Abstract
Similar to many proteins trafficking through the secretory pathway, cellular prion protein (PrP) partly retrotranslocates from the endoplasmic reticulum to the cytosol through the endoplasmic reticulum-associated degradation (ERAD) pathway in an attempt to alleviate accumulation of cellular misfolded PrP. Surprisingly, familial PrP mutants fail to retrotranslocate and simultaneously block normal cellular PrP retrotranslocation. That impairments in retrotranslocation of misfolded proteins could lead to global disruptions in cellular homeostasis prompted further investigations into PrP mutant retrotranslocation defects. A gain- and loss-of-function approach identified human E3 ubiquitin ligase, Hrd1, as a critical regulator of PrP retrotranslocation in mammalian cells. Expression of familial human PrP mutants, V210I129V and M232R129V, not only abolished PrP retrotranslocation, but also that of Hrd1-dependent ERAD substrates, transthyretin TTRD18G and α1-anti-trypsin A1ATNHK. Mutant PrP expression decreased binding immunoglobulin protein (BiP) levels by 50% and attenuated ER stress-induced BiP by increasing BiP turnover 6-fold. Overexpression of BiP with PrP mutants rescued retrotranslocation of PrP, TTRD18G and A1ATNHK. PrP mutants-induced cell death was also rescued by co-expression of BiP. These results show that PrP mutants highjack the Hrd1-dependent ERAD pathway, an action that would result in misfolded protein accumulation especially in terminally differentiated neurons. This could explain the age-dependent neuronal degeneration in familial prion diseases.
Introduction
The exact mechanism by which mutant prion proteins (PrP) induce age-dependent neuronal degeneration is unknown. Misfolded PrP is thought to be central to the degeneration of neurons in prion diseases. PrP is translated through the secretory pathway and the newly synthesized protein accumulates as a glycophosphatidylinositol (GPI)-anchored protein on the outer leaflet of the plasma membrane. While most newly synthesized PrPs escape endoplasmic reticulum-associated degradation (ERAD) (1,2), approximately 10% of newly synthesized PrP is retrotranslocated from the endoplasmic reticulum (ER) to the cytosol (CyPrP) via the ER-associated degradation (ERAD) pathway (3–7).
Most familial PrP mutants escape retrotranslocation and concomitantly prevent endogenous PrP retrotranslocation in cell lines and primary human neurons (8,9). While a specific mechanism for PrP ERAD has not yet been elucidated in mammalian cells, proteasome-dependent degradation of human unglycosylated PrP was stabilized in yeast lacking the expression of the Hrd1p–Hrd3p retrotranslocation complex (10). Yeast misfolded ER lumenal or intramembrane glycoproteins undergo ERAD through the Hrd1 complex containing transmembrane E3 ubiquitin ligase Hrd1p, three lumenal ER misfolded glycoprotein sensor proteins: Yos9 (mammalian XTP3-B and OS-9), Htmlp (mammalian EDEM 1/2/3) and Kar2 (mammalian BiP), scaffold proteins Hrd3 (mammalian SEL1L) and Usa1 (mammalian HERP) and retrotranslocon, Der1 (mammalian Derlin-1, -2 or -3) (11,12). Despite high conservation of protein quality control mechanisms, some key differences exist between human and yeast ERAD systems. Hrd1p has two functional human homologues: Hrd1 and Gp78 (13,14). Both are expressed in the central nervous system, upregulated by ER stress and have multiple substrates, but Hrd1 is the most topologically similar to Hrd1p (13,15–18). Gp78 ubiquitinates and promotes proteasomal-dependent degradation of unglycosylated PrP (19), but there are no reports on the potential role of Hrd1 in glycosylated PrP retrotranslocation.
To determine whether mammalian Hrd1 mediates the retrotranslocation of normally glycosylated human wild-type and mutant PrP, we examined CyPrP levels in human CR7 glioblastoma cells under proteasome inhibition. Overexpression of human Hrd1 enhanced PrP retrotranslocation, whereas targeted small interfering RNA (siRNA) knockdown of Hrd1 decreased CyPrP levels. Expression of familial PrP mutants, PrP-V210I129V and PrP-M232R129V, prevented translocation of endogenous PrP to the cytosol, concomitantly blocked retrotranslocation of model Hrd1-mediated ERAD substrates, mutant transthyretin TTRD18G and α1-anti-trypsin null Hong Kong variant, A1ATNHK and decreased cellular survival. While PrP mutants did not induce the UPR, they considerably depressed BiP levels and BiP transfection rescued retrotranslocation and restored cell viability. We conclude that these familial PrP mutants impede Hrd1-mediated ERAD by decreasing BiP levels, thus facilitating cellular accumulation of multiple misfolded ERAD-substrate proteins. The accumulation of multiple misfolded proteins with age could explain neuronal dysfunction and degeneration in familial prion diseases.
Results
E3 ubiquitin ligase Hrd1 is responsible for the retrotranslocation of human glycosylated PrP
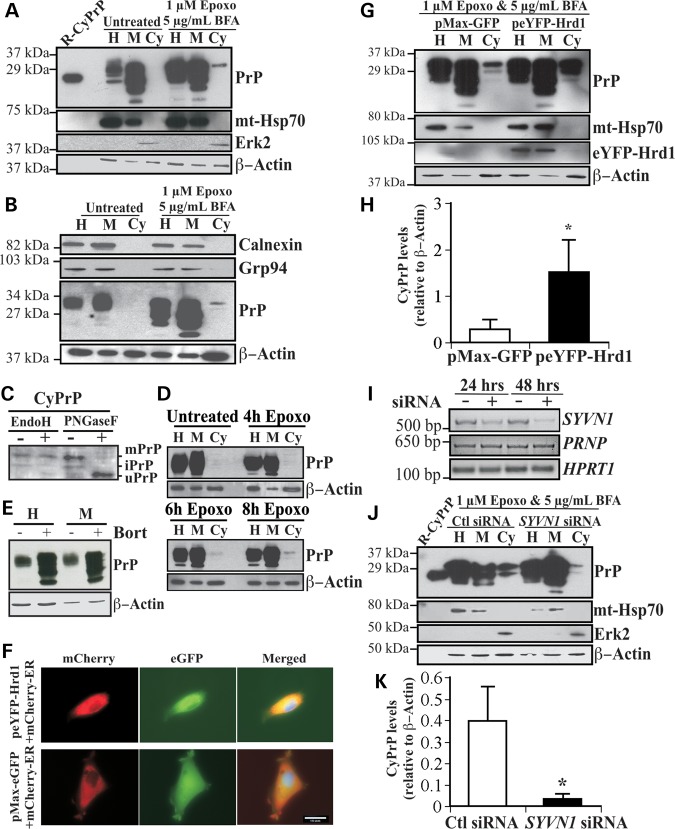
The human glioblastoma CR7 cell line was chosen because of its high endogenous expression of PrP. To assess the pre-Golgi PrP degradation expected in ERAD, CR7 cells were treated for 18 h with the potent irreversible proteasome inhibitor epoxomicin and Golgi-disaggregating brefeldin A (BFA) before submitting the cells to subcellular fractionation to separate membrane from cytosolic proteins, as previously done on human primary neurons, mouse neuroblastoma N2A cells and human breast carcinoma MCF7 cells (8). Abundant levels of non- and immature-glycosylated PrP were present in both the homogenate and membrane fractions. As expected, CyPrP was detected in epoxomicin- and BFA-treated cells, but not in untreated cells (Fig. 1A). Immunoblotting with anti-mitochondrial heat shock protein 70 (mt-Hsp70) confirmed the absence of contaminating membrane proteins in the cytosolic fraction, whereas extracellular signal-regulated kinase 2 (Erk2) was detected only in the cytosolic fractions. β-actin immunoblotting confirmed equal loading of proteins between untreated and epoxomicin–BFA-treated fractions from paired subcellular compartments. The possibility that the glycosylated CyPrP was the result of ER leakage or an impure cytosolic fraction was excluded by showing that the ER lumenal protein, Grp94 and the ER localized membrane protein, calnexin, were absent from the cytosolic fraction in untreated and BFA- and epoxomicin-treated cells (Fig. 1B). Furthermore, general leakiness from the ER can be excluded, because the cytosol from the non-epoxomicin-treated CR7 cells does not contain PrP.
Figure 1.
Overexpression of Hrd1 enhances PrP retrotranslocation while target knockdown by siRNA abolishes CyPrP in CR7 cells. (A) Western blot analysis with anti-PrP (3F4), anti-mitochondrial heat shock protein 70 (mt-Hsp70), anti-Erk2 and anti-β-Actin in CR7 cells treated for 18 h with epoxomicin (epoxo) and brefeldin A (BFA) and fractionated into homogenate (H), membrane (M) and cytosolic (Cy) fractions. Equal amounts of proteins from matched fraction pairs were loaded (example: Cy versus Cy). R-PrP; recombinant cytosolic PrP23–231. (B) Western blot with anti-calnexin, Grp94, PrP and actin of the subcellular fractions. (C) Western blot analyses of Endo H and PNGase F treated CyPrP with the 3F4 anti-PrP antibody show mature PrP (mPrP), immature PrP (iPrP) and unglycosylated PrP (uPrP). The small bar left to the panel represents the 27 kDa molecular weight marker. (D) Western blot analyses with anti-PrP 3F4 of proteins from subcellular fractionated CR7 cells treated with epoxomicin for 0, 4, 6 and 8 h. (E) Western blot analyses of the whole homogenate (H) or membrane fraction (M) of proteins from CR7 cells without or with bortezomib. (F) Fluorescent light microscopy of CR7 cells co-transfected with mCherry-ER and peYFP-Hrd1 or GFP mammalian expression vector. Nuclear DNA was stained with Hoechst 33342 staining. Scale bar represents 200 μm. (G) Western blot analysis with anti-PrP, anti-mt-Hsp70, anti-Hrd1, anti-Erk2 and anti-β-actin in CR7 cells transfected with either pMaxGFP or peYFP-Hrd1 and subjected to the subcellular fractionation. Proteins were loaded as in Figure 1A. (H) Relative CyPrP to membrane PrP levels evaluated by densitometry and depicted as fold increase over β-actin protein levels. (I) Ethidium bromide agarose gel of SYVN1, PRNP and HPRT1 amplicons obtained by RT-PCR from CR7 cells transfected for 24 or 48 h with either scrambled non-targeting control or SYVN1-targeting siRNA. (J) Western blot analysis with anti-PrP, anti-mt-Hsp70, anti-Erk2 and anti-β-actin in proteins extracted from subcellular fractions of CR7 cells transfected with 250 nm of either non-targeting scrambled or SYVN1-targeting siRNA followed by 18 h treatment with 1 µM epoxomicin and 5 µg/ml BFA. Proteins are loaded as in Figure 1A. (K) PrP in the cytosolic fraction relative to PrP in the membrane fraction of CR7 cells transfected with either non-targeting scrambled or SYVN1-targeting siRNA. CyPrP levels, evaluated by densitometry, are depicted as fold increase over β-actin. All graphed data represent the mean ± SEM of at least three independent experiments and were analyzed by independent samples t-test, *P < 0.05.
