Abstract
T-Cell acute lymphoblastic leukemia (t-all) is a malignancy of white blood cells, characterized by an uncontrolled accumulation of T-cell progenitors. During leukemic progression, immature T cells grow abnormally and crowd into the bone marrow, preventing it from making normal blood cells and spilling out into the bloodstream. Recent studies suggest that only discrete cell populations that possess the ability to recreate the entire tumour might be responsible for the initiation and propagation of t-all. Those unique cells are commonly called “cancer stem cells” or, in the case of hematopoietic malignancies, “leukemia stem cells” (lscs). Like normal hematopoietic stem cells, lscs are thought to be capable of self-renewal, during which, by asymmetrical division, they give rise to an identical copy of themselves as well as to a daughter cell that is no longer capable of self-renewal activity and represents a more “differentiated” progeny. Here, we review the main pathways of self-renewal activity in lscs, focusing on their involvement in the maintenance and development of t-all. New stem cell–directed therapies and lsc-targeted agents are also discussed.
Keywords: Leukemia stem cells, t-all, leukemia, Notch, Wnt, β-catenin, hif, mouse models, patient-derived xenografts
INTRODUCTION
Acute lymphoblastic leukemia (all) is an aggressive tumour of the hematopoietic system. Most alls (85%) originate from the B-cell lineage (so-called b-all); the rest (15%) are of T-cell lineage (or t-all)1. By definition, all is an aggressive malignancy that can be fatal if left untreated. Most human cases of t-all occur in young children between 2 and 5 years of age, but t-all can occur at any age2. The cure rate depends primarily on the age of the patient. Outcomes in infants are better than those in children more than 10 years of age; most adults succumb quickly to the disease. The long-term survival rate is 30%–40%3.
T-Cell all is the result of genetic mutations or chromosomal translocations (or both) that alter the functional pathways involved in the proliferation, survival, growth, and differentiation of hematopoietic progenitors during T-cell development4. During the period since the late 1990s, published reports have noted aberrant expression by selected groups of transcription factors in t-all as a result of various chromosomal rearrangements or small intrachromosomal deletions. More than 50% of human cases of t-all are characterized by genetic alterations in NOTCH1 or other genes related to Notch1 signalling, resulting in constitutive activation of the Notch1 pathway5,6. Moreover, more than 70% of human t-all presents loss of p16/INK4A and p14/ARF suppressor genes as result of deletions in the CDKN2A locus (chromosome band 9p21)7. Additional oncogenic transcription factors include c-Myc8, nkx2-1, nkx2-29; the lim domain only (LMO) genes, LMO1 and LMO210; the HOXA homeobox genes11; and the basic helix-loop-helix family members TAL112 and TAL213. Aberrant expression of these regulatory genes leads to a block in the differentiation process and induces uncontrolled proliferative signals in t-all.
DISCOVERY OF LSCs IN T-ALL
Leukemia stem cells were first described in the mid-1980s by John Dick’s group, when they demonstrated the existence of lsc-enriched subsets in the heterogeneous bulk populations of acute myeloid leukemia14. Those primitive cells had the capacity to be serially transplanted and could give rise to differentiated cells, recreating the complete phenotypic heterogeneity of primary leukemia14.
In t-all, cell populations with enriched lsc activity were initially reported within the CD34+CD4− or CD34+CD7− subsets. Those cell populations were able to reinitiate disease in naïve immunocompromised non-obese diabetic and severe combined immunodeficiency (nod/scid) mice—so-called leukemia-initiating cell activity15. However, in a subsequent study, it was reported that the t-all development in nod/scid mice is not strictly correlated with CD34 surface expression16. After the foregoing reports, a third study emerged from John Dick’s group. Looking at a cohort of 5 cases, they found that the CD7+ phenotype was a stronger marker of leukemia-initiating cell activity and that a lower number of CD7+CD1a− cells were transplantable in a more permissive host17. Notably, CD34 positivity was not a consistent finding across their cohort, and additional experiments suggested that those subsets performed better in a co-culture system with OP9 stroma cells expressing the Notch1 ligand delta-like 1 and were dependent on Notch1 signalling. In a very limited subset of cases, those authors also highlighted some correlation with dexamethasone resistance in vivo17.
Further experimental evidence of lsc activity in T-cell leukemia was acquired using mouse models of t-all based on the genes most commonly affected in patients. In particular, Dr. Hong Wu’s group showed that in a pten-null t-all mouse model (PTEN is mutated at a frequency of 5%–10% in t-all18), the c-KitmidCD3+Lin− subset was the most efficiently transplantable in syngeneic recipients19. In the transgenic TAL1/LMO1 mouse model of t-all, an enrichment of lsc activity was reported in CD4−CD8− CD44−CD25+ (DN3) and CD4−CD8−CD44−CD25− (DN4) fractions20,21. Finally, it was shown that in Notch1- and KrasG12D-induced t-all mouse models, lscs are enriched in the CD44+ROSlow subset22 (Table i). Those findings suggest that t-all might consist of different subpopulations, reflecting the complex heterogeneity of T-cell leukemia9. A better comprehension of lsc distribution in human t-all awaits complementary studies.
TABLE I.
Leukemia cell subsets enriched with leukemia stem cells in T-cell acute lymphoblastic leukemia
| Reference | Cell-surface markers | Tumour type | Minimal tumourigenic dose (cells) | Transplant technique | Strain of recipient mice |
|---|---|---|---|---|---|
| Cox et al., 200715 | CD34+CD4− or CD34+CD7– | Human | 104 | Intravenous injection | Immunodeficient (NSGa) |
| Guo et al., 200819 | c-KitmidCD3+Lin− | Pten−/−induced mouse leukemia | 102 | Intravenous injection | Immunodeficient (SCID) |
| Armstrong et al., 200917 | CD7+CD1a– | Human | 103 | Intrafemoral injection | Immunodeficient (NSGa) |
| McCormack et al., 201020, Tremblay et al., 201021 | CD4−CD8−CD44−CD25+ (DN3), CD4−CD8−CD44−CD25− (DN4) | TAL1/SCLand LMO1/2induced mouse model | 103 | Intravenous injection | Immunodeficient (Rag1−/−) |
| Giambra et al., 201222 | ROSlow | Human | 105 | Intravenous injection | Immunodeficient (NSGa) |
| ROSlowCD44+ | Notch1 ΔE-induced mouse leukemia | 102 | Intravenous injection | Syngeneic (C57BL/6) | |
| ROSlowCD44+ | KrasG12D-induced mouse leukemia | 103 | Intravenous injection | Syngeneic (C57BL/6) |
The Jackson Laboratory, Bar Harbor, ME, U.S.A.
