mutant forms of Hrb98DE and hnRNPA2 co-localize with stress granule marker ROX8 and sequester other RBPs
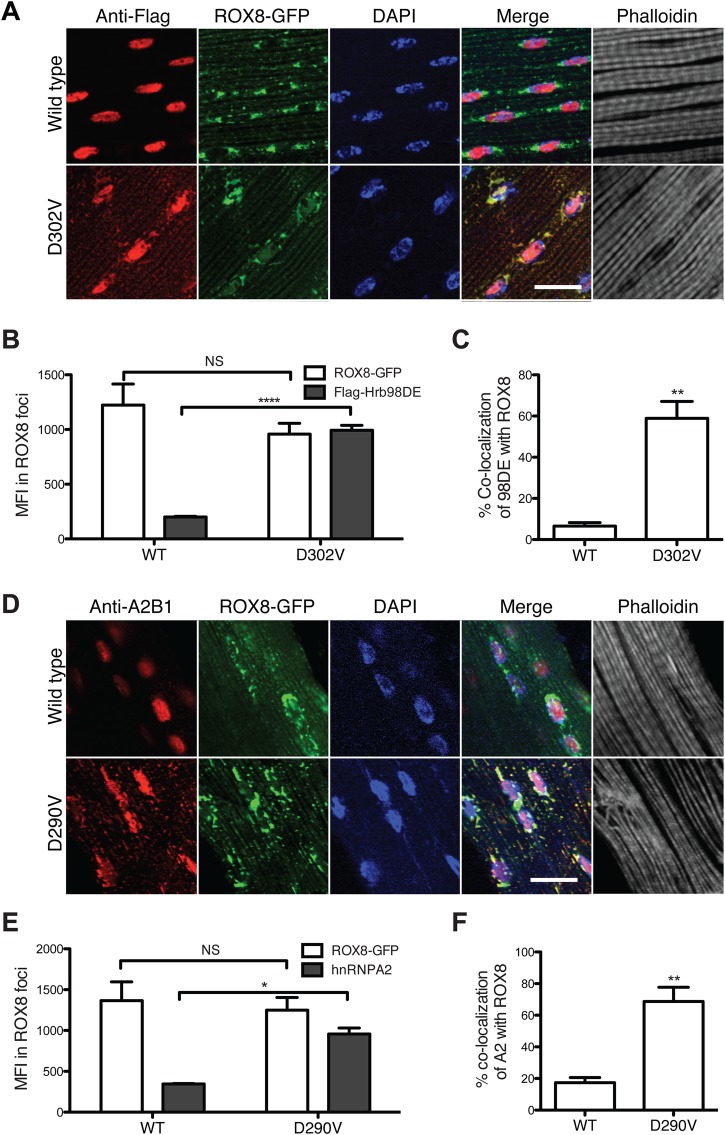
In our previous study, we found both WT and mutant hnRNPA2 located in stress granules in HeLa cells under different stress conditions (3). Therefore, we examined whether the cytoplasmic inclusions formed by mutant Hrb98DE co-localized with stress granule markers in vivo. A transgenic Drosophila carrying green fluorescence protein (GFP)-tagged ROX8 was generated. ROX8, the Drosophila ortholog of human TIAR, was reported as being a stress granule marker in the Drosophila S2 cell line (24). Ectopic expression of ROX8-GFP in Drosophila indirect flight muscles revealed a granulated staining pattern, with most granules in para-nuclear regions (Supplementary Material, Fig. S3A). Under heat shock conditions, the small granules of ROX8-GFP aggregated and formed larger puncta that were dissembled when the stress was removed (Supplementary Material, Fig. S3B). This pattern indicates that ROX8 is a good marker for studying stress granule dynamics in vivo. When ROX8-GFP was co-expressed with Hrb98DE WT protein, they remained separate. Interestingly, when Hrb98DE D302V was co-expressed with ROX8-GFP, its cytoplasmic aggregates were almost completely co-localized with the ROX8 signal (Fig. 2A). To ensure that this was an unbiased conclusion, we performed quantitative analysis to compare the localization of Hrb98DE WT or D302V from images taken from at least three individual replicates. We found that the mean fluorescence intensity (MFI) of the D302V mutant was highly increased in the ROX8-positive foci, whereas the MFI of Hrb98DE WT remained low (Fig. 2B). We also found that ∼60% of the total Hrb98DE D302V signal overlapped with ROX8-GFP signal, indicating that most of the cytoplasmic D302V co-localized with ROX8-GFP (Fig. 2C). However, <10% of Hrb98DE WT signal co-localized with ROX8-GFP, which may be due to some overlapping of signals at the nuclear envelope regions. To confirm this finding, we also examined the localization pattern relationship between human hnRNPA2 and ROX8. Similar to what we found with Hrb98DE, ROX8 only co-localized with hnRNPA2 D290V in the cytoplasm when it formed inclusions (Fig. 2D). Quantitative analysis confirmed that D290V co-localized with ROX8 more than the WT did (Fig. 2E). The percentage of D290V co-localization with ROX8 was significantly greater than that of WT (Fig. 2F). The co-localization of ROX8 to cytoplasmic inclusions of the Hrb98DE and hnRNPA2 mutant highlights the fact that the pathological inclusions may share properties similar to those of stress granules.
Figure 2.
Mutants of Hrb98DE and hnRNPA2 co-localize with stress granule marker ROX8 in cytoplasmic inclusions in Drosophila indirect flight muscle. (A) Day 2 adult Drosophila expressing ROX8-GFP (green) with either WT or D302V Flag-Hrb98DE (red) were dissected to examine the cellular localization of these proteins. Flag-Hrb98DE WT localized in nuclei, and Flag-Hrb98DE D302V co-localized with ROX8-GFP in cytoplasmic inclusions. Scale bar: 20 µm. (B) The MFI of Flag-Hrb98DE D302V in ROX8-positive foci was significantly higher than that of Flag-Hrb98DE WT. P < 0.0001, t-test. (C) The percentage of the fluorescence signal of Flag-Hrb98DE D302V co-localized with ROX8-GFP was significantly higher than that of WT. P < 0.01, t-test. (D) Day 2 adult Drosophila expressing GFP-ROX8 (green) with either WT or D290V hnRNPA2 (red) were dissected to examine the cellular localization of these proteins. hnRNPA2 WT localized in nuclei, and hnRNPA2 D290V co-localized with ROX8-GFP in cytoplasmic inclusions. Scale bar: 20 µm. (E) The MFI of hnRNPA2 D290V in ROX8-positive foci was significantly higher than that of hnRNPA2 WT. P < 0.05, t-test. (F) The percentage of the fluorescence signal of hnRNPA2 D290V co-localized with ROX8-GFP was significantly higher than that of WT. P < 0.01, t-test.