Epoxomicin inhibits cytosolic PNGase activity (20) and therefore CyPrP remained glycosylated and migrated slower than the 25 kDa unglycosylated PrP. To confirm that CyPrP is glycosylated, we treated the cytosolic proteins with EndoH, which cleaves the chitiose core of high mannose N-linked oligosaccharides, and PNGase F, which cleaves N-acetylglucosamine and asparagine in high mannose, hybrid and complex N-linked oligosaccharides from proteins. The results show a shift of a 29 kDa immature-glycosylated CyPrP into a 25 kDa CyPrP by EndoH, whereas PNGase F completely deglycosylates the 32 and 29 kDa CyPrP into a 25 kDa PrP, the size expected for unglycosylated PrP lacking both the N- and C-terminal signal peptides and the GPI anchor (Fig. 1C). The mature glycosylation of CyPrP is consistent with the fact that BFA relocalizes part of the Golgi complex into the ER resulting in more mature N-linked glycosylation than would be expected (21). The presence of N-linked glycosylation on CyPrP confirms retrotranslocation from the ER. CyPrP was apparent within 6 h of epoxomicin treatment alone (Fig. 1D), thereby excluding the possibility that long-term proteasome inhibition combined with BFA treatment led to the generation of CyPrP. Furthermore, treatment of the cells with another proteasome inhibitor, bortezomib, clearly showed an increase in membrane-associated PrP levels indicating that PrP is turned over by the proteasome (Fig. 1E).
To determine whether Hrd1 regulates mammalian PrP retrotranslocation, cells were transfected with an expression vector encoding eYFP-human Hrd1. The eYFP protein was fused to the N-terminus of Hrd1, away from the catalytic RING-finger domain and putative substrate-binding proline-rich or transmembrane regions. The eYFP-Hrd1 construct was transfected into CR7 cells with 70% efficiency. eYFP-Hrd1 co-localized with the ER-associated mCherry-ER-3 protein, as expected, and in contrast to the cytosolic distribution of GFP expressed from the pMax-GFP vector (Fig. 1F). The eYFP-Hrd1-expressing cells become more compact and elongated compared with the GFP-expressing cells. Following transfection with peYFP-Hrd1 or pMax-GFP, CR7 cells were treated with epoxomicin and BFA and subjected to subcellular fractionation. Overexpression of eYFP-Hrd1 markedly increased CyPrP (Fig. 1G). Expression of eYFP-Hrd1 was confirmed in the homogenate and membrane fractions. Immunodetection of subcellular membrane and cytosolic markers, mt-Hsp70 and Erk2, respectively, in addition to β-actin confirmed the purity and equal loading of respective fractions. The CyPrP levels were calculated relative to membrane PrP levels to exclude possible leakiness artifacts due to different expression levels of the protein. Densitometric analyses of band intensities demonstrated that overexpression of Hrd1 increased CyPrP levels 3-fold (Fig. 1H). To complement the gain-of-function observed with Hrd1 overexpression, we examined the effect of targeted knockdown of Hrd1 mRNA (referred to by the gene name, SYVN1) with siRNA. CR7 cells were transfected with an SYVN1-targeting pool of four different siRNAs or a scrambled, non-targeting pool for 24 h before 18 h of treatment with epoxomicin and BFA. A strong reduction of SYVN1 mRNA following 24 and 48 h of siRNA treatment was confirmed by RT-PCR, whereas PRNP and HPRT1 mRNA levels remained stable (Fig. 1I). Transfection with scrambled siRNAs did not affect PrP retrotranslocation, whereas SYVN1-targeting siRNA substantially decreased CyPrP levels (Fig. 1J). In three independent replications, targeted knockdown of SYVN1 decreased CyPrP levels by 11-fold (Fig. 1K). Together, these experiments confirm that the Hrd1 is intimately involved in mammalian PrP retrotranslocation.
Familial PrP mutants inhibit retrotranslocation of PrP and model Hrd1-mediated ERAD substrates, TTRD18G and A1ATNHK
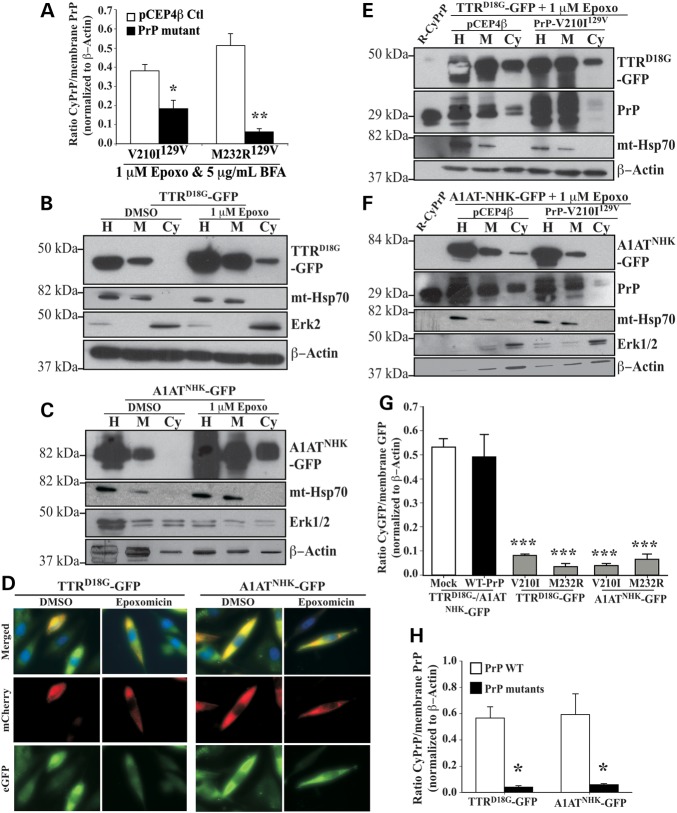
Familial PrP mutants prevent retrotranslocation of endogenously expressed PrP to varying extent and in a dominant-negative fashion (8,9). To investigate if familial PrP mutants interfere with the retrotranslocation of other Hrd1-mediated ERAD substrates, we examined the effect of M232R129V and V210I129V PrP mutants associated with familial Creutzfeldt–Jakob disease, on the retrotranslocation of two model Hrd1 substrates, the lumenal non-glycosylated mutant transthyretin TTRD18G (22) and the lumenal glycosylated α1-anti-trypsin null Hong Kong variant, A1ATNHK (23). We chose the two PRNP mutations, M232R129V and V210129V for this study because of all 20 PRNP mutants previously tested (8,9), these most strongly inhibited retrotranslocation thus providing a clear model to clearly identify the retrotranslocation mechanism. As shown previously in N2a mouse neuroblastoma cells (8), expression of both PrP mutants significantly reduced CyPrP in human CR7 cells (Fig. 2A). Expression of the PrP-V210I129V mutant reduced CyPrP levels 2-fold, whereas PrP-M232R129V reduced CyPrP levels 8-fold, compared with vector-transfected controls. Accumulation of both TTRD18G-GFP and A1ATNHK-GFP was observed in the cytosolic fraction from cells treated with epoxomicin, confirming these proteins are efficiently retrotranslocated, as previously shown in HEK293 and HeLa cells (18) (Fig. 2B and C). Furthermore, while cells expressing TTRD18G-GFP and A1ATNHK-GFP mutants and treated with DMSO alone show GFP reporter co-localized with the mCherry-ER-3 marker, inhibition of the proteasome additionally showed cytoplasmic GFP fluorescence (Fig. 2D), consistent with retrotranslocated TTRD18G-GFP and A1ATNHK-GFP accumulating in the cellular cytosol.
Figure 2.
Familial PrP mutants inhibit retrotranslocation of both endogenous cellular PrP and other ERAD substrates. (A) Quantification by densitometry of western blot analyses of CyPrP levels relative to membrane PrP levels in CR7 cells transfected with pCEP4β-empty, pCEP4β-PrP-V210I129V or -PrP-M232R129V and subjected to the subcellular fractionation. Data are depicted as a ratio of CyPrP relative to membrane PrP after normalizing to actin levels. *P < 0.05, **P < 0.01 (B, C) Western blot analysis with anti-GFP, anti-mt-Hsp70, anti-Erk1/2 and anti-β Actin in CR7 cells transfected with pcDNA3.1(-)-TTRD18G-GFP or pcDNA3.1(-)-A1ATNHK-GFP and subjected to subcellular fractionation. Proteins were loaded as in Figure 1A. (D) Fluorescence microscopy of CR7 cells co-transfected with mCherry-ER and TTRD18G-GFP or A1ATNHK-GFP treated with DMSO or epoxomicin. Nuclear DNA is identified using Hoechst staining. (E,F) Western blot analysis with anti-GFP, anti-PrP, anti-mt-Hsp70, anti-Erk1/2 and anti-β-actin in CR7 cells co-transfected with TTRD18G-GFP (E) or A1ATNHK-GFP (F) and PrP-V210I129V, followed by the subcellular fractionation assay. (G) Densitometry quantification of western blot analysis for cytosolic GFP in CR7 cells co-transfected with either TTRD18G-GFP or A1ATNHK-GFP and with PrP-V210I129V or PrP-M232R129V. Data are depicted as a ratio of cytosolic GFP relative to membrane GFP, represent the mean ± SEM of at least three independent experiments and are analyzed using a one-way ANOVA followed by Tukey's post-tests. *P < 0.05, ***P < 0.001. (H) Densitometry quantification of western blots for wild-type or mutant CyPrP relative to membrane PrP in cells transfected with TTRD18G-GFP or A1ATNHK-GFP. Statistical evaluations were done as in G.