NSG = NOD SCID gamma; SCID = severe combined immunodeficiency; DN3/4 = double-negative 3/4; TAL1 = T-cell acute lymphocytic leukemia 1; LMO1/2 = LIM domain only 1/2; ROS = reactive oxygen species.
New discoveries about the functional mechanisms of lsc maintenance and propagation and the key regulatory pathways involved in lsc self-renewal in human and mouse models of t-all are emerging. Those new findings could contribute to the development of therapies that will be more efficient in specifically targeting the malignant stem-cell population and in reducing cytotoxic effects on normal stem-cell subsets.
SELF-RENEWAL PATHWAYS OF LSC ACTIVITY
Self-renewal is the main feature that discriminates stem-cell from differentiated-cell populations. It is defined as the capacity to self-renew for indefinite periods while the entire differentiation potential is preserved.
The role of stem cells is indeed to generate differentiated progeny while maintaining the undifferentiated stem-cell pool. This unique property of stem cells results from their ability to divide asymmetrically: Of two daughter cells, one retains the stem-cell characteristics; the other is destined for a limited number of future divisions and will produce even-more-specialized cells23.
The functional mechanisms that control self-renewal in lscs are not clearly described. Increasing evidence shows that common pathways regulate the self-renewal capacity in both normal and cancer stem cells and promote cancer progression when deregulated24. The most relevant examples include the Notch1, Wnt, and hypoxia-inducible factor (hif) cellular signalling pathways that modulate the maintenance and activity of lscs in t-all16,19,22,25,26. Interestingly, acquisition of mutations within those pathways contributes to leukemic progression by promoting self-renewal and survival within supportive niches. In the subsections that follow, we review those three functional pathways, underlining their role in the activity of lscs and progression of T-cell leukemia.
Notch1 Signalling Pathway
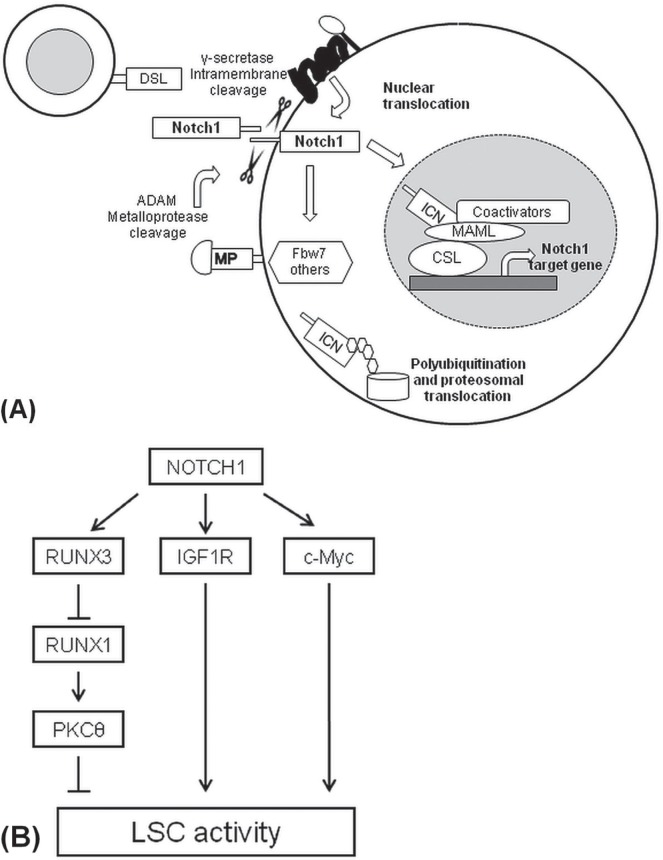
Notch1 signalling is one of main pathways involved in the proliferation, maintenance, and survival of T-cell leukemia; it is activated when the transmembrane receptor Notch1 recognizes ligands from the Jagged family (for example, jag1 and jag2) or delta-like proteins dll1, dll3, and dll4 on neighbouring cells27. The binding of the extracellular subunit of Notch1 (ecn1) to its ligand activates several proteolytic cleavages and allows for the release and translocation of the intracellular Notch1 domain (icn1) to the nucleus. The intracellular Notch1 domain acts as a transcription factor and forms part of a transcriptional complex with coactivators such as mastermind-like protein 1 and the dna-binding transcription factor csl, leading to the activation or repression of Notch1 target genes28 [Figure 1(A)].
FIGURE 1.
The Notch1 signalling pathway. (A) The binding of Notch1 ligand (DSL) with the Notch1 receptor activates two subsequent protease cleavages. Afterward, intracellular Notch1 (ICN1) translocates into the nucleus where it interacts with other coactivators in a protein complex for the induction of Notch1 target genes. ADAM = A disintegrin and metalloproteinase; MAML = mastermind-like protein; MP = metal-loproteinase; CSL = CBF1/Su(H)/Lag-1 transcription factor complex. (B) The Notch1 signalling pathway promotes leukemia stem cell (LSC) activity by upregulation of IGF1R and c-Myc target genes and the indirect downregulation of protein kinase C–theta (PKCθ). RUNX3/1 = runt-related transcription factor 3/1.