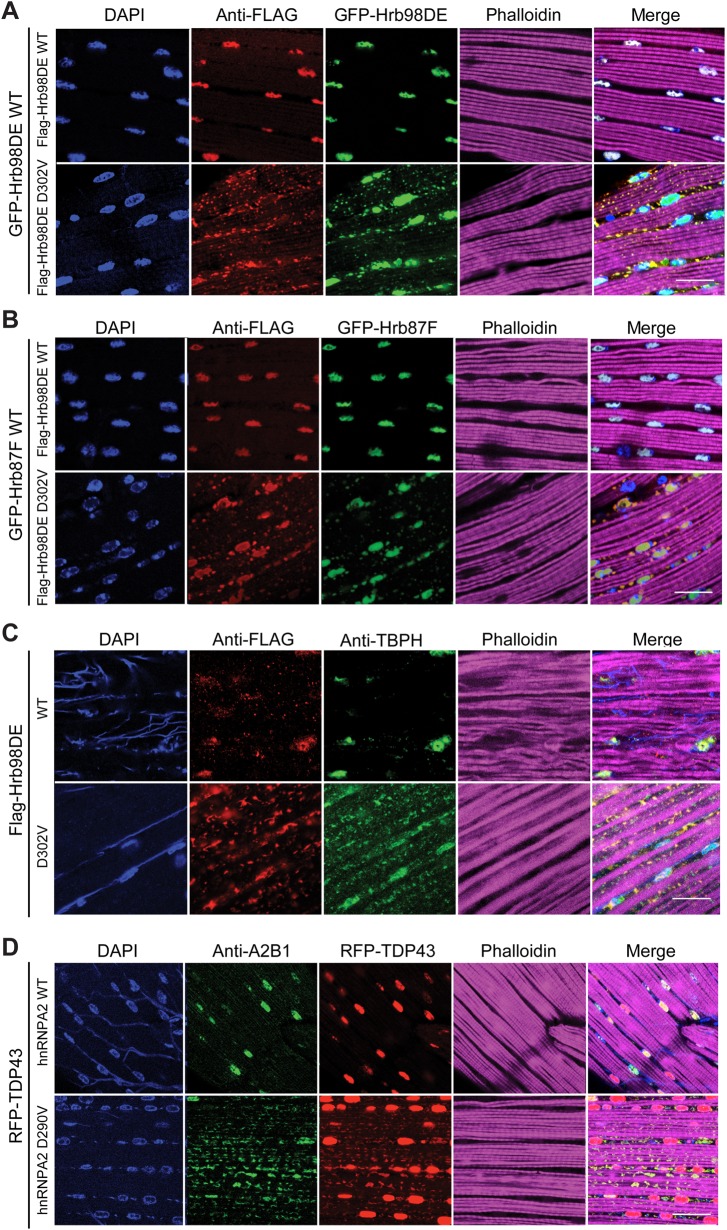
In our previous study, mutation of hnRNPA2B1 or hnRNPA1 was found to cause disease in an autosomal-dominant inheritance pattern (3). In patient muscles, hnRNPA2B1 and hnRNPA1 were often found to be excluded from the nucleus and aggregated in the cytoplasm, despite the presence of one WT copy of the gene. Furthermore, aberrant cytoplasmic accumulation of TDP-43, the pathological hallmark of amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD), was also present, suggesting that the mutant forms of hnRNPA2B1 and hnRNPA1 can affect cellular localization of other RBPs that do not carry disease-causing mutations. Therefore, we sought to examine whether our Hrb98DE D302V Drosophila model exhibits properties similar to those of hnRNPA2B1 in patients with MSP. To determine whether mutant Hrb98DE is able to sequester other RBPs, we co-expressed GFP-tagged Hrb98DE WT or Hrb87F with Flag-tagged WT or D302V form of Hrb98DE in Drosophila indirect flight muscles. As expected, GFP-Hrb98DE WT and GFP-Hrb87F were found in the nucleus, co-localizing with Flag-Hrb98DE WT (Fig. 3A and B). However, some GFP-Hrb98DE WT was translocated to the cytoplasm and co-localized with Flag-Hrb98DE D302V in the aggregates (Fig. 3A). A similar result was found when GFP-Hrb87F was co-expressed with Flag-Hrb98DE D302V (Fig. 3B). More importantly, endogenous TAR DNA-binding protein-43 homolog (TBPH), the Drosophila ortholog of human TDP-43, co-localized with cytoplasmic Flag-Hrb98DE D302V, suggesting that it was also sequestered to cytoplasmic inclusions (Fig. 3C). In agreement with this observation, co-expression of red fluorescence protein (RFP)-tagged human TDP-43 with hnRNPA2 D290V in indirect flight muscles revealed sequestration of TDP-43 to the cytoplasmic aggregates (Fig. 3D), but nuclear localization of TBPH or RFP-TDP-43 was not altered by expression of WT Hrb98DE or hnRNPA2, respectively (Fig. 3C and D). Finally, we examined whether the cytoplasmic aggregates caused by Hrb98DE D302V could be suppressed by reduction of endogenous Hrb98DE. To reduce the levels of endogenous Hrb98DE protein without affecting ectopically expressed Hrb98DE D392V, we used a deficiency line on the third chromosome, Df(3R)BSC322, in which the entire Hrb98DE gene was deleted. Endogenous Hrb98DE heterozygotes led to a marked suppression of the Flag-Hrb98DE D302V-associated cytoplasmic aggregation (Supplementary Material, Fig. S4), suggesting that the aggregate phenotype is hypermorphic rather than neomorphic. Altogether, our data suggest that expression of Hrb98DE D302V in Drosophila indirect flight muscle causes a mutation- and age-dependent formation of cytoplasmic disease protein aggregates that sequester other WT RBPs. This recapitulates the muscular pathology observed in hnRNPA2B1-related MSP.
Figure 3.
WT RBPs were sequestered in the Hrb98DE D302V-positive cytoplasmic inclusions. (A) Expression of GFP-Hrb98DE WT in indirect flight muscle co-localized with Flag-Hrb98DE D302V in cytoplasmic inclusions and with Flag-Hrb98DE WT in nuclei. (B) Expression of GFP-Hrb87F WT in indirect flight muscle co-localized with Flag-Hrb98DE D302V in cytoplasmic inclusions and with Flag-Hrb98DE WT in nuclei. (C) Endogenous TBPH was detected in Drosophila expressing either WT or D302V Flag-Hrb98DE in indirect flight muscle. TBPH was sequestered into cytoplasmic aggregates when Flag-Hrb98DE D302V, but not WT, was expressed. (D) RFP-TDP-43 expressed in indirect flight muscle was sequestered into cytoplasmic aggregates when hnRNPA2 D290V, not WT, was expressed. Scale bars represent 20 µm.
Expression of MRJ inhibits cytoplasmic inclusion formation in the Hrb98DE/hnRNPA2 mutant
Our findings demonstrating that the disease mutation in Hrb98DE and hnRNPA2B1 drives formation of cytoplasmic aggregates that co-localize with stress granule marker, ROX8-GFP, suggest that excess assembly and persistence of stress granules may be in the center of disease pathogenesis. The current hypothesis is that disease-causing mutations increase the aggregation-propensity of the target proteins, incorporating them more tightly into stress granules. These irreversibly aggregated proteins may account for the pathological inclusions. Therefore, any modulation that prevents irreversible aggregation of disease-associated proteins may reduce their toxicity.
HSPs are well-known modulators of protein aggregation. Interestingly, it was shown that mutations in a family of DNAJ/HSP40 chaperone, DNAJB6, cause LGMD1E, another type of inherited myopathy (19,20,25). LGMD1E caused by DNAJB6 mutation shares histopathological features with disease initiated by mutations in VCP, hnRNPA1 or hnRNPA2B1, including deposition of fibrillar pathology of hnRNPA1, hnRNPA2B1 and TDP-43 (19,21). Therefore we hypothesized that DNAJB6 function may intersect with RBP function, specifically in regulating the aggregation propensity of these RBPs.