The effect of familial PrP mutants on the retrotranslocation of other Hrd1-mediated ERAD substrates was assessed in CR7 cells co-transfected with TTRD18G-GFP or A1ATNHK-GFP and either the PrP-M232R129V or the PrP-V210I129V mutants. Immunoblotting proteins from subcellular fractions showed strongly reduced cytosolic TTRD18G-GFP (Fig. 2E), A1ATNHK-GFP (Fig. 2F) and CyPrP, when the HRD1 substrates were co-expressed with mutant PrPs (Fig. 2E and F). Densitometric analysis of a minimum of three independent experiments showed that PrP-V210I129V decreased cytosolic TTRD18G-GFP and A1ATNHK-GFP, by 6- and 16-fold, respectively, whereas PrP-M232R129V decreased them 14- and 8-fold, respectively (Fig. 2G). This effect appeared specific to PrP mutants, because neither TTRD18G-GFP nor A1ATNHK-GFP affected CyPrP levels in the presence of the vector control (Fig. 2E and F). Furthermore, TTRD18G-GFP and A1ATNHK-GFP retrotranslocated normally when co-expressed with WT-PrP (Fig. 2H). These results indicate that neither TTRD18G-GFP nor A1ATNHK-GFP inhibited WT-PrP retrotranslocation, whereas PrP mutants inhibited endogenous PrP, and exogenously expressed PrP, TTRD18G-GFP and A1ATNHK-GFP retrotranslocation. Overall, these results suggest that familial PrP mutants are causing a blockade of several Hrd1 protein substrates.
Familial PrP mutants are enhancing degradation of BiP protein in a proteasomal-independent manner
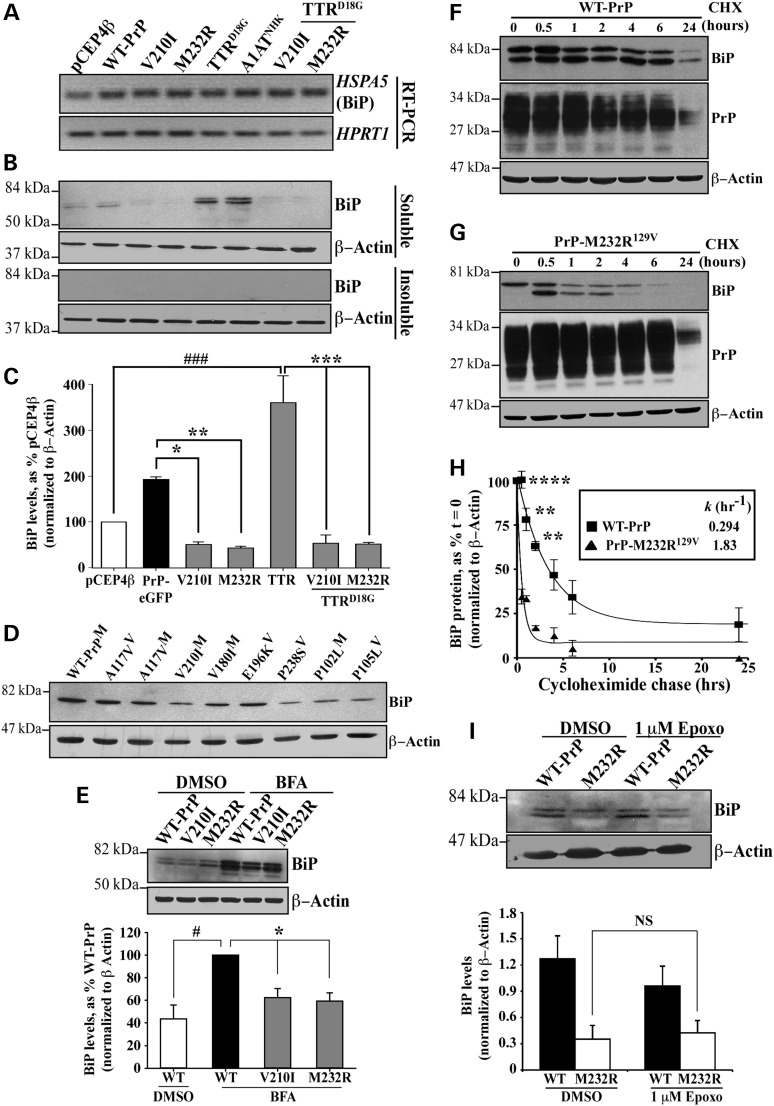
To determine whether PrP mutant expression induced ER stress, we examined the levels of the classic ER stress marker BiP in CR7 cells transfected with WT or mutant PrP, either alone or co-transfected with TTRD18G-GFP, whose turnover is known to be regulated by BiP (24). BiP mRNA levels were slightly increased in all transfected cells, consistent with an increased ER burden due to exogenously introduced proteins (Fig. 3A). BiP protein was detected in vector and WT-PrP transfected cells, but was very low in cells transfected with PrP-V210I129V and PrP-M232R129V (Fig. 3B). As expected, both TTRD18G-GFP and A1ATNHK-GFP increased BiP levels considerably. However, when either PrP mutants were co-transfected with TTRD18G-GFP, BiP levels were dramatically diminished, indicating that even in the presence of a protein known to induce ER-stress, PrP mutants suppress BiP levels. Western blot analysis did not reveal BiP in the detergent insoluble compartment (Fig. 3B). Densitometric quantification of three independent experiments confirmed a 4-fold lower level of BiP by PrP mutants, relative to WT-PrP and a 7-fold decrease relative to TTRD18G-GFP-transfected cells (Fig. 3C). Eight additional PrP mutants associated with prion diseases also decreased the levels of BiP relative to those in WT PrP-transfected cells (Fig. 3D). Furthermore, the expression of PrP mutants attenuated BFA-induced BiP upregulation by approximately 40% (Fig. 3E). The ability to suppress BiP in the presence of such a strong inductive force, indicates a very robust effect. Overall, these results suggest that misfolded PrP mutants may escape the ERAD pathway by decreasing levels of BiP protein, a sensor essential for the recognition of misfolded ER lumenal glycoproteins in the Hrd1 retrotranslocon.
Figure 3.
Familial PrP suppress the levels of unfolded protein response protein BiP. (A) Ethidium bromide-stained agarose gel of HSPA5 (BiP) and HPRT1 amplicons obtained by RT-PCR. (B) Western blot analysis with anti-BiP and anti-β-actin in CR7 cells transfected with WT-PrP, PrP-V210I129V or -M232R129V, TTRD18G-GFP and A1ATNHK-GFP either alone or co-transfected with PrP mutants (order is as depicted in A). Top and bottom panels represent detergent soluble and insoluble proteins, respectively. (C) Densitometry quantification of BiP protein levels presented in Figure 3B. ###P < 0.001, relative to pCEP4β and *P < 0.05, **P < 0.01 and ***P < 0.001, relative to WT-PrP-EGFP or TTRD18G. (D) Western blot with anti-BiP and β-actin of proteins extracted from CR7 cells transfected with WT PrP or different mutant PrPs. (E) Western blot analysis with anti-BiP and anti-β-actin in CR7 cells transfected with WT-PrP or mutant PrP and treated with either DMSO or BFA presented with corresponding quantification by densitometry of BiP protein levels. (F,G) Western blots with anti-BiP, anti-PrP 3F4 or anti-β-actin of proteins extracted from WT-PrP (F) or PrP-M232R (G) transfected cells treated with cycloheximide for the indicated times. (H) Kinetic analysis of BiP degradation following inhibition of total protein synthesis with 0.75 μg/ml cycloheximide in CR7 cells expressing WT-PrP or PrP-M232R129V PrP for 24 h. Graph represents an exponential decay plot for the degradation of BiP with respect to time. (I) Western blot analysis of anti-BiP and anti-β-actin in cells transfected with WT-PrP or PrP-M232R129V followed by treatment with 1 μM epoxomicin or DMSO. Data are presented with quantification of band intensities by densitometry. All graphed data represent the mean ± SEM of at least three independent experiments and were analyzed using one-way ANOVAs followed by Tukey's post-tests.
While BiP mRNA levels indicate that transcription is unaffected, familial PrP mutants may be interfering with other aspects of BiP regulation, chiefly mRNA translation or protein degradation. The stability of BiP protein in the presence of WT or mutant PrP was compared using cycloheximide chase experiments. Cells were transfected with WT or mutant PrP for 24 h before the addition of cycloheximide. Following inhibition of protein translation, BiP and PrP were turned over in a time-dependent manner in WT PrP-transfected cells, with a half-life of 2.3 h for BiP (Fig. 3F and H). Notably, within 6 h post-cycloheximide treatment in PrP-M232R129V-transfected cells, BiP was considerably reduced (Fig. 3G). In contrast, PrP levels were maintained, thus excluding the possibility that PrP and BiP are coordinately turned over (Fig. 3G). Furthermore, BiP was significantly less stable with a half-life of just 23 min and a rate of degradation six times faster in PrP-M232R129V-expressing cells than in WT-PrP-transfected cells (k−1= 1.83 and k−1= 0.294, respectively) (Fig. 3H).
To determine whether enhanced degradation of BiP in the presence of PrP-M232R129V was dependent on the proteasome, levels of BiP protein were examined in cells transfected with WT or mutant PrP, with or without epoxomicin. Inhibition of the proteasome failed to stabilize BiP in the presence of PrP-M232R129V (Fig. 3I). This is consistent with reports showing that both WT and mutant BiP can be turned over by proteasome-independent pathways (25,26).
PrP mutants do not activate the unfolded protein response in CR7 and SK-N-SH cells
The UPR response is initiated when chaperone BiP/glucose-recognition protein 78 (Grp78) detects and binds misfolded proteins in the lumen of the ER, thereby detaching from ER stress sensor proteins: inositol requiring enzyme 1 (IRE1α), activating transcription factor 6 (ATF6α) and PKR-like ER localized kinase (PERK), which generate increased transcription of ER chaperone and other UPR target genes via spliced X-box binding protein (sXBP1) and cleaved ATF6α, and protein translation inhibition by phosphorylation of eukaryotic translation initiation factor 2 (peIF2α) (reviewed by (27,28)). Transmissible scrapie PrP (PrPSc) increases sXBP1 and peIF2α, but only GADD34 dephosphorylation of peIF2α or pharmacological inhibition of PERK prevent neurodegeneration in infected mice brains (29–31). In contrast, familial PrP mutants do not seem to induce ER stress in brain (32).