The constitutive expression of Notch1 in hematopoietic stem cells (hscs) leads to immortalized and cytokine-dependent cell lines that are able to generate cell progenitors with lymphoid and myeloid features both in vivo and in vitro29. Using transgenic Notch1 reporter mice, it was also shown that the Notch1 signalling pathway is downregulated and less active in the differentiated hematopoietic cells of peripheral lymphoid organs. Inhibition of Notch1 signalling causes accelerated differentiation of hscs in vitro and depletion of hscs in vivo, suggesting that the Notch1 pathway is important for the maintenance of hscs in an undifferentiated state30.
In cancer, deregulation of Notch1 signalling can induce the transcriptional expression of several oncogenes and inhibit various tumour suppressors, leading to the generation of strong oncogenic signals involved in the malignant transformation of immature T progenitors and the maintenance of established leukemia. Some examples of oncogenic Notch1 targets include pre-tcra, CD28, Deltex-1, hey1, hes1, and c-Myc, that activate programs such as the mtor (mammalian target of rapamycin)– dependent pi2k–Akt/pkb pathway, supporting cell-cycle progression and growth31. More than 50% of human t-all cases carry alterations in the NOTCH1 gene itself5; 15% have mutations in FBW7/Sel-10, the F-box protein 7 involved in Notch1 turnover6. In both cases, it leads to hyperactivation of Notch1 signalling. About one quarter of human t-all cases show mutations in the extracellular heterodimerization domain of Notch1 receptor that weaken the interaction between the extracellular subunit of Notch1 and the icn1, reducing the minimal signal required by a ligand to activate the pathway32. Deletion of the pest domain in the C-terminal portion of Notch1 receptor occurs in 12% of t-all patients and increases the stability of intracellular Notch1 (or icn1) by lowering the protein turnover. Both mutations have been detected simultaneously in more than 18% of human cases5. Notably, the Notch1 signalling pathway has been instrumental in the development of mouse models of t-all5,33, which have helped achieve a better understanding of the role played by Notch1 in the self-renewal activity of lscs16,22,25.
In T-cell leukemia, the Notch1 signalling pathway promotes both the growth and the survival of bulk cells34, but it has also been shown to play a role in the self-renewal of lscs, as assayed by serial transplantation of primary human t-all into immunocompromised nod/scid mice16. Notably, in the Tal1/Lmo2 mouse model of t-all, in vivo treatment of leukemic mice with γ-secretase inhibitors (gsis) significantly reduced lsc frequency and decreased cell survival25. Both observations support the idea that the Notch1 pathway is required for lsc maintenance in vitro and in vivo.
It was also reported that several Notch1 target genes modulate the leukemia-initiating activity in t-all22,35,36 [Figure 1(B)]. The deletion of insulin-like growth factor 1 receptor in Notch1-induced mouse leukemia reduces the transplantability of cells to secondary recipients and, consequently, the lsc frequency. Moreover, pharmacologic inhibition of insulin-like growth factor 1 receptor impairs the viability and growth of human t-all cells in vitro35.
Expression of c-Myc was also correlated with lsc activity in t-all. In Notch1-induced mouse leukemia harbouring the c-MycGFP fusion allele, c-MycGFP–positive cell subsets were enriched in lscs and, in microarray gene expression profiling, showed a gene signature similar to that of embryonic and hematopoietic stem cells. Interestingly, the selective bet bromodomain inhibitors—for example, jq1—that specifically target brd4, a transcriptional c-Myc activator, impair the in vitro growth of human t-all, representing a potential therapeutic strategy for the treatment of T-cell leukemia36.
Our group demonstrated that the downregulation of protein kinase C θ expression correlates with low levels of reactive oxygen species and further modulates the lsc activity of CD44+ subsets in t-all. Protein kinase C θ is an indirect target of Notch1 in t-all and is suppressed by a Notch1-induced transcriptional circuit that involves the induced runt-related transcription factor 3 (runx3) and runx1. In particular, the Notch1 signalling pathway activates runx3 expression, which then represses runx1, a tumour suppressor that positively modulates protein kinase C θ expression22 [Figure 1(B)]. Finally, compared with wild-type NOTCH1 pediatric t-all, mutated NOTCH1 pediatric t-all has a higher leukemia-initiating cell frequency within the CD34 compartment37.
Overall, the foregoing observations demonstrate that the Notch1 signalling pathway sustains the self-renewal activity of lscs in human t-all and suggest that novel and Notch1-specific drugs might improve clinical outcomes, especially for patients with refractory or relapsed leukemia.
Wnt Signalling Pathway
The Wnt signalling pathway is a well-known regulatory mechanism of self-renewal activity in cancer and stem-cell biology.
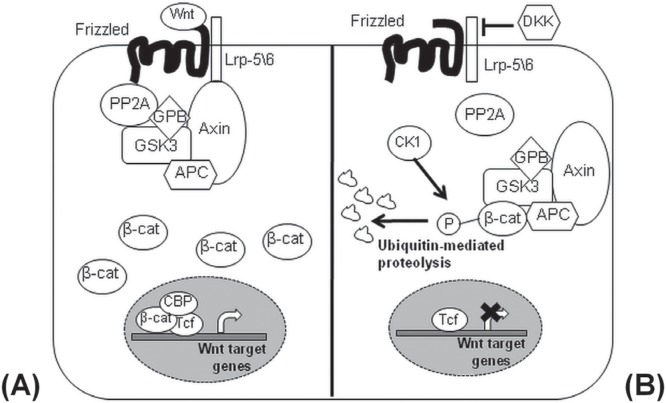
In the canonical Wnt pathway, the signal transduction cascade is started by the secretion of glycoproteins called Wnts, members of a highly conserved family of 19 ligands that further modulate various cellular processes in the receptive cell, including growth, survival, polarity, and differentiation38. Wnt ligand binding to the Frizzled or lrp-5/6 receptors blocks the degradation of β-catenin that will then translocate into the nucleus and act as a coactivator alongside transcription factors from the T-cell factor/lymphoid enhancing factor (tcf/lef) family. That complex induces the transcription of Wnt target genes such as cyclin D1 and c-Myc. When Frizzled or lrp-5/6 receptors are not engaged, Wnt signalling is not active, and β-catenin forms a protein complex with adenomatosis polyposis coli, axin, protein phosphatase 2A, casein kinase 1α, and glycogen synthase kinase 3, which promotes β-catenin phosphorylation by casein kinase 1α and glycogen synthase kinase 3 and its degradation after translocation to the proteasomal machinery39 (Figure 2).