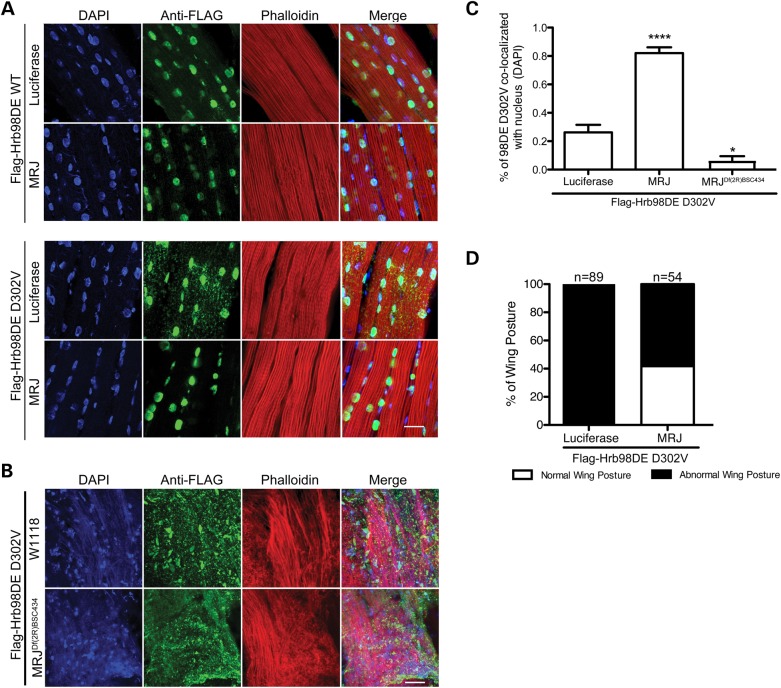
We generated a transgenic Drosophila line carrying MRJ and co-expressed either WT or D302V form of Flag-Hrb98DE in the indirect flight muscle. Although expression of MRJ had no obvious effect on Hrb98DE WT localization, the cytoplasmic aggregation of Flag-Hrb98DE D302V was significantly reduced in fly muscles expressing MRJ (Fig. 4A). Moreover, reducing the endogenous MRJ levels by half using a classical loss of function line led to a marked enhancement of cytoplasmic aggregation of Hrb98DE D302V (Fig. 4B). Quantification analysis showed that expression of MRJ significantly increased the amount of Hrb98DE D302V in the nucleus to about 80% of the total signal versus only about 20% in control. Reduction of endogenous MRJ decreased the amount of Hrb98DE D302V in the nucleus to <10% (Fig. 4C). Expression of MRJ also partially rescued abnormal wing posture phenotype caused by Hrb98DE D302V (Fig. 4D).
Figure 4.
Expression of MRJ inhibited Hrb98DE D302V-positive cytoplasmic inclusion formation and rescued abnormal wing posture phenotype. (A) Either WT or D302V Flag-Hrb98DE was co-expressed either with luciferase or with MRJ in Drosophila indirect flight muscle. The cellular localization of Flag-Hrb98DE was detected by anti-Flag antibody. (B) Cytoplasmic re-localization of Flag-Hrb98DE D302V was enhanced, following knockdown of MRJ levels using a deficiency allele. (C) The percentage of nuclear Flag-Hrb98DE D302V signals was significantly increased when it was co-expressed with MRJ (P < 0.0001, t-test). (D) Co-expression of MRJ rescued the abnormal wing posture phenotype caused by Flag-Hrb98DE D302V. P < 0.0001, χ2 test. Scale bar in (A) and (B), 20 µm.
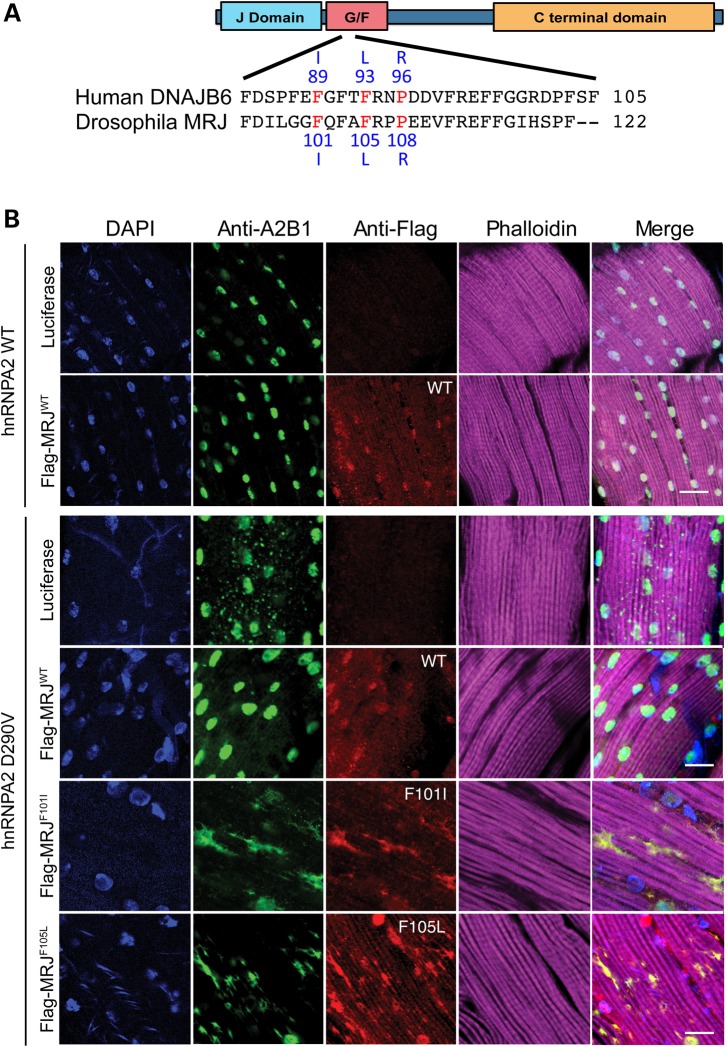
In patients with LGMD1E, multiple missense mutations in the G/F domain have been identified. These mutations cause changes in amino acid sequence, including Phe89Ile, Phe93Leu and Pro96Arg (19,20,25). Careful alignment of human DNAJB6 and Drosophila MRJ amino acid sequences identified the following conserved amino acids that correspond to the human disease mutations: Phe101, Phe105 and Pro108 (Fig. 5A). Therefore, we generated transgenic Drosophila lines with MRJ mutations to determine whether the modifier effect of MRJ WT on hnRNPA2 D290V cytoplasmic inclusion would be affected by the disease-causing mutations. Immunohistochemical analysis showed that hnRNPA2 WT localized appropriately to nuclei, whereas hnRNPA2 D290V largely accumulated in cytoplasmic inclusions as reported previously (3) (Fig. 5B). Flag-MRJ expression did not overtly affect the cellular localization of hnRNPA2 WT. However, hnRNPA2 D290V-containing cytoplasmic inclusions were significantly reduced when Flag-MRJ WT was co-expressed (Fig. 5B), confirming the genetic interaction between MRJ and Hrb98DE/hnRNPA2. Interestingly, neither Flag-MRJ F101I nor Flag-MRJ F105L mutants were able to rescue hnRNPA2 D290V-associated cytoplasmic inclusions. Rather, hnRNPA2 D290V was almost completely excluded from the nucleus and cytoplasmic aggregation was markedly enhanced, resulting in larger cytoplasmic aggregates that were both MRJ- and hnRNPA2-positive (Fig. 5B). These results highlight an indispensable role of the G/F domain in the genetic modifier effect of MRJ.
Figure 5.
WT, but not disease-causing mutants of MRJ, inhibited hnRNPA2 D290V cytoplasmic inclusion. (A) Schematics show the conserved domain of the Drosophila MRJ protein and its partial alignment with its human homolog DnaJB6. Three disease-causing point mutations in human DanJB6 are highlighted (F89I, F93L and P96R), along with the corresponding MRJ mutations used in our study (F101I and F105L). (B) Human protein hnRNPA2 was co-expressed with luciferase, Flag-MRJ WT, F101I or F105L in Drosophila indirect flight muscle. Although hnRNPA2 WT exclusively localizes to the nucleus, hnRNPA2 D290V largely accumulated in cytoplasmic inclusions. The hnRNPA2 D290V cytoplasmic inclusions were greatly reduced by WT Flag-MRJ, but not by F101I or F105L. Scale bar, 20 µm.