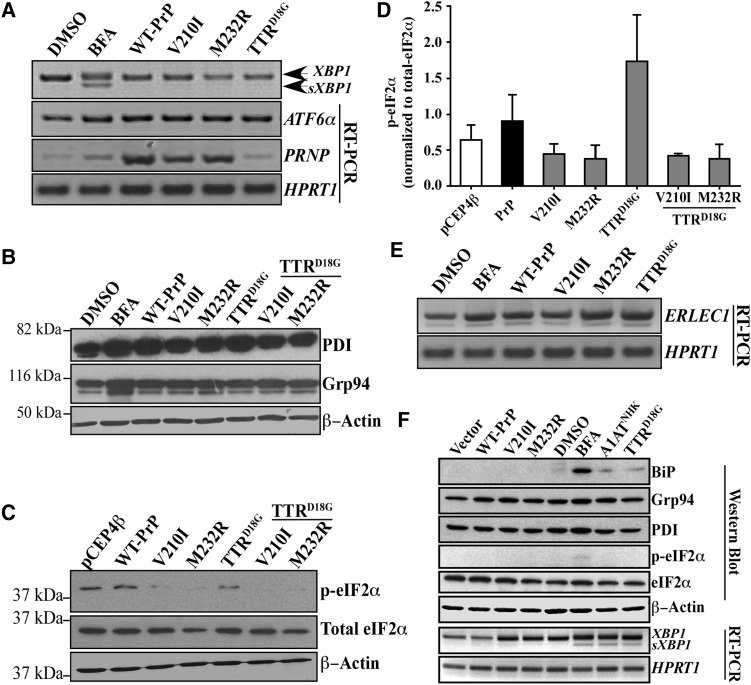
To determine whether the PrP mutants under study activated the UPR pathway, sXBP1 levels, as an indicator of active IRE1, were examined by RT-PCR. While BFA-induced sXBP1 mRNA, neither PrP mutants nor TTRD18G did (Fig. 4A). BFA, WT-PrP, PrP mutants and TTRD18G-GFP increased ATF6α mRNA levels; however, activation of the ATF6α transcriptional activity was unlikely, because neither BiP mRNA (Fig. 3A) nor Grp94 protein levels (Fig. 4B) were increased, both of which are upregulated by cleaved ATF6α (33,34). PrP mutants decreased phosphorylated, but not total eIF2α, relative to vector and WT-PrP-transfected cells (Fig. 4C), indicating that protein translation is continuing, unmitigated, in the presence of PrP mutants (Fig. 4D). These results indicate that PrP mutants do not activate the UPR in the CR7 cell line.
Figure 4.
Familial PrP mutants inhibit eIF2α phosphorylation but, otherwise do not activate the unfolded protein response. (A) Ethidium bromide agarose gel of XBP1, splice XBP1 (sXBP1), ATF6α, PRNP and HPRT1 amplicons obtained by RT-PCR from CR7 cells treated with DMSO, 5 µg/ml BFA or transfected with either WT-PrP, PrP-V210I129V, -M232R129V or mutant TTRD18G. (B) Western blot analysis with anti-PDI, anti-Grp94 and anti-β-actin. (C) Western blot analysis with anti-p-eIF2α, anti-eIF2α and anti-β-actin in transfected CR7 cells. (D) Densitometry quantitation of the levels of p-eIF2α relative to total eIF2α. Data represent the mean ± SEM of three independent experiments and are analyzed by one-way ANOVA followed by Tukey's post hoc tests. (E) Ethidium bromide-stained agarose gel of ERLEC1 and HPRT1 amplicons obtained by RT-PCR from CR7 cells transfected with WT PrP, mutant PrP or TTRD18G (no epoxomicin nor BFA in transfected cells). (F) Western blot analyses and RT-PCR from proteins and RNA extracts of SK-N-SH neuroblastoma cells transfected with WT PrP and mutant PrP, A1ATNHK, TTRD18G or treated with BFA.
Accumulation of misfolded proteins in the ER due to PrP mutant-mediated BiP degradation would likely trigger a stress–response even in the absence of BiP upregulation and a classic UPR. To determine whether PrP mutants were affecting other markers of ER stress, we examined ER stress-regulated protein prolyl disulfide isomerase (PDI) and found that it was increased with BFA, PrP and TTRD18G. Mutant PrPs slightly attenuated levels of PDI in the absence or presence of TTRD18G (Fig. 4B). In contrast, ER-resident chaperone Grp94 levels were not affected in PrP or TTRD18G-transfected cells, but were upregulated by BFA, as expected of a potent ER stressor.
ERLEC1 encodes the ER-resident lectin protein XTP3-B, which, in addition to BiP and the EDEM family, acts to discriminate terminally misfolded proteins from folding intermediates and deliver them to the Hrd1 complex for degradation (35). Treatment with BFA, or transfection with WT PrP, mutant PrP and TTRD18G, in the absence of BFA, increased levels of ERLEC1 mRNA and the increase was most prominent in PrP-M232R129V and TTRD18G-transfected cells (Fig. 4E). These results indicate that PrP mutants can upregulate proteins associated with sorting and folding, but does not induce the UPR.
To further analyze the effect of familial PrP mutants on the UPR, human neuroblastoma SK-N-SH cells were transfected with the V210I and M232R PrP mutants. BFA treatment, A1ATNHK and TTRD18G transfections served as positive controls to induce the UPR (Fig. 4F). A slight increase in BiP levels and XBP1 splicing was observed with the BFA treatments, and with A1ATNHK, and TTRD18G transfections, but not with the PrP mutant transfections. Phosphorylation of eIF2α was observed only with BFA treatment. All conditions slightly increased the levels of Grp94 and only BFA increased the PDI levels. This indicates that the failure of PrP mutants to mount the UPR in the CR7 glial cells is replicated in the SK-N-SH neuronal cell line.
Overexpression of BiP rescues PrP-mutant-induced cell death and restores retrotranslocation of misfolded Hrd1-mediated ERAD substrates
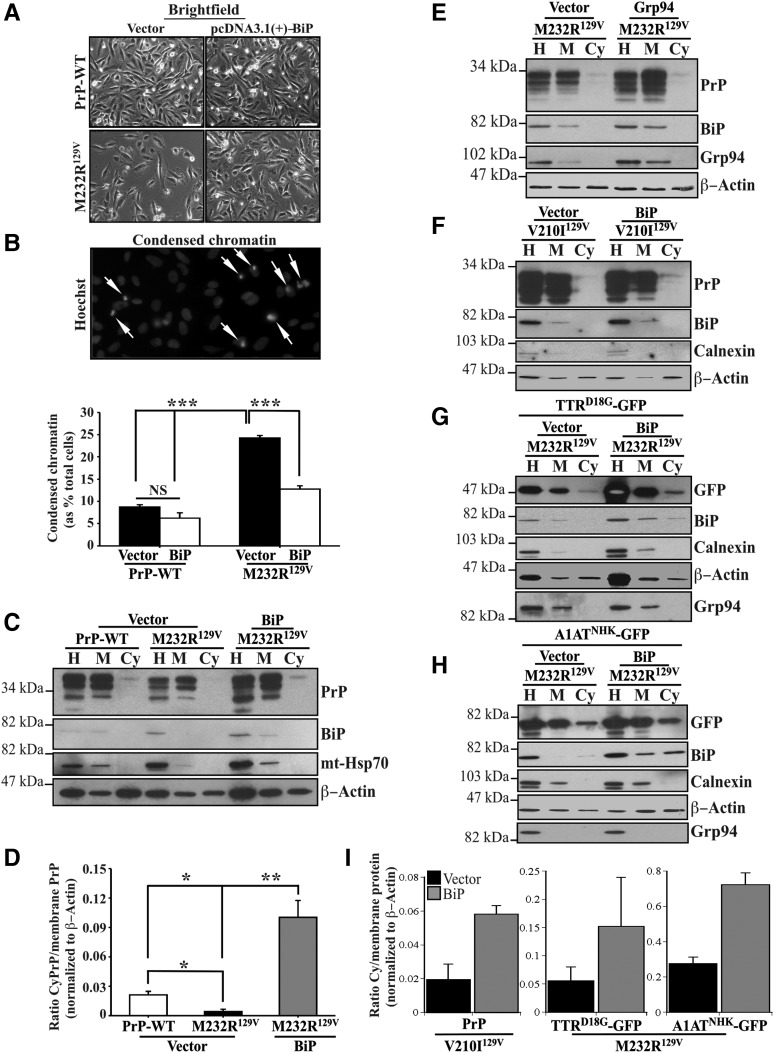
To assess the functional relationship between familial PrP-induced BiP degradation and cellular survival, cell viability was determined by examination of condensed chromatin in CR7 cells transfected with either WT-PrP or PrP-M232R129V, in the presence or absence of BiP. Equal numbers of healthy cells were observed in WT-PrP and BiP co-transfected cells. However, transfection with PrP M232R129V dramatically decreased total cell numbers, and this was rescued by co-expression with BiP (Fig. 5A). Examination of nuclear DNA by Hoechst staining showed a significant increase in the percentage of cells displaying apoptotic nuclei, characterized by clumped and condensed chromatin in mutant PrP-expressing cells, when compared with the oval, homogeneously stained nuclei of healthy cells (Fig. 5A and B). Notably, overexpression of BiP reversed PrP-M232R129V-induced apoptosis. Taken together, these data support the idea that re-introduction of BiP protein is able to reverse cellular deficits caused by familial PrP mutants.
Figure 5.
Overexpression of BiP rescues PrP-mutants-induced loss of protein retrotranslocation. (A) Brightfield of CR7 cells co-transfected with WT-PrP or PrP-M232R129V, in combination with either empty pcDNA3.1(+) vector or pcDNA3.1(+)-BiP. Scale bar represents 200 μm. (B) Fluorescent light microscopy shows normal and condensed (arrows) nuclear DNA with Hoechst 33342 staining. Quantification of condensed chromatin in CR7 cells presented in Figure 5A. Data are depicted as a percentage of total Hoechst positive nuclei displaying condensed chromatin. (C, D) Western blot analysis with anti-PrP, anti-BiP, anti-mt-Hsp70 and anti-β-actin presented with corresponding quantification by densitometry of proteins extracted from CR7 cells co-transfected with WT-PrP or PrP-M232R129V, in combination with either empty pcDNA3.1(+) vector or pcDNA3.1(+)-BiP. All graphed data represent the mean ± SEM of at least three independent experiments and are analyzed by one-way ANOVA followed by Tukey's post-tests. *P < 0.05, **P < 0.01, ***P < 0.001. (E, F) Western blot analysis with anti-PrP, anti-BiP, anti-calnexin, anti-Grp94 and anti-β-actin of proteins extracted from CR7 cells co-transfected with WT-PrP or PrP-V210I129V, in combination with either empty pcDNA3.1(+) vector or pcDNA3.1(+)-BiP. (G,H) Western blot analysis with anti-GFP, anti-BiP, anti-calnexin, anti-Grp94 and anti-β-actin of proteins extracted from CR7 cells co-transfected with TTRD18G (G) or A1ATNHK (H) and WT-PrP or PrP-M232R129V, in combination with either empty pcDNA3.1(+) vector or pcDNA3.1(+)-BiP. (I) Densitometry of cytosolic PrP relative to membrane PrP from experiments in Figure 5F–H. Data represent the mean and SD or variance of 3 (V210I) and 2 (TTRD18G or A1ATNHK) independent experiments.