FIGURE 2.
The canonical Wnt signalling pathway. (A) To activate the canonical Wnt signalling pathway, Wnt ligands bind the Frizzled and Lrp5/6 receptors and promote the stabilization of β-catenin after the recruitment and destruction of axin\APC\GSK3 complex. The activated β-catenin enters into the nucleus and induces the expression of Wnt target genes. APC = adenomatous polyposis coli; GSK3 = glycogen synthase kinase 3; PP2A = protein phosphatase 2A; GPB = GSK3 binding protein; CBP = CREB binding protein; Tcf = T-cell–specific transcription factor. (B) In the absence of Wnt ligands, the axin\APC\GSK3 complex leads to the proteasomal degradation of β-catenin after phosphorylation mediated by casein kinase 1α (CK1α). DKK = Dickkopf Wnt signalling pathway inhibitor; P = Phosphoryl group.
In normal hematopoietic development, the Wnt signalling pathway sustains the self-renewal activity of hscs. Expression of activated β-catenin preserves hscs in an undifferentiated state in long-term in vitro cultures, increasing their numbers by a factor of between 20 and 4840. Moreover, inhibition of Wnt signalling reduces the capacity of hscs to reconstitute the hematopoietic system in secondary recipients, emphasizing the role of that pathway in lymphoid and myeloid lineage commitment and in the maintenance of hscs40. Interestingly, the expression levels of HoxB4 and Notch1 increase in hscs transduced with the activated form of β-catenin, suggesting that Wnt signalling might cooperate with the HoxB4 and Notch1 pathways to mediate its own functions40,41.
Given the relevance of Wnt signalling in normal hematopoietic stem cells, the expectation is that, when deregulated, Wnt signalling might contribute to leukemia establishment and self-renewal activity by lscs26,42,43. Aberrant pathway activation leads to the malignant transformation of mouse hematopoietic cells and produces both chronic and acute myeloid leukemia39. In mouse thymocytes, constitutive stimulation of the pathway by transduction with active forms of β-catenin generates T-cell leukemia within 60–80 days44, and notably, lscs of mouse and human t-all are characterized by elevated levels of β-catenin protein19,26. In approximately 85% of human cases of t-all, abnormally high expression of β-catenin is observed at the protein level in non-leukemic thymocytes45. Moreover, recent studies have demonstrated that Cre-mediated β-catenin stabilization in CD4Cre-CtnnbΔex3 mice blocks normal T-cell development at the double-positive stage and predisposes the double-positive T-cells to malignant transformation. Notably, Notch1 activation was not required for β-catenin–induced lymphomagenesis, suggesting that activation of β-catenin might induce t-all independently of Notch144. Moreover, it was reported that, in human and mouse models of t-all, T-cell factor 1 (tcf-1) acts as a T-cell–specific tumour suppressor46,47. In particular, mice with a tcf-1–null genetic background developed T-cell leukemia and showed an aberrant upregulation of Lef1 in pre-leukemic thymocytes and in established leukemic cells, supporting the idea that tcf-1 and lef-1 have opposing but cooperative functions during the malignant transformation47. Those observations highlight the importance of tcf-1 and β-catenin as critical regulators of T-cell malignant transformation and leukemia maintenance.
The role of Wnt-signalling in leukemia stem-cell biology has been emphasized in human and mouse models of t-all19,26. In pten-null leukemia, the c-KitmidCD3+Lin– cell subsets, enriched in self-renewable lscs, are characterized by high levels of unphosphorylated β-catenin and by an aberrant overexpression of c-Myc, a well-known Wnt target oncogene48. Moreover, the conditional deletion of the β-catenin gene impairs lsc frequency, suggesting that the Wnt signalling pathway might contribute to the maintenance of lsc activity in t-all and that the loss of pten might cooperate with β-catenin for lsc transformation19.
Recently, using an integrated f luorescent reporter of Wnt signalling, it has been also reported that, in Notch1-induced mouse leukemia, only a small fraction of bulk leukemia cells shows active Wnt signalling. Those Wnt-active cells are highly enriched for lsc activity, and the deletion of β-catenin demonstrates a detrimental effect on lsc frequency26. Interestingly, blocking Wnt signalling in xenograft-expanded human t-all by transduction with lentivirus encoding dominant negative tcf49 impairs the transplantability of human cells into immunocompromised secondary recipients, demonstrating their dependence on the canonical Wnt pathway. From a therapeutic perspective, inhibitors of Wnt signalling, such as the tankyrase inhibitor XAV-93950 and indomethacin42, reduce in vitro proliferation and survival of various human t-all cell lines and xenograft-expanded patient T lymphoblasts, suggesting that pharmacologic inhibitors of the Wnt pathway could potentially be used in therapies for the treatment of patients with aggressive t-all26.