LGMD1E patients with DNAJB6 mutation display hnRNPA2B1 pathology
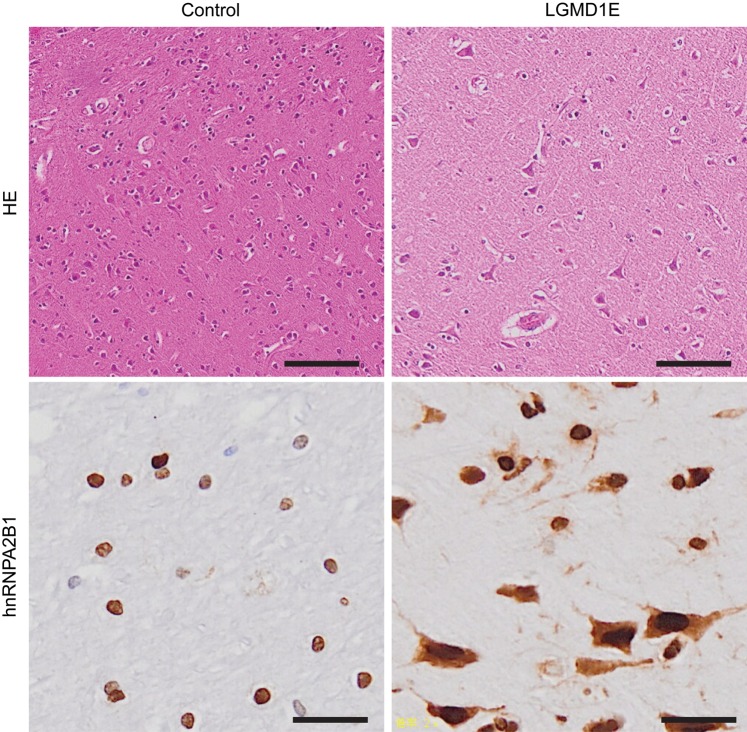
It has been previously reported that TDP-43 forms aggregates in muscle tissues from LGMD1E patients (19). A case study demonstrated that a patient with LGMD1E caused by DNAJB6 mutation also developed frontotemporal dementia (FTD). However, in this case, TDP-43 pathology was not observed in the affected brain, even though there was a severe reduction of DNAJB6 immunoreactivity in the frontal cortex when compared with samples from normal control (26). Absence of TDP-43 pathology concomitant with DNAJB6 mutation has also been reported in a recent mouse model of LGMD1E. Transgenic mice expressing the LGMD1E mutant, DNAJB6 F93L, under a muscle-specific promoter develop muscle pathology, consistent with myopathy. Immunohistochemistry of muscle tissues from these mice found that TDP-43 was present in the myonuclei of DNAJB6 WT or F93L expressing mouse muscle, but did not accumulate in the sarcoplasm. However, both hnRNPA2B1 and hnRNPA1 accumulated as small puncta in the center of myofibers of these mice (18). In addition, both hnRNPA2B1 and hnRNPA1 were found to accumulate in muscle tissues from LGMD1E patients (21), suggesting that the accumulation of hnRNPA2B1 and hnRNPA1 inclusions is a more common feature of DNAJB6-associated LGMD1E pathology. In order to determine whether hnRNPA2B1 is mislocalized in FTD and LGMD (DNAJB6-F93L mutation) patient brains, in which no TDP-43 pathology was observed (26), we performed immunostaining with anti-hnRNPA2B1 antibody. Remarkably, massive accumulation of hnRNPA2B1 was found in the cytoplasm of cells within the temporal lobe, compared with the exclusively nuclear localization of hnRNPA2B1 in the temporal lobe of control brain (Fig. 6), suggesting that hnRNPA2B1 pathology might be a prominent feature of DNAJB6-associated LGMD1E.
Figure 6.
hnRNPA2B1 was accumulated in the cytoplasm in patients with LGMD1E. Hematoxylin and eosin and hnRNPA2B1 stainings were performed in cortical tissues from patients with FTD and LGMD1E caused by DNAJB6 F93L mutation. hnRNPA2B1 staining demonstrates that temporal lobes of patients with FTD and LGMD1E largely accumulate hnRNPA2B1 in the cytoplasm. Scale bar, 50 µm.
MRJ increasingly interacts with Hrb98DE under heat shock conditions
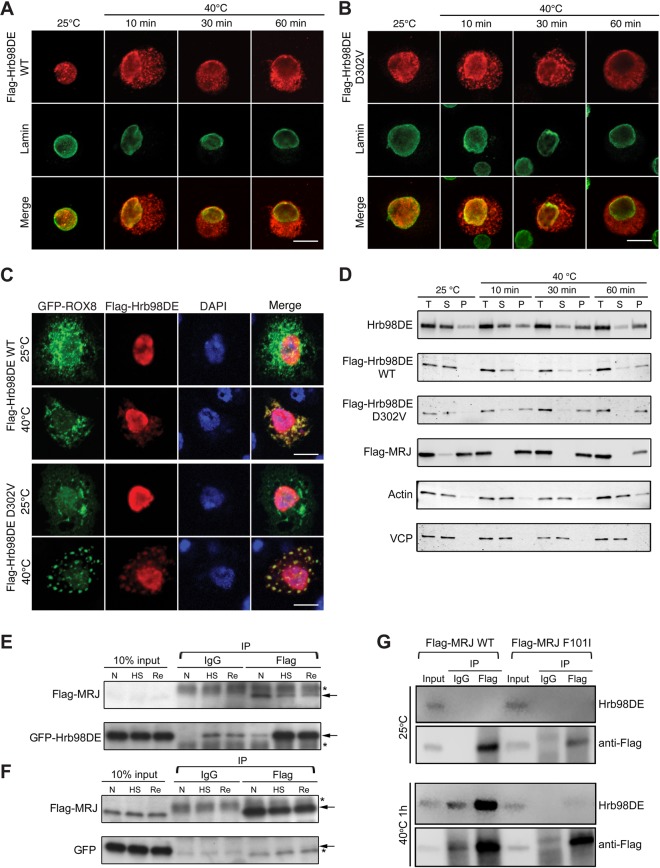
In order to further characterize the molecular mechanism by which MRJ WT, not disease-associated mutants, affects the aggregation property of Hrb98DE D302V, we sought to explore their physical interaction using co-immunoprecipitation (co-IP) assay in cultured cells. In Drosophila S2 cells, both WT and D302V mutant forms of Hrb98DE were localized to the nuclei under normal culture conditions (25°C, Fig. 7A and B). However, upon heat shock stress at 40°C, Hrb98DE translocated from the nuclei into cytoplasm (40°C, Fig. 7A and B). After 1 h of heat shock, almost all Hrb98DE protein was depleted from the nucleus. Neither WT nor D302V Flag-Hrb98DE significantly differed in the translocation rate. Heat-shock-induced cytoplasmic translocation of Hrb98DE, particularly the D302V mutant, co-localized with the stress granule marker ROX8-GFP (Fig. 7C), indicating that Hrb98DE translocates to stress granules upon heat shock stress, consistent with hnRNPA2B1 and hnRNPA1 stress granule localization in mammalian cell lines (3).
Figure 7.