To determine whether BiP overexpression was able to rescue PrP mutant-induced defective retrotranslocation, we examined retrotranslocation of total PrP in cells expressing WT or M232R129V PrP, alone or in the presence of exogenous BiP. As previously seen, M232R129VPrP dramatically decreased CyPrP levels, representing a failure of both endogenously expressed WT-PrP and transfected PrP-M232R129V to retrotranslocate from the ER (Fig. 5C). Co-expression of BiP with M232R129V PrP rescued CyPrP retrotranslocation and moreover increased significantly the levels of CyPrP relative to those in WT-PrP-transfected cells (Fig. 5C and D). In contrast, overexpression of Grp94 did not rescue the retrotranslocation of CyPrP relative to membrane PrP in PrP-M232R-transfected cells (Fig. 5E). However, BiP overexpression rescued V210I129V PrP (Fig. 5F and I), TTRD18G (Fig. 5G and I) and A1ATNHK (Fig. 5H and I), retrotranslocation. Unexpectedly, we noted the presence of cytosolic BiP in cells transfected with TTRD18G and A1ATNHK, especially in cells co-transfected with BiP (Fig. 5G and H). In contrast, Grp94, another lumenal ER protein, was not detected in the cytosol, again excluding simple leakiness of the ER lumenal proteins into the cytosol during the subcellular fractionation. It is possible that concentrated cytosol from a large number of cells reveals retrotranslocated BiP on western blots, whereas this would not be observed in single cell analyses by immunocytochemistry or fluorescently tagged proteins. These results are also consistent with reports of other ER lumenal proteins that have been observed to retrotranslocate: calreticulin and OS-9 (36–39). Nevertheless, these results strongly support the downregulation of BiP as the mechanism for the defective retrotranslocation in PrP mutant expressing cells.
Discussion
Our results highlight four novel features of PrP retrotranslocation: (1) the E3 ubiquitin ligase Hrd1 is essential for mammalian PrP retrotranslocation, (2) familial PrP mutants block the retrotranslocation of misfolded model Hrd1 protein substrates, (3) familial PrP mutants block Hrd1-dependent ERAD by decreasing expression of the ER-resident chaperone BiP and BiP overexpression rescues Hrd1 protein substrate retrotranslocation and (4) familial PrP mutants do not activate the UPR response.
E3 ubiquitin ligase, Hrd1, is essential to mammalian PrP retrotranslocation
Uncovering a mechanism for the generation of CyPrP becomes particularly important in the context of PrP's physiological role in both disease and health. The accumulation of CyPrP leads to aggregation and neurodegeneration in mouse neuroblastoma N2a cells and in CyPrP-transgenic mouse cerebellar granule neurons (40). Additionally, CyPrP protects against Bax-mediated cell death in human primary CNS neurons and breast carcinoma MCF-7 cells (5,41–43). Reduced CyPrP in cells expressing familial PrP mutants is associated with a loss of PrP's anti-Bax function, a deficit that is reversed through the re-introduction of CyPrP (8,9). Therefore, fully understanding the molecular mechanism of PrP retrotranslocation could help design therapeutic intervention to treat prion diseases.
We identify Hrd1 as a critical regulator of mammalian glycosylated PrP retrotranslocation. Overexpression of Hrd1 in CR7 human glioblastoma cells increased, whereas targeted knockdown of Hrd1, decreased CyPrP levels, consistent with the retrotranslocation of human unglycosylated PrP by hrd1p in yeast (10). Results in yeast could not be extrapolated to mammalian systems, because yeast hrd1p possesses two mammalian homologues: Hrd1 and Gp78, two ER-resident E3 ligases with distinct, yet overlapping, pools of misfolded protein substrates (14,18,44). Gp78 targets unglycosylated PrP (19), whereas here, Hrd1 targets glycosylated PrP for proteasomal degradation. It is not unprecedented for different glycosylation species of the same protein to utilize divergent retrotranslocation machinery (45).
Hrd1 appears essential for the degradation of a number of misfolded proteins associated with CNS aging and neurodegenerative conditions (18). Hrd1 is important to the developing and aging brain and an age-dependent decrease in Hrd1 may be involved in the accumulation of misfolded Hrd1 protein substrates (46). Knockout of Hrd1-stabilizing protein, SEL1, is lethal in mice (39). Hrd1 is also important for the retrotranslocation and degradation of mutant huntingtin, wild-type parkin-associated endothelin receptor-like receptor, and wild-type amyloid precursor protein; three proteins associated with Huntington's, Parkinson's and Alzheimer's disease (AD), respectively (47–49). Hrd1 levels are significantly decreased in AD brains (47). Here, we add PrP as an additional Hrd1 substrate associated with prion diseases.
Familial PrP mutants block the retrotranslocation of misfolded Hrd1 protein substrates
Not only do familial PrP mutants escape ERAD, but they simultaneously prevent the retrotranslocation and subsequent degradation of model Hrd1-substrates TTRD18G-GFP and A1ATNHK-GFP. The defect in retrotranslocation is a specific feature of mutant PrP, because neither TTRD18G-GFP nor A1ATNHK-GFP, when co-expressed with WT-PrP, prevented the generation of CyPrP. The inhibition of model Hrd1 protein substrates, TTRD18G-GFP and A1ATNHK-GFP, retrotranslocation by PrP mutants is consistent with previous work showing that endogenously expressed PrP was unable to retrotranslocate when co-expressed with PrP mutants (8). Jodoin et al. (2007) proposed that the loss of anti-Bax function by CyPrP contributes to the neuronal apoptosis observed in familial prion diseases (9,50–52). However, the disruption in dislocation of other Hrd1 substrates, pinpoint a novel mechanism of familial prion disease pathology, wherein the buildup of misfolded Hrd1-substrates could eventually result in neuronal dysfunction and degeneration. This is supported by the observation that PrP mutant expression increased cell death in CR7 cells. The inhibition of misfolded protein retrotranslocation by PrP mutants may explain why neurons are the primary target of familial PrP diseases, despite PrP being largely expressed in many other tissues of the human body. The accumulation of misfolded proteins would be expected to be most detrimental to the terminally differentiated neurons, which, unlike dividing cell types, cannot cope with deregulated proteostasis by self-renewal.
Familial PrP mutants block the retrotranslocation of Hrd1 protein substrates by enhancing turnover of misfolded protein sensor, BiP
Investigation into the broader implications of a disrupted ERAD pathway in familial prion disease revealed that PrP mutants decrease BiP protein levels through enhanced turnover. Interestingly, BiP is part of the Hrd1 complex and serves as an ER lumenal sensor protein to capture ER misfolded glycoproteins for dislocation to the cytosol (11). The direct implication of these lower BiP levels in PrP mutant-mediated inhibition of retrotranslocation was confirmed by the re-introduction of BiP, which not only rescued retrotranslocation of PrP and other Hrd1 protein substrates, A1ATNHK and TTRD18G, but also restored cell viability. Whether the rescue by BiP will apply to all other PRNP mutants will need to be empirically verified but would be expected in PRNP mutants that reduce the levels of BiP as shown in Figure 3D.
Downregulation of BiP has been associated with neurodegenerative diseases. Mice expressing the E200K or octapeptide repeat PrP mutation showed an age-dependent decrease in BiP protein levels compared with WT mice (32,53). BiP protein levels were reduced in sporadic and familial Presenilin I AD brains (54). Marinesco–Sjögren syndrome is caused by mutations in BiP cofactor SIL1, possibly leading to a loss of functional BiP in neurons (55). This syndrome was reproduced in the SIL1−/− ‘Woozy mouse’ manifesting as adult-onset ataxia accompanied by Purkinje cell loss as a result of accumulation of misfolded proteins (56). Lastly, a traditional Japanese medicine, Yokukansan, improved memory impairment in AD patients (57), and rescued mutant PS1-induced apoptosis by upregulating BiP in cells (58). Together, these results indicate a very important role of BiP in the management of misfolded proteins associated with neurodegenerative CNS conditions.
Familial PrP mutants V210I129V and M232R129V do not induce the unfolded protein response in CR7 cells
While PrP mutants dramatically reduced BiP levels, they did not induce the UPR PERK-mediated phosphorylation of eIF2α, IRE1-mediated splicing of XBP1 and cleaved ATF6 targets, Grp94, BiP (59). Lack of UPR activation was unexpected, because UPR is generally induced by misfolded proteins in order for cells to stop de novo protein translation and increase chaperones to repair the misfolded proteins. However, the results are consistent with failure to observe an ER stress response in familial prion diseases (60). In a transgenic octapeptide repeat PrP mutant mouse, sXBP1 was upregulated in liver, but never in brain, despite the presence of neurodegeneration (32). However, BiP levels were unaltered in PrP-P102L and PrP-E200K transfected neuroblastoma SH-SY5Y cells, whereas PrP-G114V and PrP-A117V mutants slightly increased BiP levels (61). Despite an increase in BiP binding, steady state, endogenous levels of full-length mHtt do not cause ER stress or mount a ‘typical’ UPR, but Htt-transfected mouse striatal cells become more sensitive to subsequent ER stressors (62).
The lack of activation of the UPR in conditions of downregulated BiP levels is unexpected nevertheless. BFA, A1ATNHK and TTRD18G are able to mount the UPR response in glioblastoma CR7 and neuroblastoma SK-N-SH cells, so the UPR response itself is intact in these cells. Because the activation of the UPR can be achieved by two different mechanisms: a downregulation of BiP and by direct interaction of the misfolded proteins with the UPR sensors (63), it is possible that not all cells activate UPR in response to downregulated BiP. In itself, the lack of UPR activation by PrP mutants may not be so surprising, because the PrP mutants were shown to traffic to and attach as GPI-anchored PrP at the cell surface rather than accumulate in the ER (8).
In contrast to familial prion disease (60), transmissible PrP diseases show UPR activation. A dramatic loss of synaptic proteins occurs in PrPSc-infected mouse brain neurons, as a result of progressive increases in phospho-PERK and phospho-eIF2α-mediated protein synthesis inhibition (31). Pharmacologically inhibiting PERK phosphorylation in PrPSc-infected mice restored synaptic protein synthesis and rescued behavioral deficits, while protecting against spongiform degeneration and neuronal cell loss (30). These compelling data fit with other reports showing PrPSc-mediated increases in PDI and ER chaperones Grp58, BiP and Grp94 (64,65). Consequently, PERK-based treatments may not apply to familial prion diseases.