HIF-1 Signalling Pathway
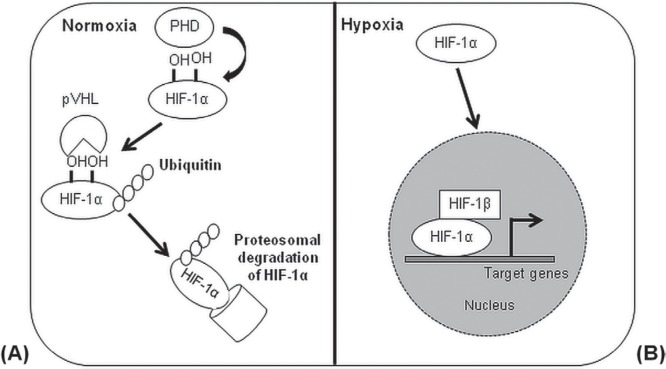
Oxygen regulates major cellular processes, including proliferation, survival, and metabolism. In both normal and malignant hematopoietic compartments, conditions of restricted oxygen modulate the self-renewal activity and differentiation of stem subsets by the transcriptional activity of hypoxia inducible factors (hifs)51. Those transcription factors are critical for the cellular response to low levels of oxygen or hypoxia (<3% O2, 20 mmHg) and ensure cell growth in hypoxic microenvironments by mediating the switch from oxidative to glycolytic metabolism. The hifs act as highly-conserved heterodimeric complexes composed of 2 subunits (α and β). The α subunits (hif-1α, hif-2α, hif-3α) are sensitive to high concentrations of oxygen and are stable only under hypoxic conditions. The β subunit, known as hif-1β or aryl hydrocarbon receptor nuclear translocator, is constitutively expressed and does not respond to oxygen levels52 (Figure 3). Of the hif-α isoforms, hif-1α is the most studied and the most broadly expressed in various tissues. In presence of high oxygen concentrations (>3% O2), hif-1α is, like the other α subunits, hydroxylated at conserved proline sites by the hif prolyl hydroxylases, triggering its degradation by the proteasome after ubiquitination by the von Hippel–Lindau ubiquitin E3 ligase complex53. The lack of oxygen under hypoxic or anoxic conditions inhibits the activity of hif prolyl hydroxylase, which uses oxygen as a co-substrate and stabilizes hif-1α, allowing the transcriptional activation of hif-1α target genes54.
FIGURE 3.
Regulation of the hypoxia-inducible factor 1α (HIF-1α) signalling pathway. (A) Under normoxic conditions, HIF-1α is degraded by the proteasomic machinery after hydroxylation and ubiquitination mediated by prolyl hydroxylases (PHDs) and the pVHL–E3–ubiquitin ligase complex. pVHL = von Hippel–Lindau tumour suppressor; E3 = ubiquitin-protein ligase. (B) In the presence of hypoxic microenvironments, HIF-1α is stable and translocates into the nucleus, where it forms a complex with the HIF-1β subunit for the activation of HIF target genes.
Since the early 2000s, hifs have been known for their importance for cancer stem cells in solid tumours54. However, emerging evidence suggests that hif signalling plays a critical role in leukemogenesis and maintenance of lscs. It was reported that the induction of hif activity can promote a stem-like phenotype and increase the number of lscs51. In acute myeloid leukemia, hif-1α is overexpressed and selectively activated in CD34+CD38− lsc–enriched subsets, and notably, the pharmacologic inhibition of hif-1α by echinomycin55 affects the transplantability of human lscs into immunocompromised mice56. In a mouse model of chronic myeloid leukemia, the deletion of hif-1α in Bcr-Abl–expressing lscs impairs the leukemogenic activity of lscs after transplant into secondary recipients and induces the expression of p16(Ink4a) and p19(Arf), affecting the cell survival and promoting apoptosis57.
In the Notch1-induced mouse model of t-all, it was recently reported that cell subsets enriched in lscs and with active Wnt signalling reside preferentially within hypoxic niches in vivo. In particular, hif-1α stabilization directly upregulates the expression of β-catenin at the transcriptional level, which potentiates Wnt signalling26. Moreover, hif-1α deletion in mouse leukemia and hif inhibition on patient-derived t-all xenografts by lentiviral small hairpin rnas reduce the lsc frequency, suggesting that the hif and Wnt/β-catenin signalling pathways cooperate together to support lsc function in t-all26.
In human t-all, hif-1α appears to be a critical regulator of the Notch1 pathway as well. The stabilization of hif-1α potentiates Notch1 signalling and so alters the expression of two matrix metalloproteinases, mmp2 and mmp9, involved in the invasion and chemoresistance of leukemic cells58. In the lsc-enriched c-Kit+Sca1+ subsets of mouse T-cell lymphoma, generated by a mutant isoform of Epm2a, hif-1α was also described to promote lsc activity by repressing Hes1, a Notch1-target gene and negative regulator of the Notch1 pathway56. Those observations indicate that hif signalling is strictly connected with relevant pathways of lsc maintenance in T-cell leukemia and lymphoma (such as Notch1 and Wnt signalling) and that blocking hif activity might improve conventional therapies by generating lsc-specific and less-detrimental treatments.
SUMMARY: TARGETING LEUKEMIA STEM CELLS
The idea that only restricted and self-renewing stem cells have the capacity to maintain and propagate malignant cell populations has strong implications about the way a tumour should be treated. Traditional therapies have been designed and applied to target the bulk of tumour mass, without any distinction between the various cell subsets. However, the failure of those therapies suggests that cancer stem cells might not be being efficiently targeted, even if the conventional remedies produce dramatic responses59. In modulating the Notch1, Wnt/β-catenin, and hif pathways, stem-cell signalling pathway modifiers could thus hold promise as targeted agents in the treatment of refractory or relapsed t-all.
The pharmacologic drugs most widely known to suppress Notch signalling are gsis that block the proteolytic cleavage mediated by γ-secretase and thus prevent the translocation of the intracellular Notch1 fragment icn1 into the nucleus60. In phase i clinical trials, the antitumoural activities of several gsis—for example, RO4929097 and MRK-003—have already been tested in mouse models of t-all and in relapsed t-all patients61–63. Those studies show that Notch1 signalling pathway inhibition by gsis increases the sensitivity of leukemia cells to radiation and cytotoxic chemotherapy64. However, adverse side effects in leukemia patients and non-selectivity have been reported with gsis60. To prevent gut toxicity, several approaches, such as treatment with glucocorticoids65 or optimal dosing of gsis, have been proposed63. Because gut toxicity is a result of Notch2 inhibition by gsi66, another alternative is to use a Notch1-specific agent67.
The effective inhibition of Wnt signalling is also a critical objective on the road to cure cancer. Negative physiologic regulators of Wnt signalling, such as the secreted Frizzled-related proteins and the Dickkopf-related protein 1, have been used as natural inhibitors of the Wnt pathway in cancer68–70. However, since about 2005, high-throughput screening of libraries containing small chemical compounds on cells transduced with Wnt reporters have identified various synthetic inhibitors of Wnt signalling—among them, the molecules X AV939 and pyrvinium. Specifically, XAV939 increases axin stability through the inhibition of tankyrase activity, and pyrvinium induces β-catenin phosphorylation through the activation of casein kinase50,71. Notably, alternative strategies such as blocking antibodies that target Wnt ligands or receptors have been also synthesized to hamper the Wnt signalling pathway72,73.