Heat shock changed the solubility and cellular localization of Hrb98DE and induced interaction between Hrb98DE and MRJ. (A) S2 cells transfected with Flag-Hrb98DE WT were fixed before or after 40°C heat shock and stained with anti-Flag (red) and anti-lamin (green, nuclear marker) to determine the cellular localization of Flag-Hrb98DE. Flag-Hrb98DE WT resided in the nucleus before heat shock and quickly translocated to the cytoplasm after heat shock. (B) Same experiment as (A), except Flag-Hrb98DE D302V was transfected into S2 cells. (C) S2 cells transfected with Flag-Hrb98DE and ROX8-GFP were treated with or without heat shock. Flag-Hrb98DE co-localized with ROX8-GFP in the cytoplasm after heat shock treatment. (D) Heat shock-induced changes in Hrb98DE's solubility in 0.2% Triton X-100 lysis buffer (Supplementary Material, Fig. S7). Equal amounts of cells were lysed either in Laemmli sample buffer to obtain a total (T) sample or in lysis buffer to obtain the soluble fraction (S) and pellet fraction (P). Endogenous Hrb98DE or transfected Flag-Hrb98DE WT, D302V or Flag-MRJ was detected by western blotting to determine their solubility to lysis buffer. Hrb98DE was detected in the soluble fraction (S) at normal condition. After heat shock, Hrb98DE became insoluble to lysis buffer and became detectable in the pellet fraction (P). The solubility of VCP and actin was not changed after heat shock and used as loading controls along with Sypro Ruby-stained gel (Supplementary Material, Fig. S8C). (E) Cells transfected with Flag-MRJ and GFP-Hrb98DE were treated at normal (25°C, N), heat shock (40°C for 1 h, HS) or recovery (30 min at 25°C after 1 h 40°C treatment and recovery). Using magnetic bead-based co-IP, GFP-Hrb98DE was found to be increasingly associated with Flag-MRJ in the heat shock (HS) and recovery (Re) conditions compared with that in normal (N) condition. In control IgG co-IP, only trace amounts of GFP-Hrb98DE are detected after heat shock, which may be due to the contamination of the insoluble GFP-Hrb98DE. (F) As a control, cells were transfected with Flag-MRJ and GFP and co-IP was performed as described in (E). GFP was not found to co-precipitate with Flag-MRJ in any condition. (G) Cells that transfected with either WT or F101I mutant form of Flag-MRJ were treated with heat shock, and co-IP was performed as described in (E) and (F). Endogenous Hrb98DE proteins were detected using a monoclonal antibody we generated (Supplementary Material, Fig. S4). Arrows in (E) and (F), band of interest. Asterisks in (E) and (F), nonspecific band. Scale bars, 10 µm.
We also examined the localization pattern of endogenous Hrb98DE by immunostaining with anti-Hrb98DE antibody (Supplementary Material, Fig. S5). Consistent with the localization of ectopically expressed Hrb98DE, endogenous protein was exclusively found in the nucleus under normal conditions, but slowly re-localized to cytoplasmic puncta upon heat shock (Supplementary Material, Fig. S6). To examine the impact of the disease-associated mutation in MRJ on the localization of endogenous Hrb98DE in S2 cells, we expressed Flag-tagged versions of WT and F101I mutant MRJ. Both WT and mutant MRJ proteins were ubiquitously expressed throughout the cells, and this localization pattern was unaffected by heat shock in cultured S2 cells (Supplementary Material, Fig. S6). Interestingly, expression of WT MRJ inhibited the translocation of endogenous Hrb98DE to the cytoplasm after heat shock (Supplementary Material, Fig. S6), whereas mutant MRJ F101I had no impact on endogenous Hrb98DE localization.
We next examined whether MRJ is able to regulate Hrb98DE solubility in non-ionic detergent-containing buffer in S2 cells under normal or heat shock conditions. In order to measure the levels of protein solubility change, we designed a fractionation assay in which proteins were sequentially extracted in the same volume of 0.2% Triton X-100 (lysis buffer) and urea buffer (Supplementary Material, Fig. S7). Under normal culture conditions, endogenous Hrb98DE dissolved in lysis buffer, while a small fraction remained in the pellet. After heat shock, the soluble protein fraction was gradually reduced, and the protein recovered in the pellet fraction was increased, suggesting that Hrb98DE becomes more insoluble upon heat shock (Fig. 7D). Flag-tagged WT and Hrb98DE D302V behaved similarly, with D302V exhibiting faster dynamics in its change of solubility (Fig. 7D, second and third rows). We also found that mutant Hrb98DE (D302V) is less soluble than WT protein in indirect flight muscle in fly. Greater amounts of mutant Hrb98DE (D302V) were recovered in the Triton X-100-insoluble fraction (Supplementary Material, Fig. S8A). The change in Hrb98DE solubility was very similar to that of ROX8 (Supplementary Material, Fig. S8B), supporting our initial finding that the cytoplasmic aggregates of Hrb98DE or hnRNPA2 represent a stress-granule-like structure. Consistent with its localization pattern, Flag-MRJ solubility was not changed upon heat shock. Flag-MRJ was detected primarily in the pellet fraction under normal conditions and remained insoluble during heat shock (Fig. 7D, fourth row).
To overcome the challenge of co-IP within the insoluble fraction, we used magnetic beads conjugated to a Flag antibody in order to collect the protein of interest and then performed magnetic separation of the bound protein complexes from the input lysate. This prevented precipitation of non-specific insoluble contaminants by centrifugal force. S2 cells transfected with Flag-MRJ and GFP-Hrb98DE were lysed followed by co-IP using an anti-Flag antibody. Normal mouse immunoglobulin G (IgG) was used as a negative control for pull-down. As shown in Figure 7E, there was very little association between Flag-MRJ and GFP-Hrb98DE under normal conditions. However, the interaction significantly increased after the cells were treated with heat shock at 40°C. The interaction was maintained even when cells were recovered for 30 min at normal temperature after the heat shock. To verify that the interaction between Flag-MRJ and GFP-Hrb98DE is independent of the GFP tag, the same co-IP assay was performed using lysate from S2 cells transfected with GFP and Flag-MRJ. No interaction was detected between GFP and Flag-MRJ (Fig. 7F).
The disease-associated mutant MRJ failed to rescue cytoplasmic inclusion phenotype caused by Hrb98DE D302V in fly and also failed to inhibit cytoplasmic re-localization of Hrb98DE upon heat shock in cultured S2 cells (Fig. 5B and Supplementary Material, Fig. S6). Therefore, we next examined whether mutant MRJ F101I has the ability to interact with Hrb98DE, similar to WT MRJ. Remarkably, despite comparable levels of expression, MRJ F101I was not able to interact with endogenous Hrb98DE, although WT MRJ was found to interact with endogenous Hrb98DE as efficiently as with overexpressed Hrb98DE (Fig. 7G). Taken together, these results suggest that MRJ plays a regulatory role in the subcellular localization of Hrb98DE either through direct interaction or in complex and that this role is impaired by a disease-causing mutation of the MRJ G/F domain.
Discussion
Although accumulation of aberrant protein aggregates occurs in many muscular and neuronal degenerative diseases, very little is known about the underlying molecular mechanisms leading to aggregate formation. The identification of TDP-43 as the major protein constituent in such aggregates (27), and discovery of mutations in TDP-43 and FUS in familial ALS (28–30), highlights the contributing role of RBPs in neurodegenerative diseases. Furthermore, mutations in other RNPs including hnRNPA1, hnRNPA2B1, hnRNPDL and TIA-1 have been recently characterized to play a causal role in myopathies and bone diseases (3–5,31,32), implicating RNPs in the degeneration of several post-mitotic tissues.