In conclusion, our data supporting a novel mechanism by which familial prion mutants alter neuronal homeostasis. An impaired ability to recognize misfolded proteins and restore balance through the UPR and ERAD could explain the age-dependent neurodegeneration seen in familial prion disease patients.
Materials and Methods
Cell cultures and pharmacological treatments
CR7 human glioblastoma cells (obtained, with ethical approval, from Dr Melinda Estes, Cleveland Clinic, Cleveland, OH) were maintained in Opti-MEM reduced serum media (Invitrogen, Burlington, Ontario, Canada) supplemented with 2.4 g/l NaHCO3 and 20% fetal bovine serum (FBS; Thermo Fisher Scientific, Mississauga, Ontario, Canada). The sex of the donor is unknown. SK-N-SH cells were obtained from ATCC and maintained in DMEM (Invitrogen) supplemented with 3.7 g/l NaHCO3 and 10% FBS (Thermo Fisher Scientific). To examine retrotranslocation, CR7 cells were treated for 18 h with 1 μM of irreversible proteasome inhibitor epoxomicin (Enzo Life Sciences, Farmingdale, NY). The minimal concentration of epoxomicin required to fully inhibit proteasomal activity on CR7 cells was empirically determined by testing the efficiency of 0.15–1 µM epoxomicin on CR7 cells. Proteasomal activity was assessed in a fluorogenic assay using Succ-LLVY-AMC as a substrate. While 50% inhibition was achieved with 0.2 µM, strong inhibition of proteasomal activity in CR7 cells required 1 µM epoxomicin (7% activity remaining). The 1 µM concentration of epoxomicin was not toxic to the CR7 cells. In initial experiments, 5 μg/ml of Golgi-disaggregating BFA (Sigma, Oakville, Ontario, Canada) was used to ensure that CyPrP was generated from the endoplasmic reticulum via ERAD and to replicate our previous observations (8) in the CR7 cell line. Alternatively, where indicated, cells were treated with epoxomicin alone or 75 μM of reversible proteasome inhibitor Bortezomib (Calbiochem, Billerica, MA) for 18 h prior to subcellular fractionation.
DNA transfections and RNA interference
Both DNA transfection and RNA interference by siRNA were carried out using the Amaxa Nucleofection system, Kit V, program T-030 for CR7 cells and program P-020 for SK-N-SH cells, with the Nucleofector 2b device (Lonza, Mississauga, Ontario, Canada). These cell-specific parameters were optimized using Amaxa Cell Line Optimization Nucleofector Kit (Lonza). Transfection of 1 × 106 cells (on average, passages 9–18) was achieved using 1 μg of peYFP-Hrd1 (a generous gift from Dr Fang, University of Maryland, College Park, MA), control pMax-eGFP (included in the Amaxa Nucleofection Kit, Lonza), PrP mutants (all in the pCEP4β expression vector) (8), pcDNA3.1(+)-BiP (deposited by Richard C. Austin, McMaster U. Hamilton, ON, Canada in Addgene, Cambridge, MA) and pcDNA3.1(-)-TTRD18G-GFP or pcDNA3.1(-)-A1ATNHK-GFP (both a kind gift from Dr Kopito and Dr Christianson, Stanford University, Stanford, CA) (18). Double transfections were carried out using 0.5 μg of either PrP mutant, or pCEP4β-Empty combined with 1.5 μg of TTRD18G-GFP or A1ATNHK-GFP. Alternatively, rescue experiments were carried out using a 50:50 ratio of WT or mutant PrP to BiP or a 1:2:2 ratio of TTRD18G-GFP/A1ATNHK-GFP to mutant PrP to BiP. For siRNA interference, 250 nm of ON-TARGET Plus Human SYVN1 or Non-Targeting SMART pool siRNA (Dharmacon, Thermo Fisher Scientific) were transfected into 1 × 106 CR7 cells, using the Amaxa nucleofection system described above. SK-N-SH cells were either lysed in NP40 buffer (150 mm NaCl, 50 mm Tris–HCl pH 8.0, 5 mm EDTA, 1% NP40) prior to western blot analysis or placed in TRIZOL to extract RNA.
Isolation of total RNA and reverse-transcriptase polymerase chain reaction
Total RNA was extracted from CR7 cells using the TRIZOL reagent (Invitrogen, Life Technologies, Burlington, ON) following the manufacturer's instructions. cDNA was prepared using avian myeloblastosis reverse transcriptase (AMV-RT, Roche, Laval, Quebec, Canada), following the manufacturer's protocol. Briefly, 2 μg total RNA was used for cDNA synthesis by AMV-RT with poly dT primers. PCR amplification of PRNP, SYVN1, HSPA5 (BiP), XBP1, ATF6α, ERLEC1 or HPRT1 cDNAs was done with the following primers: forward 5′-ACGCGGATCCCA AGAAGCGCCCGAAGCCT-3′ and reverse 5′-GCCGCTCGAGGCTCGATCCTCTCTGGTA-3′ for PRNP, forward 5′-CTGGCTCTGCCCCAGAGGCTGGCCCT-3′ and reverse 5′-GTGGGCAACAGGA GACTCCAGCTTCTGCAGG-3′ for SYVN1, forward 5′-TCAAGTTCTTGCCCTTC AAGG-3′ and reverse 5′-AAATAAGCCTCAGCGGTTT CTT-3′ for HSPA5 (BiP), forward 5′-GGGTCCAAGTTGTCCAGAATGC-3′ and reverse 5′-TTACGAGAGAAAACTCATGGC-3′ for XBP1, forward 5′-TGGGGGAGTC ACACAGCTCCC-3′ and reverse 5′-AGCTGCCGCTTCAGTGTTCCA-3′ for ATF6α, forward 5′-GCTCACTGTTGGGACAACCCACA-3′ and reverse 5′-CCATGTCCCGAC AACCACAGAGG for ERLEC1 and forward 5′-CCTGGCGTCGTGATTAGTGAT-3′ and reverse 5′-AGACGTTCAGTCCTGTCCATAA-3′ for HPRT1.
Subcellular fractionation assay
Subcellular fractionations were performed as described previously (8) with modifications. Forty-two hours post-transfection and 18 h post-treatment, approximately 3 × 106 CR7 cells were collected in 0.25% trypsin–EDTA (Invitrogen) and spun at 500g for 5 min at 37°C. Cells were washed three times in 10 ml of ice-cold PBS and suspended in 1 ml of fractionation buffer (8% sucrose (w/v), 20 mm HCl–Tricine, pH 7.8 and 1 mm EDTA) supplemented with protease inhibitors (38 μg/ml 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), 0.5 μg/ml leupeptin, 0.1 μg/ml pepstatin and 0.1 μg/ml Nα-Tosyl-Lys Chloromethyl Ketone, all from Sigma, St Louis, MI). Cells were lysed with 18 strokes in a 2 ml Dounce homogenizer (Kontes, VWR, Mississauga, Ontario, Canada). The homogenate was spun at 2000g for 10 min at 4°C to pellet nuclei and unbroken cells. The resultant supernatant was collected and further spun at 100 000g for 1 h at 4°C. The supernatant containing cytosolic proteins was removed, and the pellet containing membrane proteins was washed two times in fractionation buffer and resuspended in 200 μL of Triton X-100 lysis buffer (150 mm NaCl, 2 mm EDTA, 0.5% Triton-X (v/v), 0.5% sodium deoxycholate (w/v), 50 mm Tris–HCl, pH 7.5). Proteins were quantified using the Bradford reagent (BioRad, Mississauga, Ontario, Canada), and equal amounts of proteins from matched fraction pairs (for example, 100 μg of untreated versus 100 μg of epoxomicin-treated cytosolic proteins) were precipitated overnight in four volumes of ice-cold methanol and re-suspended in Lammeli buffer (66) prior to western blotting.
Endoglycosidase H and peptide-N-glycosidase F digestion
To assess glycosylation of cytosolic PrP, CR7 cells were treated with 1 μM of epoxomicin and 5 μg/ml BFA for 18 h followed by subcellular fractionation to obtain cytosolic proteins (described above). Glycosidase treatments were done with New England Biolab enzymes and supplied buffers as recommended (New England Biolabs, Pickering, Ontario, Canada). Two hundred μιχρογραμσ of total proteins were precipitated overnight in methanol and re-suspended in 1% NP-40 buffer supplemented with one time protein denaturing buffer. Proteins were boiled for 10 min followed by digestion with 1500 U of either endoglycosidase H (EndoH) or peptide-N-glycosidase F (PNGaseF) for 4 h at 37°C in the presence, respectively, of the provided G5 or G7 reaction buffers. Digested proteins were precipitated overnight in methanol and re-suspended in Lammeli buffer prior to western blotting.
Western blot analysis
All proteins were separated on 10% SDS-PAGE with the exception of those probed for PrP or eYFP-Hrd1, which were separated on 15% and 8%, respectively. Proteins were transferred to polyvinylidene fluoride membranes at 15 V over 10 min using the Trans-blot Turbo Transfer System (1.5 mm gel, ‘High MW’ program, BioRad). PrP was detected with 1:10 000 anti-PrP109–112 3F4 antibody (67). Antibodies specific for the C-terminal of Hrd1 (1:250; Abgent, San Diego, CA), mitochondrial Hsp70 (1:500; Thermo Fisher Scientific, Waltham, MA), ERK 1/2 (1:1000; Cell Signaling, Danvers, MA) or anti-ERK-2 (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA), GFP (1:2500; Clone B-2, Santa Cruz Biotechnology), anti-eIF2α (1:500, 9722; Cell Signaling), anti-phospho-eIF2α (1:500, 9721; Cell Signaling), anti-BiP (rabbit monoclonal C50B12), anti-PDI (C81H6), anti-Grp94 (2104), anti-calnexin (2679) (all 1:1000; Cell Signaling ER stress antibody sampler kit) and anti-β-actin (1:2500; Sigma, Clone AC-15) were also used for western blotting. Immunoreactivity was detected with 1:5000 anti-mouse (GE Healthcare, Baie d'Urfe, Quebec, Canada) or 1:5000 anti-rabbit (Dako, Burlington, Ontario, Canada) IgG secondary antibodies conjugated to horseradish peroxidase or 1:5000 anti-mouse IgG secondary antibodies conjugated to alkaline phosphatase (Jackson ImmunoResearch, West Grove, PA) and chemiluminescence (GE Healthcare, Mississauga, Ontario, Canada) or NBT/BCIP (Promega, Madison, WI).