A novel class of therapeutic agents, presently in clinical trials as anticancer agents, consists of drugs that specifically target hif signalling74. Some effective inhibitors of the hif pathway are echinomycin, a peptide antibiotic that blocks the binding of hif-1α to dna55; chetomin, which prevents the interaction of hif-1α with the transcriptional coactivator p300–cbp75; the heat shock protein 90 inhibitor 17-allyl-aminogeldanamycin, which promotes a von Hippel–Lindau–independent degradation of hif-1α76; and 2-methoxyestradiol, an inhibitor of microtubule polymer ization77. Other small molecules, such as YC-1 [3-(5′-hydroxymethyl-2′-furyl)-1-benzylindazole], thioredoxin, and topoisomerase i inhibitors, have also been show n to reduce hif-1α expression and xenograf t growth78,79. However, ongoing screens are still in progress and might lead to the characterization of more selective and effective hif-1 inhibitors in the near future.
In conclusion, observations suggest that leukemia stem cells might not be targeted using a single universal strategy. To achieve complete patient remission and to improve clinical outcomes by preventing disease relapse, future research must focus on the mechanisms of crosstalk by the self-renewal signalling pathways and the activity of other oncogenes and tumour suppressors.
ACKNOWLEDGMENTS
This review was supported by research funding from the Terry Fox Foundation and the Canadian Institutes of Health Research.
CONFLICT OF INTEREST DISCLOSURES
We have read and understood Current Oncology’s policy on disclosing conflicts of interest, and we declare that we have none.
REFERENCES
- 1.United States Department of Health and Human Services, National Institutes of Health, National Cancer Institute (nci) Childhood Acute Lymphoblastic Leukemia Treatment—for health professionals (PDQ) [Web page] Bethesda, MD: NCI; 2015. [Available at: http://www.cancer.gov/types/leukemia/hp/child-all-treatment-pdq; cited 11 December 2015] [Google Scholar]
- 2.Coustan-Smith E, Song G, Clark C, et al. New markers for minimal residual disease detection in acute lymphoblastic leukemia. Blood. 2011;117:6267–76. doi: 10.1182/blood-2010-12-324004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moricke A, Zimmermann M, Reiter A, et al. Prognostic impact of age in children and adolescents with acute lymphoblastic leukemia: data from the trials all-bfm 86, 90, and 95. Klin Padiatr. 2005;217:310–20. doi: 10.1055/s-2005-872515. [DOI] [PubMed] [Google Scholar]
- 4.Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J Clin Invest. 2012;122:3398–406. doi: 10.1172/JCI61269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 6.O’Neil J, Grim J, Strack P, et al. FBW7 mutations in leukemic cells mediate Notch pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–24. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrando AA, Neuberg DS, Staunton J, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/S1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 8.Erikson J, Finger L, Sun L, et al. Deregulation of c-Myc by translocation of the alpha-locus of the T-cell receptor in T-cell leukemias. Science. 1986;232:884–6. doi: 10.1126/science.3486470. [DOI] [PubMed] [Google Scholar]
- 9.Homminga I, Pieters R, Langerak AW, et al. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell. 2011;19:484–97. doi: 10.1016/j.ccr.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 10.Boehm T, Foroni L, Kaneko Y, Perutz MF, Rabbitts TH. The rhombotin family of cysteine-rich lim-domain oncogenes: distinct members are involved in T-cell translocations to human chromosomes 11p15 and 11p13. Proc Natl Acad Sci U S A. 1991;88:4367–71. doi: 10.1073/pnas.88.10.4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soulier J, Clappier E, Cayuela JM, et al. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (t-all) Blood. 2005;106:274–86. doi: 10.1182/blood-2004-10-3900. [DOI] [PubMed] [Google Scholar]
- 12.Begley CG, Aplan PD, Davey MP, et al. Chromosomal translocation in a human leukemic stem-cell line disrupts the T-cell antigen receptor delta-chain diversity region and results in a previously unreported fusion transcript. Proc Natl Acad Sci U S A. 1989;86:2031–5. doi: 10.1073/pnas.86.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia Y, Brown L, Yang CY, et al. TAL2, a helix-loop-helix gene activated by the (7;9)(q34;q32) translocation in human T-cell leukemia. Proc Natl Acad Sci U S A. 1991;88:11416–20. doi: 10.1073/pnas.88.24.11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–7. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 15.Cox CV, Martin HM, Kearns PR, Virgo P, Evely RS, Blair A. Characterization of a progenitor cell population in childhood T-cell acute lymphoblastic leukemia. Blood. 2007;109:674–82. doi: 10.1182/blood-2006-06-030445. [DOI] [PubMed] [Google Scholar]
- 16.Armstrong F, Brunet de la Grange P, Gerby B, et al. Notch is a key regulator of human T-cell acute leukemia initiating cell activity. Blood. 2009;113:1730–40. doi: 10.1182/blood-2008-02-138172. [DOI] [PubMed] [Google Scholar]
- 17.Chiu PP, Jiang H, Dick JE. Leukemia-initiating cells in human T-lymphoblastic leukemia exhibit glucocorticoid resistance. Blood. 2010;116:5268–79. doi: 10.1182/blood-2010-06-292300. [DOI] [PubMed] [Google Scholar]
- 18.Gutierrez A, Sanda T, Grebliunaite R, et al. High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood. 2009;114:647–50. doi: 10.1182/blood-2009-02-206722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo W, Lasky JL, Chang CJ, et al. Multi-genetic events collaboratively contribute to pten-null leukaemia stem-cell formation. Nature. 2008;453:529–33. doi: 10.1038/nature06933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCormack MP, Young LF, Vasudevan S, et al. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science. 2010;327:879–83. doi: 10.1126/science.1182378. [DOI] [PubMed] [Google Scholar]