An interesting common feature of many RNPs is the presence of an aggregation-prone PrLD that allows them to rapidly self-associate. The PrLD of TIA-1 was shown to be required for the assembly of stress granules, providing the first evidence connecting the PrLD of an RNP to stress granules (33). Moreover, the majority of disease-linked mutations in RNPs occur in the PrLD, leading to increased propensity of protein aggregation and stress granule formation (34). Together with the finding that TDP-43 pathology occurring in ALS and FTLD co-localizes with stress granule markers (35), stress granule biology has been suggested to be linked to the pathogenesis of several disorders. Therefore, we have hypothesized that mutations perturbing normal stress granule dynamics may lead to prolonged periods of stable stress granules, resulting in the evolution into the pathological aggregates (36).
Using Drosophila models of MSP, we have successfully recapitulated the pathological changes observed in muscles of MSP patients with hnRNPA2B1 mutation. Flies expressing a disease-causing mutation in either hnRNPA2 or Hrb98DE develop the accumulation of many RBPs in cytoplasmic aggregates (Figs 1–3). These mutant proteins show remarkable levels of co-localization with the stress granule marker, Rox8, demonstrating that pathological inclusions may share properties similar to those of stress granules (Fig. 2). Although ectopically expressed ROX8 also forms cytoplasmic granules in muscle, it did not sequester other RBPs, as disease-linked Hrb98DE or hnRNPA2 mutant proteins did. The distinct property of RNP sequestration may underlie hnRNPA2 or Hrb98DE pathogenesis by causing disruption of the normal function (hypomorph), gain of WT function (hypermorph) or even gain of toxic function (neomorph) of WT RNPs. Our data showing that reduction of endogenous Hrb98DE protein rescues Hrb98DE D302V mutant-associated aggregation (Supplementary Material, Fig. S4) demonstrate that mutant RNPs engage normal WT RNPs to confer cellular toxicity, providing evidence that the disease mutation in Hrb98DE results in a hypermorphic allele.
Notably, we identified a Hsp40 chaperone, DNAJB6/MRJ, as a genetic modifier of mutant hnRNPA2- or Hrb98DE-associated myopathy. Expression of WT MRJ, but not disease-associated MRJ mutants, rescues mutant Hrb98DE and hnRNPA2B1-associated cytoplasmic mislocalization and abnormal wing phenotype and interacts with Hrb98DE after heat shock. Moreover, reduction of the endogenous MRJ levels by MRJ heterozygous deficiency line markedly enhanced mutant protein-associated cytoplasmic aggregation (Figs. 4 and 5). Our results provide the first evidence of the genetic interaction between two functionally unrelated, yet myopathy-related, genes, hnRNPA2B1 and DNAJB6. DNAJB6 protein is composed with three distinct domains: an N-terminal J domain in which HSP70 binds (37,38), a G/F domain and a C-terminal substrate-binding domain (39). Remarkably, all known mutations linked to LGMD1E localize to an eight amino acid stretch within the G/F domain (Fig. 5A), highlighting the functional importance of this domain. A recent report demonstrated that the G/F domain of DNAJB6 is required for resolving nuclear TDP-43 stress granules. LGMD1E-associated mutations on the G/F domain of DNAJB6 abrogate this ability of DNAJB6, resulting in the persistence of TDP-43 aggregates (18). In fact, our results also demonstrate that the intact function of the G/F domain is essential for reducing mutation-dependent Hrb98DE or hnRNPA2 cytoplasmic aggregates. Our co-IP data demonstrate that the LGMD1E-associated MRJ mutation remarkably reduces Hrb98DE interaction (Fig. 7G), suggesting that the dysfunction of DNAJB6 mutants might be attributed to the inability of mutant proteins to bind RNP substrates.
Finally, we have analyzed patient samples with FTD and LGMD caused by a DNAJB6 mutation. Although the muscle from the patient revealed TDP-43 pathology, TDP-43 pathology was absent in the brain tissue from the same patient (26). However, we found that this patient developed well-defined hnRNPA2B1 pathology in the affected brain area (Fig. 6), linking previously overlooked hnRNPA2B1 pathology to LGMD. The association of hnRNPA2B1 with pathology in LGMD implies that aberrant function of hnRNPA2B1 occurs in LGMD patients, giving us insight into explaining the common pathology identified from two genetically unrelated diseases.
Overall, our data emphasize the potential shared mechanism in the pathogenesis of different clinical diseases. Future work investigating the contribution of individual factors will help improve understanding of the pleiotropic effects of these disease genes, leading to the development of a targeted approach for therapeutic intervention of inherited myopathy.
Materials and Methods
Generation of phylogram for hnRNPA2B1 gene family
TBPH of D. melanogaster was used to BLAST (Basic Local Alignment Search Tool) against the RefSeq protein database, including species of Mus musculus, D. melanogaster, Anopheles gambiae, H. sapiens, Danio rerio (zebrafish), Caenorhabditis elegans and Gallus gallus (junglefowl). The top 100 sequences (according to BLAST E values) were obtained. Sequences sharing >98% similarity were filtered by UCLUST (40) because they likely represent alternatively spliced variants of the same genes. Eighty-one sequences remaining after this process were aligned using the MAFFT multiple sequence alignment program (41). The DNA-coding sequences of all proteins were extracted and threaded onto the protein sequences to form an in-frame codon alignment. Squint (42) was used to extract the DNA-coding alignment of the two RRM domains present in each protein using the protein alignment as a guide. Phylogenetic trees of the codon alignment including 21 genes from only human and Drosophila were generated using the genetic algorithm-based GARLI (43) method.
DNA constructs
The complementary DNA sequences encoding Drosophila Hrb98DE (accession number NM_170373), Hrb87F (accession number NM_057459), mrj (accession number NM_166153) and ROX8 (accession number NM_ 170117.2) were obtained from Drosophila Genomics Resource Center (clone numbers LD38464, LD32727, LD10701 and LD31194, respectively). Full-length sequences were then amplified by performing polymerase chain reaction (PCR) assays and subcloned into pUASTattB vectors. In some cases, an epitope tag of FLAG, GFP or RFP was added to either the 5′ end or the 3′ end of the gene via PCR. Missense mutations of Hrb98DE (Hrb98DE D302V) and MRJ (MRJ F101I, F105L) were generated by using a QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, St. Jude Hartwell Center). All plasmid constructs were confirmed by Sanger DNA sequencing.
Fly stocks and culture
Flies carrying transgenes in pUASTattB vectors were generated by performing a standard injection and φC31 integrase-mediated transgenesis technique (BestGene Inc.). To express a transgene in muscles, Mhc-Gal4 was used (from G. Marqués). All Drosophila stocks were maintained in a 25°C incubator with a 12 h day/night cycle.
Antibodies
The following commercial antibodies were used in this study: mouse monoclonal anti-Flag (M2, Sigma), anti-hnRNPA2B1 (EF-67, Santa Cruz Biotechnology), anti-GFP (B-2, Santa Cruz Biotechnology) and anti-LAMIN (ADL84.12c, Developmental Studies Hybridoma Bank). Mouse anti-Hrb98E antibodies were generated as described. The rabbit anti-TBPH (N) antibody was a generous gift from D. Morton.
Preparation of adult fly muscle for immunofluorescence
Adult flies were embedded in a drop of OCT compound (Sakura Finetek) on a glass slide, frozen in liquid nitrogen and bisected sagittally by using a razor blade. After being fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS), fly tissues were permeabilized in PBS containing 0.2% Triton X-100, and indiscriminant binding was blocked by adding 5% normal goat serum in PBS. The hemithoraces were stained by using corresponding antibodies, Alexa Fluor 647 phalloidin (Life Technologies) and 4′,6-diamidino-2-phenylindole (DAPI), according to manufacturer's instructions. Stained hemithoraces were treated in 80% glycerol for 30 min. The indirect flight muscle was then removed and mounted in ProLong Gold Antifade Mountant (Life Technologies) for immunofluorescence photographing. All photographs were captured on a confocal microscope (Zeiss LSM 780 NLO Meta) with Zeiss ZEN software. To assess the co-localization of Hrb98DE or hnRNPA2 with ROX8 (Fig. 2B and E), photographs captured from corresponding Drosophila were imported into SlideBook 5 (Intelligent Imaging Innovations Inc.). A mask of ROX8 signal was generated by using the automatic default setting and then applied to the channel of either Hrb98DE or hnRNPA2. MFI inside or outside of the mask was measured by using the analysis function of the software. Photographs of at least three individual flies were analyzed, and data were then exported to Prism 6 (GraphPad software) to generate bar graphs. The data were also analyzed with DAPI signal used to generate the mask.