Fluorescence microscopy and cell viability assays
To assess subcellular distribution, eGFP, eYFP-Hrd1, A1ATNHK- or TTRD18G-coding plasmids were co-transfected in CR7 cells by nucleofection (described above) with mCherry-ER-3, a kind gift from Dr Micheal Davidson (Florida State University, FL) obtained through Addgene (#55041) and plated on poly-d-lysine coated coverslips or glass bottom plate. eGFP and eYFP-Hrd1-transfected cells were imaged after 24 h, whereas cells transfected with A1ATNHK- or TTRD18G- were treated after 24 h with DMSO or 1 µM epoxomicin for 18 h before imaging. Chromatin was stained using Hoechst 33342 (1 μg/ml, ImmunoChemistry Technologies, Bloomington, MN) before fixing in phosphate buffer saline containing 2% glutaraldehyde (Sigma G-5882) and 0.2% formaldehyde (Thermo Scientific #28908) and mounting on slide. To assess cell viability, condensed chromatin was examined in CR7 cells following Hoechst 33342 staining in cells transfected with WT or familial PrP mutants, with or without pcDNA3.1(+)-BiP. A minimum of 15 visual fields were quantified for each condition.
Cycloheximide chase
CR7 cells were transfected with either WT-PrP or PrP-M232R129V. Twenty-four hours post-transfection, cells were incubated with 75 μg/ml cycloheximide (Sigma) and harvested at indicated times over a 24-h period in NP-40 lysis buffer supplemented with protease inhibitor cocktail. One hundred μg of proteins from each time-point was precipitated overnight in four volumes of ice-cold methanol and analyzed by western blotting. Using densitometry of non-saturated X-ray films, arbitrary values were generated for BiP protein bands, which were subsequently normalized to β-actin. Values were converted to percent time 0 for each respective protein. The data were fit to a nonlinear, one-phase decay equation (y = (A– B) × Exp((−1) × k × x) + B), where A and B represent the highest and lowest values, respectively, and k represents the rate constant. Half-lives and rates of decay were generated using Prism software (GraphPad Software, version 6.0, San Diego, CA).
Densitometry and statistical analysis
Quantification of western blot analysis was carried out by evaluating relative density of cytosolic and membrane protein bands with respect to corresponding β-actin loading controls using ImageJ (National Institutes of Health, version 1.47t (64-bit), Bethesda, MD). Statistical significance of the results was analyzed, where indicated, by either independent samples t-test or one-way ANOVA followed by Tukey's multiple comparisons test in Prism (GraphPad Software, version 6.0, San Diego, CA). A P-value of <0.05 was used to determine statistical significance.
Funding
This work was supported by the Canadian Institutes of Health Research operating grants MOP89376 and MOP102738 to A.L.B.
Acknowledgements
We are grateful to Dr Shengyun Fang (University of Maryland, MA) for generously providing the peYFP-Hrd1 construct and to both Dr John Christianson and Dr Ron Kopito (Stanford University, CA) for the pcDNA3.1(-)-TTRD18G-GFP and pcDNA3.1(−)-1plasmids. We also thank Dr Gyanesh Sharma for his assistance with the kinetics analysis and Dr Raphael Rouget for generating all recombinant PrP proteins used in these experiments. Lastly, we thank the entire LeBlanc laboratory team for all technical advice and discussion provided through the process. M.A.D. is a recipient of Canadian Institutes of Health Research Frederick Banting and Charles Best doctoral scholarship.
Conflict of Interest statement. None of the authors have any conflict of interest.
References
- 1.Ashok A., Hegde R.S. (2009) Selective processing and metabolism of disease-causing mutant prion proteins. PLoS Pathog., 5, e1000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Satpute-Krishnan P., Ajinkya M., Bhat S., Itakura E., Hegde R.S., Lippincott-Schwartz J. (2014) ER stress-induced clearance of misfolded GPI-anchored proteins via the secretory pathway. Cell, 158, 522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zanusso G., Petersen R.B., Jin T., Jing Y., Kanoush R., Ferrari S., Gambetti P., Singh N. (1999) Proteasomal degradation and N-terminal protease resistance of the codon 145 mutant prion protein. J. Biol. Chem., 274, 23396–23404. [DOI] [PubMed] [Google Scholar]
- 4.Yedidia Y., Horonchik L., Tzaban S., Yanai A., Taraboulos A. (2001) Proteasomes and ubiquitin are involved in the turnover of the wild-type prion protein. EMBO J., 20, 5383–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roucou X., Guo Q., Zhang Y., Goodyer C.G., LeBlanc A.C. (2003) Cytosolic prion protein is not toxic and protects against Bax-mediated cell death in human primary neurons. J. Biol. Chem., 278, 40877–40881. [DOI] [PubMed] [Google Scholar]
- 6.Ma J., Lindquist S. (2001) Wild-type PrP and a mutant associated with prion disease are subject to retrograde transport and proteasome degradation. Proc. Natl. Acad. Sci. USA, 98, 14955–14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin T., Gu Y., Zanusso G., Sy M., Kumar A., Cohen M., Gambetti P., Singh N. (2000) The chaperone protein BiP binds to a mutant prion protein and mediates its degradation by the proteasome. J. Biol. Chem., 275, 38699–38704. [DOI] [PubMed] [Google Scholar]
- 8.Jodoin J., Laroche-Pierre S., Goodyer C.G., LeBlanc A.C. (2007) Defective retrotranslocation causes loss of anti-Bax function in human familial prion protein mutants. J. Neurosci., 27, 5081–5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jodoin J., Misiewicz M., Makhijani P., Giannopoulos P.N., Hammond J., Goodyer C.G., LeBlanc A.C. (2009) Loss of anti-Bax function in Gerstmann–Sträussler–Scheinker syndrome-associated prion protein mutants. PloS ONE, 4, e6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Apodaca J., Kim I., Rao H. (2006) Cellular tolerance of prion protein PrP in yeast involves proteolysis and the unfolded protein response. Biochem. Biophys. Res. Commun., 347, 319–326. [DOI] [PubMed] [Google Scholar]
- 11.Ruggiano A., Foresti O., Carvalho P. (2014) Quality control: ER-associated degradation: protein quality control and beyond. J. Cell Biol., 204, 869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mehnert M., Sommer T., Jarosch E. (2014) Der1 promotes movement of misfolded proteins through the endoplasmic reticulum membrane. Nat. Cell Biol., 16, 77–86. [DOI] [PubMed] [Google Scholar]
- 13.Kaneko M., Ishiguro M., Niinuma Y., Uesugi M., Nomura Y. (2002) Human HRD1 protects against ER stress-induced apoptosis through ER-associated degradation. FEBS Lett., 532, 147–152. [DOI] [PubMed] [Google Scholar]
- 14.Fang S., Ferrone M., Yang C., Jensen J.P., Tiwari S., Weissman A.M. (2001) The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA, 98, 14422–14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nadav E., Shmueli A., Barr H., Gonen H., Ciechanover A., Reiss Y. (2003) A novel mammalian endoplasmic reticulum ubiquitin ligase homologous to the yeast Hrd1. Biochem. Biophys. Res. Commun., 303, 91–97. [DOI] [PubMed] [Google Scholar]
- 16.Kaneko M., Yasui S., Niinuma Y., Arai K., Omura T., Okuma Y., Nomura Y. (2007) A different pathway in the endoplasmic reticulum stress-induced expression of human HRD1 and SEL1 genes. FEBS Lett., 581, 5355–5360. [DOI] [PubMed] [Google Scholar]
- 17.Shen Y., Ballar P., Apostolou A., Doong H., Fang S. (2007) ER stress differentially regulates the stabilities of ERAD ubiquitin ligases and their substrates. Biochem. Biophys. Res. Commun., 352, 919–924. [DOI] [PubMed] [Google Scholar]
- 18.Christianson J.C., Olzmann J.A., Shaler T.A., Sowa M.E., Bennett E.J., Richter C.M., Tyler R.E., Greenblatt E.J., Harper J.W., Kopito R.R. (2012) Defining human ERAD networks through an integrative mapping strategy. Nat. Cell Biol., 14, 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shao J., Choe V., Cheng H., Tsai Y.C., Weissman A.M., Luo S., Rao H. (2014) Ubiquitin ligase gp78 targets unglycosylated prion protein PrP for ubiquitylation and degradation. PLoS ONE, 9, e92290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostankovitch M., Altrich-Vanlith M., Robila V., Engelhard V.H. (2009) N-glycosylation enhances presentation of a MHC class I-restricted epitope from tyrosinase. J. Immunol., 182, 4830–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lippincott-Schwartz J., Yuan L.C., Bonifacino J.S., Klausner R.D. (1989) Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell, 56, 801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vidal R., Garzuly F., Budka H., Lalowski M., Linke R.P., Brittig F., Frangione B., Wisniewski T. (1996) Meningocerebrovascular amyloidosis associated with a novel transthyretin mis-sense mutation at codon 18 (TTRD 18G). Am. J. Pathol., 148, 361–366. [PMC free article] [PubMed] [Google Scholar]
- 23.Christianson J.C., Shaler T.A., Tyler R.E., Kopito R.R. (2008) OS-9 and GRP94 deliver mutant α1-antitrypsin to the Hrd1–SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol., 10, 272–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Susuki S., Sato T., Miyata M., Momohara M., Suico M., Shuto T., Ando Y., Kai H. (2009) The endoplasmic reticulum-associated degradation of transthyretin variants is negatively regulated by BiP in mammalian cells. J. Biol. Chem., 284, 8312–8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donoso G., Herzog V., Schmitz A. (2005) Misfolded BiP is degraded by a proteasome-independent endoplasmic-reticulum-associated degradation pathway. Biochem. J., 387, 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi Y., Porter K., Parameswaran N., Bae H.K., Pestka J.J. (2009) Role of GRP78/BiP degradation and ER stress in deoxynivalenol-induced interleukin-6 upregulation in the macrophage. Toxicol. Sci., 109, 249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J., Kaufman R.J. (2006) From acute ER stress to physiological roles of the unfolded protein response. Cell Death Differ., 13, 374–384. [DOI] [PubMed] [Google Scholar]