- 21.Tremblay M, Tremblay CS, Herblot S, et al. Modeling T-cell acute lymphoblastic leukemia induced by the SCL and LMO1 oncogenes. Genes Dev. 2010;24:1093–105. doi: 10.1101/gad.1897910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giambra V, Jenkins CR, Wang H, et al. Notch1 promotes T cell leukemia-initiating activity by runx-mediated regulation of pkc-theta and reactive oxygen species. Nat Med. 2012;18:1693–8. doi: 10.1038/nm.2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- 24.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 25.Tatarek J, Cullion K, Ashworth T, Gerstein R, Aster JC, Kelliher MA. Notch1 inhibition targets the leukemia-initiating cells in a Tal1/Lmo2 mouse model of t-all. Blood. 2011;118:1579–90. doi: 10.1182/blood-2010-08-300343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giambra V, Jenkins C, Lam SH, et al. Leukemia stem cells in t-all require active Hif1α and Wnt signaling. Blood. 2015;125:3917–27. doi: 10.1182/blood-2014-10-609370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guruharsha KG, Kankel MW, Artavanis-Tsakonas S. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet. 2012;13:654–66. doi: 10.1038/nrg3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lobry C, Oh P, Aifantis I. Oncogenic and tumor suppressor functions of Notch in cancer: it’s NOTCH what you think. J Exp Med. 2011;208:1931–5. doi: 10.1084/jem.20111855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varnum-Finney B, Xu L, Brashem-Stein C, et al. Pluripotent, cytokine-dependent, hematopoietic stem cells are immortalized by constitutive Notch1 signaling. Nat Med. 2000;6:1278–81. doi: 10.1038/81390. [DOI] [PubMed] [Google Scholar]
- 30.Duncan AW, Rattis FM, DiMascio LN, et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat Immunol. 2005;6:314–22. doi: 10.1038/ni1164. [DOI] [PubMed] [Google Scholar]
- 31.Aster JC, Pear WS, Blacklow SC. Notch signaling in leukemia. Annu Rev Pathol. 2008;3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malecki MJ, Sanchez-Irizarry C, Mitchell JL, et al. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol. 2006;26:4642–51. doi: 10.1128/MCB.01655-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pear WS, Aster JC, Scott ML, et al. Exclusive development of T cell neoplasms in mice transplanted w ith bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–91. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weng AP, Nam Y, Wolfe MS, et al. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of Notch signaling. Mol Cell Biol. 2003;23:655–64. doi: 10.1128/MCB.23.2.655-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medyouf H, Gusscott S, Wang H, et al. High-level igf1r expression is required for leukemia-initiating cell activity in t-all and is supported by Notch signaling. J Exp Med. 2011;208:1809–22. doi: 10.1084/jem.20110121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King B, Trimarchi T, Reavie L, et al. The ubiquitin ligase fbxw7 modulates leukemia-initiating cell activity by regulating Myc stability. Cell. 2013;153:1552–66. doi: 10.1016/j.cell.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma W, Gutierrez A, Goff DJ, et al. Notch1 signaling promotes human T-cell acute lymphoblastic leukemia initiating cell regeneration in supportive niches. PLoS One. 2012;7:e39725. doi: 10.1371/journal.pone.0039725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–50. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 39.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 40.Reya T, Duncan AW, Ailles L, et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–14. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 41.Hayward P, Kalmar T, Arias AM. Wnt/Notch signalling and information processing during development. Development. 2008;135:411–24. doi: 10.1242/dev.000505. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Krivtsov AV, Sinha AU, et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–3. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao C, Blum J, Chen A, et al. Loss of beta-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer Cell. 2007;12:528–41. doi: 10.1016/j.ccr.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo Z, Dose M, Kovalovsky D, et al. Beta-catenin stabilization stalls the transition from double-positive to single-positive stage and predisposes thymocytes to malignant transformation. Blood. 2007;109:5463–72. doi: 10.1182/blood-2006-11-059071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ng OH, Erbilgin Y, Firtina S, et al. Deregulated Wnt signaling in childhood T-cell acute lymphoblastic leukemia. Blood Cancer J. 2014;4:e192. doi: 10.1038/bcj.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tiemessen MM, Baert MR, Schonewille T, et al. The nuclear effector of Wnt-signaling, Tcf1, functions as a T-cell–specific tumor suppressor for development of lymphomas. PLoS Biol. 2012;10:e1001430. doi: 10.1371/journal.pbio.1001430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu S, Zhou X, Steinke FC, et al. The tcf-1 and lef-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity. 2012;37:813–26. doi: 10.1016/j.immuni.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He TC, Sparks AB, Rago C, et al. Identification of c-Myc as a target of the apc pathway. Science. 1998;281:1509–12. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 49.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–6. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 50.Huang SM, Mishina YM, Liu S, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–20. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 51.Gezer D, Vukovic M, Soga T, Pollard PJ, Kranc KR. Concise review: genetic dissection of hypoxia signaling pathways in normal and leukemic stem cells. Stem Cells. 2014;32:1390–7. doi: 10.1002/stem.1657. [DOI] [PubMed] [Google Scholar]