Generation and characterization of anti-Hrb98DE mouse monoclonal antibody
Full-length Hrb98DE was cloned into the pET28a vector to generate recombinant protein. The recombinant 6XHis-tagged Hrb98DE was expressed in Escherichia coli BL21(DE3) cells (Novagen), grown in Turbo Prime Broth medium at 37°C and induced by adding 1 mM IPTG for 4 h at 25°C before harvesting. The cell pellet was resuspended in 25 mM Tris–HCl, pH 8.0, 500 mM NaCl, 10% glycerol, 1 mM β-mercaptoethanol and 1 mM 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride and was disrupted in a high-pressure homogenizer. After the cell lysate was centrifuged at 20 000g for 40 min, the supernatant was subjected to immobilized metal-affinity chromatography, and the protein was eluted with 150 mM imidazole after a wash with 40 mM imidazole. After purification, recombinant Hrb98DE was confirmed by matrix-assisted laser desorption/ionization with tandem time-of-flight mass analyzer. The recombinant Hrb98DE was sent to GenScript for monoclonal antibody production. The final anti-Hrb98DE Clone 15C9 was selected on the basis of enzyme-linked immunosorbent assay activity against the immunogen and validated by performing western blotting and immunofluorescence assays (Supplementary Material, Fig. S5).
Drosophila S2 cell culture, transfection and immunofluorescence
S2 cells purchased from ATCC were cultured in modified Schneider medium (Sigma), supplemented with 10% heat-inactivated fetal bovine serum (HyClone, Thermo Fisher) and 10% Pen Strep (Gibco, Life Technologies) in a 25°C incubator. Cells were transferred into 1.5 ml tubes and incubated in a 40°C heat block for transient heat shock treatment. For transient expression of proteins, Lipofectamine 2000 (Life Technologies) was used, according to the manufacturer's instructions. A pAC-GAL4 plasmid (from Dr S. Ogden, St Jude Children's Research Hospital) was co-transfected with pUASTattB constructs carrying the gene-of-interest. Cells were harvested 48 h after transfection and processed for further treatment. For immunofluorescence, S2 cells were fixed in 4% paraformaldehyde in PBS and then applied to slides by cytospin. Cells were then permeabilized with 0.5% Triton X-100 in PBS for 10 min, blocked with 5% goat serum in PBS for 30 min and incubated with primary antibody for 2 h at room temperature (20–25°C) or overnight at 4°C. Primary antibodies were visualized with secondary antibodies conjugated to Alexa Fluor 488 or Alexa Fluor 555 (Life Technologies), and nuclei were detected by DAPI staining. All photographs were captured on a confocal microscope (Zeiss LSM 780 NLO Meta) with Zeiss ZEN software. The same capturing condition was applied when signals from different samples were needed for comparison.
Analysis of detergent solubility and co-IP
Cell lysate was prepared by lysing the S2 cells in 1× (150 mM NaCl, 20 nm phosphate buffer, 10% glycerol and 0.2% Triton X-100) with Halt protease inhibitor cocktail (Thermo Scientific). Cell lysates were treated twice with 30 s sonication (Misonix S-4000, Misonix Inc.) to ensure the complete homogenization of samples, which is important to reduce the likelihood of non-specific co-IP products forming due to the remaining cell debris. Without removing the insoluble fraction, the whole-cell lysate was then added with 2–5 µg of corresponding antibody and incubated at 4°C for 2 h. Then, 10–60 µl of Protein G Dynabeads (Life Technologies) was added to each sample, according to the manufacturer's protocol to precipitate the protein of interest. The beads were washed three to five times with 1× lysis buffer to remove non-specific binding. Samples were eluted by boiling the Dynabeads in 1× Laemmli sample buffer and collecting the supernatants.
Western blotting
Samples were resolved by electrophoresis on an AnyKD Mini-PROTEAN precast gel (Bio-Rad) and then transferred to PVDF membrane by using the wet transfer method. After transfer, samples were blocked with either 5% milk or 1% gelatin (Sigma) in TBS buffer with 0.05% Tween-20 (Thermo Fisher). After incubation of primary antibodies, samples were incubated with either IRDye fluorescence-labeled secondary antibodies (LI-COR) or TrueBlot horseradish peroxidase-conjugated secondary antibodies, following the manufacturers’ protocols. Protein bands were visualized by using the Odyssey Fc system (LI-COR) and Image Studio (LI-COR).
Immunohistochemistry of human brain tissues
Six-micron cryostat sections of muscle biopsies were fixed first in 4% paraformaldehyde for 5 min and then in acetone for 5 min. The slides were rinsed and washed in PBS before 30 min incubation with Image-iTFX signal enhancer reagent (Life Technologies). After serial washes in PBS, the slides were covered with 10% normal goat serum in PBS/BSA buffer. Overnight incubation with primary antibodies was carried out at 4°C. The slides then underwent serial washes in PBS before 60 min incubation with the secondary antibody Alexa Fluor 488 (1:400, Life technologies). The nuclei were detected using DAPI (Wako). The slides then underwent serial washes in PBS and were coverslipped. Images were acquired using a fluorescence microscope (BZ-X170, Keyence) with BZ-X Analyzer software.
Supplementary Material
Funding
This work was supported by a grant from Muscular Dystrophy Association (MDA) to H.J.K., from the ALS Association to R.-M.K., grants from the NIH and MDA to E.D.R. and from Target ALS, The Packard Center for ALS Research at the Johns Hopkins University and The ALS Association to J.P.T.
Supplementary Material
Acknowledgements
We thank the Bloomington Drosophila Stock Center, the VDRC Stock Center, Drs G. Marqués, D. Morton and S. Ogden for fly lines and sharing reagents, as well as Drs J. Tanboon, I. Nishino, H. Yaguchi and Y. Hayshi for the help of providing and helping with immunohistochemistry of samples from human patients. We also thank the Cell and Tissue Imaging Core at St Jude Children's Research Hospital for assistance.
Conflict of Interest statement. None declared.