- 28.Hetz C. (2012) The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol., 13, 89–102. [DOI] [PubMed] [Google Scholar]
- 29.Hetz C., Lee A.H., Gonzalez-Romero D., Thielen P., Castilla J., Soto C., Glimcher L.H. (2008) Unfolded protein response transcription factor XBP-1 does not influence prion replication or pathogenesis. Proc. Natl. Acad. Sci. USA, 105, 757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno J.A., Halliday M., Molloy C., Radford H., Verity N., Axten J.M., Ortori C.A., Willis A.E., Fischer P.M., Barrett D.A. (2013) Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci. Transl. Med., 5, 206ra138. [DOI] [PubMed] [Google Scholar]
- 31.Moreno J., Radford H., Peretti D., Steinert J., Verity N., Martin M., Halliday M., Morgan J., Dinsdale D., Ortori C. et al. (2012) Sustained translational repression by eIF2a-P mediates prion neurodegeneration. Nature, 485, 507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quaglio E., Restelli E., Garofoli A., Dossena S., De Luigi A., Tagliavacca L., Imperiale D., Migheli A., Salmona M., Sitia R. et al. (2011) Expression of mutant or cytosolic PrP in transgenic mice and cells is not associated with endoplasmic reticulum stress or proteasome dysfunction. PLoS ONE, 6, e19339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee K., Tirasophon W., Shen X., Michalak M., Prywes R., Okada T., Yoshida H., Mori K., Kaufman R.J. (2002) IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev., 16, 452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H., Harada A., Mori K. (2007) Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev. Cell, 13, 365–376. [DOI] [PubMed] [Google Scholar]
- 35.Fujimori T., Kamiya Y., Nagata K., Kato K., Hosokawa N. (2013) Endoplasmic reticulum lectin XTP3-B inhibits endoplasmic reticulum-associated degradation of a misfolded α1-antitrypsin variant. FEBS J., 280, 1563–1575. [DOI] [PubMed] [Google Scholar]
- 36.Afshar N., Black B.E., Paschal B.M. (2005) Retrotranslocation of the chaperone calreticulin from the endoplasmic reticulum lumen to the cytosol. Mol. Cell Biol., 25, 8844–8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cumming R.C., Andon N.L., Haynes P.A., Park M., Fischer W.H., Schubert D. (2004) Protein disulfide bond formation in the cytoplasm during oxidative stress. J. Biol. Chem., 279, 21749–21758. [DOI] [PubMed] [Google Scholar]
- 38.Petris G., Vecchi L., Bestagno M., Burrone O.R. (2011) Efficient detection of proteins retro-translocated from the ER to the cytosol by in vivo biotinylation. PLoS ONE, 6, e23712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun S., Shi G., Han X., Francisco A.B., Ji Y., Mendonça N., Liu X., Locasale J.W., Simpson K.W., Duhamel G.E. (2014) Sel1L is indispensable for mammalian endoplasmic reticulum-associated degradation, endoplasmic reticulum homeostasis, and survival. Proc. Natl. Acad. Sci. USA, 111, E582–E591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma J., Wollmann R., Lindquist S. (2002) Neurotoxicity and neurodegeneration when PrP accumulates in the cytosol. Science, 298, 1781–1785. [DOI] [PubMed] [Google Scholar]
- 41.Bounhar Y., Zhang Y., Goodyer C.G., LeBlanc A. (2001) Prion protein protects human neurons against Bax-mediated apoptosis. J. Biol. Chem., 276, 39145–39149. [DOI] [PubMed] [Google Scholar]
- 42.Roucou X., Giannopoulos P., Zhang Y., Jodoin J., Goodyer C., LeBlanc A. (2005) Cellular prion protein inhibits proapoptotic Bax conformational change in human neurons and in breast carcinoma MCF-7 cells. Cell Death Differ., 12, 783–795. [DOI] [PubMed] [Google Scholar]
- 43.Lin D.T., Jodoin J., Baril M., Goodyer C.G., LeBlanc A.C. (2008) Cytosolic prion protein is the predominant anti-Bax prion protein form: exclusion of transmembrane and secreted prion protein forms in the anti-Bax function. Biochim. Biophys. Acta (BBA)-Mol. Cell Res., 1783, 2001–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kikkert M., Doolman R., Dai M., Avner R., Hassink G., van Voorden S., Thanedar S., Roitelman J., Chau V., Wiertz E. (2004) Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J. Biol. Chem., 279, 3525–3534. [DOI] [PubMed] [Google Scholar]
- 45.Shenkman M., Groisman B., Ron E., Avezov E., Hendershot L.M., Lederkremer G.Z. (2013) A shared endoplasmic reticulum-associated degradation pathway involving the EDEM1 protein for glycosylated and nonglycosylated proteins. J. Biol. Chem., 288, 2167–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gray D.A., Tsirigotis M., Woulfe J. (2003) Ubiquitin, proteasomes, and the aging brain. Sci. SAGE KE, 2003, 6. [DOI] [PubMed] [Google Scholar]
- 47.Kaneko M., Koike H., Saito R., Kitamura Y., Okuma Y., Nomura Y. (2010) Loss of HRD1-mediated protein degradation causes amyloid precursor protein accumulation and amyloid-β generation. J. Neurosci., 30, 3924–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Omura T., Kaneko M., Okuma Y., Orba Y., Nagashima K., Takahashi R., Fujitani N., Matsumura S., Hata A., Kubota K. (2006) A ubiquitin ligase HRD1 promotes the degradation of Pael receptor, a substrate of Parkin. J. Neurochem., 99, 1456–1469. [DOI] [PubMed] [Google Scholar]
- 49.Yang H., Zhong X., Ballar P., Luo S., Shen Y., Rubinsztein D.C., Monteiro M.J., Fang S. (2007) Ubiquitin ligase Hrd1 enhances the degradation and suppresses the toxicity of polyglutamine-expanded huntingtin. Exp. Cell Res., 313, 538–550. [DOI] [PubMed] [Google Scholar]
- 50.Dorandeu A., Wingertsmann L., Chrétien F., Delisle M.B., Vital C., Parchi P., Montagna P., Lugaresi E., Ironside J.W., Budka H. (1998) Neuronal apoptosis in fatal familial insomnia. Brain Pathol., 8, 531–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferrer I. (2002) Synaptic pathology and cell death in the cerebellum in Creutzfeldt-Jakob disease. Cerebellum, 1, 213–222. [DOI] [PubMed] [Google Scholar]
- 52.Gray F., Chrétien F., Adle-Biassette H., Dorandeu A., Ereau T., Delisle M.-B., Kopp N., Ironside J.W., Vital C. (1999) Neuronal apoptosis in Creutzfeldt-Jakob disease. J. Neuropathol. Exp. Neurol., 58, 321–328. [DOI] [PubMed] [Google Scholar]
- 53.Cohen E., Avrahami D., Frid K., Canello T., Lahad E.L., Zeligson S., Perlberg S., Chapman J., Cohen O.S., Kahana E. (2013) Snord 3A: a molecular marker and modulator of prion disease progression. PLoS ONE, 8, e54433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katayama T., Imaizumi K., Sato N., Miyoshi K., Kudo T., Hitomi J., Morihara T., Yoneda T., Gomi F., Mori Y. (1999) Presenilin-1 mutations downregulate the signalling pathway of the unfolded-protein response. Nat. Cell Biol., 1, 479–485. [DOI] [PubMed] [Google Scholar]
- 55.Senderek J., Krieger M., Stendel C., Bergmann C., Moser M., Breitbach-Faller N., Rudnik-Schöneborn S., Blaschek A., Wolf N.I., Harting I. (2005) Mutations in SIL1 cause Marinesco-Sjögren syndrome, a cerebellar ataxia with cataract and myopathy. Nat. Genet., 37, 1312–1314. [DOI] [PubMed] [Google Scholar]
- 56.Zhao L., Longo-Guess C., Harris B.S., Lee J.-W., Ackerman S.L. (2005) Protein accumulation and neurodegeneration in the woozy mutant mouse is caused by disruption of SIL1, a cochaperone of BiP. Nat. Genet., 37, 974–979. [DOI] [PubMed] [Google Scholar]
- 57.Mizukami K., Asada T., Kinoshita T., Tanaka K., Sonohara K., Nakai R., Yamaguchi K., Hanyu H., Kanaya K., Takao T. (2009) A randomized cross-over study of a traditional Japanese medicine (kampo), yokukansan, in the treatment of the behavioural and psychological symptoms of dementia. Int. J. Neuropsychopharmacol., 12, 191–199. [DOI] [PubMed] [Google Scholar]
- 58.Hiratsuka T., Matsuzaki S., Miyata S., Kinoshita M., Kakehi K., Nishida S., Katayama T., Tohyama M. (2010) Yokukansan inhibits neuronal death during ER stress by regulating the unfolded protein response. PLoS ONE, 5, e13280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoshida H., Haze K., Yanagi H., Yura T., Mori K. (1998) Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J. Biol. Chem., 273, 33741–33749. [DOI] [PubMed] [Google Scholar]
- 60.Unterberger U., Hoftberger R., Gelpi E., Flicker H., Budka H., Voigtlander T. (2006) Endoplasmic reticulum stress features are prominent in Alzheimer disease but not in prion diseases in vivo. J. Neuropathol. Exp. Neurol., 65, 348–357. [DOI] [PubMed] [Google Scholar]
- 61.Wang X., Shi Q., Xu K., Gao C., Chen C., Li X.L., Wang G.R., Tian C., Han J., Dong X.P. (2011) Familial CJD associated PrP mutants within transmembrane region induced Ctm-PrP retention in ER and triggered apoptosis by ER stress in SH-SY5Y cells. PLoS ONE, 6, e14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lajoie P., Snapp E.L. (2011) Changes in BiP availability reveal hypersensitivity to acute endoplasmic reticulum stress in cells expressing mutant huntingtin. J. Cell Sci., 124, 3332–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maly D.J., Papa F.R. (2014) Druggable sensors of the unfolded protein response. Nat. Chem. Biol., 10, 892–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hetz C., Russelakis-Carneiro M., Maundrell K., Castilla J., Soto C. (2003) Caspase-12 and endoplasmic reticulum stress mediate neurotoxicity of pathological prion protein. EMBO J., 22, 5435–5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoo B.C., Krapfenbauer K., Cairns N., Belay G., Bajo M., Lubec G. (2002) Overexpressed protein disulfide isomerase in brains of patients with sporadic Creutzfeldt–Jakob disease. Neurosci. Lett., 334, 196–200. [DOI] [PubMed] [Google Scholar]
- 66.Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 67.Kascsak R., Rubenstein R., Merz P., Tonna-DeMasi M., Fersko R., Carp R., Wisniewski H., Diringer H. (1987) Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J. Virol., 61, 3688–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]