- 52.Lee KE, Simon MC. From stem cells to cancer stem cells: hif takes the stage. Curr Opin Cell Biol. 2012;24:232–5. doi: 10.1016/j.ceb.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 53.Maxwell PH, Wiesener MS, Chang GW, et al. The tumour suppressor protein vhl targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–5. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 54.Heddleston JM, Li Z, Lathia JD, Bao S, Hjelmeland AB, Rich JN. Hypoxia inducible factors in cancer stem cells. Br J Cancer. 2010;102:789–95. doi: 10.1038/sj.bjc.6605551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kong D, Park EJ, Stephen AG, et al. Echinomycin, a small-molecule inhibitor of hypoxia-inducible factor-1 dna-binding activity. Cancer Res. 2005;65:9047–55. doi: 10.1158/0008-5472.CAN-05-1235. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y, Liu Y, Malek SN, Zheng P. Targeting hif1α eliminates cancer stem cells in hematological malignancies. Cell Stem Cell. 2011;8:399–411. doi: 10.1016/j.stem.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang H, Li H, Xi HS, Li S. hif1alpha is required for survival maintenance of chronic myeloid leukemia stem cells. Blood. 2012;119:2595–607. doi: 10.1182/blood-2011-10-387381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zou J, Li P, Lu F, et al. Notch1 is required for hypoxia-induced proliferation, invasion and chemoresistance of T-cell acute lymphoblastic leukemia cells. J Hematol Oncol. 2013;6:3. doi: 10.1186/1756-8722-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huff CA, Matsui WH, Smith BD, Jones RJ. Strategies to eliminate cancer stem cells: clinical implications. Eur J Cancer. 2006;42:1293–7. doi: 10.1016/j.ejca.2006.01.045. [DOI] [PubMed] [Google Scholar]
- 60.Previs RA, Coleman RL, Harris AL, Sood AK. Molecular pathways: translational and therapeutic implications of the Notch signaling pathway in cancer. Clin Cancer Res. 2015;21:955–61. doi: 10.1158/1078-0432.CCR-14-0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luistro L, He W, Smith M, et al. Preclinical profile of a potent gamma-secretase inhibitor targeting Notch signaling with in vivo efficacy and pharmacodynamic properties. Cancer Res. 2009;69:7672–80. doi: 10.1158/0008-5472.CAN-09-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kolb EA, Gorlick R, Keir ST, et al. Initial testing (stage 1) by the pediatric preclinical testing program of RO4929097, a gamma-secretase inhibitor targeting Notch signaling. Pediatr Blood Cancer. 2012;58:815–18. doi: 10.1002/pbc.23290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tammam J, Ware C, Efferson C, et al. Down-regulation of the Notch pathway mediated by a gamma-secretase inhibitor induces anti-tumour effects in mouse models of T-cell leukaemia. Br J Pharmacol. 2009;158:1183–95. doi: 10.1111/j.1476-5381.2009.00389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Z, Li Y, Ahmad A, et al. Targeting Notch signaling pathway to overcome drug resistance for cancer therapy. Biochim Biophys Acta. 2010;1806:258–67. doi: 10.1016/j.bbcan.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Real PJ, Tosello V, Palomero T, et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med. 2009;15:50–8. doi: 10.1038/nm.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Riccio O, van Gijn ME, Bezdek AC, et al. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of cdk inhibitors p27Kip1 and p57Kip2. EMBO Rep. 2008;9:377–83. doi: 10.1038/embor.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu Y, Cain-Hom C, Choy L, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–7. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 68.Lavergne E, Hendaoui I, Coulouarn C, et al. Blocking Wnt signaling by sfrp-like molecules inhibits in vivo cell proliferation and tumor growth in cells carrying active beta-catenin. Oncogene. 2011;30:423–33. doi: 10.1038/onc.2010.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang H, Wang X, Hu R, et al. Methylation of SFRP5 is related to multidrug resistance in leukemia cells. Cancer Gene Ther. 2014;21:83–9. doi: 10.1038/cgt.2013.87. [DOI] [PubMed] [Google Scholar]
- 70.Zhu Y, Sun Z, Han Q, et al. Human mesenchymal stem cells inhibit cancer cell proliferation by secreting dkk-1. Leukemia. 2009;23:925–33. doi: 10.1038/leu.2008.384. [DOI] [PubMed] [Google Scholar]
- 71.Thorne CA, Hanson AJ, Schneider J, et al. Small-molecule inhibition of Wnt signaling through activation of casein kinase 1alpha. Nat Chem Biol. 2010;6:829–36. doi: 10.1038/nchembio.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ettenberg SA, Charlat O, Daley MP, et al. Inhibition of tumorigenesis driven by different Wnt proteins requires blockade of distinct ligand-binding regions by lrp6 antibodies. Proc Natl Acad Sci U S A. 2010;107:15473–8. doi: 10.1073/pnas.1007428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang J, Baskar S, Kwong KY, Kennedy MG, Wiestner A, Rader C. Therapeutic potential and challenges of targeting receptor tyrosine kinase ror1 with monoclonal antibodies in B-cell malignancies. PLoS One. 2011;6:e21018. doi: 10.1371/journal.pone.0021018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Semenza GL. Targeting hif-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–32. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 75.Kung AL, Zabludoff SD, France DS, et al. Small molecule blockade of transcriptional coactivation of the hypoxiainducible factor pathway. Cancer Cell. 2004;6:33–43. doi: 10.1016/j.ccr.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 76.Isaacs JS, Jung YJ, Mimnaugh EG, Martinez A, Cuttitta F, Neckers LM. Hsp90 regulates a von Hippel Lindau–independent hypoxia-inducible factor-1 alpha–degradative pathway. J Biol Chem. 2002;277:29936–44. doi: 10.1074/jbc.M204733200. [DOI] [PubMed] [Google Scholar]
- 77.Mabjeesh NJ, Escuin D, LaVallee TM, et al. 2me2 inhibits tumor growth and angiogenesis by disrupting microtubules and dysregulating hif. Cancer Cell. 2003;3:363–75. doi: 10.1016/S1535-6108(03)00077-1. [DOI] [PubMed] [Google Scholar]
- 78.Yeo EJ, Chun YS, Cho YS, et al. YC-1: a potential anticancer drug targeting hypoxia-inducible factor 1. J Natl Cancer Inst. 2003;95:516–25. doi: 10.1093/jnci/95.7.516. [DOI] [PubMed] [Google Scholar]
- 79.Welsh SJ, Williams RR, Birmingham A, Newman DJ, Kirkpatrick DL, Powis G. The thioredoxin redox inhibitors 1–methylpropyl 2–imidazolyl disulfide and pleurotin inhibit hypoxia-induced factor 1alpha and vascular endothelial growth factor formation. Mol Cancer Ther. 2003;2:235–43. [PubMed] [Google Scholar]