References
- 1.Cardamone M., Darras B.T., Ryan M.M. (2008) Inherited myopathies and muscular dystrophies. Semin. Neurol., 28, 250–259. [DOI] [PubMed] [Google Scholar]
- 2.Taylor J.P. (2015) Multisystem proteinopathy: intersecting genetics in muscle, bone, and brain degeneration. Neurology, 85, 658–660. [DOI] [PubMed] [Google Scholar]
- 3.Kim H.J., Kim N.C., Wang Y.D., Scarborough E.A., Moore J., Diaz Z., MacLea K.S., Freibaum B., Li S., Molliex A. et al. (2013) Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature, 495, 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vieira N.M., Naslavsky M.S., Licinio L., Kok F., Schlesinger D., Vainzof M., Sanchez N., Kitajima J.P., Gal L., Cavacana N. et al. (2014) A defect in the RNA-processing protein HNRPDL causes limb-girdle muscular dystrophy 1G (LGMD1G). Hum. Mol. Genet., 23, 4103–4110. [DOI] [PubMed] [Google Scholar]
- 5.Klar J., Sobol M., Melberg A., Mabert K., Ameur A., Johansson A.C., Feuk L., Entesarian M., Orlen H., Casar-Borota O. et al. (2013) Welander distal myopathy caused by an ancient founder mutation in TIA1 associated with perturbed splicing. Hum. Mutat., 34, 572–577. [DOI] [PubMed] [Google Scholar]
- 6.King O.D., Gitler A.D., Shorter J. (2012) The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res., 1462, 61–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J., Mittag T., Taylor J.P. (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell, 163, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M. et al. (2015) A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell, 162, 1066–1077. [DOI] [PubMed] [Google Scholar]
- 9.Lin Y., Protter D.S., Rosen M.K., Parker R. (2015) Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell, 60, 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchan J.R., Kolaitis R.M., Taylor J.P., Parker R. (2013) Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell, 153, 1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michels A.A., Kanon B., Konings A.W., Ohtsuka K., Bensaude O., Kampinga H.H. (1997) Hsp70 and Hsp40 chaperone activities in the cytoplasm and the nucleus of mammalian cells. J. Biol. Chem., 272, 33283–33289. [DOI] [PubMed] [Google Scholar]
- 12.Ahmad A., Bhattacharya A., McDonald R.A., Cordes M., Ellington B., Bertelsen E.B., Zuiderweg E.R. (2011) Heat shock protein 70 kDa chaperone/DnaJ cochaperone complex employs an unusual dynamic interface. Proc. Natl Acad. Sci. USA, 108, 18966–18971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clare D.K., Saibil H.R. (2013) ATP-driven molecular chaperone machines. Biopolymers, 99, 846–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greene M.K., Maskos K., Landry S.J. (1998) Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc. Natl Acad. Sci. USA, 95, 6108–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cummings C.J., Mancini M.A., Antalffy B., DeFranco D.B., Orr H.T., Zoghbi H.Y. (1998) Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat. Genet., 19, 148–154. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez-Funez P., Nino-Rosales M.L., de Gouyon B., She W.C., Luchak J.M., Martinez P., Turiegano E., Benito J., Capovilla M., Skinner P.J. et al. (2000) Identification of genes that modify ataxin-1-induced neurodegeneration. Nature, 408, 101–106. [DOI] [PubMed] [Google Scholar]
- 17.Kakkar V., Prins L.C., Kampinga H.H. (2012) DNAJ proteins and protein aggregation diseases. Curr. Top. Med. Chem., 12, 2479–2490. [DOI] [PubMed] [Google Scholar]
- 18.Stein K.C., Bengoechea R., Harms M.B., Weihl C.C., True H.L. (2014) Myopathy-causing mutations in an HSP40 chaperone disrupt processing of specific client conformers. J. Biol. Chem., 289, 21120–21130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harms M.B., Sommerville R.B., Allred P., Bell S., Ma D., Cooper P., Lopate G., Pestronk A., Weihl C.C., Baloh R.H. (2012) Exome sequencing reveals DNAJB6 mutations in dominantly-inherited myopathy. Ann. Neurol., 71, 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarparanta J., Jonson P.H., Golzio C., Sandell S., Luque H., Screen M., McDonald K., Stajich J.M., Mahjneh I., Vihola A. et al. (2012) Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy. Nat. Genet., 44, 450–455, S451–S452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bengoechea R., Pittman S.K., Tuck E.P., True H.L., Weihl C.C. (2015) Myofibrillar disruption and RNA-binding protein aggregation in a mouse model of limb-girdle muscular dystrophy 1D. Hum. Mol. Genet., 24, 6588–6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freibaum B.D., Chitta R.K., High A.A., Taylor J.P. (2010) Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Proteome Res., 9, 1104–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toombs J.A., Petri M., Paul K.R., Kan G.Y., Ben-Hur A., Ross E.D. (2012) De novo design of synthetic prion domains. Proc. Natl Acad. Sci. USA, 109, 6519–6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khong A., Jan E. (2011) Modulation of stress granules and P bodies during dicistrovirus infection. J. Virol., 85, 1439–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Couthouis J., Raphael A.R., Siskind C., Findlay A.R., Buenrostro J.D., Greenleaf W.J., Vogel H., Day J.W., Flanigan K.M., Gitler A.D. (2014) Exome sequencing identifies a DNAJB6 mutation in a family with dominantly-inherited limb-girdle muscular dystrophy. Neuromuscul. Disord., 24, 431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yabe I., Tanino M., Yaguchi H., Takiyama A., Cai H., Kanno H., Takahashi I., Hayashi Y.K., Watanabe M., Takahashi H. et al. (2014) Pathology of frontotemporal dementia with limb girdle muscular dystrophy caused by a DNAJB6 mutation. Clin. Neurol. Neurosurg., 127, 10–12. [DOI] [PubMed] [Google Scholar]
- 27.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M. et al. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science, 314, 130–133. [DOI] [PubMed] [Google Scholar]
- 28.Sreedharan J., Blair I.P., Tripathi V.B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J.C., Williams K.L., Buratti E. et al. (2008) TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science, 319, 1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vance C., Rogelj B., Hortobagyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P. et al. (2009) Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science, 323, 1208–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwiatkowski T.J. Jr, Bosco D.A., Leclerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T. et al. (2009) Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science, 323, 1205–1208. [DOI] [PubMed] [Google Scholar]
- 31.Couthouis J., Hart M.P., Shorter J., DeJesus-Hernandez M., Erion R., Oristano R., Liu A.X., Ramos D., Jethava N., Hosangadi D. et al. (2011) A yeast functional screen predicts new candidate ALS disease genes. Proc. Natl Acad. Sci. USA, 108, 20881–20890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Couthouis J., Hart M.P., Erion R., King O.D., Diaz Z., Nakaya T., Ibrahim F., Kim H.J., Mojsilovic-Petrovic J., Panossian S. et al. (2012) Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum. Mol. Genet., 21, 2899–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L.M., Anderson P. (2004) Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell, 15, 5383–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y.R., King O.D., Shorter J., Gitler A.D. (2013) Stress granules as crucibles of ALS pathogenesis. J. Cell Biol., 201, 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu-Yesucevitz L., Bilgutay A., Zhang Y.J., Vanderweyde T., Citro A., Mehta T., Zaarur N., McKee A., Bowser R., Sherman M. et al. (2010) Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS ONE, 5, e13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramaswami M., Taylor J.P., Parker R. (2013) Altered ribostasis: RNA-protein granules in degenerative disorders. Cell, 154, 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szyperski T., Pellecchia M., Wall D., Georgopoulos C., Wuthrich K. (1994) NMR structure determination of the Escherichia coli DnaJ molecular chaperone: secondary structure and backbone fold of the N-terminal region (residues 2–108) containing the highly conserved J domain. Proc. Natl Acad. Sci. USA, 91, 11343–11347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corsi A.K., Schekman R. (1997) The lumenal domain of Sec63p stimulates the ATPase activity of BiP and mediates BiP recruitment to the translocon in Saccharomyces cerevisiae. J. Cell Biol., 137, 1483–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiu X.B., Shao Y.M., Miao S., Wang L. (2006) The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci., 63, 2560–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar R.C. (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26, 2460–2461. [DOI] [PubMed] [Google Scholar]
- 41.Katoh K., Asimenos G., Toh H. (2009) Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol., 537, 39–64. [DOI] [PubMed] [Google Scholar]
- 42.Goode M.G., Rodrigo A.G. (2007) SQUINT: a multiple alignment program and editor. Bioinformatics, 23, 1553–1555. [DOI] [PubMed] [Google Scholar]
- 43.Zwickl D.J. (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Ph.D. dissertation, The University of Texas at Austin. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.